Abstract
Macrophages within adipose tissue play a key role in mediating inflammatory responses in adipose tissue that are associated with obesity-related metabolic complications. In an effort to identify novel proteins secreted from adipocytes that may negatively regulate macrophage inflammation, we found that peroxiredoxin (PRX)-like 2 activated in M-CSF stimulated monocytes (PAMM), a CXXC-type PRX-like 2 domain-containing redox regulatory protein, is a novel secreted protein with potent anti-inflammatory properties. PAMM is secreted from mature human adipocytes but not preadipocytes. Overexpression of PAMM significantly attenuated lipopolysaccharide (LPS)-induced macrophage inflammation. Incubation of macrophages with adipocyte-conditional medium treated with anti-PAMM antibody significantly enhanced LPS-induced interleukin-12 (IL-12) expression in Raw264.7 cells. In addition, incubation of Raw264.7 cells with purified PAMM protein had a similar anti-inflammatory effect. Moreover, forced expression of PAMM in Raw264.7 cells resulted in decreased LPS-induced ERK1/2, p38 and c-Jun N-terminal kinase (JNK) phosphorylation, suggesting that PAMM exerted the anti-inflammatory function probably by suppressing the mitogen-activated protein kinase (MAPK) signalling pathway. Mutations in the CXXC motif of PAMM that suppressed its anti-redox activity were still able to suppress production of inflammatory cytokines in LPS-stimulated macrophages, suggesting that PAMM’s anti-inflammatory properties may be independent of its antioxidant properties. Finally, PAMM was highly expressed in both white (WAT) and brown adipose tissues (BAT) and further increased in obesity status. Our results suggest that adipocyte-derived PAMM may suppress macrophage activation by inhibiting MAPK signalling pathway.
Keywords: adipocyte, anti-inflammation, macrophage, peroxiredoxin-like 2 activated in M-CSF-stimulated monocytes (PAMM), redox, secreted protein
INTRODUCTION
Adipose tissue expansion during obesity is associated with increased macrophage infiltration [1,2]. Cross-talk between adipocytes and macrophages significantly contributes to the development of obesity, insulin resistance and diabetes [3,4]. Adipocyte-secreted factors such as monocyte chemotactic protein-1 (MCP-1, also known as CCL2) and tumour necrosis factor α (TNFα) promote macrophage infiltration and activation [5]. On the other hand, factors derived from infiltrating macrophages alter adipocyte function and insulin sensitivity [6]. As adipose tissue is an active endocrine organ, we hypothesized that adipose tissues may also secrete proteins that negatively regulate macrophage-mediated inflammation. To identify novel proteins secreted by adipocytes that exert anti-inflammatory effects on macrophages, we studied the differentiation of preadipocytes (3T3-L1 cells) with a combination of gene expression profiling and bioinformatic techniques to search for highly induced genes that encoded proteins with putative signal peptides in mature adipocytes but not preadipocytes. We identified 13 genes that were of interest. One gene, which encodes a protein called PRX-like 2 activated in M-CSF stimulated monocytes (PAMM), was further explored in the current study.
PAMM, a protein that is expressed in bone marrow monocytes on stimulation with macrophage-colony stimulating factor (M-CSF) and receptor activator of nuclear factor kappa-B ligand (RANKL), contains a CXXC motif-type peroxiredoxin (PRX)-like 2 domain [7]. PRX, which is a scavenger of H2O2 and alkyl hydroperoxides in living organisms, exerts a protective antioxidant role in cells through its peroxidase activity, whereby H2O2, peroxynitrite and a wide range of organic hydroperoxides are reduced and detoxified [8,9]. Mammalian PRX family members can be divided into six distinct groups (type 1 through type VI) [8]. PRX II is of particular interest because it appears to provide selective, specific and localized control of receptor-mediated signal transduction [9]. PRX II deficient mice have been shown to exhibit an over-responsiveness of macrophage inflammatory responses and enhanced sensitivity to lipopolysaccharide (LPS)-induced lethal shock. This suggests that PRX II, through modulation of reactive oxygen species (ROS) synthesis via NADPH oxidase activity, is an essential negative regulator of LPS-induced inflammatory signalling that is crucial for the prevention of excessive host responses to microbial products [10].
PAMM is a member of the PRX-like 2 containing protein family whose function has not been well characterized. Previously, we have reported that overexpression of PAMM in human embryonic kidney (HEK293) cells resulted in an increased GSH–GSSG ratio indicating a shift toward a more reduced environment [7]. Moreover, expression of PAMM in Raw264.7 cells protected them from hydrogen peroxide-induced oxidative stress, indicating that PAMM regulates cellular redox status. Furthermore, M-CSF and RANKL can induce up-regulation of PAMM in Raw264.7 cells and induces the cells to differentiate into osteoclasts [7]. In the present study, we found that PAMM is a novel secreted protein with potent anti-inflammatory properties. In contrast with PRX II, whose anti-inflammatory properties are highly dependent on its antioxidant activity, PAMM’s ability to inhibit production of inflammatory cytokines in LPS-stimulated macrophages appears to be due to the suppression of mitogen-activated protein kinase (MAPK) signalling pathways.
EXPERIMENTAL
Cell culture
The murine macrophage cell line Raw264.7 was obtained from the A.T.C.C. and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 units/ml of penicillin and streptomycin and 10% FBS (endotoxin < 1 ng/ml; Sigma). 3T3-L1 preadipocytes were cultured in DMEM supplemented with 10%FBS, 100 units/ml penicillin, 100 µg/ml streptomycin in a 5% CO2 humidified atmosphere and allowed to reach confluence. Differentiation of 2-day postconfluent preadipocytes was induced by incubation with a cocktail of 5 µg/ml insulin, 1 µM dexamethasone and 0.5 mM 3-isobutyl-1-methylxanthine for 10 days. Human primary preadipocytes and adipocyte differentiation medium were purchased from Cell Applications. HEK293 cells were cultured in DMEM medium supplemented with 100 units/ml of penicillin and streptomycin and 10% FBS (Sigma).
Reagents
Antibodies against β-actin, phospho-ERK1/2, ERK1/2, phosphor-p38, p38, phospho-c-Jun N-terminal kinases (JNK), were purchased from Cell Signaling Technology. Anti-C10orf58 (PAMM) antibody (HPA009025), anti-Flag antibody and LPS were purchased from Sigma. PAMM ELISA kit was purchased from MyBiosource.
Microarray hybridization
Genome-wide expression screening was conducted to identify genes up-regulated during 3T3-L1 differentiation. In brief, total RNA was extracted from 3T3-L1 cells and differentiated adipocytes. The RNA was used as a template to generate mixed cDNA probes by reverse transcription. These probes were hybridized to the mouse MG-U74Av2 chip from Affymetrix, according to the manufacturer’s instructions, at the Microarray Center in University of Texas Southwestern Medical Center.
