Abstract
One dimensional selective TOCSY experiments have been shown to be advantageous in providing improved data inputs for principle component analysis (PCA) (Sandusky and Raftery 2005a, b). Better subpopulation cluster resolution in the observed scores plots results from the ability to isolate metabolite signals of interest via the TOCSY based filtering approach. This report reexamines the quantitative aspects of this approach, first by optimizing the 1D TOCSY experiment as it relates to the measurement of biofluid constituent concentrations, and second by comparing the integration of 1D TOCSY read peaks to the bucket integration of 1D proton NMR spectra in terms of precision and accuracy. This comparison indicates that, because of the extensive peak overlap that occurs in the 1D proton NMR spectra of biofluid samples, bucket integrals are often far less accurate as measures of individual constituent concentrations than 1D TOCSY read peaks. Even spectral fitting approaches have proven difficult in the analysis of significantly overlapped spectral regions. Measurements of endogenous taurine made over a sample population of human urine demonstrates that, due to background signals from other constituents, bucket integrals of 1D proton spectra routinely overestimate the taurine concentrations and distort its variation over the sample population. As a result, PCA calculations performed using data matrices incorporating 1D TOCSY determined taurine concentrations produce better scores plot subpopulation cluster resolution.
Keywords: 1D TOCSY, Metabolomics, Metabolite profiling, Biofluids, PCA, Quantitative analysis, NMR
Introduction
The field of metabolomics combines high resolution analytical methods such as NMR and/or mass spectrometry with the multivariate statistical analysis of populations of complex matrix samples such as biofluids, foods, etc. (Nicholson et al. 1999; Fiehn et al. 2000; Kell 2004; Fernie et al. 2004; Van der Greef and Smilde 2005; Van Dien and Schilling 2006; Serkova and Niemann 2006; Pan and Raftery 2007; Gowda et al. 2008; Zhang et al. 2010). The principal objective of the statistical analysis of biofluid populations, as it is currently being applied for forensic and diagnostic purposes, is to detect subpopulations within the parent population in such a way that “unknown” samples may be assigned to one or another of the scores plot subpopulations. Standard metabolomic studies often fail because of inadequate subpopulation cluster resolution, as can often be seen in the scores plots of principal component analysis (PCA) or even supervised multivariate methods (such as partial least squares discriminant analysis, PLSDA). Thus, any technical innovation that routinely increases the subpopulation cluster resolution constitutes a significant advance in the field.
In previous publications the authors have demonstrated that using discrete concentration measurements of selected biofluid constituents, made using the 1D TOCSY experiment, as data inputs for PCA calculations will improve scores plot subpopulation cluster resolution over what can be obtained using bucket integrals of 1D proton NMR spectra (Sandusky and Raftery 2005a, b). In a subsequent publication it was demonstrated that using discrete biofluid constituent concentrations made using a spectral library software as PCA data inputs has a very similar effect, in that it will also improve scores plot subpopulation cluster resolution over what can be obtained using bucket integrals of 1D proton NMR spectra as PCA data inputs (Weljie et al. 2006).
Though these studies demonstrated that the use of discrete component measurements as data inputs will improve the PCA scores plot subpopulation cluster resolution for metabolomics studies of biofluid populations, they did not investigate the origins of this effect in a quantitative manner. This work reexamines this phenomenon. After a discussion on optimizing the 1D TOCSY experiment as it applies to the measurement of biofluid chemical constituents, this report compares the precision and accuracy of constituent measurements made in a complex biofluid matrix (human urine) via the 1D TOCSY experiment with those obtained by the bucket integration of 1D proton NMR spectra, and by analysis using Chenomx spectral library software. The utility of adding even limited 1D TOCSY data to multivariate statistical analysis is also demonstrated.
Experimental
NMR samples
Taurine, lactate, TMAO, histidine, and TSP (sodium 3-trimethylsilyl (2,2,3,3 2H4) 1-propionate) were purchased from Sigma–Aldrich (St. Louis, MO) and used without further purification. Metabolite stock solutions were prepared in 100 mM phosphate buffer at pH 7. Human urine samples were collected from healthy volunteers in accordance with the Institutional Review Board at Purdue University. For NMR analysis, urine samples were prepared by the addition of 120 µl of 0.5 M phosphate buffer, pH 7, to 480 µl of neat urine. All NMR samples were run in 5 mm tubes with 10% added D2O (Cambridge Isotope Laboratories Inc., Andover, MA) and 100 µM TSP.
NMR spectroscopy
NMR spectra were acquired on a Bruker AVANCE DRX 500 MHz spectrometer (Bruker-Biospin, Fremont, CA), using a 5 mm inverse HCN triple resonance probe equipped with XYZ axis gradient coils. All spectra were acquired at 25°C, and were referenced to the TSP methyl peak at 0.000 ppm. All pulse sequences were performed and spectra were acquired using the Bruker XWINNMR software package, release 3.5. Spectral data were processed and integrated using the Bruker Topspin software package, release 2.0. Chenomx measurements of endogenous taurine concentrations were made in Chenomx Profiler 5.1 (Chenomx, Edmonton, Canada) using the 500 MHz pH 6–8 metabolite spectral library, and the 100 µM TSP peak (0.000 ppm) as a concentration reference.
1D proton NMR spectra
1D proton NMR spectra were acquired using a 1D NOESY pulse sequence incorporating presaturation for water suppression during the relaxation delay and mixing time (Nicholson et al. 1995; Belton et al. 1998). The relaxation delay and mixing times were set to 2 s and 300 ms, respectively, and the presaturation power used was the minimum needed to effect complete suppression of the water peak. The sweep width was 10,330 Hz. In order to achieve high signal-to-noise ratios for minor components, sixty-four FID transients (of 64 k points) were averaged, resulting in a total acquisition time of 7 min. The FIDs were zero filled once, and 0.3 Hz line broadening was used in processing the spectra. A “qfil” background correction with a spectral width of 0.2 ppm was used to remove any remaining water peak.
1D TOCSY spectra
The 1D TOCSY pulse sequences described in Fig. 1 were written as modifications of those found in the Bruker XWINNMR pulse program library to match descriptions of the sequences found in (Kessler et al. 1986; Stott et al. 1995; and Facke and Berger 1995). Gaussian-shaped pulsed z-field gradients were 1 ms in duration. The 100% strength for the gradient pulses was 50 gauss per cm. The Gaussian, Secant and IBURP1 shaped selective pulses used were generated using the XWINNMR “Shape Tool” utility incorporated in the Bruker XWINNMR software package (Bauer et al. 1984; Geen et al. 1989; Geen and Freeman 1991). The Shape Tool utility was also used to calculate the duration of the chosen shaped pulses corresponding to the desired excitation band width that was typically 10–25 Hz, depending on the multiplet structure of the excited peak (Sandusky and Raftery 2005a, b). Two Hz was added to these excitation bandwidths to allow for small variations in chemical shift between samples. Typical shaped pulse lengths were 40–100 ms. The power levels of the shaped pulses effecting selective 90° or 180° rotations were determined and optimized independently for each pulse shape and excitation band width used. Typical pulse power levels for the B1 field were in the range of 58 to 68 dB (Bruker).
