Abstract
Lysine acetylation (AcK), a posttranslational modification wherein a two-carbon acetyl group binds covalently to a lysine residue, occurs prominently on mitochondrial proteins and has been linked to metabolic dysfunction. An emergent theory suggests mitochondrial AcK occurs via mass action rather than targeted catalysis. To test this hypothesis we performed mass spectrometry-based acetylproteomic analyses of quadriceps muscles from mice with skeletal muscle-specific deficiency of carnitine acetyltransferase (CrAT), an enzyme that buffers the mitochondrial acetyl-CoA pool by converting short-chain acyl-CoAs to their membrane permeant acylcarnitine counterparts. CrAT deficiency increased tissue acetyl-CoA levels and susceptibility to diet-induced AcK of broad-ranging mitochondrial proteins, coincident with diminished whole body glucose control. Sub-compartment acetylproteome analyses of muscles from obese mice and humans showed remarkable overrepresentation of mitochondrial matrix proteins. These findings reveal roles for CrAT and L-carnitine in modulating the muscle acetylproteome and provide strong experimental evidence favoring the nonenzymatic carbon pressure model of mitochondrial AcK.
The recent epidemic surge in the rates of obesity and closely related metabolic diseases has sparked intense research aimed at understanding the cellular and molecular consequences of persistent overnutrition (Ogden et al., 2014). Among many adverse outcomes of chronic positive energy imbalance is a steady decay in mitochondrial performance (Lowell and Shulman, 2005). These organelles are increasingly recognized as a key regulatory hub for processes such as nutrient sensing, retrograde signaling, autophagy and cell survival, in addition to their well-established roles in ATP production and cellular bioenergetics (Pagliarini and Rutter, 2013). Accordingly, disease-associated perturbations in mitochondrial quality and function have broad-ranging clinical and therapeutic implications.
While many disease states are characterized by perturbed expression of multiple genes involved in respiratory function (Mootha et al., 2003), dysregulation at the genomic level does not fully explain the changes in mitochondrial bioenergetics frequently associated with obesity and diabetes (Holloszy, 2009). Also contributing to obesity-induced perturbations in mitochondrial performance are several posttranslational modifications (PTMs) that modulate stability, turnover, and/or function of mitochondrial proteins. Recent applications of mass spectrometry has drawn attention to lysine acetylation as a prominent mitochondrial PTM that is increasingly recognized as a marker of cellular energy stress (Dittenhafer-Reed et al., 2015; Hebert et al., 2013; Kendrick et al., 2011; Rardin et al., 2013; Still et al., 2013). Protein acetylation is a reversible modification in which a two-carbon acetyl group is covalently bound to the ε-amino group of a lysine residue (Anderson and Hirschey, 2012). A growing number of reports provide evidence that acetylation of certain lysines can affect mitochondrial protein interactions, function and/or enzymatic activities (Bharathi et al., 2013; Hirschey, 2011; Hirschey et al., 2010; Hirschey et al., 2011; Jing et al., 2011; Still et al., 2013). The strongest evidence that these PTMs can impart adverse physiologic consequences comes from mice lacking sirtuin 3 (SIRT3), a NAD+-dependent deacetylase that removes acetyl groups from specific lysine residues (Hebert et al., 2013; Newman et al., 2012; Rardin et al., 2013). SIRT3-deficient mice display varying degrees of increased mitochondrial protein acetylation within key metabolic tissues and develop symptoms reminiscent of the metabolic syndrome when challenged by high fat feeding (Dittenhafer-Reed et al., 2015; Hirschey et al., 2011; Lantier et al., 2015).
Whereas this field has been steadily gaining knowledge about the enzymes and physiological circumstances that regulate mitochondrial protein deacylation, the biological factors that influence the addition of acetyl groups to lysine side chains remain poorly understood. One idea gaining increasing traction suggests that unlike acylation reactions in other subcellular compartments, acetylation of mitochondrial proteins occurs largely through non-enzymatic mechanisms as a consequence of mass action, rather than targeted catalysis (Ghanta et al., 2013; Wagner and Payne, 2013). This model predicts that physiological and nutritional conditions that raise mitochondrial concentrations of acetyl-CoA “push” these protein modifications by expanding the local pool of acetyl donors. Relevant to this hypothesis is evidence that overfeeding results in incomplete oxidation of carbon fuels, reflected by increased accumulation of mitochondrial-derived acylcarnitine species that originate from their corresponding acyl-CoA precursors (Koves et al., 2008). Taken together, these findings imply that chronic energy surplus results in a mismatch between substrate supply and demand, which in turn increases mitochondrial carbon load (Muoio, 2014). Also related to this general model are recent studies showing that mitochondrial acetyl-CoA balance can be nutritionally regulated via the carnitine-dependent enzyme, carnitine acetyltransferase (CrAT). This enzyme is most abundant in skeletal muscle and heart and localizes to the mitochondrial matrix (Muoio et al., 2012; Noland et al., 2009; Seiler et al., 2015). The freely reversible CrAT reaction interconverts short chain acyl-CoAs and their corresponding carnitine conjugates. Importantly, unlike their acyl-CoA precursors, acylcarnitine metabolites can traverse the inner mitochondrial membrane and thereby permit mitochondrial efflux of excess acyl moieties. These findings raise the intriguing possibility that this acetyl group buffering system plays a key role in mitigating nutrient-induced acetylation of mitochondrial proteins.
The goal of the current study was to test the hypothesis that macronutrient load and carnitine-mediated acetyl group buffering directly impact mitochondrial protein acetylation in skeletal muscle. To this end, we employed a label-free quantitative mass spectrometry-based acetylproteomics approach to examine the consequences of skeletal muscle-specific CrAT deficiency on diet-induced acetylation of mitochondrial proteins. Our findings not only establish important roles for CrAT and L-carnitine in modulating the muscle acetylproteome, but also provide strong experimental evidence that advances the “carbon pressure” concept of mitochondrial lysine acetylation.
RESULTS
Carnitine and CrAT mitigate acetyl-CoA-induced acetylation of mitochondrial proteins in vitro
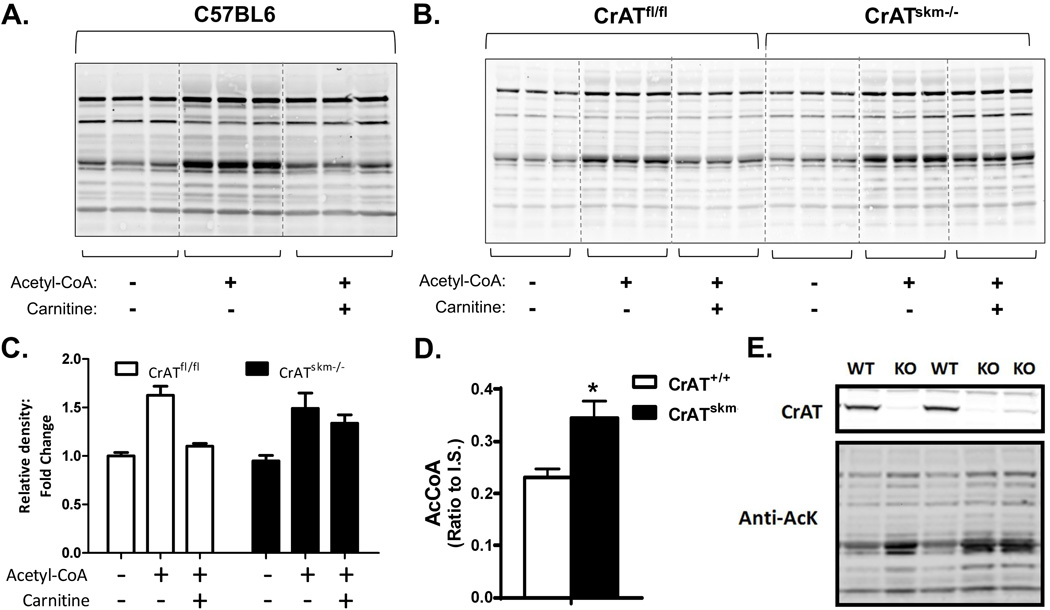
Provision of L-carnitine to muscle mitochondria permits CrAT-mediated conversion of acetyl-CoA to acetylcarnitine (Noland et al., 2009). The potential role of L-carnitine in offsetting acetyl-CoA-induced lysine acetylation of skeletal muscle mitochondrial proteins was initially tested in vitro using a permeabilized mitochondrial incubation system combined with immunoblot analysis using an anti-acetyllysine antibody. Consistent with previous reports (Wagner and Payne, 2013), total acetylation of proteins in isolated mitochondria increased when the concentration of acetyl-CoA in the incubation buffer was raised from 0 to 2.5 mM, the latter of which is within the estimated physiologic range (Hansford and Johnson, 1975; Wagner and Payne, 2013) (Fig. 1a). Addition of 5 mM L-carnitine resulted in a near-complete block of detectable acetyl-CoA-induced acetylation of several mitochondrial proteins, whereas other bands were unaffected (Fig. 1a). To determine whether this effect of L-carnitine depends on the acetyl-CoA-buffering activity of CrAT, we repeated similar experiments using mitochondria prepared from CrATskm−/− mice, which lack the enzyme specifically in skeletal muscle, as well as CrATfl/fl littermate control animals (Seiler et al., 2015). As anticipated, L-carnitine failed to defend against protein acetylation when provided to mitochondrial lysates prepared from CrAT-deficient muscles (Fig. 1b–c). Abundance of the mitochondrial electron transferring flavoprotein (ETF), used as a loading control, was unaffected by genotypes and carnitine treatment.
