Abstract
Here, we present the basic concept and theoretical framework of a scientific hypothesis called Cancer Evolution–Development (“Cancer Evo-Dev”), based on our recent studies of the molecular mechanisms by which chronic infection with the hepatitis B virus induces hepatocarcinogenesis, together with related advances in that field. Several aspects central to our hypothesis are presented:
■ Immune imbalance—caused by the interaction of genetic predispositions and environmental exposures such as viral infection—is responsible for the maintenance of chronic non-resolving inflammation. Non-resolving inflammation promotes the occurrence and progression of cancers, characterized by an evolutionary process of “mutation–selection–adaptation” for both viruses and host cells.
■ Under a microenvironment of non-resolving inflammation, proinflammatory factors promote mutations in viral or host genomes by transactivation of the expression of cytidine deaminases and their analogues. Most cells with genomic mutations and mutated viruses are eliminated in the competition for survival in the inflammatory microenvironment. Only a small percentage of the mutated cells that alter their survival signal pathways and exhibit the characteristics of “stem-ness” can survive and function as cancer-initiating cells.
■ Cancers generally develop with properties of “backward evolution” and “retro-differentiation,” indicating the indispensability of stem-like signal pathways in the evolution and development of cancers.
The hypothesis of Cancer Evo–Dev not only lays the theoretical foundation for understanding the mechanisms by which inflammation promotes the development of cancers, but also plays an important role in specific prophylaxis, prediction, early diagnosis, and targeted treatment of cancers.
Keywords: Hepatocarcinogenesis, evolution, inflammation, hepatitis B virus, mutations
INTRODUCTION
Cancers are usually caused by the generation and accumulation of somatic mutations—a process that abides by the Darwinian Theory of Evolution: mutation–selection–adaptation. That evolutionary theory of cancer has profound theoretic and clinical implications for the investigation of carcinogenic mechanisms, cancer prevention, and targeted therapy. Decades of research have provided broad evidence to support the idea that inflammation caused by infection is an important cause of cancer. However, few efforts have been made to apply evolutionary biology in understanding and controlling inflammation-induced malignant transformation.
Chronic infection is responsible for more than 25% of human cancers. Rous sarcoma virus, a retrovirus, was the first reported cancer-inducing virus. It was isolated by Francis Peyton Rous in 1910 and was named after the investigator1. Hepatitis B virus (hbv) was discovered by Baruch Samuel Blumberg in 19662. The causative relationships between human papillomavirus and cervical carcinoma, Epstein–Barr virus and nasopharyngeal carcinoma, hbv and hepatocellular carcinoma (hcc), and Helicobacter pylori and gastric cancer were established in 1985, 1990, 1995, and 2000 respectively3. In the 1960s, David Baltimore found that rna viruses can integrate into chromosomes, revealing the link between viral infection and somatic mutations1. The strong association between hbv infection and hcc has been reported in many observational studies, with odds ratios (ors) ranging from 5 to 49 in case–control studies and relative risks ranging from 7 to 98 in cohort studies4.
Population genetic studies, including genome-wide association studies, have shown that the associations between single-nucleotide polymorphisms and most human cancers are generally weak, with ors in the 1.0–1.3 range5. Those epidemiologic data highlight the importance to carcinogenesis of infection exposures, consistent with the results of cancer genome research. Mutation patterns related to chronic inflammation have been identified in most cancers6. In some cancers, the inflammation-related mutations increase with time, accompanied by a decline in the mutations related to the initial exposure7. Those distribution characteristics and the switch in mutation domination can be analyzed from an evolutionary perspective, suggesting that inflammation induced by chronic infection might not only cause somatic mutations, but also play an important role in micro-environmental selection.
Here, we focus on the scientific hypothesis of Cancer Evolution–Development (“Cancer Evo-Dev”) and summarize the basic concepts and theoretical framework of that theory. Based on prior research, and especially our own experience in hbv-induced hcc, we present a theoretical framework of Cancer Evo-Dev to elucidate the mechanisms by which cancer develops and the most cost-effective ways to control malignancies.
FRAMEWORK OF CANCER EVO-DEV
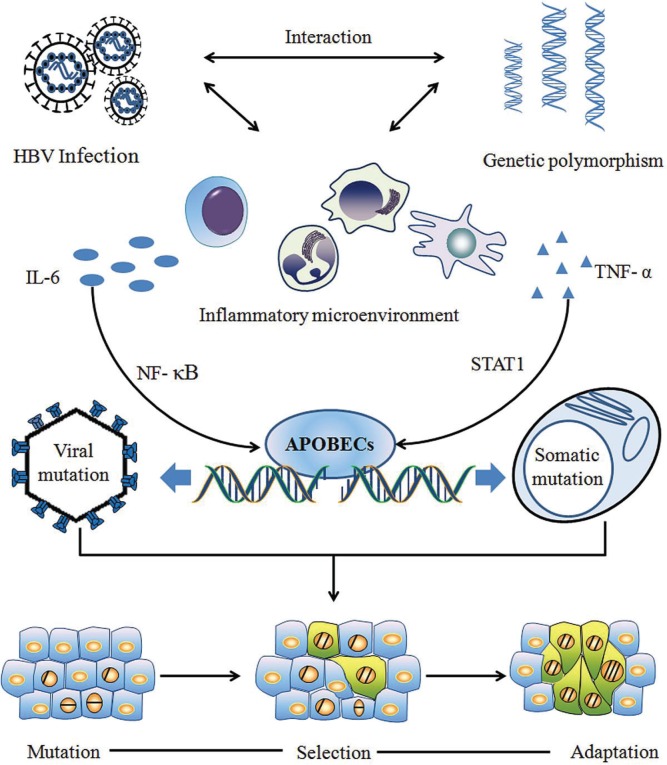
Figure 1 depicts the framework of Cancer Evo-Dev as exemplified by hepatocarcinogenesis. A series of important molecular events—from inf lammatory precancerous lesions to carcinogenesis, postoperative recurrence, and metastasis—can occur in the evolutionary process. Those events include, but are not limited to, epigenetic modification, generation and selection of somatic mutations, and alteration of dynamic signal pathway networks. The synergetic effects of genetic and environmental factors contribute to imbalance in the immune system, resulting in the activation and maintenance of non-resolving inflammation, thus providing a fertile microenvironment for the process of Cancer Evo–Dev.