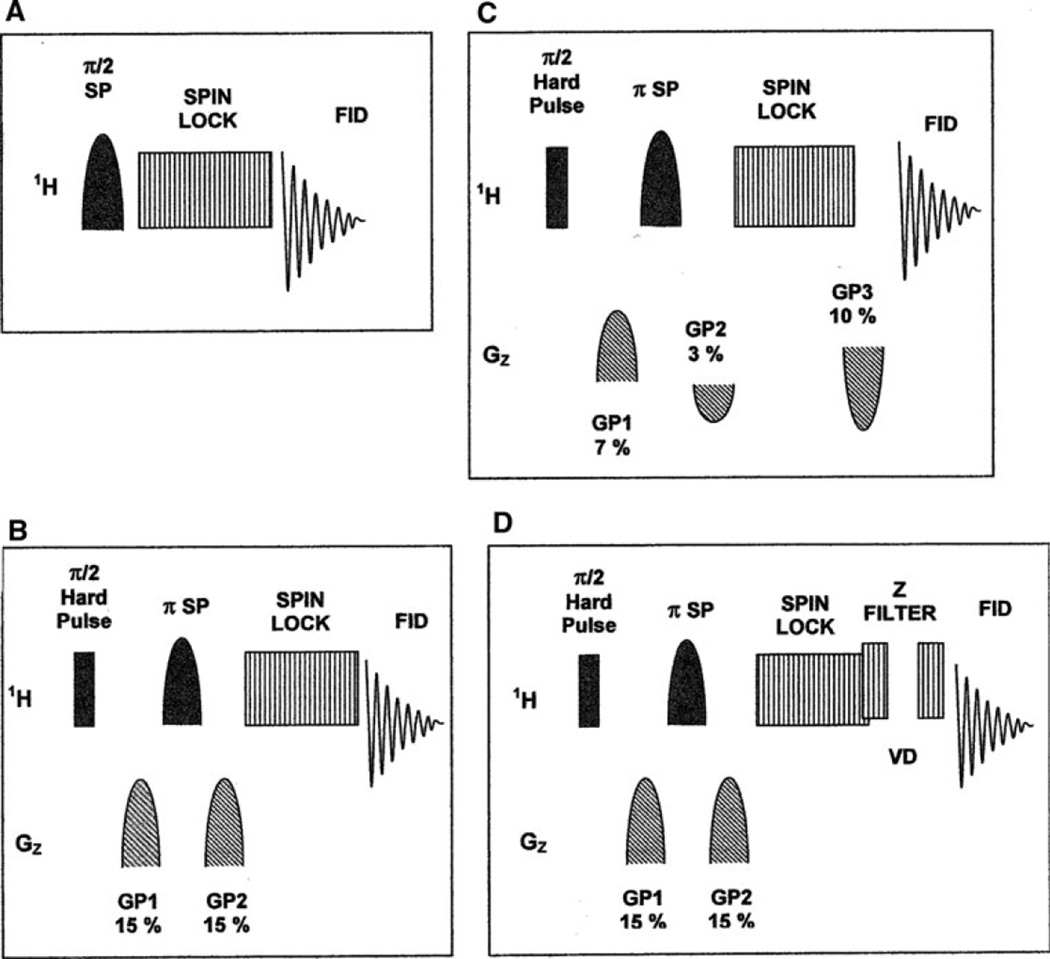
Fig. 1.
Various forms of the Selective TOCSY pulse sequence: a Original form of the experiment as described by Kessler et al. (1986); b experiment with pulsed field gradient spin echo module (PFGSE) for improved suppression outside the target excitation band (Stott et al.1995); c experiment using asymmetric field gradients (Facke and Berger 1995); d experiment incorporating both a PFGSE module for selective excitation, and a z filter for read peak rectification (Sorensen et al. 1984). SP shaped RF pulse; Gaussian, Secant, or IBURP 1. GP pulsed z field gradient. SPINLOCK MLEV 17, DIPSI 2, DIPSI 3, or FLOPSY 8. VD z filter variable delay
The MLEV 17 TOCSY spinlock sequence used in the experiment was the same as that for pulse sequences found in the Bruker XWINNMR pulse program library (Bax and Davis 1985). The DIPSI 2, DIPSI 3 and FLOPSY 8 spinlock sequences were written to match the descriptions given in (Shaka et al. 1988; Kadkhodaie et al. 1991). The z-filter used in pulse sequence D consisted of two spinlock power level 90° pulses separated by a variable delay (VD) (Sorensen et al. 1984).The z-filter VD list was made using the first ten positive values of a random number list that was generated for a mean value of 10 ms with a standard deviation of 5 ms, as calculated using the Microsoft Excel random number generator. The sweep width used for the 1D TOCSY experiments was 5,000 Hz. Eight 64 k point FID transients were averaged in each 1D TOCSY experiment, resulting in a total acquisition time of 85 s if two dummy scans are included. Line broadening of 0.3 Hz was used in processing the spectra.
Statistical analysis
Populations of 1D proton urine spectra were prepared as data matrices for PCA calculations by bucket integration in AMIX 2.1 (Bruker-Biospin, Fremont, CA). Before bucket integration, the spectra were aligned by setting the TSP peaks to a value of 0.000 ppm. Simple rectangular buckets with widths of 0.04 ppm were employed, and the sum of the integrals for each spectrum was scaled to a value of 1.0. Exclusion regions were employed in those cases, as described in the Results Section below, where integrals arising from the spectral features of ethanol, hippurate and creatinine were excluded. The urea peak appearing between 5.0 and 6.2 ppm was excluded in all cases. The bucket integral tables generated by AMIX were exported into Microsoft Office EXCEL 2003 for analysis (Microsoft Corp., Redmond, WA). Substitutions of the taurine 1D TOCSY read peak integrals for the corresponding taurine 1D proton bucket integrals, and renormalization of the resulting data matrices, were also performed in EXCEL. PCA calculations were performed using both Minitab 13 (Minitab Inc., State College, PA) and MATLAB R2007a (The Mathworks Inc., Natick, MA) with equivalent results. PCA calculations were performed using mean centered data and unit variance scaling. Pearson product moment correlation coefficients were calculated using the “PEARSON” function in Microsoft Office EXCEL 2003. ANOVA p values and F-numbers for the PC1 and PC2 scores were calculated using the “anova” function in MATLAB R2007a.
Results
TOCSY optimization
The 1D TOCSY experiment was originally described in the mid 1980s (Kessler et al. 1986) and various modifications have since then been presented in the literature. These include modifications to the basic pulse sequence (Fig. 1), (Stott et al. 1995; Facke and Berger 1995) different types of frequency selective pulses (Bauer et al. 1984; Geen et al. 1989; Geen and Freeman 1991), and various TOCSY spin lock sequences (Bax and Davis 1985; Shaka et al. 1988; Kadkhodaie et al. 1991). The effectiveness of these variations, as they relate to the measurement of biofluid constituent concentrations, were examined in experiments using human urine as a representative biofluid matrix. Four common urine constituents, hippurate, histidine, taurine and lactate were used as target species. For each combination of pulse sequence, selective pulse shape and TOCSY spinlock, the experimental parameters were optimized so as to obtain the highest possible target read peak signal-to-noise ratio. The best results were obtained using a sequence incorporating a pulse field gradient spin echo (PFGSE) module for selective band excitation (sequence B in Fig. 1). The IBURP shaped pulse used in the PFGSE module for selective inversion provides a more uniform excitation across the target excitation bandwidth, and thus produces a 10–15% improvement in the read peak intensity over that produced using a Gaussian or Secant shaped pulse (data not shown) (Bauer et al. 1984; Geen et al. 1989; Geen and Freeman 1991). It was also found that FLOPSY 8 performed best as the TOCSY spinlock, except when the target species has smaller J couplings, in which case DIPSI 2 or DIPSI 3 can be used (Table 1) (Shaka et al. 1988; Kadkhodaie et al. 1991). A z-filter modification to pulse sequence B is also sometimes useful to remove negative components from the read peaks (sequence D in Fig. 1) (Sorensen et al. 1984).