Plasmids
Flag-tagged human PAMM expression plasmid was generated by inserting human PAMM coding fragment into pCMV-MAT-Tag-FLAG-1 vector (Sigma) at HindIII and BamHI sites. PAMMC88G mutant plasmid was previously generated [7]. Flag-MCPIP1 plasmid was generated in the laboratory as described previously [11].
Transfection
Transient transfection into Raw264.7 cells was performed by electroporation following the manufacturer’s instruction (Amaxa, MD). Briefly, Raw264.7 cells were grown to confluence in DMEM medium supplemented with 10% FBS. Cells were collected and washed once with DMEM medium, and re-suspended with the electroporation buffer (Amaxa). After electroporation, the cells were plated on 6-well plates and the transfection efficiency was monitored by fluorescent microscopy. Over 60% of cells were GFP positive.
Northern blot
Total RNA was isolated from mouse tissues using RNA STAT-60 reagent (Tel-Test) following the manufacturer’s instruction [11]. Fifteen micograms of total RNA was denatured and electrophoresed on 1% agarose-formaldehyde gels. Uniformity of sample loading was verified by UV visualization of ethidiumbromide-stained gels before transfer to Nylon membranes. The cDNA probes for PAMM were amplified by PCR using a cDNA clone from A.T.C.C. as templates. 32P-labelled cDNA was prepared using the random priming method (Invitrogen). Hybridization was performed using QuckHyb buffer (Strategene) at 65°C for 2 h or overnight. Then membranes were washed once with 2 × SSC and once with 0.1 × SSC, 1% SDS for 20 min at 65°C.
Quantitative real-time PCR
After removing genomic DNA using DNAse I (Ambion), 2.4 µg of total RNA from RAW264.7 cells was reverse transcribed to cDNA using a commercially available kit (Applied Biosystems). Quantitative real-time PCR was performed with a 7900HT fast real-time PCR system (ABI) using 2 × SYBR Green master mixture (Bio-Rad). Forty cycles were conducted as follows: 95°C for 30 s, 60°C for 30 s, preceded by 1 min at 95°C for polymerase activation. Primer sequences for all genes we measured in this report are available upon request. Quantification was performed by the dCT method, with β-actin used for normalization.
Protein isolation and Western blot
After washing twice with PBS, cells were gently scraped with a rubber policeman into 5 ml of ice-cold PBS, and centrifuged at 1000 g for 5 min at 4°C. Cells from each 10-cm dish were then resuspended and lysed in 0.5 ml of lysis buffer containing 50 mM NaH2PO4, pH7.6, 250 mM NaCl, 50 mM NaF, 10 mM imidazole, 0.5% Nonidet P-40, 1 µg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride. The cell lysate was left on ice for approximately 20 min and then sonicated and centrifuged at 10000 g for 10 min at 4 °C. Protein concentrations were determined by the Bradford method (Bio-Rad Laboratories), with bovine serum albumin (BSA) as the standard. For Western blotting, proteins (50 µg) were separated by SDS/PAGE and transferred onto nitrocellulose membranes in transfer buffer containing 0.1% SDS. The membranes were blocked with 5% nonfat dry milk in 0.05% Tween 20 in TBS (TTBS) for 2 h and incubated with the primary antiserum at a 1:1000 dilution in the blocking buffer for 1 h. After being washed with TTBS three times for 10 min each, the membranes were incubated with a 1:2000 dilution of secondary antibody in TTBS for 1 h. Following three 10-minwashes with TTBS, membranes were incubated with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and exposed to X-ray film.
Measurement of GSH–GSSG ratio
Raw264.7 cells were transiently transfected with control plasmid or PAMM or PAMMC88G. The GSH–GSSG ratio was measured with the glutathione reductase/5,5′-dithiobis-(2-nitrobenzoic acid; DTNB) assay kit (Bioxytech GSH-412, OXIS International) according to the manufacturer’s instructions. In brief, total GSH (GSHt) and GSSG (oxidized GSG) concentrations were derived from GSH and GSSG standard curves and converted to nanomoles per milligram of protein. Reduced GSH concentrations were found by subtracting GSSG from GSHt. Finally, the GSH–GSSG ratio was calculated by dividing the difference between GSHt and GSSG concentrations by the GSSG concentration [ratio = GSHt − 2 (GSSG)/GSSG]. A decreased GSH–GSSG ratio is an indicator of oxidative stress.
Purification of PAMM from BL21 E. coli
pET28a-human PAMM plasmid was transformed into competent BL21 Escherichia coli cells, and transfectants were isolated on agar plates containing kanamycin (30 µg/ml). Cells were grown to log phase (D = 0.6–0.7) in LB broth with kanamycin. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 µg/ml) was then added to the medium to induce the T7 promoter. Cells were harvested after 2 h and the human PAMM protein was purified by the His-bind purification kit. The protein was eluted with imidazole (300 mM, pH 6.0) and dialysed against water. Cell lysates and purified protein were subjected to examination by Coomassie Blue staining and Western blot analysis.
Statistics
Data are expressed as mean ± S.D. Statistical analysis between two groups was performed by an unpaired Student’s t-test. Comparison between three or more groups was performed by one-way ANOVA followed by an unpaired Student’s t-test. A value of P < 0.05 was considered statistically significant.