Figure 1. Carnitine and CrAT mitigate acetyl-CoA-induced acetylation of mitochondrial proteins in vitro.
Representative anti-acetyllysine Western blots of permeabilized mitochondria from quadriceps muscle from (a) C57BL6 mice or (b) Control (CrATfl/fl) and CrAT-deficient (CrATskm−/−) mice, incubated ±2.5 mM acetyl-CoA and ±5 mM L-carnitine (60 min, 32°C, pH 8.0). (c) Corresponding quantification of the band intensities (entire lane) for panel b. Quadriceps muscles from CrATfl/fl and CrATskm−/− mice fed a high fat diet for 12 weeks were used for (d) measurement of tissue acetyl-CoA levels normalized to an internal standard (IS) (data are mean±SEM; n=5) and (e) Western blot analysis of CrAT protein (upper panel) and lysine acetylation (lower panel) in mitochondria isolated from CrATskm−/− (KO) and CrATfl/fl mice (WT).
Results of the in vitro assays strongly implied a role for the CrAT/L-carnitine system in regulating mitochondrial protein acetylation. To assess this possibility in vivo, we evaluated total lysine acetylation by Western blot analysis of isolated muscle mitochondria harvested from CrATfl/fl and CrATskm−/− mice fed a high fat (HF) diet for 12 weeks. Notably, CrAT deficiency increased tissue levels of acetyl-CoA (Fig. 1d) as well as total abundance of lysine-acetylated proteins (Fig. 1e). Considering that CrAT resides in the mitochondrial matrix, we presume the increase in tissue acetyl-CoA content reflects changes in this compartment specifically.
High fat feeding expands the detectable acetyl-proteome in skeletal muscle
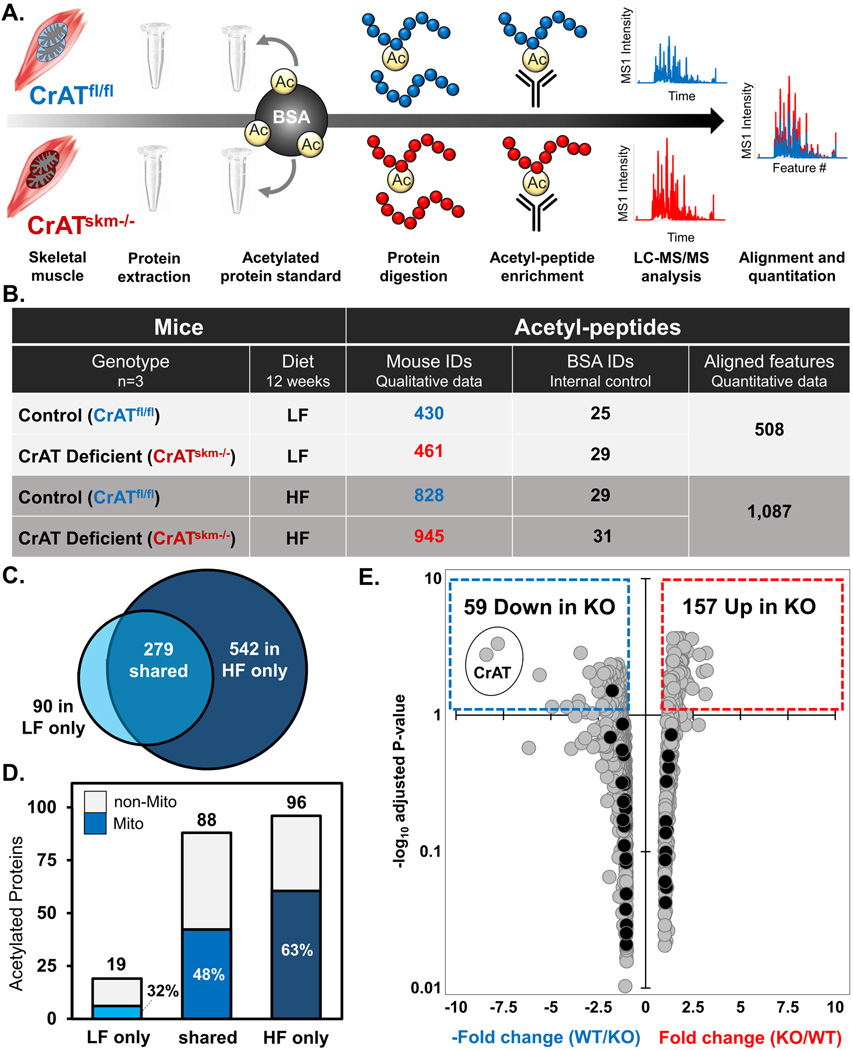
To gain further insights into the impact of nutrient load and CrAT activity on the muscle acetylome, we began by evaluating the inventories of acetylated peptides detected in quadriceps tissue from CrATfl/fl control mice fed either a low fat (LF) or HF diet. Our proteomics strategy (Fig. 2a) combined enzymatic digestion of the whole-tissue lysate, antibody-based acetylpeptide enrichment, and nanoflow reverse-phase liquid chromatography (LC)-MS/MS. The acetylpeptides identified at a 1% false discovery rate (FDR) mapped to a total of 911 non-redundant sites of acetylation, of which 279 were identified in both diets, 542 were identified only in mice fed the HF diet, and 90 were identified only in the LF group (Fig. 2b–c). This qualitative analysis suggested that the size of the acetylproteome, defined by the number of acetylation sites identified, more than doubled (222% increase) in response to chronic HF feeding; whereas the identification of internal standard acetylated BSA peptides, spiked at 5 ppm into all samples, was similar between diets (Fig. 1b). Additionally, when represented by proteins containing at least one identified acetylation site, the identified protein acetylome expanded by nearly this same amount (172%) in the HF compared to LF diet condition (Fig. 2d). Thus, in addition to increasing the number of acetylation sites identified at a fixed FDR, the HF diet appeared to augment the total number of proteins harboring at least one detectable acetyl-lysine modification. To begin assessing characteristics shared among muscle proteins with diet-induced alterations in acetylation, we used the opensource web tool db2db (Mudunuri et al., 2009) to quantify the percentage of proteins that localize to the mitochondrial, cytosolic, and/or nuclear compartments in both the LF and HF conditions. Interestingly, 63% of the acetylated proteins identified exclusively in the HF diet group are resident mitochondrial proteins, whereas those specific to the LF group were comprised of only 32% mitochondrial proteins (Fig. 2d).
Figure 2. High fat feeding expands the detectable acetyl-proteome in skeletal muscle.