Table 1.
Effect of TOCSY Spinlock sequences on the intensity of read peaks (data obtained using pulse sequence B and the IBURP1 shaped pulse)
| Target | Excitation peak | Read peak | ΔHz | J(Hz) | Mixing time (ms) | Read peak signal to noise ratio | |||
|---|---|---|---|---|---|---|---|---|---|
| MLEV-17 | DIPSI-2 | DIPSI-3 | FLOPSY-8 | ||||||
| Taurine | Taurine 1 | Taurine 2 | 90 | 7 | 80 | 42 | 45 | 45 | 48 |
| Histidine | Taurine 1 | Hist α | 370 | 7 | 40 | 67 | 78 | 74 | 90 |
| Lactate | CH3 | CH | 1,450 | 7 | 35 | 26 | 30 | 30 | 40 |
| Histidine | Hist β | Hist 2(4) | 1,989 | 2–3 | 210 (140)a | 8 | 16 | 17 | 10a |
Histidine mixing time for maximum read peak intensity was 140 ms for FLOPSY 8 and 210 ms for other spinlocks sequences
Quantitation
Application of this basic 1D TOCSY experiment to any particular biofluid constituent of interest is very straight forward, and involves three steps. First, the target peak excitation frequency relative to the center of the spectrum, or “offset,” and target peak width are measured from a 1D proton spectrum. For many common biofluid constituents, hippurate, citrate, lactate and creatinine would be examples in the case of urine, this can be done using the endogenous concentrations. In other cases, where peak overlap completely obscures the target peak, it may be necessary to “spike” the constituent of interest into the first sample of the sample population set. Second, for each constituent target peak three 1D TOCSY parameters (selective pulse length, selective pulse power, and TOCSY mixing time) should be adjusted so as to optimize the read peak intensity. The selective pulse length can be calculated from the target excitation peak width using utilities such as the VNMR “PBOX” or XWINNMR “Shape Tool.” However, we strongly recommend the addition of ~2 Hz to the observed target excitation peak width when performing this calculation (Sandusky and Raftery 2005a, b). This “loose fit” will avoid the potential problem of small peak shifts that can occur as a result of pH or ion concentration variations in the samples. The selective pulse power is adjusted to give the largest excitation peak with the TOCSY power completely attenuated. The TOCSY is then turned back on, and the optimal TOCSY mixing time is determined. Third, if measurements of absolute concentrations are needed, as opposed to relative concentrations, the response of the 1D TOCSY experiment for each particular constituent of interest should be calibrated using a spiked sample. Of course, in analyzing a set of samples for a metabolomics study of a biofluid population, this parameter optimization and calibration procedure need be done on only one sample.
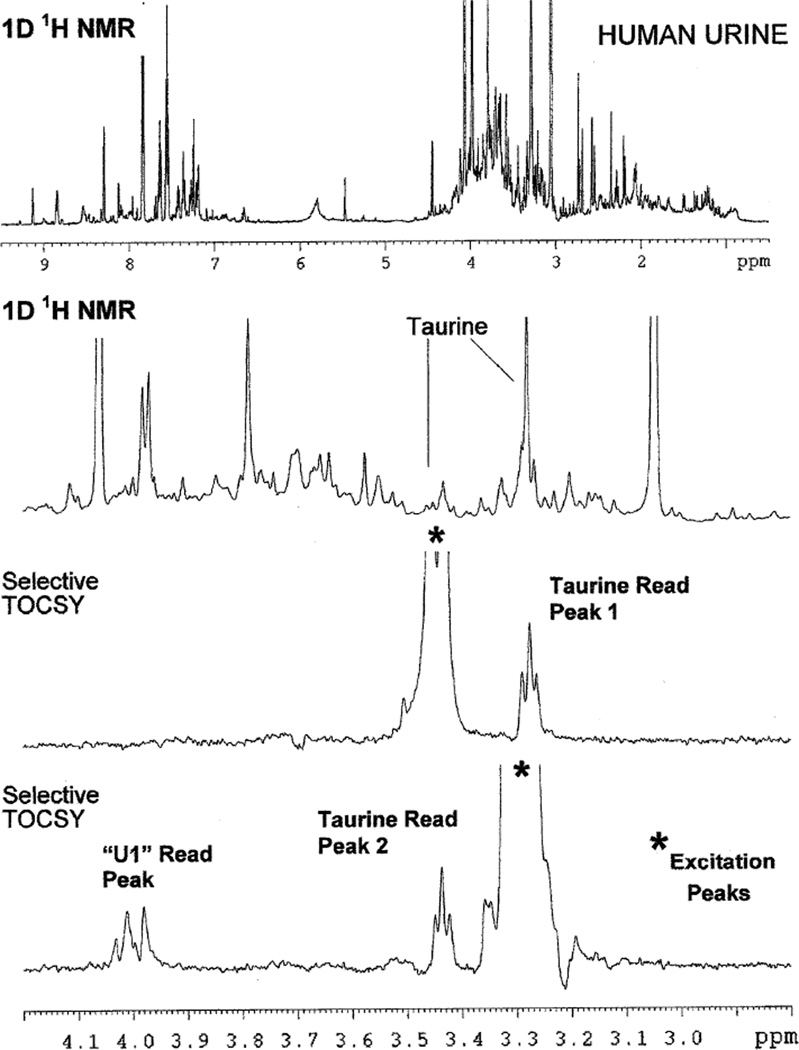
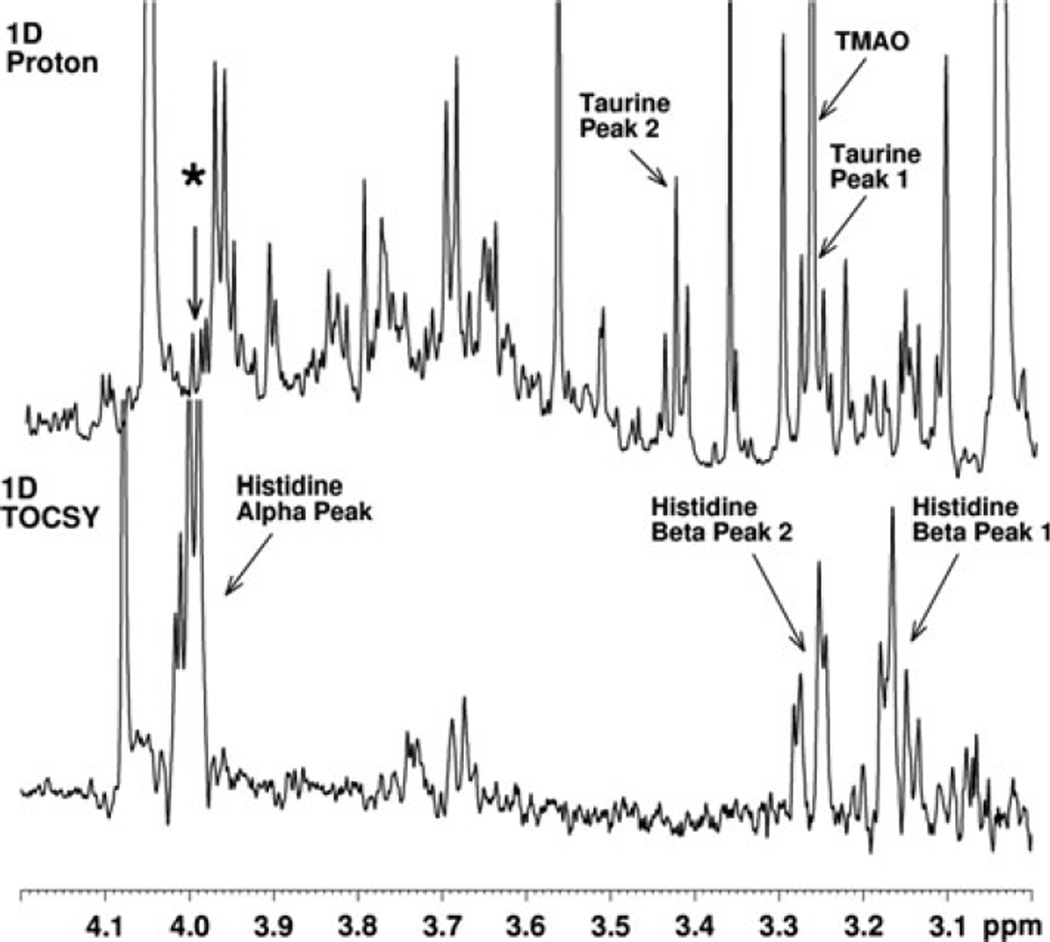