RESULTS
Identification of PAMM as a novel secreted protein from adipocytes
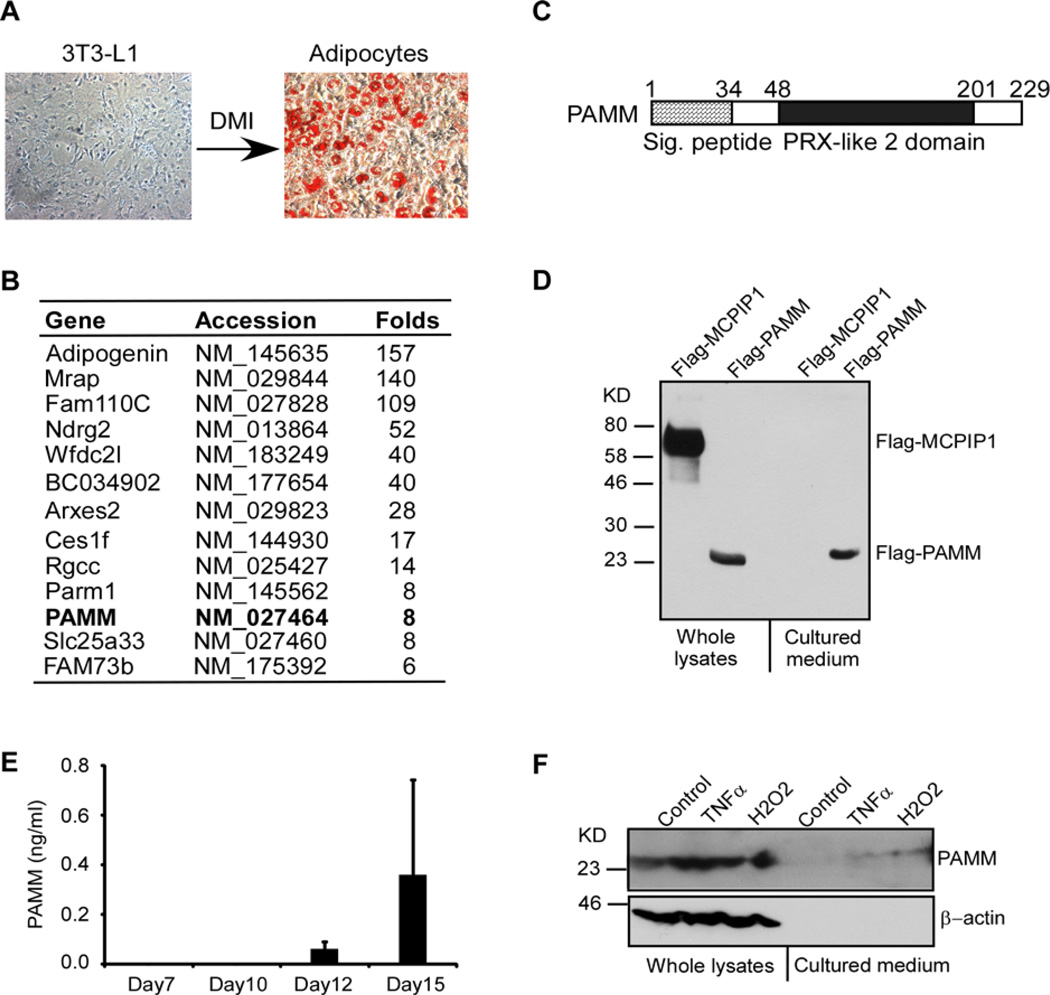
3T3-L1 is a well-established preadipocyte cell line. After incubation with a cocktail of DMI (dexamethasone, methylxanthine and insulin), 3T3-L1 cells can be fully differentiated into adipocytes (Figure 1A). To identify the putative novel proteins secreted by adipocytes, we first performed microarray analysis to compare differences in gene expression profiles in differentiated adipocytes compared with undifferentiated 3T3-L1 cells. Genes that were up-regulated by more than 5-fold in adipocytes compared with 3T3-L1 cells were initially identified. We then restricted our analysis to genes that encoded proteins containing putative signal peptides, indicating that these proteins are likely to be secreted (Figure 1B). PAMM, a protein that contains a CXXC type PRX-like 2 domain, was selected for further study. Human PAMM protein has 229 amino acids with a putative signal peptide at its N-terminal (Figure 1C). The predicted molecular mass of PAMM is ~24 kDa. To determine if PAMM is a secreted protein, we generated an expression plasmid encoding Flag-PAMM fusion protein. We transiently transfected HEK293 cells with this plasmid and a control plasmid encoding Flag-MCPIP1. Flag-MCPIP1 was selected as a control because we have extensively studied this cellular protein, which is not secreted [12]. We collected whole cell lysates and cultured medium from transfected cells for Western blot analysis. As shown in Figure 1D, both Flag-PAMM and Flag-MCPIP1 appeared in whole cell lysates, but only Flag-PAMM appeared in concentrated conditional cultured medium, suggesting that PAMM is a secreted protein. To further determine if endogenous PAMM protein is secreted from human adipocytes, human primary preadipocytes were differentiated into mature adipocytes by incubation with the differentiation medium for 15 days. The PAMM levels in the cultured medium were measured by ELISA. As shown in Figure 1(E), PAMM was not detectable in undifferentiated preadipocyte cultured medium (data not shown), but significantly increased in the cultured medium after 12 days and 15 days differentiation. Furthermore, PAMM was found at low levels in cultured medium from TNFα and H2O2-stimulated Raw264.7 cells (Figure 1F). Taken together, these results suggest that PAMM is a novel secreted protein from mature adipocytes but not preadipocytes.
Figure 1. Identification of PAMM as a secreted protein from adipocytes.
(A) 2-day post-confluent 3T3-L1 cells were incubated with a cocktail of DMI for 10 days to induce differentiation into adipocytes. The cells were stained by Oil O and the images were taken by Nikon microscope system. (B) Affymatrix microarray analysis of gene expression in 3T3-L1 cells and adipocytes. Uncharacterized genes that were up-regulated by more than 5-fold in adipocytes and encoded proteins containing putative signal peptides are listed in this table. (C) Scheme of human PAMM protein structure. Human PAMM is a 229 amino acid protein containing a putative signal peptide and a PRX-like 2 domain. (D) HEK293 cells were transiently transfected with Flag-PAMM or Flag-MCPIP1. Whole cell lysates and culture medium were analysed for Flag-PAMM and Flag-MCPIP1 by Western blot analysis with anti-Flag antibody. The results are representative of three independent experiments, which showed similar results. (E) Human primary preadipocytes were incubated with adipocyte differentiation medium for different days as indicated. The cultured media were collected and concentrated for ELISA analysis of PAMM protein. Data are presented as mean ± S.D. (F) Raw264.7 cells were stimulated with or without 10 ng/ml TNFα or 400 µM H2O2 for 4 h. Whole cell lysates and culture medium were collected for Western blot analysis with PAMM and β-actin antibodies.