(a) Schematic of the experimental workflow. After 12 weeks of feeding either a low fat (LF) or high fat (HF) diet, mouse quadriceps muscles were excised and protein extracted. Acetylated BSA standard was added to each sample prior to protein digestion. Peptides were immunoprecipitated (IP) with anti-acetyl lysine antibody and acetylpeptides were analyzed using high resolution LC-MS/MS to evaluate the acetylpeptide inventory and perform label-free quantitation. (b) Qualitative summary and data metrics for acetylpeptides identified at 1% FDR in quadriceps muscles from CrATskm−/− and CrATfl/fl mice. (c) Venn diagram showing overlap between non-redundant sites of acetylation mapped by acetylpeptides identified at 1% FDR in quadriceps muscles from CrATfl/fl mice fed a LF compared to HF diet. (d) Corresponding bar graph indicating the number of skeletal muscle proteins from CrATfl/fl mice containing at least one acetylation site, with the dark-shaded region indicating the fraction localizing to the mitochondrion. (e) Volcano plot of fold-change vs. significance of quantified acetylpeptides in CrATskm−/− compared to CrATfl/f mice fed a HF diet. The x-axis indicates magnitude (arithmetic scale) and directionality (negative numbers indicate a decrease in KO) of abundance changes and the y-axis shows the adjusted P-values in −log10 space. Significantly down-regulated and up-regulated peptides (adjusted P<0.05, or −log10 adjusted P>1.3) in CrATskm−/− quadriceps are contained within the dotted boxes in blue and red, respectively.
CrAT deficiency exacerbates nutrient-induced acetylation of mitochondrial proteins
To gain further insights into the role of CrAT in influencing mitochondrial protein acetylation we next performed a label-free quantitative protein acetylomic comparison of quadriceps muscles from CrATfl/fl and CrATskm−/− animals, fed either a LF or HF diet for 12-weeks. This label-free quantitation strategy included the CrATfl/fl animals used for the forgoing qualitative diet comparison (Fig. 2c–d), but now with the addition of genotype comparisons (Fig. 2a–b). Alignment of mass spectral features between runs was performed informatically for quantitative assessment of the area under the MS1-extracted ion chromatograms for each feature (i.e., acetylpeptide). Preliminary analysis showed that the impact of CrAT deletion was much more pronounced in the context of diet-induced obesity; thus we proceeded to analyze the dataset from mice consuming the HF diet in greater detail (see Supplemental Table 1 for in-depth analysis of the HF diet acetylproteomic data).
In mice fed the HF diet, we identified 1,087 aligned peptide features containing at least one acetylated lysine residue at a 1% FDR (Fig. 2b,e, Supplemental Table 1). These acetylpeptides mapped to a total of 236 acetylated proteins. For quantitative comparison between genotypes, acetylpeptides were selected and robust mean normalization (top and bottom 10% signals excluded) was performed to reduce quantitative bias. Whereas 116 proteins were assigned only 1 quantified acetylpeptide, an additional 100 proteins had multiple acetylpeptides. A small number (16 proteins) had more than 10 sites quantified, while 138 quantified acetylpeptides were assigned to a single structural protein, myosin 4. Considering a predicted quantitative FDR of less than 5% (P<0.05, after adjusting for multiple hypotheses), 157 acetypeptides increased with loss of CrAT (Fig. 2e). Of the 59 acetylpeptides that decreased with loss of CrAT expression, two mapped to the CrAT enzyme itself (circled in Fig. 2e). Only one internal standard acetylpeptide (mapping to the yeast alcohol dehydrogenase internal control added post-IP) was among the 216 acetylpeptides passing our significance threshold, whereas the other 35 internal standard acetylpeptides (all from acetylated BSA added prior to digestion, see black dots in Fig. 2e) were unchanged, yielding a calculated quantitative FDR of less than 1% (1 out of 216), well below our predicted quantitative FDR (5%).
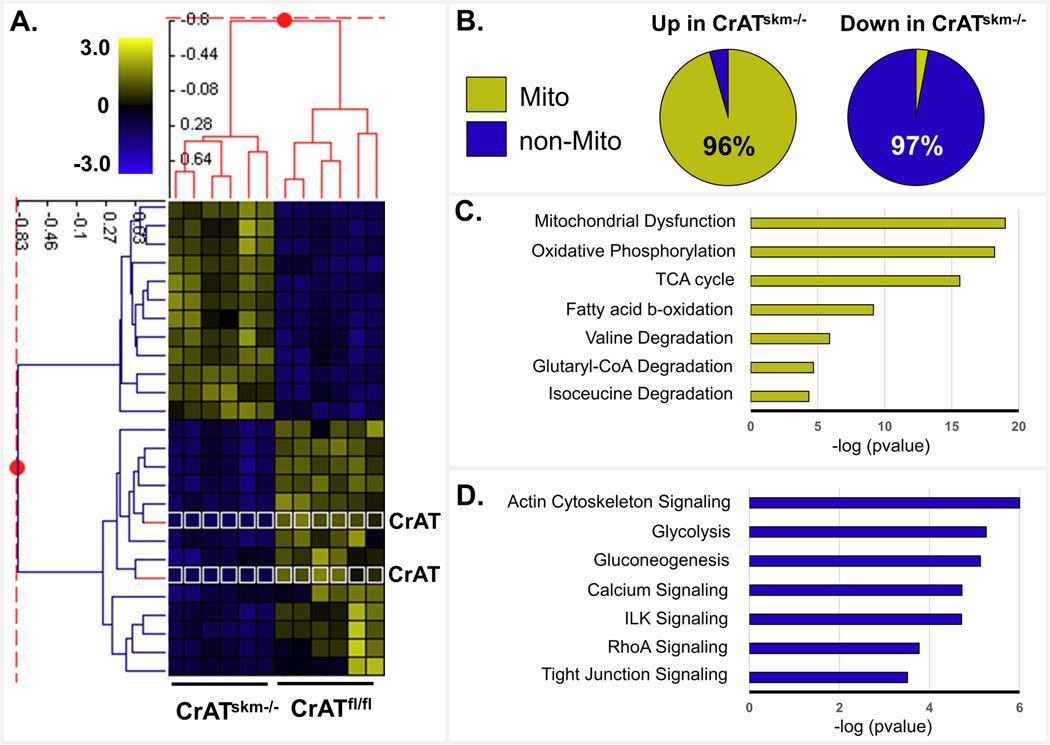
Using z-score transformed data we performed a two-dimensional agglomerative cluster analysis of 32 acetypeptides that differed in abundance between genotypes by at least 2.0-fold (adjusted P<0.05). This analysis revealed a robust and perfectly sorted genotype-specific acetylation pattern wherein acetylpeptides increased (yellow) in the CrATskm−/− muscles belonged exclusively to mitochondrial proteins (Fig. 3a). Acetylpeptides that decreased (blue) in response to CrAT deficiency belonged exclusively to cytosolic proteins, with the single exception of CrAT itself (highlighted by a white outline around the heat maps pixels in Fig. 3a), which was present only in the CrATfl/fl control group and found to be acetylated on two lysine residues. To investigate the possibility of coordinated changes within common cellular pathways, we lowered the differential abundance threshold to 1.5 fold (adjusted P<0.05), which revealed 91 acetylpeptides—belonging to 55 proteins—that were more abundant in the CrATskm−/− compared to CrATfl/fl muscles (see Supplemental Table 1). Of those 55 proteins, 96% were assigned to the mitochondrial compartment (Fig. 3b) and encompassed a broad range of bioenergetic processes as determined by Ingenuity Pathway Analysis (www.Ingenuity.com) (Fig. 3c). A much a smaller subset of 37 acetylated peptides were less abundant in the CrATskm−/− muscles. Disregarding the genetically-engineered decrease in CrAT and the internal standard that was added (both highlighted in Fig. 2e), 97% of the acetylpeptides that decreased with CrAT deficiency belonged to non-mitochondrial proteins (Fig. 3b) involved in muscle contraction, structural components, glucose metabolism, and vesicle trafficking (Fig. 3d). Western blot analysis of total abundance of several mitochondrial proteins, including citrate synthase and multiple oxidative phosphorylation complex subunits, were unremarkable, suggesting that loss of CrAT did not alter relative mitochondrial mass (Supplemental Fig. 1a–b). Protein levels of SIRT3 were likewise unaffected by genotype (Supplemental Fig. 1c).