Figure 2 illustrates the use of the 1D TOCSY experiment applied to the concentration measurement of a single chemical constituent present in a complex biofluid mixture. In this particular example the target species is taurine. It should be noted that when observed as a pure species, taurine gives a classic A2X2 proton NMR spectrum with two triplets of equal intensity appearing at 3.45 and 3.28 ppm. Significantly, in the proton NMR spectrum of the human urine sample shown in Fig. 2, both of these taurine triplets are largely obscured by the presence of other species. The application of the 1D TOCSY experiment to this sample allows the clean observation of taurine triplets without interference from the other metabolites present. Thus, the use of the lower field triplet at 3.45 ppm as the “target peak” for selective pulse excitation produces a taurine triplet TOCSY “read peak” at 3.28 ppm, while the use of the high field triplet at 3.25 ppm as the “target peak” produces a TOCSY “read peak” for taurine at 3.45 ppm and an additional read peak resulting from a second metabolite species designated here as “U1,” or Unknown 1. The endogenous taurine concentration in this sample was 400 µM. Subsequently, using a urine sample spiked with an aliquot of standard histidine, the major constituent species interfering with the observation of the taurine high field peak were determined to be trimethylamine oxide (TMAO), which produces a singlet at 3.28 ppm, and histidine, which has peaks at 3.17, 3.28 and 4.0 ppm (Fig. 3).
Fig. 2.
Use of the Selective TOCSY experiment to isolate the peaks of the metabolite taurine from obscuring peaks in a human urine sample. Note that taurine peaks are completely obscured in 1D 1H NMR spectrum (top two spectra), but cleanly isolated in the Selective TOCSY spectra. Selective TOCSY experiments used pulse sequence B shown in Fig. 1, employing an IBURP1 shaped pulse to effect a 25 Hz selective excitation centered at the frequency positions indicated by an asterisk (*), and a 35 ms DIPSI 3 spinlock. Experiments were performed at an endogenous taurine concentration of 0.4 mM. The unknown species labeled here as “U1” was subsequently identified as histidine (see Fig. 3)
Fig. 3.
Selective TOCSY experiment performed on another human urine sample with target excitation at 4.0 ppm. Note that the experiment establishes that one of the histidine beta peaks contributes, along with TMAO, to the peaks obscuring the high field taurine triplet peak at 3.28 ppm. The experimental set up was similar to that used for the experiment shown in Fig. 2. An IBURP 1 pulse was used to effect a 20 Hz selective excitation centered at 4.0 ppm indicated by the asterisk (*). A DIPSI 2 spinlock was applied for 40 ms. Histidine observed was endogenous in this sample
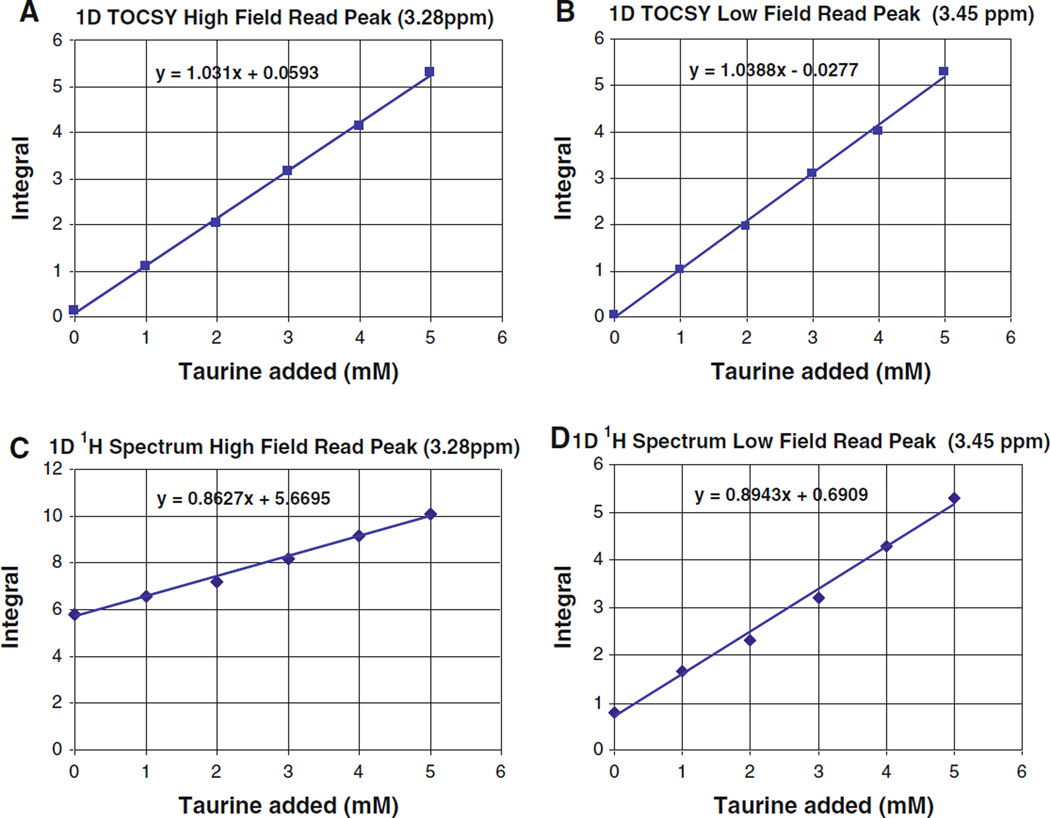
Figure 4 presents a titration of taurine into a human urine sample. Comparing the plots of the integrals of the taurine TOCSY read peaks and to those of corresponding segments of the 1D proton spectra indicates that, as measurements of biofluid constituent concentrations, the TOCSY read peaks are equivalent to bucket integrals of 1D proton spectra in terms of precision. The slopes of least squares linear fits for the low field and high field taurine 1D TOCSY read peak integration plots are in agreement to within ±1%.