PAMM is a negative regulator of macrophage inflammation
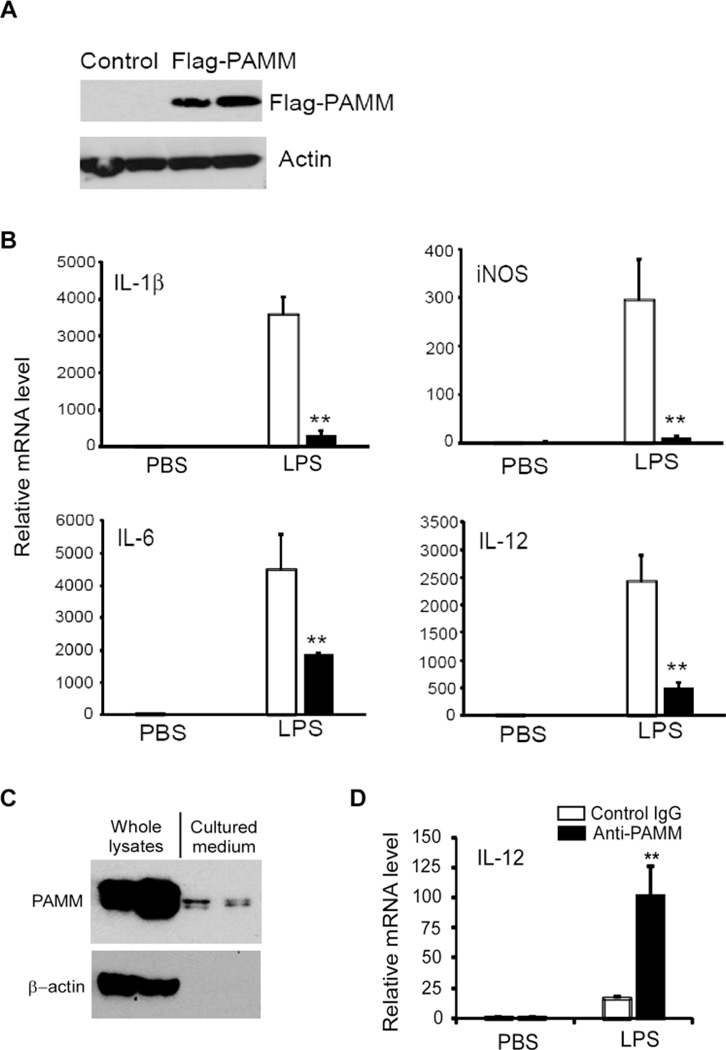
Previously, Xu et al. [7] reported that PAMM is a redox regulatory protein that modulates osteoclast differentiation. However, the role of PAMM in macrophage-mediated inflammation remains unknown. Consequently, we examined the effects of PAMM overexpression on the expression of inflammatory cytokines in LPS stimulated Raw264.7 cells. Raw264.7 cells were transiently transfected with Flag-PAMM expression plasmid or control plasmid. After 24 h, transfected cells were stimulated with 1 µg/ml of LPS for 8 h. RNA and cell lysates were harvested from the treated cells for Q-PCR and Western blot analysis. PAMM overexpression in Raw264.7 cells was confirmed by Western blot with anti-Flag antibody (Figure 2A). As determined by Q-PCR (Figure 2B), overexpression of PAMM markedly attenuated LPS-induced mRNA expression for IL-1β, interleukin-6 (IL-6), IL-12 and inducible nitric oxide synthase (iNOS). These results suggest that PAMM functions as a potent negative regulator of inflammatory responses in LPS stimulated macrophages. To further determine if adipocyte-derived PAMM negatively regulates macrophage inflammation, we first collected the conditional cultured medium from differentiated 3T3-L1 adipocytes and then treated the medium with anti-PAMM or control IgG to neutralize PAMM protein in the medium. Raw264.7 cells were incubated with the treated medium for 30 min and then stimulated with or without LPS for 8 h. As shown in Figure 2(C), Western blot showed that PAMM protein could be detected in the cultured medium from mature 3T3-L1 adipocytes. As shown in Figure 2(D), neutralized PAMM in the cultured medium by anti-PAMM significantly increased LPS-induced IL-12 expression in Raw264.7 cells, suggesting that secreted PAMM exerted an inhibitory effect on LPS-induced macrophage activation.
Figure 2. Overexpression of PAMM inhibited inflammatory gene expression in Raw264.7 cells.
(A) Raw264.7 cells were transiently transfected with Flag-PAMM expression plasmid or control plasmid. Cell lysates from transfected cells were analysed by Western blot with anti-Flag antibody. The same membrane was probed by anti-actin antibody and served as a loading control. (B) Transfected Raw264.7 cell were stimulated with or without 1.0 µg/ml LPS for 8 h. Relative mRNA levels of IL-1β, IL-6, IL-12 and iNOS were detected by QPCR. Data represent mean ± S.D., n = 3, **P <0.001 compared with control group. (C) 2-day post-confluent 3T3-L1 cells were incubated with a cocktail of DMI for 10 days to induce differentiation into adipocytes. The cell lysates and cultured media were collected for Western blot analysis of PAMM protein with anti-PAMM. (D) The conditional cultured medium from above experiments was pre-incubated with anti-PAMM (1.0 ng/ml) or control IgG for 1 h, then the medium was added into Raw264.7 cells and incubated for 1 h. Then the cells were stimulated with or without 1.0 µg/ml LPS for 8 h. Relative mRNA level of IL-12 was detected by QPCR. Data represent mean± S.D., n = 3, **P <0.001 compared with control IgG group.
Purified PAMM protein inhibited inflammatory genes expression in Raw264.7 cells
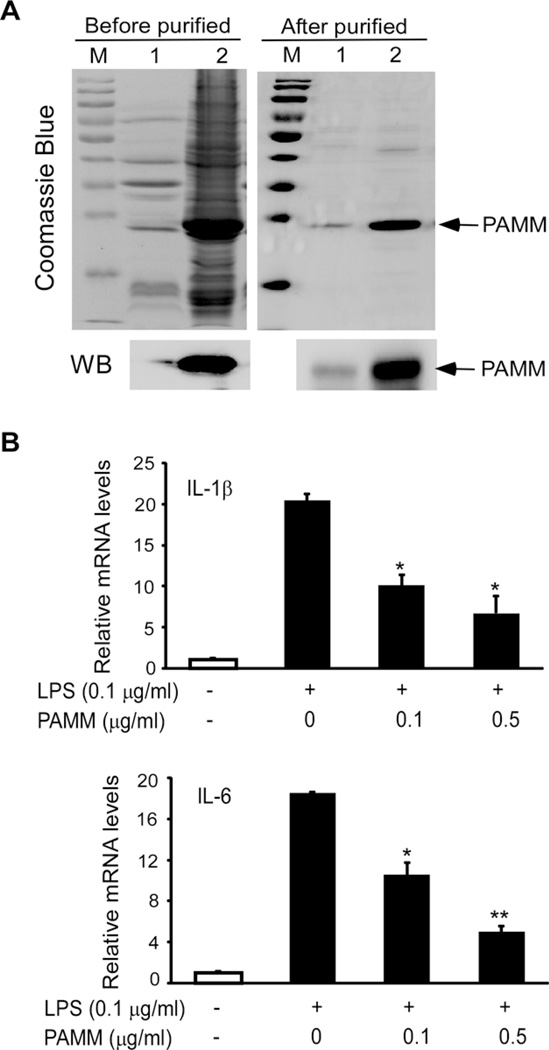
To further confirm that extracellular PAMM exerts anti-inflammatory effect on macrophages, PAMM protein was purified using the BL21 E. coli system, and purified PAMM was confirmed by both Coomassie Blue staining and Western blot analysis with anti-PAMM antibody. As shown in Figure 3(A), human PAMM protein was purified from both supernatants and cell pellets from BL21 E. coli broth cultures. Next, Raw264.7 cells were pretreated with 0, 0.1 or 0.5 µg/ml of purified PAMM protein for 30 min, then stimulated with 0.1 µg/ml of LPS or PBS (negative control) for 8 h. Total RNA was isolated from treated cells and controls for QPCR analysis. As shown in Figure 3(B), purified PAMM protein dose-dependently suppressed the mRNA expression of IL-1β and IL-6 in LPS-stimulated Raw264.7 cells. These results suggest that the inflammatory response of macrophages can be suppressed by exposure to PAMM secreted by other cell types (e.g. adipocytes).