Figure 3. CrAT deficiency exacerbates nutrient-induced acetylation of mitochondrial proteins.
Acetylproteomic analyses were performed on quadriceps muscles from CrATskm−/− and CrATfl/f mice fed a high fat diet for 12 weeks. (a) Two-dimensional agglomerative cluster analysis using z-score transformed data showing 32 acetypeptides that differed in abundance between genotypes by at least 2.0-fold (adjusted P<0.05). Yellow indicates acetylpeptides increasing up to 3-fold, and blue indicates acetylpeptides decreasing up to 3 fold in the CrATskm−/− mice relative controls. Pixels outlined in white indicate CrAT acetylpeptides. (b) Localization annotation (Gene ontology) was assessed for 154 acetylpeptides (mapping to 69 proteins) that were increased at least 1.5-fold (Up) and 27 acetylpeptides (mapping to 18 proteins) that were decreased at least 1.5-fold (Down) in CrATskm−/− mice relative to controls. For both pie charts, the yellow area represents the percentage of peptides mapping to mitochondrial-localized proteins (96% of those increased), with blue indicating the percentage of peptides mapping to non-mitochondrial proteins (97% of those decreased). (c) Pathway assessment (Ingenuity algorithm) for acetylation events that increase in CrATskm−/− mice relative to controls. The bar graph indicates the significance (−log pvalues) of each pathway as being enriched for changing acetylation. (d) Pathway analysis (as in panel c) for acetylation events that decrease in CrATskm−/− mice relative to controls.
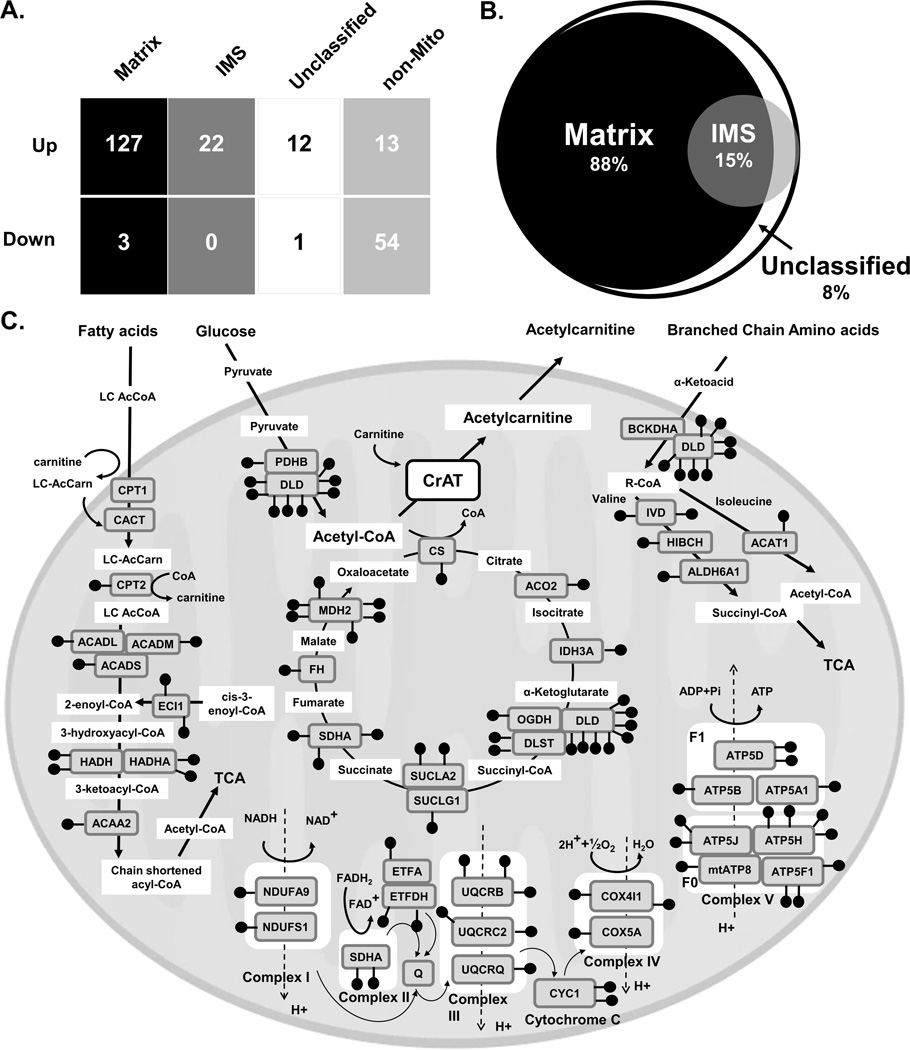
To evaluate sub-organelle localization of acetylated proteins we analyzed our list of acetylated peptides against the Broad Institute’s MitoCarta compendium (Pagliarini et al., 2008)—an expansive inventory of mitochondrial proteins and their sub-compartment localization within the matrix and inner membrane space (IMS) (Hung et al., 2014; Rhee et al., 2013) (Fig. 4a, Supplemental Table 1). Interestingly, whereas only 42% of proteins in MitoCarta map to the matrix (Supplemental Fig. 2a), 81% of the acetylpeptides identified in the present study were assigned to matrix proteins (Supplemental Fig. 2b), along with 88% of the mitochondrial acetylpeptides (127/144) that increased significantly (adjusted P<0.05) with CrAT deficiency (Fig. 4b). Only three matrix acetylpeptides decreased in CrATskm−/− mice (Fig. 4a) (two belonging to CrAT). Although 15% of mitochondrial-localized acetylpeptides that increased with CrAT deletion were assigned to IMS proteins, this subset was enriched for proteins that dually localize to the matrix, presumably proteins that span the inner mitochondrial membrane. Manual inspection of acetylpeptides increased with CrAT deletion but currently lacking sub-mitochondrial classification revealed that these too are likely to reside in the matrix. Figure 4c provides a map of mitochondrial acetylation sites in pathways that were found to enriched for hyperacetylation in CrAT deficient muscles, illustrating the broad range of metabolic pathways affected. To determine whether a similar acetylproteomic profile would emerge from humans, we analyzed quadriceps biopsies from nine overweight subjects (assessed in three pooled samples of three), all with BMI≥29. We identified a total of 515 acetylpeptides at a 1% FDR, 16 of which were generated from our acetylated BSA internal standard (Supplemental Table 2). A subset of 198 acetylpeptides mapped to mitochondrial proteins, of which 163 belong to proteins that localize to the mitochondrial matrix (Supplemental Fig. 2d), consistent with the animal models. Moreover, analysis of both mouse (Supplemental Fig. 2a–b) and human (Supplemental Fig. 2c–d) muscle tissues showed that the relative number of acetylation events on matrix (compared to non-matrix) targets exceeds the relative distribution of proteins comprising these compartments by a factor of two (i.e. 81–82% of acetylated mitochondrial peptides map to matrix proteins whereas the total mitochondrial proteome consists of only 41–42% matrix proteins). Taken together, these results supports the notion that mitochondrial matrix proteins are particularly susceptible to non-enzymatic lysine acetylation in response to chronic caloric surplus (Ghanta et al., 2013; Wagner and Payne, 2013).
Figure 4. Mitochondrial matrix proteins are highly susceptible to lysine acetylation induced by diet and CrAT deficiency.
(a) Table showing number of acetylated peptides significantly changing in CrATskm−/− mice relative to CrATfl/f controls in the context of the HF diet, binned by mitochondrial localization. Analysis was performed by mapping the current dataset to the Broad Institute’s MitoCarta proteome inventories of the mammalian mitochondrion, the mitochondrial matrix (Matrix) and the mitochondrial inner membrane space (IMS). Acetylpeptides mapping to proteins with overlapping localization to both the Matrix and IMS were considered for both categories. MitoCarta proteins that were not found to be Matrix or IMS were considered “Unclassified” and proteins not contained in the MitoCarta were considered non-mitochondrial (non-Mito). (b) Venn Diagram showing the submitochondrial localization of acetylation events increasing in CrATskm−/− relative to CrATfl/f controls. The white circle represents the relative number of acetylpeptides across the entire mitochondrion, black represents the matrix, and grey represents the IMS, with compartmental overlap depicted. (c) Map of mitochondrial metabolic pathways with increased abundance of acetylated proteins in muscle from CrATskm−/− mice. Black “lollypops” depict acetylpeptides within pathways detected by the Ingenuity algorithm as having increased acetylation in CrATskm−/− mice (Fig. 3c), with each symbol representing an individual acetylated lysine.