Fig. 4.
Titration of taurine into a human urine sample: a integral of 1D TOCSY spectrum around 3.28 ppm; b integral of 1D proton spectrum around 3.28 ppm; c integral of 1D TOCSY spectrum around 3.45 ppm; and d integral of 1D proton spectrum around 3.45 ppm. Experimental parameters were similar to those used for Fig. 2. Note that ordinate offsets in (c) and (d) are due to contributions from histidine, TMAO and other species
More important however is that, because peak overlap adds an “integral background” to the bucket integrals of the 1D proton spectra, the integrals of the TOCSY read peaks provide a much more accurate, and internally consistent, measurement of constituent concentrations. This effect can readily be seen as the large offsets or intercept values in Fig. 4c and d. Thus, the bucket integral of the 1D proton spectrum at the high field taurine triplet implies an endogenous taurine concentration of 6.6 mM (Fig. 4c), while bucket integration of the same 1D proton spectrum at the low field taurine triplet indicates an endogenous taurine concentration of 770 µM (Fig. 4d). The actual endogenous taurine concentration in this sample, as measured using 1D TOCSY, is below 100 µM.
Tables 2 and 3 presents 1D TOCSY and bucket integral measurements of the endogenous taurine concentration in a population of six human urine samples collected from six different individuals. In all six samples the bucket integration of the 1D proton spectrum significantly overestimates the taurine concentration, and gives internally inconsistent measurements. Clearly in the case of the high field taurine triplet region around 3.28 ppm the major contributions to the bucket integral background are TMAO and one of the histidine beta peaks (Fig. 3). However, the presence of a significant integral background in the low field taurine triplet bucket integral around 3.45 ppm (column #4 in Table 2) suggests that, in crowded regions of the 1D proton spectra of biofluids, there is an aggregate contribution to bucket integral background coming from many minor constituents. These minor constituents would presumably be present at low concentrations, and would thus be undetected and unrecognized as individual biofluid constituents. It is also important to note that the 1D TOCSY measurements of the two taurine read peaks show a very high degree of statistical correlation over the sample population, giving a Pearson Product Moment Correlation Coefficient of 0.997 (see Table 3). In contrast, the bucket integrals of the 1D proton spectra in the corresponding regions give a Pearson product moment correlation coefficient of −0.408, indicating that these two regions are dominated by contributions from species other than taurine.
Table 2.
Measurement of endogenous taurine in a population of six human urine samples
| Column no. | 1D TOCSY read peak | 1D proton spectrum peaks | ||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Urine sample | 3.28 ppm | 3.45 ppm | 3.28 ppm | 3.45 ppm |
| a | 0.786 | 0.831 | 3.784 | 1.670 |
| b | 0.466 | 0.455 | 2.733 | 1.443 |
| c | 0.915 | 0.918 | 4.186 | 1.859 |
| d | 0.436 | 0.387 | 2.767 | 1.019 |
| e | 0.157 | 0.130 | 4.584 | 1.353 |
| f | 0.058 | 0.028 | 6.572 | 0.773 |
Peak integration comparison for 1D TOCSY and 1D proton spectra (mM Taurine)
Table 3.
Pearson correlation product moments for integral columns
| Columns | Type | Pearson correlation product moment |
|---|---|---|
| 1 × 2 | TOCSY × TOCSY | 0.997 |
| 1 × 3 | TOCSY × 1D | − 0.494 |
| 1 × 4 | TOCSY × 1D | 0.848 |
| 2 × 3 | TOCSY × 1D | 0.067 |
| 2 × 4 | TOCSY × 1D | 0.777 |
| 3 × 4 | 1D × 1D | − 0.408 |
Measurement of the endogenous taurine concentrations in the set of six urine samples was also made using the Chenomx Profiler spectral library software, Chenomx Inc., Edmonton Canada. In these Chenomx measurements the low field taurine triplet at 3.45 ppm was used as the indicator of taurine concentration, and the 100 µM TSP peak at 0.000 ppm was used as the concentration reference. The resulting taurine concentrations, measured by applying the Chenomx approach to the 1D 1H NMR spectra, were in general intermediate between the values measured using 1D TOCSY and those determined by integration of the 1D proton spectra. Also in two cases, samples e and f, the endogenous taurine concentrations were too low relative to the obscuring species to allow for identification and measurement using the Chenomx software. A newer version of the software (Chenomx 7.0) was also used to verify the concentration determination, and resulted in essentially the same value.