Figure 3. Purified PAMM protein attenuated inflammatory gene expression in Raw264.7 cells.
(A) PAMM was purified from BL21 E. coli by Akata wash. Purified PAMM was examined by Coomassie Blue staining and Western blot analysis with anti-PAMM antibody. M: marker; 1: supernatant; 2: pellet. (B) Raw264.7 cells were incubated with 0, 0.1 or 0.5 µg/ml of purified PAMM protein for 30 min and then stimulated with or without 0.1 µg/ml of LPS for 8 h. Relative mRNA levels of IL-1β and IL-6 were detected by QPCR. Data represent mean± S.D., n = 3, *P <0.05, **P <0.001 compared with LPS without PAMM group.
Overexpression of PAMM inhibited the MAPK signal pathways
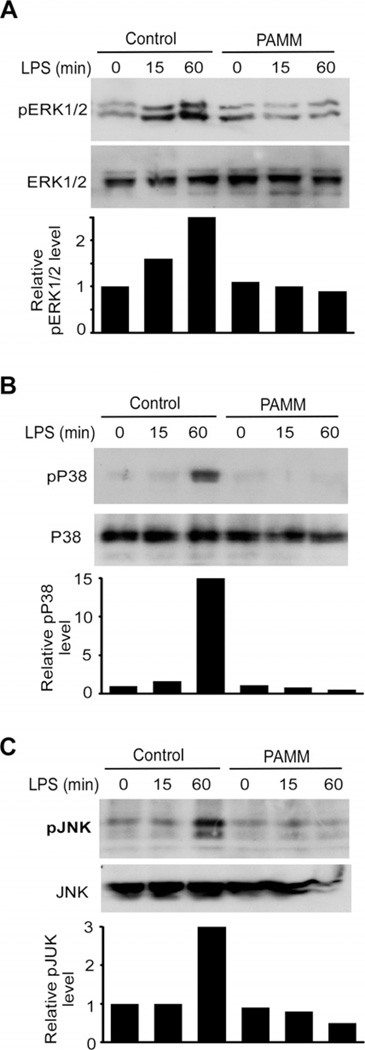
To define the mechanisms by which PAMM suppresses LPS-induced expression of inflammatory cytokines in Raw264.7 cells, we analysed the effect of overexpression of PAMM on LPS-induced activation of NF-κB and MAPK signal pathways. Raw264.7 cells were transiently transfected with Flag-PAMM expression plasmid or control plasmid. After 24 h, transfected cells were stimulated by 1 µg/ml of LPS for different time points as indicated in Figure 3, and cell lysates were harvested for Western blot analysis. As shown in Figure 3, overexpression of PAMM significantly attenuated LPS-induced phosphorylation of ERK1/2, p38 and JNK, but not IKKβ and p65 (data not shown) in Raw264.7 cells. These results suggest that PAMM suppressed the LPS-induced MAPK signalling pathways. Given the critical role of MAPK signalling in responses of macrophages to various inflammatory stimuli, it appears that PAMM suppresses inflammatory responses in LPS-stimulated macrophages by suppressing the MAPK signalling pathways (Figure 4).
Figure 4. Overexpression of PAMM inhibited the MAPK signalling pathway.
Raw264.7 cells were transiently transfected with PAMM expression plasmid or control plasmid. After 24 h, the transfected cells were incubated overnight, then stimulated with 100 ng/ml LPS for 0, 15 and 60 min as indicated. Cell lysates were extracted and Western blot was performed to detect the phosphorylation of ERK1/2 (A), p38 (B) and pJNK (C) with specific antibodies as indicated. Total protein levels of ERK1/2, p38 and JNK were also detected to serve as loading controls. Three independent experiments showed similar results.
The anti-inflammatory property of PAMM is not dependent on its anti-redox activity
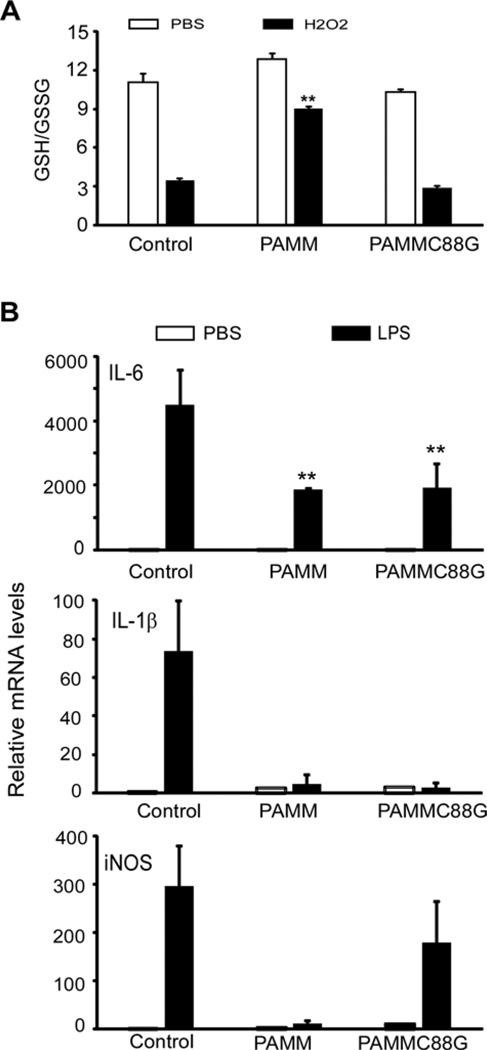
As previously reported, PAMM contains a CXXC-type PRX-like 2 domain [7]. Overexpression of PAMM, shifts the redox status of transfected cells towards a more reduced environment [7]. In contrast, overexpression of a PAMMC88G mutant, in which a cysteine residue was substituted by glycine at position 88 in the CXXC motif, failed to shift the redox status of transfected cells towards a more reduced environment [7]. These results suggest that PAMM functions as a redox regulatory protein and that the CXXC motif is essential for its anti-redox activity [7]. To further define the mechanisms by which PAMM suppresses macrophage inflammation, we transiently transfected Raw264.7 cells with a PAMM expression plasmid, a PAMMC88G mutant plasmid or a control plasmid. Transfected cells were stimulated with H2O2 for 24 h, then GSH–GSSG ratios were measured. GSH is a major tissue antioxidant that is converted to GSSG under conditions of oxidative stress. Thus, reduced GSH–GSSG ratios indicate oxidative stress. As shown in Figure 5(A), overexpression of either PAMM or PAMMC88G failed to affect the GSH–GSSG ratio in normal cells. H2O2 treatment, in contrast, significantly decreased the ratio of GSH–GSSG, indicating oxidative stress. Overexpression of PAMM, but not PAMMC88G, significantly prevented H2O2-caused decreases in the GSH–GSSG ratio, suggesting that PAMM has the capacity to decrease oxidative stress. To examine if PAMM’s anti-inflammatory property is dependent on its anti-redox activity, we transiently transfected Raw264.7 cells with PAMM, PAMMC88G or control plasmids. 24 h later, the transfected cells were stimulated with 1 µg/ml of LPS for 8 h, and total RNA was harvested for QPCR analysis. As shown in Figure 5(B), overexpression of either PAMM or the PAMMC88G mutant, inhibited LPS-induced expression of IL-6 and IL-1β. Since the C → G mutation in PAMMC88G abolishes PAMM’s antioxidant properties, the capacity of PAMMC88G to suppress IL-6 and IL-1β expression in LPS stimulated macrophages suggests that PAMM’s ability to suppress inflammatory cytokine production in LPS-stimulated macrophages is unrelated to its antioxidant properties.