In silico structural analysis and modeling of acetylpeptides and proteins increased by CrAT deficiency
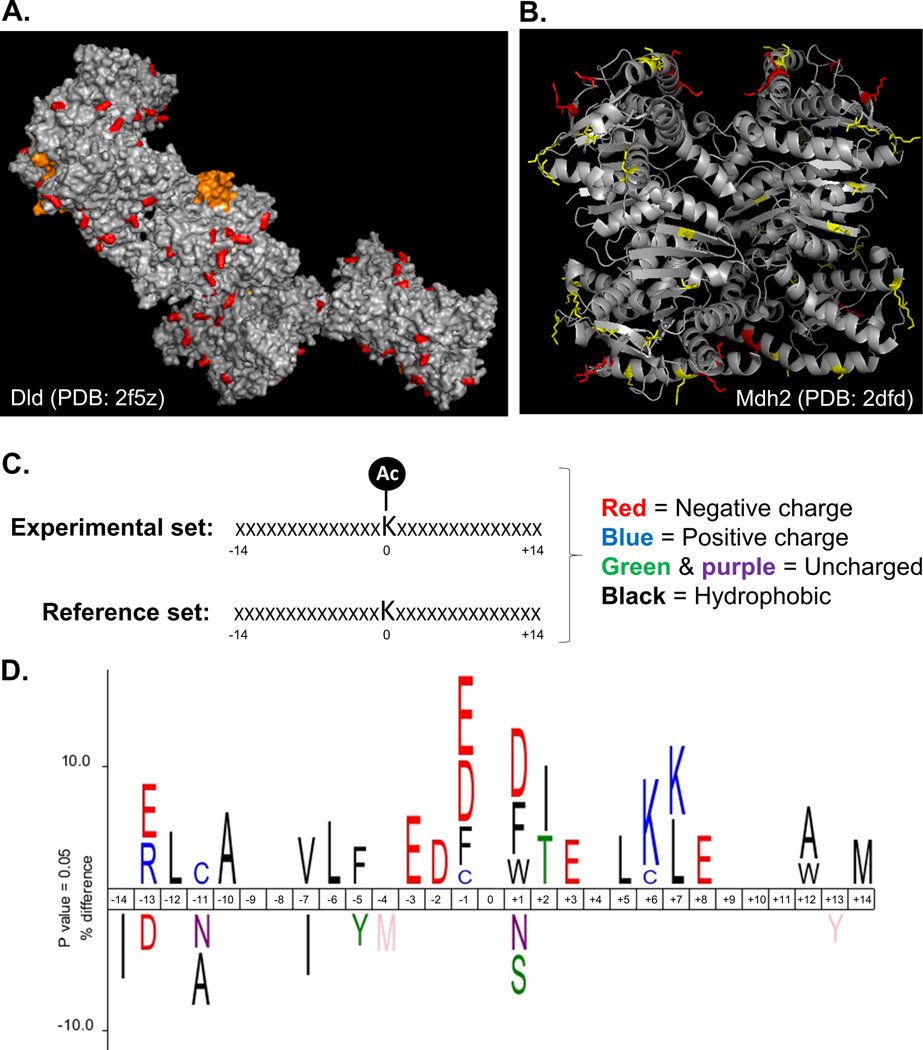
We next sought to gain insight into the tertiary environments of the acetylated mitochondrial residues affected by CrAT deficiency, presuming that acetylation events driven by mass action would tend to occur on exposed sites that are more likely to encounter an acetyl donor. To this end, we performed in silico homology-based three-dimensional structure modeling using the opensource SWISS-Model Repository (Kiefer et al., 2009), which was applied to only those peptides (40 total) belonging to mouse proteins (23 total) with a sequence identity of ≥90% when compared to the protein used to generate the crystal structure. Interestingly, 100% of the 40 acetyl-lysine residues meeting the above criteria were predicted as solvent exposed sites that protrude away from the surface of the protein. For example, Figure 5a shows a pymol-generated image of the acetyl sites (in red) identified on the 11-subunit crystal structure of dihydrolipoamide dehydrogenase (Dld), which functions as the E3 subunit of several dehydrogenase complexes, including PDH, αKDH and BCKD. Figure 5b provides a model of the secondary structure locations of acetyl sites identified within the tetrameric structure of Mdh2, the mitochondrial isoform of malate dehydrogenase.
Figure 5. In silico structural analysis and modeling of acetylpeptides and proteins increased by CrAT deficiency.
(a) Structure (PDB: 2f5z) of the E3 complex of pyruvate dehydrogenase (Dld) showing acetylated lysine residues (AcK) that increased in the CrATskm−/− (highlighted in red). Residues in orange represent the FAD-linked lipoamide arm of the enzyme. Modeling was performed using PyMOL. (b) Structure (PDB: 2dfd) of mitochondrial malate dehydrogenase 2 (Mdh2) showing localization of acetylated residues. AcK highlighted in red represent differentially abundant PTMs identified from CrATskm−/− mice, whereas those in yellow were unchanged between genotypes. (c) Icelogo analysis was performed by comparing the amino acid landscape surrounding AcKs identified in CrATskm−/− muscle to a mouse reference set containing information about the identity of the amino acids in the same 28 positions neighboring lysine residues in all mouse proteins. (d) The top results show the enrichment of amino acids seven residues N- (negative) or C- (positive) terminal to acetylated lysines (centered at position “0”) identified to be increased in CrATskm−/− muscle compared to controls after 12 weeks of high fat diet feeding. The bottom results show the enrichment of amino acids surrounding the average lysine present in mouse proteins. Letters are color coded according to amino acid type using the scheme shown in panel c.
The opensource Icelogo web application (Colaert et al., 2009) was then used to visualize conserved sequence patterns in the acetyl peptides identified in CrAT null muscles as compared to those detected in the control group. First, the primary amino acid sequence of the 14 residues flanking each mitochondrial acetyl site was compared against all non-acetylated lysine residues belonging to the same mitochondrial proteins (i.e. reference set) (Fig. 5c). Frequencies of residues that were significantly (adjusted P<0.05) overrepresented (positive y-axis) or underrepresented (negative y-axis) in each position are represented in Figure 5d by the height of the single-letter amino acid abbreviations. As compared to the reference set, analysis of the set of acetyl peptides that were increased in CrAT null muscles revealed a clear overrepresentation of acidic residues (mainly glutamate and aspartate) in the immediate neighborhood of the acetylated lysine, along with a duplex of adjacent lysine residues at positions +6 and +7. A similar but less pronounced pattern emerged when the analysis was performed using mitochondrial acetyl sites that were unchanged between genotypes (not shown). In sum, these observations are consistent with the premise that some lysine residues are more vulnerable to non-enzymatic acetylation than others due to their primary, secondary and tertiary protein structures (Ghanta et al., 2013; Hebert et al., 2013).