FIGURE 1.
Schematic of the basic framework of Cancer Evolution–Development (Evo-Dev). Interactions between genetically predisposed inflammatory signalling molecules (such as human leucocyte antigen) and chronic infection [such as with hepatitis B virus (HBV)] contribute to the activation and persistence of chronic inflammation. Cytokines in the nuclear factor kappa B (NF-κB) or STAT1 signal pathways stimulate expression of members of the family of cytidine deaminases and their analogues called “apolipoprotein B messenger RNA editing enzyme, catalytic polypeptide-like” (APOBECs), which induce somatic and viral genomic alterations by C-to-U hypermutation. Distinct mutant lineages are selected by the inflammatory microenvironment. Mutants with characteristics of stem cells live through the survival selection and evolve into tumour-initiating cells by altering the signalling network. The process of Cancer Evo-Dev—characterized by “backward evolution” and “retro-differentiation”—is consequently triggered.
Under conditions of non-resolving inflammation, activated nuclear factor κB (nf-κb) and proinflammatory molecules can transactivate the expression of nucleic acid editing enzymes such as the family of cytidine deaminases and their analogues called “apolipoprotein B messenger rna editing enzyme, catalytic polypeptide-like” (apobecs) in humans, thus promoting viral and somatic mutations. Viral mutants facilitate the malignant transformation of normal cells. Most mutant cells are eliminated by selective pressures, such as those exerted by the inflammatory immune response, although a small proportion of mutant cells survive in precancerous lesions. Those surviving mutant clones later evolve into tumour-initiating cells (also known as stem-like tumour-initiating cells) by altering the original cell signal patterns and promoting epithelial-to-mesenchymal transition (emt) through epigenetic regulation. Some established cancer markers such as alpha-fetoprotein and carcinoembryonic antigen are usually expressed at the embryonic stage, silenced after birth, and re-expressed in cancer patients. That evidence implies that the process of Cancer Evo-Dev can be characterized as “backward evolution” and “retro-differentiation.”
THE EVOLUTIONARY CHARACTERISTICS OF CARCINOGENESIS
Retro-differentiation and Backward Evolution
The development of the human embryo repeats the process of human evolution within 10 months. Most genes expressing at this stage will be silenced after delivery, but some genes, such as that for alpha-fetoprotein, will re-express for a short period of time when an organism is injured.
Most cells in tumour tissue are highly differentiated and have limited proliferative activity. However, some cells with stem-cell-like characteristics become the main malignant subgroup in tumour tissues. SALL4, an oncofetal protein, is one of the key factors for self-renewal and maintenance of embryo stem-cell pluripotency. SALL4 is expressed in the human fetal liver and silenced in the adult liver, but it can be detected in a subgroup of hccs. The re-expression of SALL4 is related to the “stem function” of tumour cells and indicates invasion and unfavourable prognosis. Furthermore, the cell aging process is accompanied by shortening of its telomeres, which does not occur in tumour cells.
In terms of morphology, emt is the process by which epithelial cells lose their epithelial characteristics and acquire mesenchymal characteristics, structure, and biologic function. An emt usually occurs at a critical stage of embryonic development, and it is equally important for tumour metastasis. In the process of tumour invasion through emt, epithelial cells acquire “stem-ness,” including self-renewal and antiapoptotic capacities. Most tumour cells are differentiated, with limited amplification ability. However, a small proportion of cells with the “stem-ness” feature become the main malignant cell subsets in cancer and are known as cancer-initiating cells, responsible for the disease’s malignant nature and chemoresistance.
“Dead-End” Evolution of HBV
In cancers induced by chronic viral infection, not only host cells, but also the virus, conduct the process of evolution. Hepatocarcinogenesis induced by hbv is a typical Cancer Evo-Dev process, and hbv evolution serves as a valuable clue for investigating the mechanism underlying that process. Hepatitis B virus belongs to the Hepadnaviridae family and is evolutionarily conservative in the long-term evolution of species. Genetic evolution analysis indicates that human hbv and hepadnaviruses isolated from non-human primates are in the same evolutionary branch8. However, the evolution of the hbv genome is evident in a subset of infected individuals during chronic infection.