Figure 5. The anti-inflammatory property of PAMM is not dependent on its antioxidant activity.
(A) Raw264.7 cells were transiently transfected with PAMM or PAMMC88G expression plasmids or control plasmid. After 24 h, transfected cells were treated with or without 400 µM of H2O2 for 2 h. The GSH–GSSG ratio was measured in the cytosolic fraction from these cells. Data represent mean± S.D., n = 3, **P <0.001 compared with control group. (B) The transfected cells were stimulated with or without 1 µg/ml of LPS for 8 h. Relative mRNA levels of IL-6, IL-1β and iNOS were detected by QPCR. Data represent mean ± S.D., n = 3, **P <0.001 compared with control group.
PAMM was broadly expressed but highly enriched in adipose tissues
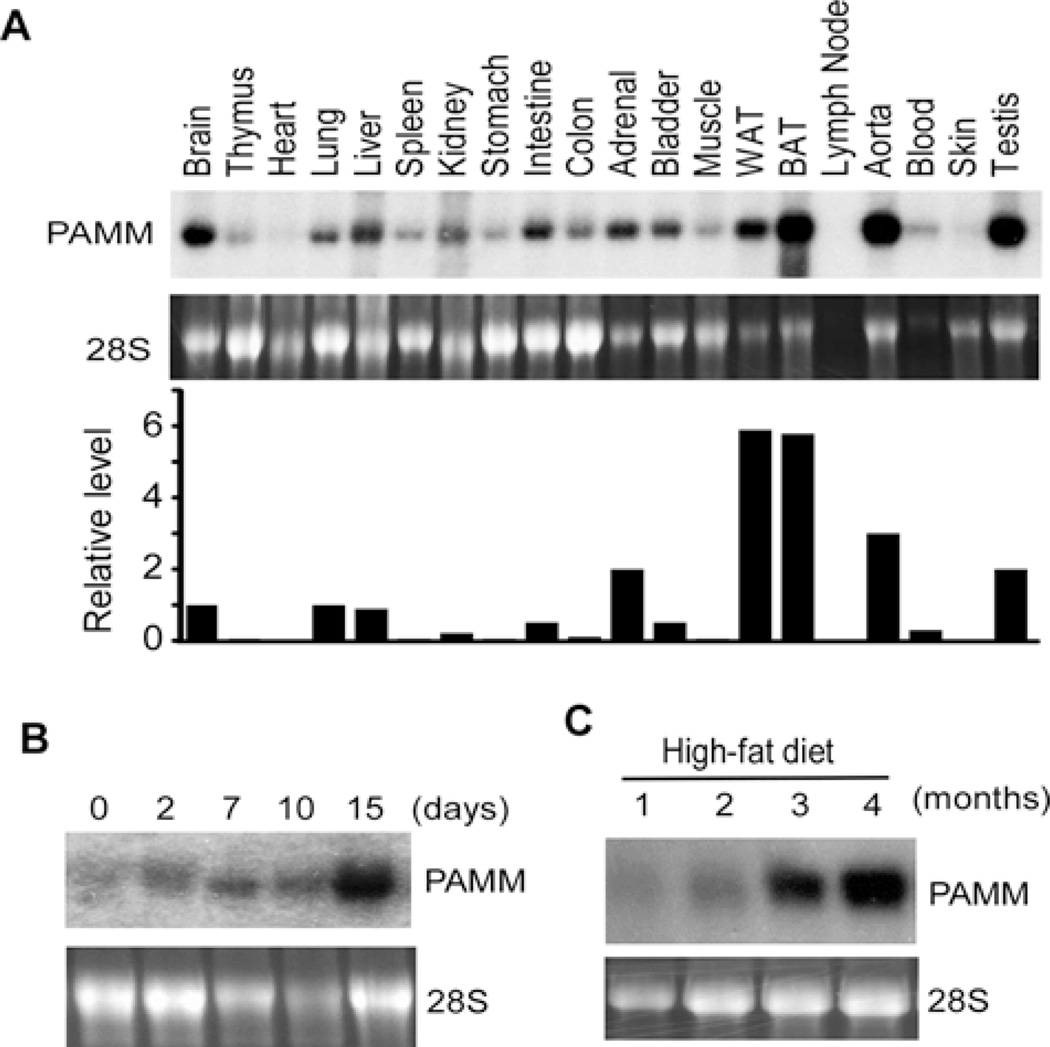
To examine the expression pattern of PAMM mRNA in mouse tissues, we isolated total RNA from adult mouse tissues. Northern blot analysis showed that PAMM mRNA was also broadly expressed in virtually all tissues examined from mice, but was most highly expressed in adipose tissues (Figure 6A). In addition, we observed that PAMM mRNA was up-regulated during 3T3-L1 differentiation (Figure 6B). To examine the expression of PAMM mRNA in adipose tissues under obese status, wild-type C57/BL6 mice at the age of one month were fed with high-fat diet (D12492, containing 60 kcal%fat) for four months [13]. The white adipose tissues (WAT) were collected for Northern blot analysis. As shown in Figure 6(C), PAMM mRNA was significantly increased during high-fat diet-induced obesity, suggesting that PAMM expression is associated with obesity.