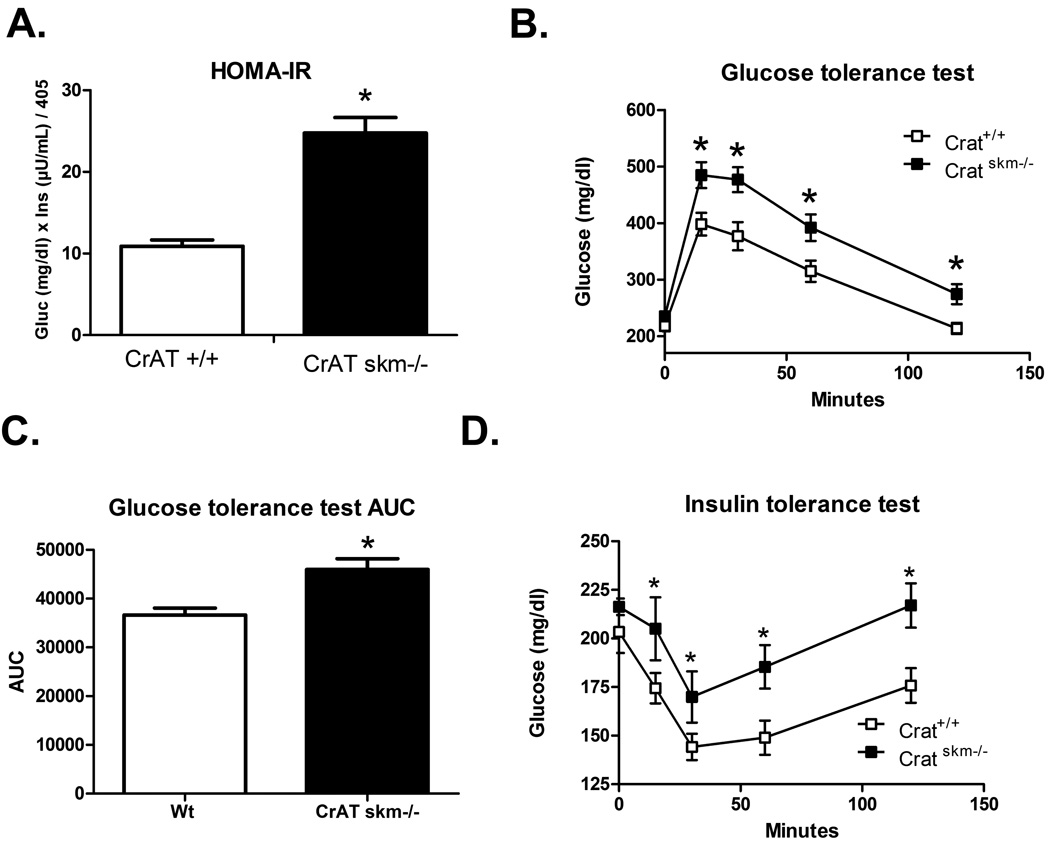
Hyperacetylation of mitochondrial proteins in CrATskm−/− mice is accompanied by impaired whole body glucose homeostasis
Because studies in Sirt3 KO mice have linked hyperacetylation of mitochondrial proteins to increased susceptibility to diet-induced glucose intolerance and muscle insulin resistance (Lantier et al., 2015), we sought to determine whether a similar phenotype would manifest in the CrATskm−/− mice. Indeed, following 12 weeks of HF feeding, the diet-induced decline in whole body glucose homeostasis was more severe in CrATskm−/− as compared to their CrATfl/fl littermates (Fig. 6a–d). Both the HOMA index of insulin resistance (Fig. 6a) and blood glucose excursions during an i.p. glucose tolerance test (Fig. 6b–c) were elevated in CrAT deficient mice, whereas lowering of blood glucose in response to an insulin tolerance test was blunted (Fig. 6d). Although these observations do not prove causation, they are consistent with mounting evidence that an upward shift in the acetylation state of mitochondrial proteins is often accompanied by perturbations in insulin action and whole body metabolic control (Hirschey et al., 2011; Jing et al., 2011; Lantier et al., 2015).
Figure 6. Hyper-acetylation of mitochondrial proteins in CrATskm−/− mice is accompanied by glucose intolerance.
CrATfl/fl and CrATskm−/− mice were fed a high fat diet for 8 weeks prior to assessment of glucose control obtain in the morning after a 2 hour fast. (a) HOMA insulin resistance (IR) index (n = 5–8 per group, *p<0.05 versus CrAT fl/fl by Student’s t-test). (b) Glucose tolerance test performed using i.p. injection of 2g glucose per Kg bodyweight (n = 5–8 per group, *p<0.001 by Student’s t-test). (c) Area under the curve from the tests shown in panel b (*p<0.005 by Student’s t-test). (d) Insulin tolerance test performed using i.p. injection of 0.75U humulin per Kg bodyweight (n = 5–8 per group, *p<0.001 by Student’s t-test).
DISCUSSION
The current study deployed mass spectrometry-based acetylproteomics to examine the interplay between nutrient load and protein acetylation in skeletal muscle, and to test the hypothesis that lysine acetylation of mitochondrial proteins fluctuates in response to local concentrations of acetyl-CoA. This carbon load concept of mitochondrial protein acetylation is gaining increasing support, but has thus far relied on circumstantial evidence. Here, we used CrAT ablation in skeletal muscle as a targeted genetic maneuver to induce a compartment-specific rise in tissue acetyl-CoA levels in the context of contrasting nutrient conditions. We report two major findings. First, HF feeding resulted in an approximate doubling of the number of unique acetylation sites and acetylated proteins identified in whole muscle tissue, and the subset of acetylpeptides detected only in the context of HF feeding belong to proteins that reside predominately in the mitochondrion. Secondly, genetic disruption of mitochondrial acetyl group buffering capacity due to deletion of the CrAT enzyme increased muscle abundance of a large number of lysine acetylpeptides belonging to mitochondrial proteins. Further, while our acetyl proteomic analysis was conducted with whole-tissue lysates, the effect of CrAT deficiency was largely specific to the mitochondrial matrix compartment and detected principally under conditions of chronic overfeeding. These results provide strong in vivo experimental evidence directly linking perturbations in mitochondrial acetyl group balance to broad-scale changes in the mitochondrial matrix acetylproteome. The findings also establish a role for the L-carnitine/CrAT buffering system in regulating protein acetylation within the mitochondrial matrix, which may help to explain why total acetylation of mitochondrial proteins trends lower in skeletal muscle as compared to other metabolic tissues (e.g. liver) when measured in the context of a standard chow diet (Dittenhafer-Reed et al., 2015; Overmyer et al., 2015).
The importance of acetyl modifications in regulating subcellular localization, stability and activity of cytoplasmic and nuclear proteins is well established (Glozak et al., 2005; Ozden et al., 2011). Acetylation events occurring in these compartments are catalyzed by several known protein acetyltransferase (KATs) that are counteracted by proteins belonging to the histone deacetylase and sirtuin deacetylase families. Interest in mitochondrial protein acetylation has escalated with the discovery that a subset of the sirtuin deacylases resides in the mitochondrial matrix (SIRT3–5), implying important regulatory roles for these PTMs in oxidative metabolism. Studies of mitochondrial protein acetylation have centered on characterizing overall coverage of acetyl-occupied lysine residues and their potential regulation by SIRT3. Notably however, SIRT3 acts only to counterbalance the acetylation side of the equation. Delineating the factors that influence the on-rate of these reactions could be equally important. Because compelling evidence of a bona fide mitochondrial-localized KAT has yet to emerge, mounting speculation posits that mitochondrial lysine acetylation occurs non-enzymatically via mass action when a deprotonated lysine side-chain reacts with the thioester of acetyl-CoA. Fitting with this prediction, the present study showed that CrAT loss-of-function increased tissue levels of acetyl-CoA and exacerbated the impact of HF feeding on the mitochondrial protein acetylome.
Interestingly, genotype-dependent differences in muscle acetyl-CoA content were statistically significant, but relatively modest. However, it is important to consider that measurements made in whole tissue at a single time point probably underestimate dynamic physiological fluctuations occurring specifically within the mitochondrial matrix, and/or the microenvironments surrounding the affected metabolic enzymes. Additionally, transient spikes in the availability of acetyl donors might be quickly buffered by the mitochondrial proteome. For example, a recent study that quantified site-specific stoichiometry of the entire E.coli proteome estimated that the amount of acetate on proteins is approximately 2 mM (Baeza et al., 2014). We therefore suspect that much of the acetyl-CoA surplus in CrATskm−/− mice is sequestered by mitochondrial proteins, in essence trapping carbons within this compartment. This explanation could account for the finding that CrAT deficiency resulted in reduced acetylation of proteins residing outside the mitochondria.