Previous research by our group established the wild-type (“standard”) hbv sequences based on the whole hbv genome sequenced in 2000 asymptomatic carriers seropositive for hepatitis B surface antigen from community-based epidemiologic surveys. Based on the wild-type hbv sequences, hcc-related mutations and their development patterns were subsequently identified. We also observed that hbv mutations posing a significant hcc risk are located mainly within the enhancer ii/basal core promoter/precore and preS regions9–11. During the hbv-induced carcinogenic “trilogy” (chronic hepatitis, liver cirrhosis, hcc), the species and frequencies of those mutations often accumulate consecutively and can be used to predict the occurrence and development of liver cirrhosis and hcc12–15. Retrospective and prospective cohort studies have both identified a combination of hbv mutations (C1653T, A1762T/G1764A, and T1753V) that have significant predictive value12,16,17. Among them, the A1762T/G1764A mutation usually appears in the early stage; other mutations, including T1753V, C1653T, G1899A, and preS deletion, are evident only in a late stage of the evolution18. Reaction to chronic hbv infection (characterized by immune response–induced hepatocyte injury and release of transaminase) is usually accompanied by hbv e-antigen (HBeAg) seroconversion and an increase in hbv mutations, indicating the selective effect of immune cells on viral mutants.
Hepatitis B virus acquired during infancy or early childhood, or at an early stage of the infection in adults, is usually of the wild type12–14. During the chronic inflammation process, especially after a HBeAg shift from HBeAg-positive to HBeAg-negative, mutant hbv subgroups gradually increase. One of the main features of mutant hbv genotypes is a deficiency of the CD8+ T-cell epitope, a consequence of immune selection. Although the hcc-related hbv mutants are present in fetal cord blood, neonatal infection is usually caused by wild-type hbv rather than by mutant subgroups. In hbv-infected children 1–15 years or age, the frequencies of hcc-related mutations increased as age increased. However, compared with their mothers, who had been exposed to chronic infection for at least 25 years, the children had fewer hcc-related hbv mutations18.
The foregoing results are based on analyses of serum hbv. In individuals with chronic hbv infection, almost all hbv is synthesized in hepatocytes and released into the circulation at a pace of up to 1011 viral particles daily. Because hbv is cleared from plasma with a half-life of approximately 1.2 days, functional hepatocytes should be responsible for the persistence of hbv mutants in peripheral blood19,20. The immune microenvironment of liver tissue is therefore necessary for the co-evolution of hbv and the host genome. Although tumour-adjacent tissues are pathologically categorized as “normal,” they typically contain precancerous lesions and have already entered the middle stage of the cancer evolution process. The hccs that relapse more than 2 years after resection are considered to be recurrent hcc and are not a result of the initial hcc cell diffusion into remnant liver tissue21.
The species and frequencies of certain hbv mutations in adjacent tissues are distinct in different populations. Together with immune markers and expression levels of inflammatory genes, they can therefore be used to predict prognosis in hcc patients receiving curative surgery. For example, hbv mutations in the enhancer ii/basal core promoter/precore region, such as A1762T/G1764A, can serve as predictive markers for survival and recurrence21, indicating that hbv evolution in adjacent tissues continues until the patient dies. Antiviral therapy can block hbv evolution in adjacent tissues by relieving inflammation, thus notably prolonging survival in hcc patients22.
Earlier studies by our group explained why Hepadnaviridae family members, including hbv, are highly conservative across species18. Wild-type hbv has advantage of infecting hepatocytes, facilitating viral spread from one individual to another, and contributing to the maintenance of its viral species. The hcc-related mutants can cause malignant transformation, but have lost the advantage of person-to-person infection. Those mutants are therefore usually eliminated at the death of the carriers, which is termed “dead-end” evolution.
EVOLUTIONARY EFFECT OF NON-RESOLVING INFLAMMATION
Providing Microenvironmental Pressure
As a defense mechanism responding to exogenous infection and injury, most short-term acute inflammation is beneficial to humans. However, chronic inflammation—a wound does not heal—is closely associated with many chronic diseases, including cancer and diabetes. It is widely accepted that most solid tumours and some hematologic malignancies are associated with non-resolving inflammation. Analogous to the Darwinian Theory of Evolution and Origin of Species, the process of cancer evolution is based on two conditions: the continuous acquisition of somatic mutations, and natural selection acting on the resultant phenotypic diversity23. A chronic inflammatory microenvironment serves as a niche for that process by inducing endogenous mutagenic factors such as apobecs and provides selection pressure.
During the process of carcinogenesis, cells must overcome four barriers:
■ The cell-cycle checkpoint that regulates cell division
■ Apoptosis, which limits cell proliferation
■ Telomere length, which determines the total number of cell divisions
■ The cell adhesion barrier that prevents cell migration
The induction and sustenance of inflammation can alter the “ecologic” conditions in local or systematic tissues (or both), weaken the functions of the foregoing barriers, induce abnormal expression of genes, cause genomic instability, and provide opportunities for backward evolution into cancer-initiating cells in mesenchymal tissues. Chronic inflammation can boost the secretion of inflammatory mediators such as eicosanoids (prostaglandin E2, leukotrienes)24, cytokines, and chemokines in stromal cells, resulting in abnormal transformation of the tissue microenvironment, infiltration of dysfunctional immune cells, and decreased epithelial integrity, all of which play an important role in Cancer Evo-Dev.