PCA
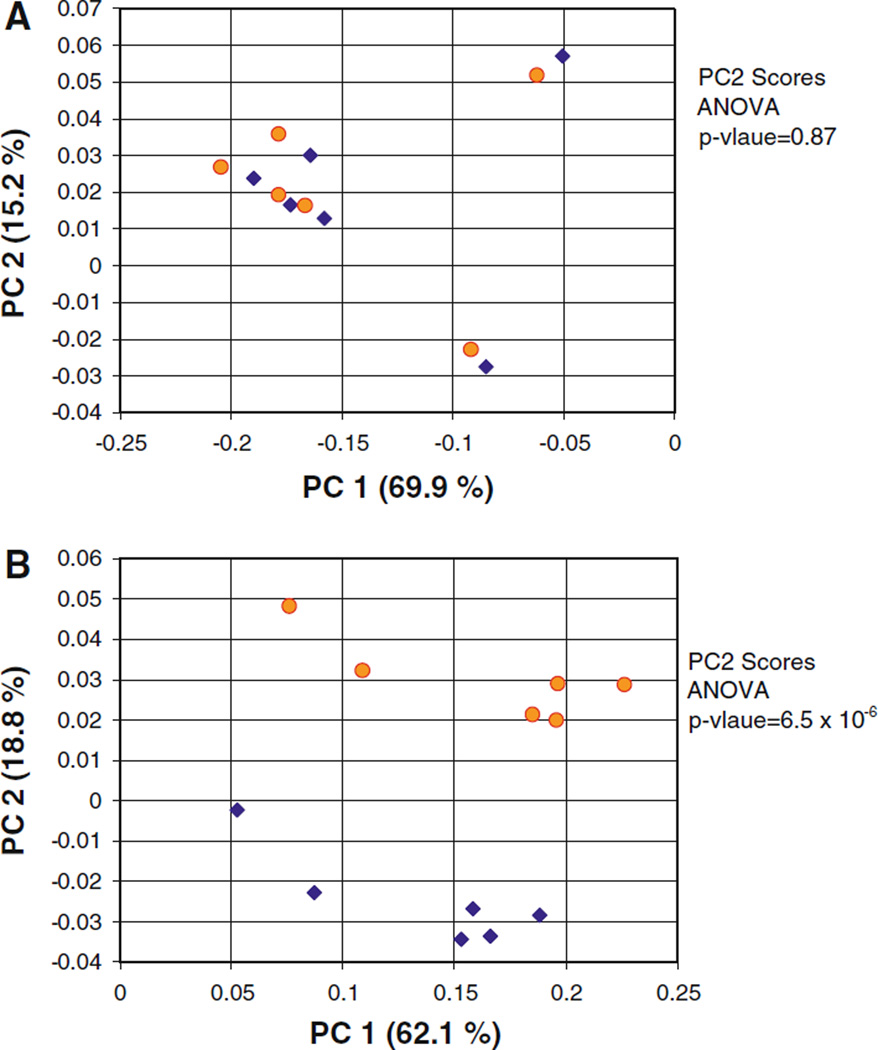
In order to examine the effect of including the more accurate 1D TOCSY measurements as PCA data inputs, a population of samples was generated by spiking taurine into each of the six samples of human urine discussed above. This resulted in a population of twelve samples with two subpopulations; a “low taurine” subpopulation with taurine concentrations between 0 and 1 mM, and a “high taurine” subpopulation with taurine concentrations between 4 and 5 mM. Interestingly, the PC1 versus PC2 scores plot from PCA calculations performed using bucket integrated 1D proton spectra fail to resolve the “high taurine” and “low taurine” subpopulations to any degree (Fig. 5a). However when 1D TOCSY read peak integrals were substituted into the bucket integral matrix in the place of the corresponding taurine 1D proton bucket integrals, PCA calculations produced complete cluster resolution of the two populations along PC2 (Fig. 5b). The ANOVA p-value for the PC2 scores drops from 0.87, for the 1D proton spectrum based calculation, to 6.5 × 10−6, for the calculation performed with a data matrix including the taurine 1D TOCSY read peak integrals. Similarly, PCA calculations performed using simulated bucket integral data indicate that inclusion of “integral background” in bucket integral based PCA calculations will generally have deleterious effects on scores plot subpopulation cluster resolution (data not shown).
Fig. 5.
PC1 versus PC2 PCA scores plots calculated from the 1D proton spectra of a set of human urine samples with two subpopulations. Yellow circles “low taurine” subpopulation (0–1 mM taurine). Blue diamonds “high taurine” subpopulation (4–5 mM taurine). Spectral features from urea (6.5–5 ppm) have been excluded. a Calculation based on data matrix of bucket integrated 1D proton spectra. b Repeat of calculation from A, but with taurine 1D TOCSY read peak integrals substituted for taurine 1D proton bucket integrals
It should be noted that in the analysis of the original 1D proton spectra, the sum of bucket integrals for each spectrum were scaled to a numerical value of 1.0. This is a standard procedure in NMR-based metabolomics, especially for urine spectra, and is performed to correct for the variation in total metabolite concentration over the sample population due to dilution effects (Zhang et al. 2009). In order to make certain that the substitution of 1D TOCSY read peak integral data into the bucket integral matrix would not grossly distort this normalization, the highest 1D TOCSY read peak integral (that from the high field triplet of sample “c” spiked with taurine), was set numerically equal to the value found in the corresponding bucket integral for the 3.28 ppm peak in the original 1D proton spectra bucket integral matrix, and the remaining 1D TOCSY read peak integrals were scaled accordingly. Lastly, before the PCA calculations were performed, the data matrix, now incorporating the scaled 1D TOCSY read peak integral data, was renormalized such that the integrals for each spectrum summed to 1.
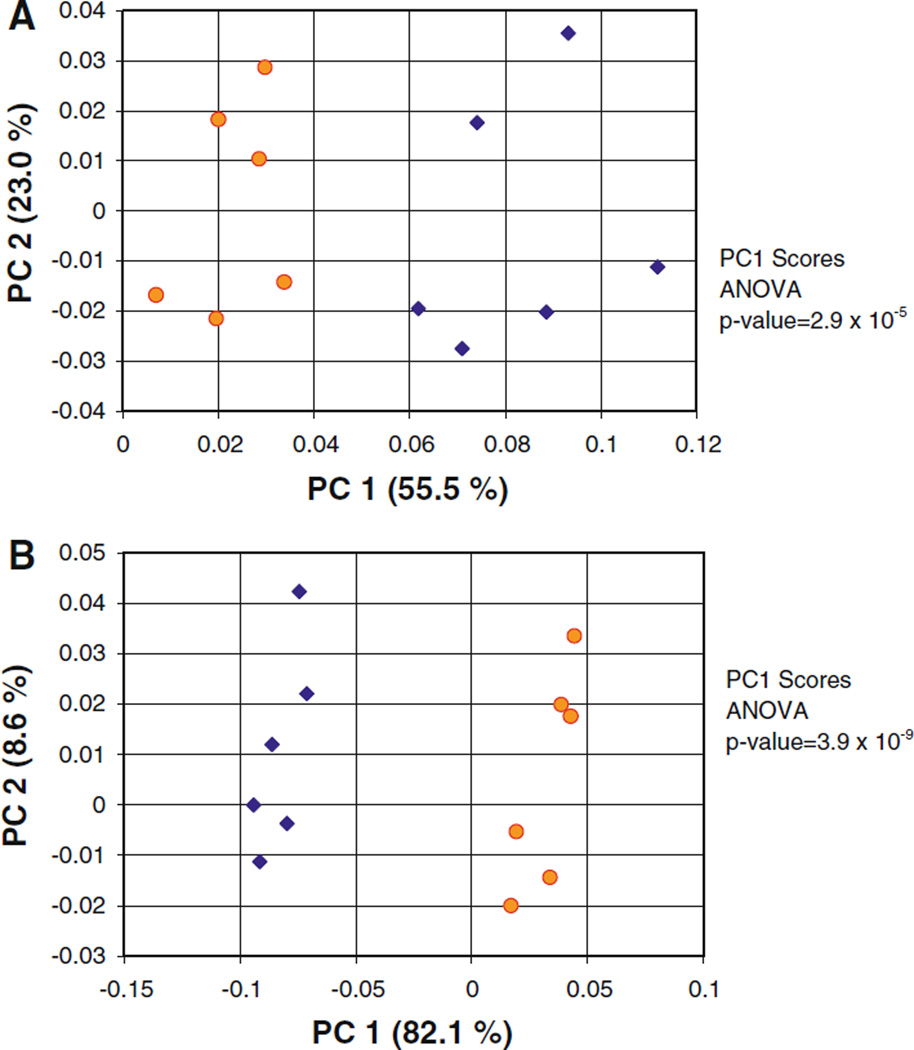
Examination of the loadings plots from the 1D proton bucket integral based PCA calculation (scores plot shown in Fig. 5a), indicated that the calculation failed to resolve “low taurine” and “high taurine” subpopulations because the variance in the sample population was dominated by variations in 3 endogenous urine metabolites: ethanol, hippurate, and creatinine. When these metabolites were excluded during the generation of the bucket integral matrix using AMIX software, the scores plot of the 1D proton spectrum PCA calculation did now resolve the “low taurine” and “high taurine” subpopulations along PC1 (Fig. 6a). However, when the taurine TOCSY read peak integrals were substituted into the same data set (i.e., excluding the ethanol, hippurate and creatinine signals) there nevertheless was a significant improvement in the cluster resolution, and the p-value for the PC1 scores decreased from 2.9 × 10−5 to 3.9 × 10−9 (Fig. 6).