Figure 6. PAMM is broadly expressed but highly enriched in adipose tissues.
(A) Northern blot analysis of PAMM mRNA expression in mouse tissues. BAT, brown adipose tissue. (B) Northern blot analysis of PAMM mRNA expression during DMI-induced 3T3-L1 differentiation. (C) C57/BL6 mice at 1-month age were fed with a high-fat diet. The white adipose tissues were collected at different time points as indicated for Northern blot analysis of PAMM mRNA.
DISCUSSION
It is now broadly accepted that low-grade, chronic inflammation associated with obesity leads to the onset of insulin resistance and Type 2 diabetes mellitus [14,15]. Obesity-associated inflammation is characterized by an increased abundance of macrophages in adipose tissue along with production of inflammatory cytokines [1,2]. Macrophages in adipose tissue secrete inflammatory mediators such as TNFα and IL-6 that interfere with adipocyte function by inhibiting responses to insulin [5]. Adipocytes also secrete many inflammatory or anti-inflammatory mediators, such as resistin and adiponectin, that interfere with macrophage function [16,17]. The focus of this study was to search for novel proteins secreted by adipocytes that negatively regulate responses of macrophages to inflammatory stimuli. To accomplish this, we first analysed alterations in gene expression between 3T3-L1 preadipocytes and differentiated 3T3-L1 adipocytes. Genes that had not been previously characterized and that were up-regulated by more than 5-fold in adipocytes compared with undifferentiated 3T3-L1 cells were selected for analysis by SignalP 3.0 and SMART programs. Genes that met these criteria were then examined to determine whether they encoded proteins with putative signal peptides, indicating that they are likely to be secreted by adipocytes. Several genes meeting all of these criteria were identified, but one in particular, attracted our interest, and the remainder of the study focused on the product of this gene. PAMM is a poorly characterized protein with a putative signal peptide and a PRX-like 2 domain. Microarray analysis demonstrated that PAMM mRNA expression was up-regulated by 8-fold in differentiated 3T3-L1 adipocytes compared with undifferentiated 3T3-L1 cells, and Northern blots confirmed that PAMM was significantly up-regulated during 3T3-L1 cell differentiation. PAMM mRNA was expressed in virtually all mouse tissues examined, but it was particularly abundant in adipose tissue. The present study demonstrated that PAMM is a secreted protein that can inhibit the production of inflammatory cytokines in LPS-stimulated macrophages.
There are five CXXC-motif containing PRX isoforms recognized in mammals [18]. On the basis of the peptide regions that harbour a conserved cysteine residue, they can be classified into typical and atypical CXXC motif-containing PRXs. Peroxiredoxin I, II, III and IV are typical 2-Cys PRXs, whereas PRX V is an atypical 2-Cys PRX [19]. Both PRX I and II lack a putative signal sequence and localize in the cytosol. However, they can be released from lysed cells and presented to natural killer (NK) cells through an unknown receptor to enhance the cytotoxic activity of NK cells [20]. Unlike PRX I and PRX II, PRX IV has a distinct hydrophobic N-terminal signal sequence that distinguishes it from the other mammalian PRXs and suggests that it is actively secreted from cells and functions in the extracellular space [21]. In addition, proteomic and molecular studies suggest that 2-Cys PRXs are secreted by many helminth pathogens [22–26]. In the present study, we first identified PAMM as a novel secreted protein that contains a PRX-like 2 domain. In addition to its anti-redox activity, PAMM also has the capacity to potently inhibit production of inflammatory cytokines by LPS-stimulated macrophages. We hypothesize that PAMM, secreted by adipocytes, may play an important role in controlling the activation and function of macrophages that infiltrate adipose tissue in vivo. Since it is broadly expressed in other, non-adipose tissues, though at substantially lower levels, PAMM may also provide an anti-inflammatory function in these tissues, but this will require additional investigation.
The mechanisms by which PAMM inhibits the expression of inflammatory cytokines in macrophages had been unknown. It is well established that H2O2 can function as a second message to inflammatory stimuli, mediating receptor-initiated signal transduction including nuclear factor-κB (NF-κB) and MAPK signal pathways [27,28]. However, we demonstrated that PAMM’s ability to inhibit production of inflammatory cytokines by LPS-stimulated macrophages is independent of its antioxidant properties. Specifically, we observed that overexpression of PAMM inhibited the phosphorylation of ERK1/2, p38 and JNK, key molecules in the MAPK signalling pathway, which is critical to the ability of macrophages to respond to various inflammatory stimuli. The mechanism by which extracellular PAMM actually inhibits the MAPK pathway is uncertain. It has previously been reported that the 2-Cys PRX from Plasmodium berghei was shown to interact with the toll-like receptor-4 on macrophages, leading to the production of pro-inflammatory cytokines [29]. Consequently, it is reasonable to speculate that PAMM binds directly to specific membrane receptors such as toll-like receptor 4 to suppress the MAPK pathway.
In summary, our present work identified PAMM as a novel secreted protein with potent anti-inflammatory properties. PAMM inhibits production of inflammatory cytokines by LPS stimulated macrophages, and we have evidence that this occurs through inhibition of the MAPK signalling pathway, rather than via its antioxidant properties. PAMM is highly expressed in adipose tissue, and is broadly expressed in all other mouse tissues investigated, suggesting that it may have essential roles in other tissues. In addition, PAMM expression in adipose tissues is highly associated with obesity, suggesting that the inflammatory environment of obese adipose may increase PAMM expression. However, why increased PAMM expression fails to turn off the macrophage inflammation needs to be further studied. Also, although we have shown that PAMM was an inhibitor of the MAPK pathway, the exact molecular mechanisms by which PAMM inhibited MAPK remain to be discovered.
Acknowledgments
FUNDING
This work was supported by the School of Medicine, University of Missouri – Kansas City [grant number DSF2014]; National Institute of Health [grant number AI103618 to (M.F.)]; and National Natural Foundation of China [(grant numbers 91339113, 81270202 and 81070095 to (H.X.), 81470435, 81170277 to (Z.J.)].
Abbreviations
- JNK
c-Jun N-terminal kinase
- DMEM
Dulbecco’s modified Eagle’s medium
- DMI
dexamethasone, methylxanthine and insulin
- HEK293
human embryonic kidney
- IL-6
interleukin-6
- iNOS
inducible nitric oxide synthase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MCPIP1
monocyte chemotactic protein induced protein 1
- M-CSF
macrophage-colony stimulating factor
- NK
natural killer
- PAMM
PRX-like 2 activated in M-CSF-stimulated monocytes
- PRX
peroxiredoxin
- RANKL
receptor activator of nuclear factor kappa-B ligand
- TNFα
tumour necrosis factor α
- TTBS
0.05% Tween 20 in TBS
- WAT
white adipose tissue.