In hcc, for instance, chronic inflammation promotes carcinogenesis by activating some evolutionarily conserved signal pathways, such as phosphoinositide 3-kinase (pi3k)/Akt/mtor (mammalian target of rapamycin), nf-κb/tumour necrosis factor α (tnf-α), Raf/mitogen-activated protein kinase (mapk)/erk, transforming growth factor β1 (tgf-β1), jak, Wnt/β-catenin, and signal transducer and activator of transcription 3 (stat3)/interleukin 6 (il-6), which play critical roles in hbv-hcc development and invasion21. By activating stat3 and nf-κb, il-6 and tnf-α can induce hepatocytes to lose their epithelial characteristics (emt) and initiate backward evolution. Transforming growth factor β1 can facilitate emt, which can be enhanced by il-6 and tnf-α. The synergistic effect of those three cytokines can promote the transformation of normal hepatocytes into stem-like cells. By relieving hepatic inflammation, antiviral therapy can significantly lower the risks of hcc occurrence and postoperative recurrence22,25. The same data also support the notion that elimination of inflammation can destroy the fertile environment for cancer evolution.
In a severe inflammatory microenvironment, a process of continuous necrosis and proliferation can help somatic mutations to accumulate, and tumour-initiating cell clones with strong viability are selected. In a broader sense, the tissue and organ reshaping that occurs under inflammatory conditions is an important way in which organisms adapt to the environment. The interaction of genetic susceptibility with environmental factors induces non-resolving inflammation and consequently promotes Cancer Evo-Dev.
APOBECs Bridge Inflammation and Cancer
The apobecs, a family of cytidine deaminases, are powerful endogenous mutagenic factors that play critical roles in many biologic processes, especially immune defense. This group of enzymes can specifically catalyze irreversible cytidine and deoxycytidine deamination to convert bases from cytosine to uracil, creating a cytosine-to-uracil mismatch in minus-stranded dna and reverse-transcript guanosine-to-adenosine transitions in plus-stranded dna.
Activation-induced cytidine deaminase and apobec3 were found in the pathways of both the acquired and the innate immune system21,22. Mutagenesis mediated by apobecs can increase the viral mutation load to a level that exceeds the threshold for viral viability. Accordingly, apobec family members can similarly increase the number of somatic mutations to a threshold that exceeds the host’s repair ability and starts the Cancer Evo-Dev process. In activation-induced cytidine deaminase transgenic animal models, mutations induced in the TP53 and β-catenin genes by constitutive expression of activation-induced cytidine deaminase can generate hcc (13.75%), lung cancer (8.75%), and gastric cancer (1.25%)26.
Three mechanisms prevent the induction of somatic mutations by the apobec family. First, apobecs rarely express in normal tissues, and short-term activation of apobecs is beneficial for eliminating pathogens. Second, the cytidine deaminase activity of the apobecs is applied almost exclusively to single-stranded nucleotides, in which mutagenesis is 200–300 times more efficient than it is in double-stranded dna. Third, the uracil-induced mutagenesis of apobecs is counteracted by uracil–dna glycosylase (ung)27. However, genetic susceptibility, viral mutations, and an unbalanced immune system interact with each other to prevent the absolute elimination of pathogens such as hbv, resulting in chronic inflammation accompanied with apobec expression.
In the inflammatory microenvironment, the proinflammatory cytokine/chemokine and nf-κb/tnf-α signal pathways are persistently activated, which can significantly increase the expression of apobecs at the transcription level28. The resulting persistent high levels of apobec expression can edit the single-stranded dnas that are temporarily generated during the transcription and replication process, consequently leading to mutagenesis in the human genome. If the balance between apobecs and ung is broken, or if the overall mutagenesis level exceeds the reserve capacity of the downstream repair pathways, somatic mutations will be generated, thus promoting the process of Cancer Evo-Dev. The apobec family therefore provides a link between high-risk infection, inflammation, oncogenes, and somatic mutation, forming the early-stage framework of Cancer Evo-Dev.
GENETIC ALTERATION IS THE BASIS OF CANCER EVO-DEV
Host Somatic Mutations
The continuous generation and selection of somatic mutations are two basic mechanisms underlying Cancer Evo-Dev. One of the classic characteristics of cancer cells is their high mutation rate. Somatic mutations can be classified according to their effects on Cancer Evo-Dev. A small proportion of the mutations can lead to malignant phenotypes that are positively selected during the evolution process and thus are called “driver” mutations. The remaining mutations are “passengers” that contribute very little to carcinogenesis29.
The spontaneous rate of somatic mutations is not high enough to trigger the evolution process. There must be some mutagenesis-driving forces—such as defective dna repair capacity, exogenous or endogenous mutagen exposures, and intrinsic mistakes of dna replication—that can increase the mutation rate in the cancer genome. A distinct mutagenic process generates various mutation combinations that are called “signatures.” Those apobec-related signatures are widely prevalent in more than half of all cancer types being investigated for a connection with inflammation, suggesting that the inflammatory immune response is the common mechanism by which mutations are generated6.