Fig. 6.
PC1 versus PC2 PCA scores plots calculated from the 1D proton spectra of a population of human urine samples with two subpopulations. Repeat of calculations presented in Fig. 5, but with spectral features from urea, ethanol, hippurate, and creatinine excluded. Yellow circles “low taurine” subpopulation (0–1 mM taurine). Blue diamonds “high taurine” subpopulation (4–5 mM taurine). a Calculation based on data matrix of bucket integrated 1D proton spectra. b Repeat of calculation from A, but with taurine 1D TOCSY read peak integrals substituted for taurine 1D proton bucket integrals
Discussion
The selective 1D TOCSY experiment can be relied upon to produce quantifiable read peaks for target metabolites present at concentrations of ~50 µM or better within a 1 min acquisition time using a room temperature 5 mm inverse probe at 500 MHz. If a 5 mm inverse cryoprobe on an 800 MHz instrument were employed the threshold sensitivity of the method could be reduced to 10 µM. Furthermore, quantifiable 1D TOCSY read peaks may be obtained on constituent species even when the peaks of these species are completely obscured in the 1D proton spectrum by the presence of other biofluid constituents.
Because the integral intensities of the 1D TOCSY read peaks are proportional to the concentration of the target metabolite present (Fig. 4; Bauer et al. 1984) they can be used as data inputs, in the place of 1D proton spectra bucket integrals, for chemometric calculations such as ANOVA, PCA or supervised multivariate methods. Furthermore, because 1D TOCSY read peaks make measurements free of the integral background inherent in bucket integration of crowded regions of 1D proton NMR biofluid spectra, using 1D TOCSY read peak integrals as input data for PCA calculations should routinely produce better scores plot subpopulation cluster resolution than that obtained using 1D proton spectra bucket integrals.
The use of discrete component concentrations in the place of 1D proton NMR bucket integrals as data input matrices for PCA calculations on biofluid sample populations has previously been shown to produce better scores plot subpopulation cluster resolution (Sandusky and Raftery 2005a, b; Weljie et al. 2006). The PCA calculation results presented in this paper indicate that two separate factors contribute to this improvement. First, constructing a PCA data matrix from discrete component concentrations allows the analyst to limit the data inputs to metabolite species that are statistically significant in resolving the subpopulations of interest, in effect excluding metabolite species whose presence in the data matrix may be deleterious to subpopulation cluster resolution. This effect is observed in comparing Fig. 6a and b. Second, the use of accurately measured discrete component concentrations, specifically concentration values that are not distorted by integral background, such as those obtained by 1D TOCSY, sharpens the statistical “signal” of the significant metabolite species as they vary over the sample population. This effect is observed in comparing Fig. 5a and b. Certainly other factors, such as the reduction or elimination of noise contributions found in 1D NMR proton bucket integral matrices (Halouska and Powers 2006), also contribute as well, though probably to a lesser extent.
The occurrence of significant integral background in the 1D proton NMR spectrum bucket integral of the low field taurine peak at 3.45 ppm suggests, not surprisingly, that an aggregate of minor constituents will contribute to the integral background in crowded regions of the spectrum, even when these minor constituents are not present at concentrations high enough for them to be detected as individual constituents. 1D proton NMR spectra of urine samples routinely contain detectable peaks from only 30–40 detectable constituent species (Foxall et al. 1993), whereas HSQC spectra of urine samples prepared using 15N ethanolamine demonstrates that there are nearly 200 carboxyl-containing constituents alone present at NMR detectable concentrations (Ye et al. 2009). Mass spectrometry indicates that perhaps as many as 1,400 constituent species are detectable in typical biofluid samples (Fischer 2010).
Chenomx, Inc. (Edmonton, Canada) has developed a method for data extraction based on an NMR spectral library and software that allows the determination of concentration measurements for constituent chemical species from 1D proton NMR spectra of biofluids. The use of this spectral library software corrects for the problems of peak overlap, and gives more accurate measurements of biofluid constituents than bucket integrals. It has been demonstrated that using these spectral library extracted concentrations in PCA calculations can increase the scores plot subpopulation cluster resolution over that which can be obtained using bucket integrated 1D proton NMR spectra as data inputs (Weljie et al. 2006). The Chenomx spectral library software is a very valuable tool for the analysis of biofluid populations. However, when significant spectral overlap or broad baseline components are present, this approach is more challenging. In such cases, the 1D TOCSY approach may find application. As demonstrated above, quantifiable 1D TOCSY read peaks may be obtained on metabolite species even when the peaks of these species are completely obscured in the 1D proton spectrum by the presence of other biofluid constituents. It seems unlikely that the use of spectral library based software can adequately address the problem presented by the contributions of an aggregate of minor constituents to the integral background as discussed above.
The authors believe that the use of discrete concentration measurements made using 1D TOCSY provides an improved technique in the forensic and diagnostic applications of metabolomics. In passing, we note that other types of TOCSY NMR experiments discussed in the literature may or may not be useful in similar contexts. Statistical TOCSY, or STOCSY, developed by the Nicholson and coworkers, uses statistical covariation over a population of biofluid samples to resolve the proton NMR spectra for certain individual metabolites (Cloarec et al. 2005). The covariation TOCSY approach developed by Bruschweiler and Zhang uses the covariation inherent in an individual molecule’s chemical shift frequencies and J couplings, as they evolve over a sampled time domain, to separate the spectra of the individual chemical species in a mixture using a reduced number of increments in the 2D spectra (Zhang and Bruschweiler 2004, 2007). Recent work by the Emsley group has shown the utility of intraspectral correlation to better define peak integral limits for improved data analysis (Holmes et al. 2007). Each of these approaches has its own strengths, and is being applied in a variety of metabolomics research studies (Cloarec et al. 2005; Blaise et al. 2009; Maher et al. 2007; Holmes et al. 2006). However, in the present case, the high degree of overlapping peaks and aggregate integral background found in many biofluid samples challenges the statistical methodologies. We note in this regard the Pearson Product Moment analysis of taurine in a population of human urine samples described in Tables 2 and 3. Reduction of this background signal prior to sophisticated statistical analysis appears to be highly useful.
Conclusions
In this paper we have shown that 1D selective TOCSY measurements remove the integral background that is intrinsically present in the bucket integrals of 1D proton NMR spectra due to the overlap of unresolved peaks from the great many chemical constituents that occur in biofluids. Thus, 1D TOCSY read peak integrals are more accurate measures of the true variances of statistically significant metabolite concentrations in a biofluid sample population than the bucket integrals of 1D proton spectra. While the use of modeling using standard compound spectra can improve quantitation, the presence of such background signals still complicates the analysis and causes errors. We have also shown that using 1D TOCSY data as inputs to PCA calculations performed on a population of human urine samples demonstrates that substituting 1D TOCSY read peak integrals for the corresponding 1D proton bucket integrals produces better subpopulation cluster resolution, even when this substitution is limited to only one statistically significant metabolite.