Footnotes
AUTHOR CONTRIBUTION
Mingui Fu conceived and coordinated the study and wrote the paper. Fang Guo, Hui He, Shengping Huang performed and analysed the experiments shown in Figures 2, 4 and 5. Zhi-Chao Fu and Tingtao Chen designed, performed and analysed the experiments shown in Figure 3. Yan Xu and Leslie Morse designed, performed and analysed the experiments shown in Figure 1E. Mingui Fu designed, performed and analysed the experiments shown in Figures 1 and 6. Christopher Papasian, Ricardo Battaglino, Xiao-Feng Yang, Zhisheng Jiang and Hong-Bo Xin helped to revise the manuscript and provided advice. All authors reviewed the results and approved the final version of the manuscript.
REFERENCES
- 1.McNelis JC, Olefsky JM. Macrophages, immunity, and metabolic disease. Immunity. 2014;17:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Lee J. Adipose tissue macrophages in the development of obesity-induced inflammation, insulin resistance and Type 2 diabetes. Arch. Pharm. Res. 2013;36:208–222. doi: 10.1007/s12272-013-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han JM, Levings MK. Immune regulation in obesity-associated adipose inflammation. J. Immunol. 2013;191:527–532. doi: 10.4049/jimmunol.1301035. [DOI] [PubMed] [Google Scholar]
- 4.Chmelar J, Chung KJ, Chavakis T. The role of innate immune cells in obese adipose tissue inflammation and development of insulin resistance. Thromb. Haemost. 2013;109:399–406. doi: 10.1160/TH12-09-0703. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Santibañez G, Lumeng CN. Macrophages and the regulation of adipose tissue remodeling. Annu. Rev. Nutr. 2014;34:57–76. doi: 10.1146/annurev-nutr-071812-161113. [DOI] [PubMed] [Google Scholar]
- 6.Ohashi K, Shibata R, Murohara T, Ouchi N. Role of anti-inflammatory adipokines in obesity-related diseases. Trends Endocrinol. Metab. 2014;25:348–355. doi: 10.1016/j.tem.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Morse LR, da Silva RA, Odgren PR, Sasaki H, Stashenko P, Battaglino RA. PAMM: a redox regulatory protein that modulates osteoclast differentiation. Antioxid. Redox Signal. 2010;13:27–37. doi: 10.1089/ars.2009.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid. Redox Signal. 2011;15:781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 9.Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med. 2005;11:571–578. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang CS, Lee DS, Song CH, An SJ, Li S, Kim JM, Kim CS, Yoo DG, Jeon BH, Yang HY, et al. Roles of peroxiredoxin II in the regulation of proinflammatory responses to LPS and protection against endotoxin-induced lethal shock. J. Exp. Med. 2007;204:583–594. doi: 10.1084/jem.20061849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi D, Huang S, Miao R, She ZG, Quinn T, Chang Y, Liu J, Fan D, Chen YE, Fu M. Monocyte chemotactic protein-induced protein 1 (MCPIP1) suppresses stress granule formation and determines apoptosis under stress. J. Biol. Chem. 2011;286:41692–41700. doi: 10.1074/jbc.M111.276006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Zhang Y, Sun T, Chandalia M, Abate N, Fan D, Xin HB, Chen YE, Fu M. Dietary obesity induced Egr-1 in adipocytes facilitates energy storage via suppression of FOXC2. Sci. Rep. 2013;3:1476. doi: 10.1038/srep01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNelis JC, Olefsky JM. Macrophages, immunity, and metabolic disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Mraz M, Haluzik M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J. Endocrinol. 2014;222:R113–R127. doi: 10.1530/JOE-14-0283. [DOI] [PubMed] [Google Scholar]
- 16.Lazar MA. Resistin- and obesity-associated metabolic diseases. Horm. Metab. Res. 2007;39:710–716. doi: 10.1055/s-2007-985897. [DOI] [PubMed] [Google Scholar]
- 17.Anderson EK, Gutierrez DA, Hasty AH. Adipose tissue recruitment of leukocytes. Curr. Opin. Lipidol. 2010;21:172–177. doi: 10.1097/MOL.0b013e3283393867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 19.Wood ZA, Schröder E, Robin-Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 20.Shau H, Gupta RK, Golub SH. Identification of a natural killer enhancing factor (NKEF) from human erythroid cells. Cell Immunol. 1993;147:1–11. doi: 10.1006/cimm.1993.1043. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto A, Okado A, Fujii T, Fujii J, Egashira M, Niikawa N, Taniguchi N. Cloning of the peroxiredoxin gene family in rats and characterization of the fourth member. FEBS Lett. 1999;443:246–250. doi: 10.1016/s0014-5793(98)01736-0. [DOI] [PubMed] [Google Scholar]
- 22.Cass CL, Johnson JR, Califf LL, Xu T, Hernandez HJ, Stadecker MJ, Yates JR, 3rd, Williams DL. Proteomic analysis of Schistosoma mansoni egg secretions. Mol. Biochem. Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gourbal BE, Guillou F, Mitta G, Sibille P, Thèron A, Pointier JP, Coustau C. Excretory–secretory products of larval Fasciola hepatica investigated using a two-dimensional proteomic approach. Mol. Biochem. Parasitol. 2008;161:63–66. doi: 10.1016/j.molbiopara.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Wu XJ, Sabat G, Brown JF, Zhang M, Taft A, Peterson N, Harms A, Yoshino TP. Proteomic analysis of Schistosoma mansoni proteins released during in vitro miracidium-to-sporocyst transformation. Mol. Biochem. Parasitol. 2008;164:32–44. doi: 10.1016/j.molbiopara.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Cui SJ, Hu W, Feng Z, Wang ZQ, Han G. Excretory/secretory proteome of the adult developmental stage of human blood fluke, Schistosoma japonicum. Mol. Cell Proteomics. 2009;8:1236–1251. doi: 10.1074/mcp.M800538-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar R, Tsuji N, Morales TH, Carmody AB, Ozols VO, Welton J, Tang L. Removal of hydrogen peroxide by a 1-cysteine peroxiredoxin enzyme of the filarial parasite Dirofilaria immitis. Parasitol. Res. 2000;86:200–206. doi: 10.1007/s004360050032. [DOI] [PubMed] [Google Scholar]
- 27.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 29.Furuta T, Imajo-Ohmi S, Fukuda H, Kano S, Miyake K, Watanabe N. Mast cell-mediated immune responses through IgE antibody and Toll-like receptor 4 by malarial peroxiredoxin. Eur. J. Immunol. 2008;38:1341–1350. doi: 10.1002/eji.200738059. [DOI] [PubMed] [Google Scholar]