Driver mutations are selected at certain phases during the Cancer Evo-Dev process, but might not be detectable at all stages. At the early stage of carcinogenesis, cells with initial driver mutations can survive and multiply rapidly. However, at later stages of Cancer Evo-Dev, cells with other driver mutations can gain more advantages in the survival competition and can replace the cells that have only initial mutations, becoming the dominant subset. For example, in lung cancer patients who continued to be exposed to cigarettes, the signatures of the initial mutations (smoke-related) showed a relative decline over time, accompanied by an increase in the apobec-related mutations30. Tracing the positive selection of drivers and the patterns of cancer genome alteration can therefore help to demonstrate the lineage of the malignancy clones and the major mutagenic factors. Exome-sequencing data from solid tumours and hematologic neoplasms confirmed the clonal heterogeneity of primary tumours and metastases, supporting the evolution model at the genetic level31.
Somatic mutations participate in Cancer Evo-Dev by altering signal pathways. Despite the tremendous effect of genetic alteration, the incidences of specific mutations in a single gene are not high in the patient population. For example, the mutation rates for ARID1A and ARID2, two genes with classic hcc-related genetic variations, in tumour tissue are 16.8% and 5.6% respectively32. Given such a low detection rate, clinical and public health experts are not really in a position to use the frequency of a single somatic mutation for the prediction, prevention, early diagnosis, and treatment of cancer; however, combinations of genetic variations might be useful in that regard. Somatic mutations with a similar function in different individuals might influence a specific signal pathway that is related to the “stem-ness” of cancer, thereby significantly promoting carcinogenesis. Using critical molecules on the signal pathway network, novel diagnostic biomarkers and therapeutic targets for those mutation combinations could potentially be discovered.
Giving the huge amount of mutation data, complex dynamic patterns, and various interactions with genetic and environmental factors, the application of systems biology is fundamental to signal-pathway mutation investigations. For instance, the pi3k/Akt/mtor, nf-κb/tnf-α, Raf/mapk/erk, tgf-β1, jak, Wnt/β-catenin, and stat3/il-6 signal pathways have been identified as playing important roles in hepatocellular carcinogenesis33. In colorectal carcinoma, a regulation network based on high-throughput data about molecule groups on signal pathways was constructed, and its prospective value was validated by cohort study34.
Tumour Heterogeneity
Tumour heterogeneity refers to two concepts. First, patients with tumours of the same pathologic type show distinct clinical manifestations, including occurrence, metastasis, therapeutic response to drugs and radiation therapies, and postoperative prognosis. Second, tumour cells in a single individual show significant differences in genomic mutation profile, evolution pathway, and gene expression. The first (“classical”) concept of tumour heterogeneity is the basis for the development of biomarkers and therapeutic targets that can predict cancer occurrence, metastasis, and therapeutic response, contributing to personalized medicine. The second concept of tumour heterogeneity was discovered and subsequently validated by the application of next-generation sequencing, which allows genomic alterations and exome evolution to be traced by multipoint deep sequencing.
Tumours in different microenvironments (tissues harbouring primary foci compared with metastatic foci) and at different treatment stages (before chemotherapy compared with after chemotherapy) have distinct mutation spectra, thus demonstrating, within a solid tumour, an obvious heterogeneity that is the result of continuously unbalanced evolution that persists under the selection pressure of the microenvironment. Therapies can also serve as a selection pressure, bring their own changes in malignant clones, and that evolution-induced heterogeneity will complicate treatment of the cancer. Cancer therapy should therefore be designed as sequential treatments with the specific purpose of targeting critical pathways during the Cancer Evo-Dev process.
Somatic Mutations in HCC
A variety of hcc-associated mutated genes—mainly including the chromatin remodelling genes (ARID1A, ARID1B, ARID2), the p53/rb tumour suppression pathway (IRF2, TP53, CDKN2A), the Wnt/β-catenin signal pathway (RPS6KA3–AXIN1, NFE2L2–CTNNB1), and the Ras/pi3k pathway (PTEN, PIK3CA, KRAS, NRAS)—were discovered by next-generation sequencing27,32,35. Most of those mutated genes were identified in hcc samples at the late phase of their evolution; the somatic mutations of precancerous lesions have received little attention. Interestingly, a close connection has been observed between some cancer-related mutations and specific environmental exposures. For instance, a deactivation-inducing mutation of IRF2, a tumour suppression gene, exists only in hbv-hcc and can cause p53 dysfunction32. In alcoholic hcc patients, the major somatic mutations usually occur within the chromatin remodelling genes, with higher detection rates36. Mutations induce oxidative stress and inactivate RPS6KA3, which can both interact with the Wnt/β-catenin pathway, suggesting a synergistic effect of those pathways35. Mutations in the promotor region of telomerase exist in 25% of liver cirrhosis and play a role in the activation of CTNNB133.
The foregoing preliminary studies have demonstrated the existence of somatic mutations that can drive hcc evolution in precancerous lesions. As studies of the cancer genome have indicated, genetic alteration is more complicated in hcc than in many other cancer types6. The long evolutionary process of hcc is partly responsible for that complexity, with its multiphasic carcinogenesis that leads to frequent shifts of mutation dominance. Another important contributor is the interaction between hbv mutation and host genetic variation.