Acknowledgments
This work was supported by the NIH (NIGMS R01GM085291-02 and 3R01GM085392-02S1). DR is a member of the Purdue University Center for Cancer Research and the Oncological Sciences Center in Discover Park at Purdue University.
Contributor Information
Peter Sandusky, Department of Chemistry, Eckerd College, St. Petersburg, FL 33711, USA.
Emmanuel Appiah-Amponsah, Department of Chemistry, Purdue University, West Lafayette, IN 47907, USA.
Daniel Raftery, Email: raftery@purdue.edu, Department of Chemistry, Purdue University, West Lafayette, IN 47907, USA.
References
- Bauer C, Freeman R, Frenkiel T, Keeler J, Shaka AJ. Gaussian pulses. J Magn Res. 1984;58:442–457. [Google Scholar]
- Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Res. 1985;65:355–360. [Google Scholar]
- Belton PS, Colquhoun IJ, Kemsley EK, Delgadillo I, Roma P, Dennis MJ, Sharman M, Holmes E, Nicholson JK, Spraul M. Application of chemometrics to the 1H NMR spectra of apple juices: discrimination between apple varieties. Food Chem. 1998;61:207–213. [Google Scholar]
- Blaise BJ, Shintu L, Elena B, Emsley L, Dumas ME, Toulhoat P. Statistical recoupling prior to significance testing in nuclear magnetic resonance based metabonomics. Anal Chem. 2009;81:6242–6251. doi: 10.1021/ac9007754. [DOI] [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson JK. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal Chem. 2005;77:1282–1289. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- Facke T, Berger S. Application of pulsed field gradients in an improved selective TOCSY experiment. J Magn Res Ser A. 1995;113:257–259. [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nature Rev Mol Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fischer S. Agilent Technologies. Personal communication. 2010 [Google Scholar]
- Foxall PJD, Parkinson JA, Sadler IH, Lindon JC, Nicholson JK. Analysis of biological fluids using 600 MHz proton NMR spectroscopy: application of homonuclear two-dimensional J-resolved spectroscopy to urine and blood plasma for spectral simplification and assignment. J Pharm Biomed Anal. 1993;11:21–31. doi: 10.1016/0731-7085(93)80145-q. [DOI] [PubMed] [Google Scholar]
- Geen H, Freeman R. Band-selective radiofrequency pulses. J Magn Res. 1991;93:93–141. [Google Scholar]
- Geen H, Wimperis S, Freeman R. Band-selective pulses without phase distortion. A simulated annealing approach. J Magn Res. 1989;85:620–627. [Google Scholar]
- Gowda GAN, Zhang SC, Gu HW, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics: a review. Expert Rev Mol Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halouska S, Powers R. Negative impact of noise on the principal component analysis of NMR data. J Magn Res. 2006;178:88–95. doi: 10.1016/j.jmr.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Holmes E, Cloarec O, Nicholson JK. Probing latent biomarker signatures and in vivo pathway activity in experimental disease states via statistical total correlation spectroscopy (STOCSY) of biofluids: application to HgCl2 toxicity. J Proteome Res. 2006;5:1313–1320. doi: 10.1021/pr050399w. [DOI] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Cloaree O, Coen M, Tang H, Maibaum E, Bruce S, Bruce S, Chan Q, Elliott P, Stamler J, Wilson ID, Lindon JC, Nicholson JK. Detection of urinary drug metabolite (xenometabolome) signatures in molecular epidemiology studies via statistical total correlation (NMR) spectroscopy. Anal Chem. 2007;79:2629–2640. doi: 10.1021/ac062305n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadkhodaie M, Rivas O, Tan M, Mohebbi A, Shaka AJ. Broadband homonuclear cross polarization using flip-flop spectroscopy. J Magn Res. 1991;91:437–443. [Google Scholar]
- Kell DB. Metabolomics and systems biology: making sense of the soup. Curr Opin Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Kessler H, Oschkinat H, Griesinger C. Transformation of homonuclear two-dimensional NMR techniques into one-dimensional techniques using Gaussian pulses. J Magn Res. 1986;70:106–113. [Google Scholar]
- Maher AD, Zirah SFM, Holmes E, Nicholson JK. Experimental and analytical variation in human urine in 1H NMR spectroscopy-based metabolic phenotyping studies. Anal Chem. 2007;79:5204–5211. doi: 10.1021/ac070212f. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Foxall PJD, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H–13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, Holmes E. “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- Sandusky P, Raftery D. Use of selective TOCSY NMR experiments for quantifying minor components in complex mixtures: application to the metabonomics of amino acids in honey. Anal Chem. 2005a;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- Sandusky P, Raftery D. Use of semiselective TOCSY and the pearson correlation for the metabonomic analysis of biofluid mixtures: application to urine. Anal Chem. 2005b;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev Mol Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- Shaka AJ, Lee CJ, Pines A. Iterative scheme for bilinear operators; application to spin decoupling. J Magn Res. 1988;77:274–293. [Google Scholar]
- Sorensen OW, Rance M, Ernst RR. z filter for purging phase—or multiplet-distorted spectra. J Magn Res. 1984;56:527–534. [Google Scholar]
- Stott K, Stonehouse J, Keeler J, Hwang TL, Shaka AJ. Excitation sculpting in high-resolution nuclear magnetic resonance spectroscopy: application to selective NOE experiments. J Am Chem Soc. 1995;117:4199–4200. [Google Scholar]
- Van der Greef J, Smilde AK. Symbiosis of chemometrics and metabolomics: past, present and future. J Chemometr. 2005;19:376–386. [Google Scholar]
- Van Dien S, Schilling CH. Bringing metabolomics data into the forefront of systems biology. Mol Syst Biol. 2006;2:1–2. doi: 10.1038/msb4100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- Ye T, Mo H, Shanaiah N, Gowda GAN, Zhang S, Raftery D. Chemoselective 15N tag for sensitive and high resolution nuclear magnetic resonance profiling of carboxyl-containing metabolome. Anal Chem. 2009;81:4882–4888. doi: 10.1021/ac900539y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Bruschweiler R. Indirect covariance NMR spectroscopy. J Am Chem Soc. 2004;126:13180–13181. doi: 10.1021/ja047241h. [DOI] [PubMed] [Google Scholar]
- Zhang F, Bruschweiler R. Robust deconvolution of complex mixtures by covariance TOCSY spectroscopy. Angew Chem Int Ed. 2007;46:2639–2642. doi: 10.1002/anie.200604599. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zheng C, Lanza IR, Nair KS, Raftery D, Vitek O. Interdependence of signal processing and analysis of urine 1H NMR spectra for metabolic profiling. Anal Chem. 2009;81:6080–6088. doi: 10.1021/ac900424c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gowda GAN, Asiago V, Ye T, Raftery D. Advances in NMR-based biofluid analysis and metabolite profiling. Analyst. 2010;135:1490–1498. doi: 10.1039/c000091d. [DOI] [PMC free article] [PubMed] [Google Scholar]