Acetyl-CoA holds a prominent position in mitochondrial metabolism as the universal oxidative end product of glucose, fatty acid, and amino acid catabolism. Additionally, pumping of protons from the mitochondrial lumen to the inner membrane space, which is mediated by the electron transport chain, maintains the matrix pH within an alkaline range closer to the pKa of lysine residues (Ghanta et al., 2013; Wagner and Payne, 2013). Thus, in theory, the chemical conditions of the mitochondrial matrix is particularly conducive to non-enzymatic lysine acetylation, which requires a deprotonated lysine primary amine. A recent in vitro study found that lysine reactivity for non-enzymatic acetylation is site-specific, depending on the primary, secondary and tertiary structure of the protein (Baeza et al., 2015). In the current investigation, analysis of structural and chemical features of the acetylation sites found to be more abundant in CrAT deficient muscles revealed some interesting trends. First, structural predictions based on in silico modeling of mitochondrial protein acetylation sites found to be more abundant in CrAT deficient muscles universally positioned these events on solvent exposed lysine residues that protrude away from the surface of the protein. We interpret this finding to suggest that this subset of acetyl modifications occurred after proper protein folding and complex assembly, which could have implications for understanding the stoichiometry and functional relevance of these events. Secondly, Icelogo analysis of primary sequence motifs revealed an overrepresentation of negatively charged residues, mainly glutamate and aspartate, in the immediate neighborhood of the mitochondrial protein acetylation sites responsive to CrAT inactivation, as well as a slightly more distant cluster of positively charged lysine residues. As recently suggested (Ghanta et al., 2013), this motif could reflect a local environment that effectively lowers the pKa of the targeted lysine, increasing the likelihood of deprotonation and thereby rendering the site susceptible to nucleophilic attack and acyl modification.
An important and yet unanswered question in this field is whether widespread acetylation of mitochondrial proteins represents a bona fide regulatory mechanism, and/or a form of protein damage that comes as a cost of the permissive metabolic environment. As discussed previously, these two possibilities are not mutually exclusive (Ghanta et al., 2013). Thus, high occupancy acetylation sites with perturbed pKa values might have evolved to permit macronutrient-induced modification and regulation of proteins in a manner that optimizes energy partitioning and storage during periods of carbon surplus (Ghanta et al., 2013). Also uncertain is whether global shifts in lysine acetylation have a physiologically relevant impact on cellular function, especially considering that many of these events appear to occur at low stoichiometry (Baeza et al., 2014; Weinert et al., 2015). Nonetheless, a growing number of studies using site-directed mutagenesis strategies show that at least some of these modifications alter activity of mitochondrial proteins, mostly in a negative direction (Hallows et al., 2006; Hallows et al., 2011; Hirschey et al., 2010; Still et al., 2013), although gain-of-function acetylation events have been reported (Fernandes et al., 2015; Lu et al., 2015). Lysine acetylation has also been linked to nutrient control of autophagy and mitophagy (Webster et al., 2014). Although these processes appear to be regulated by acetylation of specific proteins that reside outside the mitochondrial matrix, including some that localize to the outer mitochondrial membrane, the current findings suggest hyperacetylation of the matrix proteome might have a reciprocal effect on AcK events in other compartments. Still, the list of functionally relevant AcK targets is small in comparison to the breadth of sites that have been identified (Choudhary et al., 2009; Dittenhafer-Reed et al., 2015; Hebert et al., 2013; Kim et al., 2006; Rardin et al., 2013). Herein, perturbations in the acetylproteome of the CrATskm−/− mice coincided with impaired glucose tolerance, which aligns with a number of observational studies linking heightened mitochondrial acetylation and/or SIRT3 insufficiency to metabolic dysregulation. Also noteworthy is that obesity and HF feeding have been shown to diminish CrAT activity in skeletal muscle (Lindeboom et al., 2014; Seiler et al., 2014) and L-carnitine supplementation increases tissue acetylcarnitine production and efflux while also improving metabolic control when administered to obese rodents and humans (Noland et al., 2009; Ringseis et al., 2012). Taken together with the current findings, these results prompt speculation that the therapeutic benefits of L-carnitine supplementation might stem from its actions to combat stress-induced acetylation of mitochondrial proteins, including CrAT itself. This is an intriguing possibility now deserving of further investigation.
EXPERIMENTAL PROCEDURES
Animal Studies
All procedures were approved by the Duke University Institutional Animal Care and Use Committee. Mice had free access to food and water and were fed a standard chow (Purina Rodent Chow no. 5001, Purina Mills, St. Louis, MO, USA) prior to being fed a semi-purified 10% fat diet (Research Diets, D12450B) or a 45% fat diet (Research Diets, D03021303) for 12-weeks. The genetic strategy used for generating CrATfl/fl and CrATskm−/− mice was described previously (Seiler et al., 2015). Glucose tolerance and insulin tolerance tests were performed in the morning 1 hour after food withdrawal. Mice were singly housed with access to water (gel pack). After taking glucose reading using a glucometer at the 0 minute time point, mice were given an intraperitoneal injection of 2.0 g glucose per Kg body or 0.08 U/mouse Humulin, both dissolved in saline. Blood for ELISA insulin measurements was collected at 0, 15, and 60 min. At the time of harvest for proteomics analysis, weights of mice fed the high fat diet averaged 38.6 g (1.1 STE, n=5) for CrATfl/fl and 41.1g (1.8 STE, n=8) for CrATskm−/−. At time of the glucose tolerance test, animals weighed 27.5 g (STE 0.8, n=5) for CrATfl/fl and 37.5g (1.6 STE, n=8) for CrATskm−/−.
In Vitro Mitochondrial Incubations
Mitochondria were enriched from CrATfl/fl and CrATskm−/− quadriceps and gastrocnemius as described in the Supplemental Methods. Mitochondrial lysates (100 µg protein) were incubated in CrAT activity buffer (50 mM Tris-HCl, 1 M EDTA, 1 mM Nicatinamide (NAM), 1 µM Trichstatin A (TSA), protease inhibitor cocktail (Sigma #P8340); pH 8.0) at 1ug/uL. Treatments included 2.5 mM Acetyl-CoA and 5 mM L-Carnitine. Mitochondrial lysates were incubated at 32 °C for 1 hr. Reactions were stopped by placing lysates on ice and transferred to the −80 °C.
Metabolic Profiling
Acyl-CoAs were extracted from quadriceps tissue and analyzed by flow injection tandem mass spectrometry as described previously (An et al., 2004; Seiler et al., 2015).
Quantitative proteomics
Pulverized quadriceps tissue samples were solubilized in 8M urea-containing buffer as described in the Supplemental Methods. After 30 ng acetylated-BSA serum albumin (Ac-BSA) was added as an internal standard to each protein sample (5 mg), cysteine residues were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin overnight. Peptide samples were desalted, lyophilized, and subjected to immunoprecipitation with Acetyl-K PTMScan enrichment beads (Cell Signaling Technology). Peptides were eluted, desalted, lyophilized, and re-suspended along with 10 fmol/µL of yeast aldehyde dehydrogenase (ADH1_YEAST) MassPrep Standard (Waters). Samples were subjected to label-free quantitative LC/MS/MS using a nanoAcquity UPLC system (Waters Corp) coupled to a LTQ-Orbitrap XL high resolution accurate mass tandem mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ionization source. The raw data was analyzed using Rosetta Elucidator v3.3 (Rosetta Biosoftware, Inc). All LC-MS/MS runs were aligned using the PeakTeller algorithm (Elucidator) and relative peptide abundances were calculated based on area-under-the-curve (AUC) of aligned features across all runs. This MS/MS data was searched against a custom database of “target” sequences consisting of NCBI RefSeq mus musculus proteins along with internal standards and “decoy” reversed-sequences for false positive rate determination (Elias and Gygi, 2007). Database searching was performed with the Elucidator software package using Mascot search engine (v2.2), assigning precursor ion tolerance of 5 ppm and product ion tolerance of 0.8 Da. Searching allowed variable modification of N and Q (deamidation, +1 Da), M (oxidation, +16 Da), and K (acetylation, +42 Da). After aggregating all search results, annotation (assignment of peptide sequence to the quantitative MS signal) was performed for peptides whose ion score was greater than 30. For the high fat diet cohorts, this resulted in a total of 4204 peptides mapping to 932 proteins, including 1111 peptides containing at least one acetylated K residue, at 1% peptide FDR. For quantitative processing, the data was first curated to contain only high quality peptides with appropriate chromatographic peak shape (1,087 acetylpeptide features in the high fat fed mice). See the Supplemental Methods for more details. Analyzed data for the high fat diet cohorts are included in Supplemental Table 1. Analyzed data for the human pilot study are provided as Supplemental Table 2. All raw mass spectrometry files for the acetylproteomics work described herein are available online through Chorus (https://chorusproject.org, Public Project ID989) via the following links: LF and HF diet mouse acetylproteomics (Public Experiment ID2047): https://chorusproject.org/anonymous/download/experiment/22e926ba037a45d79abb3dd51e24d3df Human pilot study acetylproteomics (Public Experiment ID2048): https://chorusproject.org/anonymous/download/experiment/50216a62d2284df1a991eb2cc96fa1c3
In Silico Analysis
The three dimensional structure analysis was performed using the SWISS-MODEL repository program (Kiefer et al., 2009). “The SWISS-MODEL Repository is a database of annotated three-dimensional comparative protein structure models generated by the fully automated homology-modelling pipeline”. The sequence identity threshold was set at 90% sequence homology. The crystal structure imaging incorporating the acetyl sites was performed using PyMOL. Assigned PDB names: Dld=2f5z, MDH=2dfd The amino acid sequence motif analysis was performed using the Icelogo web analysis tool (Colaert et al., 2009) as described in the figure legends. Frequency analysis was plotted as percent difference with an adjusted P<0.05. For pathway analysis, a list of increased acetylpeptides (1.5-fold increase, adjusted P<0.05) was analyzed through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.Ingenuity.com).