Occurrence of HBV Mutations
The occurrence of hcc-related hbv mutations also strictly abides by the Darwinian model: mutation–selection–adaptation. The generation of hbv mutations follows two patterns.
The first pattern is one of absolutely random viral mutations. During viral replication, the partially double-stranded hbv dna is generated from an intermediate rna through the reverse transcription activity of the viral polymerase. The hbv reverse transcriptase lacks proofreading capacity, resulting in mutation rates in the range of 1.5×10−5 to 5×10−5 nucleotide substitutions per site per year, which can increase after HBeAg seroconversion37. Because of overlapping open reading frames, mutations can alter the genes that are necessary to viral replication. The random natural mutations are therefore constrained to specific regions of the hbv genome.
The second viral mutation pattern is induced by host cytidine deaminases; it imparts directional properties27. During chronic hbv infection, inflammatory cytokines and chemokines—including nf-κb, tnf-α, il-1, and il-6—can transactivate the apobec family of cytidine deaminases. The apobec family has a dual effect on hbv: reduction of hbv and induction of hbv mutations. Although many hbv genome fragments, including the precore/basal core promoter/enhancer ii region and the S region, are generally sensitive to editing by members of apobec338–41, the sequence encoding hbv X protein (HBx) is more vulnerable. The HBx region is the preferred editing target of apobec3, which generates carboxylic acid–terminal truncated HBx, the main form of hbv dna that is integrated into the host genome41. That apobec-induced mutagenesis is counteracted by ung, which plays an important role in the base-excision repair mechanism42. In human cells, the nuclear form of ung is far more active than its mitochondrial counterpart43. The mutagenic effect of inflammatory factors on the hbv genome depends on the degree of the damage to the apobec–ung balance27.
Although most hbv mutations are random, directional evolution of hbv occurs under immune selection pressures during chronic infection. In the initial immune tolerance phase of chronic infection, the immune pressure is weak, and most of the individual viruses are wild-type. Immune pressure increases with the progression of chronic inflammation33,44, which facilitates the gradual occurrence of viral mutations45, especially in HBeAg-negative individuals.
Most hcc-related hbv mutations are selected by the immune microenvironment before the occurrence of hcc and can be used as predictive markers. Single-nucleotide polymorphisms of the inf lammatory signal pathway genes, including STAT3, nf-κb, HLA-DP, and HLA-DQ have been demonstrated to maintain the chronic infection and to facilitate the selection of hcc-related hbv mutations that contribute to the risk of liver cancer12–14,46. In the inflammation–immune response microenvironment, most hbv mutants are eliminated by the antiviral immune response. Only a tiny fraction of mutant viruses that facilitate the regeneration of hepatocytes can survive and gradually develop into the hcc-promoting clones that adapt to the inflammatory environment. However, those viral mutants that affect the pre-cancer hepatocytes are less infectious to normal liver cells.
HBV Mutation Promotes the Evolution of HCC
The hcc-related hbv mutations promote hepatocarcinogenesis. It has been reported that, after being transfected into primary human hepatocytes and hcc cells, the HBx gene fragment and full-length hbv with A1762T/G1764A–based combination mutations can significantly facilitate the degradation of p21, a tumour suppressor protein, and increase the expression of S-phase kinase-associated protein 2, thus promoting the proliferation and metastasis of cells46,47. Mutations in the preS and S regions also notably facilitate carcinogenesis11,21. The hbv large envelope protein gene fragment (preS1/preS2/S), with F141L mutation in the preS2 region, can significantly promote the proliferation of hepatocytes by downregulating the p53 and p21 pathways and upregulating the expression of cyclin-dependent kinase 4 and cyclin A. Colony-forming rates are about twice as high for hepatocytes expressing the F141L–large envelope protein than for those expressing the wild-type hbv large envelope protein48. In prospective epidemiologic studies, hbv-infected patients with viral mutations are more likely to develop hcc49–52. Those data suggest that hbv mutations selected by the immune system can promote hcc evolution.
From the perspective of evolution biology, hbv mutations can participate in the alteration of the host genome both directly and indirectly. First, hcc-related viral mutants can cause somatic mutations by directly integrating into the human genome. Carboxylic acid–terminal truncated HBx mutation is the main form of viral integration and, compared with full-length HBx, usually demonstrates a stronger pro-carcinogenic effect22,53,54. Furthermore, postoperative antiviral therapy cannot reduce the recurrence rate and extend the lifespan of patients with carboxylic acid–terminal truncated HBx integration22. Second, the hbv mutations that contribute to the maintenance of chronic infection elicit non-resolving inflammation, which not only can increase the frequency of somatic mutations by inducing expression of the apobec family, but also can serve as a selection pressure in the microenvironment27.
SUMMARY AND FUTURE DIRECTIONS
Based on studies of hbv-induced hepatocarcinogenesis (a typical evolutionary process), we put forward the hypothesis of Cancer Evo-Dev. Under conditions of genetic predisposition, exogenous factors such as viral infection can induce chronic inflammation. The elimination of chronic infection can relieve inflammation, reducing the incidence of cancer and subsequently extending effective survival. As the hypothesis describes, tumour-initiating cells obtain a survival advantage during the evolutionary process of mutation–selection–adaptation by activating a “stem-ness” pathway and simultaneously causing evolutionary heterogeneity. Critical molecules in a functional subnetwork that maintains and promotes the Cancer Evo-Dev process can be demonstrated using systems biology approaches. The development of high-efficiency inhibitors that will target these critical molecules and block corresponding signal pathways could be a powerful treatment strategy in advanced cancers.
The hypothesis of Cancer Evo-Dev will serve two purposes: first, early prevention can reduce the cancer incidence and delay its onset; second, targeted therapy can reduce morbidity and mortality rates. Our hypothesis can therefore contribute to the realization of “P4” medicine (predictive, preventive, personalized, and participatory)55, and promote the prophylaxis and control of cancer.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Basic Research Program (grant no. 2015CB554000) and the National Natural Scientific Foundation of China (grant nos. 91529305, 81520108021). The study sponsors had no role in the writing of the manuscript and in the decision to submit the manuscript for publication.
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare that we have none.
REFERENCES
- 1.Baltimore D. rna-dependent dna polymerase in virions of rna tumour viruses. Nature. 1970;226:1209–11. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Stehelin D, Varmus HE, Bishop JM, Vogt PK. dna related to the transforming gene(s) of avian sarcoma viruses is present in normal avian dna. Nature. 1976;260:170–3. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 3.Ewald PW. 99th Dahlem Conference on infection, inflammation and chronic inflammatory disorders: symbionts and immunopathology in chronic diseases: insights from evolution. Clin Exp Immunol. 2010;160:27–34. doi: 10.1111/j.1365-2249.2010.04127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen VT, Law MG, Dore GJ. Hepatitis B–related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453–63. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- 5.Du Y, Xie J, Chang W, Han Y, Cao G. Genome-wide association studies: inherent limitations and future challenges. Front Med. 2012;6:444–50. doi: 10.1007/s11684-012-0225-3. [DOI] [PubMed] [Google Scholar]
- 6.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuong KJ, Loeb LA. apobec3b mutagenesis in cancer. Nat Genet. 2013;45:964–5. doi: 10.1038/ng.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin J, Xie J, Liu S, et al. Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am J Gastroenterol. 2011;106:81–92. doi: 10.1038/ajg.2010.399. [DOI] [PubMed] [Google Scholar]
- 10.Yin J, Xie J, Zhang H, et al. Significant association of different preS mutations with hepatitis B–related cirrhosis or hepatocellular carcinoma. J Gastroenterol. 2010;45:1063–71. doi: 10.1007/s00535-010-0253-1. [DOI] [PubMed] [Google Scholar]
- 11.Liu S, Xie J, Yin J, et al. A matched case–control study of hepatitis B virus mutations in the preS and core promoter regions associated independently with hepatocellular carcinoma. J Med Virol. 2011;83:45–53. doi: 10.1002/jmv.21829. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Yin J, Zhang Y, et al. HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J Virol. 2013;87:12176–86. doi: 10.1128/JVI.02073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie J, Zhang Y, Zhang Q, et al. Interaction of signal transducer and activator of transcription 3 polymorphisms with hepatitis B virus mutations in hepatocellular carcinoma. Hepatology. 2013;57:2369–77. doi: 10.1002/hep.26303. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Ji XW, Hou XM, et al. Effect of functional nuclear factor-κB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Ann Oncol. 2014;25:2413–19. doi: 10.1093/annonc/mdu451. [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Zhang H, Gu C, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–82. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji X, Zhang Q, Li B, et al. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol. 2014;28:201–9. doi: 10.1016/j.meegid.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 17.Yin J, Wang J, Pu R, et al. Hepatitis B virus combo mutations improve the prediction and active prophylaxis of hepatocellular carcinoma: a clinic-based cohort study. Cancer Prev Res (Phila) 2015;8:978–88. doi: 10.1158/1940-6207.CAPR-15-0160. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Xie Z, Ni H, et al. Mother-to-child transmission of hepatitis B virus: evolution of hepatocellular carcinoma–related viral mutations in the post-immunization era. J Clin Virol. 2014;61:47–54. doi: 10.1016/j.jcv.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci U S A. 1996;93:4398–402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewin SR, Ribeiro RM, Walters T, et al. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology. 2001;34:1012–20. doi: 10.1053/jhep.2001.28509. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Zhang Q, Chang W, Du Y, Zhang H, Cao G. Viral and host inflammation-related factors that can predict the prognosis of hepatocellular carcinoma. Eur J Cancer. 2012;48:1977–87. doi: 10.1016/j.ejca.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Yin J, Li N, Han Y, et al. Effect of antiviral treatment with nucleotide/nucleoside analogs on postoperative prognosis of hepatitis B virus–related hepatocellular carcinoma: a two-stage longitudinal clinical study. J Clin Oncol. 2013;31:3647–55. doi: 10.1200/JCO.2012.48.5896. [DOI] [PubMed] [Google Scholar]
- 23.Gatenby RA, Gillies RJ, Brown JS. Of cancer and cave fish. Nat Rev Cancer. 2011;11:237–8. doi: 10.1038/nrc3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–93. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LP, Zhao J, Du Y, et al. Antiviral treatment to prevent chronic hepatitis B or C–related hepatocellular carcinoma. World J Virol. 2012;1:174–83. doi: 10.5501/wjv.v1.i6.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morisawa T, Marusawa H, Ueda Y, et al. Organ-specific profiles of genetic changes in cancers caused by activation-induced cytidine deaminase expression. Int J Cancer. 2008;123:2735–40. doi: 10.1002/ijc.23853. [DOI] [PubMed] [Google Scholar]