Analyses of human muscle
Acetylproteomic analysis of human muscle was conducted on biopsy specimens obtained from nine individuals, eight female and one male with a mean age of 57, ranging from 55–67 years, and a mean body mass index of 32.8, ranging from 29–35, with fasting blood glucose levels ranging from 82–105 mg/dL.
Supplementary Material
Acknowledgments
The authors thank Muoio laboratory members Dorothy Slentz, April Hawkins, and Dr. Timothy Koves for technical assistance and help with manuscript preparation. We also thank Dr. Randy Mynatt (Pennington Biomedical Research Center) for providing the CrATfl/fl mice. We gratefully acknowledge Drs. David Pagliarini and Brendan Floyd (University of Wisconsin, Department of Biochemistry) for assistance with annotation of sub-mitochondrial protein localization, and Dr. Matthew Hirschey (Duke University, Department of Medicne) for critical review of the manuscript. This work was supported by NIH grants R01-DK089312 (D.M.M), P01-DK058398 (D.M.M), and F32-DK093256 to (M.N.D.), and Pfizer’s Investigator-Initiated Research Program (D.M.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, M.N.D. and D.M.M; Methodology, M.N.D., L.K., J.W.T, L.G.D., R.D.S., O.R.I., P.A.G and D.M.M; Investigation, M.N.D., L.K., J.W.T, L.G.D., P.A.G., and D.M.M; Writing, M.N.D., L.K., J.W.T, P.A.G and D.M.M; Funding Acquisition, M.J.B., T.P.R., and D.M.M; Supervision, J.W.T and D.M.M
REFERENCES
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nature medicine. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Anderson KA, Hirschey MD. Mitochondrial protein acetylation regulates metabolism. Essays in biochemistry. 2012;52:23–35. doi: 10.1042/bse0520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. The Journal of biological chemistry. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS chemical biology. 2015;10:122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, et al. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. The Journal of biological chemistry. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nature methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Dittenhafer-Reed KE, Richards AL, Fan J, Smallegan MJ, Fotuhi Siahpirani A, Kemmerer ZA, Prolla TA, Roy S, Coon JJ, Denu JM. SIRT3 Mediates Multi-Tissue Coupling for Metabolic Fuel Switching. Cell metabolism. 2015;21:637–646. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Weddle A, Kinter CS, Humphries KM, Mather T, Szweda LI, Kinter M. Lysine Acetylation Activates Mitochondrial Aconitase in the Heart. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanta S, Grossmann RE, Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: chemical and metabolic logic of acetyl-lysine modifications. Critical reviews in biochemistry and molecular biology. 2013;48:561–574. doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Molecular cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford RG, Johnson RN. The steady state concentrations of coenzyme A-SH and coenzyme A thioester, citrate, and isocitrate during tricarboxylate cycle oxidations in rabbit heart mitochondria. The Journal of biological chemistry. 1975;250:8361–8375. [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Molecular cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD. Old enzymes, new tricks: sirtuins are NAD(+)-dependent de-acylases. Cell metabolism. 2011;14:718–719. doi: 10.1016/j.cmet.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloszy JO. Skeletal muscle "mitochondrial deficiency" does not mediate insulin resistance. The American journal of clinical nutrition. 2009;89:463S–466S. doi: 10.3945/ajcn.2008.26717C. [DOI] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Molecular cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. The Biochemical journal. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic acids research. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell metabolism. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Lantier L, Williams AS, Williams IM, Yang KK, Bracy DP, Goelzer M, James FD, Gius D, Wasserman DH. SIRT3 is crucial for maintaining skeletal muscle insulin action and protects against severe insulin resistance in high fat fed mice. Diabetes. 2015 doi: 10.2337/db14-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom L, Nabuurs CI, Hoeks J, Brouwers B, Phielix E, Kooi ME, Hesselink MK, Wildberger JE, Stevens RD, Koves T, et al. Long-echo time MR spectroscopy for skeletal muscle acetylcarnitine detection. The Journal of clinical investigation. 2014;124:4915–4925. doi: 10.1172/JCI74830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Lu Z, Chen Y, Aponte AM, Battaglia V, Gucek M, Sack MN. Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate: elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. The Journal of biological chemistry. 2015;290:2466–2476. doi: 10.1074/jbc.M114.606228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mudunuri U, Che A, Yi M, Stephens RM. bioDBnet: the biological database network. Bioinformatics. 2009;25:555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell. 2014;159:1253–1262. doi: 10.1016/j.cell.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, Noland RC, Kovalik JP, Seiler SE, Davies MN, DeBalsi KL, Ilkayeva OR, Stevens RD, Kheterpal I, Zhang J, et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell metabolism. 2012;15:764–777. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. The Journal of biological chemistry. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland RC, Koves TR, Seiler SE, Lum H, Lust RM, Ilkayeva O, Stevens RD, Hegardt FG, Muoio DM. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. The Journal of biological chemistry. 2009;284:22840–22852. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. Jama. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer KA, Evans CR, Qi NR, Minogue CE, Carson JJ, Chermside-Scabbo CJ, Koch LG, Britton SL, Pagliarini DJ, Coon JJ, et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell metabolism. 2015;21:468–478. doi: 10.1016/j.cmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging. 2011;3:102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Rutter J. Hallmarks of a new era in mitochondrial biochemistry. Genes & development. 2013;27:2615–2627. doi: 10.1101/gad.229724.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringseis R, Keller J, Eder K. Role of carnitine in the regulation of glucose homeostasis and insulin sensitivity: evidence from in vivo and in vitro studies with carnitine supplementation and carnitine deficiency. European journal of nutrition. 2012;51:1–18. doi: 10.1007/s00394-011-0284-2. [DOI] [PubMed] [Google Scholar]
- Seiler SE, Koves TR, Gooding JR, Wong KE, Stevens RS, Ilkayeva OR, Wittmann AH, DeBalsi KL, Davies MN, Lindeboom L, et al. Carnitine Acetyltransferase Mitigates Metabolic Inertia and Muscle Fatigue During Exercise. Cell metabolism. 2015;22:65–76. doi: 10.1016/j.cmet.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler SE, Martin OJ, Noland RC, Slentz DH, DeBalsi KL, Ilkayeva OR, An J, Newgard CB, Koves TR, Muoio DM. Obesity and lipid stress inhibit carnitine acetyltransferase activity. Journal of lipid research. 2014;55:635–644. doi: 10.1194/jlr.M043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, et al. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. The Journal of biological chemistry. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. The Journal of biological chemistry. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BR, Scott I, Traba J, Han K, Sack MN. Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochimica et biophysica acta. 2014;1841:525–534. doi: 10.1016/j.bbalip.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO Journal. 2015;34(21):2620–2632. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.