- 27.Deng Y, Du Y, Zhang Q, Han X, Cao G. Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation and hepatocellular carcinoma. Cancer Lett. 2014;343:161–71. doi: 10.1016/j.canlet.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 28.Endo Y, Marusawa H, Kinoshita K, et al. Expression of activation-induced cytidine deaminase in human hepatocytes via nf-κb signaling. Oncogene. 2007;26:5587–95. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- 29.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldas C. Cancer sequencing unravels clonal evolution. Nat Biotechnol. 2012;30:408–10. doi: 10.1038/nbt.2213. [DOI] [PubMed] [Google Scholar]
- 32.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–8. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han YF, Zhao J, Ma LY, et al. Factors predicting occurrence and prognosis of hepatitis-B-virus-related hepatocellular carcinoma. World J Gastroenterol. 2011;17:4258–70. doi: 10.3748/wjg.v17.i38.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang W, Gao X, Han Y, et al. Gene expression profiling–derived immunohistochemistry signature with high prognostic value in colorectal carcinoma. Gut. 2014;63:1457–67. doi: 10.1136/gutjnl-2013-305475. [DOI] [PubMed] [Google Scholar]
- 35.Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa H, Shibata T. Comprehensive genome sequencing of the liver cancer genome. Cancer Lett. 2013;340:234–40. doi: 10.1016/j.canlet.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 37.Orito E, Mizokami M, Ina Y, et al. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci U S A. 1989;86:7059–62. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vartanian JP, Henry M, Marchio A, et al. Massive apobec3 editing of hepatitis B viral dna in cirrhosis. PLoS Pathog. 2010;6:e1000928. doi: 10.1371/journal.ppat.1000928. [Available online at: http://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1000928; cited 21 June 2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus dna strands by apobec3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:8321–6. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi C, Ishino H, Tsuge M, et al. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626–33. doi: 10.1002/hep.20580. [DOI] [PubMed] [Google Scholar]
- 41.Henry M, Guétard D, Suspène R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of hbv dna by monodomain human apobec3 cytidine deaminases and the recombinant nature of apobec3G. PLoS One. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [Available online at: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0004277; cited 21 June 2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pérez-Durán P, Belver L, de Yébenes VG, Delgado P, Pisano DG, Ramiro AR. ung shapes the specificity of aid-induced somatic hypermutation. J Exp Med. 2012;209:1379–89. doi: 10.1084/jem.20112253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kavli B, Sundheim O, Akbari M, et al. hung2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded dna, with hsmug1 as a broad specificity back up. J Biol Chem. 2002;277:39926–36. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 44.Hannoun C, Horal P, Lindh M. Long-term mutation rates in the hepatitis B virus genome. J Gen Virol. 2000;81:75–83. doi: 10.1099/0022-1317-81-1-75. [DOI] [PubMed] [Google Scholar]
- 45.Tran A, Kremsdorf D, Capel F, et al. Emergence of and take-over by hepatitis B virus (hbv) with rearrangements in the pre-S/S and pre-C/C genes during chronic hbv infection. J Virol. 1991;65:3566–74. doi: 10.1128/jvi.65.7.3566-3574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, Tai AW, Tong S, Lok AS. hbv core promoter mutations promote cellular proliferation through e2f1-mediated upregulation of S-phase kinase-associated protein 2 transcription. J Hepatol. 2013;58:1068–73. doi: 10.1016/j.jhep.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Tong S, Tai AW, Hussain M, Lok AS. Hepatitis B virus core promoter mutations contribute to hepatocarcinogenesis by deregulating skp2 and its target, p21. Gastroenterology. 2011;141:1412–21. doi: 10.1053/j.gastro.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–21. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 49.Yuan JM, Ambinder A, Fan Y, Gao YT, Yu MC, Groopman JD. Prospective evaluation of hepatitis B 1762(T)/1764(A) mutations on hepatocellular carcinoma development in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2009;18:590–4. doi: 10.1158/1055-9965.EPI-08-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh CT, So M, Ng J, et al. Hepatitis B virus-dna level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology. 2010;52:1922–33. doi: 10.1002/hep.23898. [DOI] [PubMed] [Google Scholar]
- 51.Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–43. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang ZL, Sabin CA, Dong BQ, et al. hbv A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103:2254–62. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sze KM, Chu GK, Lee JM, Ng IO. C-Terminal truncated hepatitis B virus X protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57:131–9. doi: 10.1002/hep.25979. [DOI] [PubMed] [Google Scholar]
- 54.Yip WK, Cheng AS, Zhu R, et al. Carboxyl-terminal truncated HBx regulates a distinct microrna transcription program in hepatocellular carcinoma development. PLoS One. 2011;6:e22888. doi: 10.1371/journal.pone.0022888. [Available online at: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0022888; cited 21 June 2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan X, Zhai Y, Chang W, et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123:1080–8. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]