Abstract
The composition of the gut microbiome with use of nonsteroidal anti-inflammatory drugs (NSAIDs) has not been fully characterized. Drug use within the past 30 days was ascertained in 155 adults and stool specimens were submitted for analysis. Area under the receiver operating characteristic curve (AUC) was calculated in logit models to distinguish relative abundance of operational taxonomic units (OTUs) by medication class. The type of medication had a greater influence on the gut microbiome than the number of medications. NSAIDs were particularly associated with distinct microbial populations. Four OTUs (Prevotella spp., Bacteroides spp, family Ruminococaceae, Barnesiella spp.) discriminated aspirin users from no medication (AUC=0.96; 95% CI 0.84, 1.00). The microbiome profile of celecoxib users was similar to ibuprofen users, with both showing an enrichment of Acidaminococcaceae and Enterobacteriaceae. Bacteria from families Propionibacteriaceae, Pseudomonadaceae, Puniceicoccaceae, and Rikenellaceae were more abundant in individuals who were taking ibuprofen than in controls or in individuals who were taking naproxen. Bacteroides spp. and Erysipelotrichaceae spp. discriminated use of NSAIDs with proton pump inhibitors versus NSAIDs alone (AUC=0.96; 95% CI: 0.87, 1.00). Bacteroides spp. and a bacterium of family Ruminococcaceae discriminated individuals who were taking NSAIDs in combination with antidepressants and laxatives versus individuals who were taking NSAIDs alone (AUC=0.98; 95% CI: 0.93, 1.00). In conclusion, bacteria in the gastrointestinal tract reflect the combinations of medications that people ingest. The bacterial composition of the gut varied with the type of NSAID ingested.
Keywords: microbiota, bacteria, drug, gastrointestinal tract, nonsteroidal anti-inflammatory drugs
INTRODUCTION
Nearly half of Americans are current users of prescription drugs and 10.6% took five or more prescription drugs within the past 30 days [1]. Over-the-counter (OTC) drugs are used more frequently, with 84% of Americans taking OTC medications to treat colds, influenza, coughs or sinus problems, 82% for pain, 77% for gastrointestinal upset, and 75% for allergies [2]. Nonsteroidal anti-inflammatory drugs (NSAIDs) are among the most widely used medications. It was recently estimated that 19.0% of U.S. adults (43.6 million) ingested aspirin at least three times a week on a chronic basis, while an estimated 12.8% (29.4 million) took NSAIDs at least three times a week for at least 3 months [3].
While medication use is common, the gastrointestinal effects of drug combinations have not been fully characterized. We hypothesized that the gut microbiome is responsive to the entirety of medications used at a given time and we were particularly interested in the microbiome after ingestion of NSAIDs because of their frequent use in adults and their capacity to induce gastrointestinal effects, some mediated through the inhibition of prostaglandin formation [4]. There is considerable evidence to show that NSAIDs cause bleeding, inflammation, and ulceration in the stomach and small intestine [5]. To minimize the upper gastrointestinal effects, often proton pump inhibitors or H2 receptor antagonists are prescribed for patients taking NSAIDs. Yet, the NSAID-induced enteropathy in the distal small intestine remains a concern for long-term users of NSAIDs, as well as other side effects such as the increased risk of stroke and cardiovascular disease [6].
Although NSAID-induced gastropathy and enteropathy have been recognized for some time [7], investigators more recently suggested that such effects may originate with dysbiosis, that is, perturbations in the gut microbiome [8,9]. NSAID enteropathy does not occur in germ-free mice but when Eubacterium limosum or Escherichia coli are introduced, intestinal ulceration occurs [10,11]. Other animal studies indicate that, through changes in enteric bacterial content, NSAID effects may be potentiated when administered with proton pump inhibitors [12].
In this study, we explored the biodiversity and composition of the gut microbiome after medication use in community volunteers.
METHODS
Community residents, 18 years of age or older, were recruited from southeastern Michigan in the United States and were enrolled in the study from January 2011 through January 2012 [13]. Exclusion criteria were: diarrhea within the previous 7 days, pregnancy, hospitalization, and residence in a skilled nursing facility. The study was approved by the University of Michigan Institutional Review Board.
Subjects completed a questionnaire regarding demographic characteristics, medical history, diet, and all medications (both OTC and prescription) used within the past 30 days. Vitamins, minerals and other supplements were also recorded. For purposes of this study, we considered use of any prescription drug, OTC drug, herbal supplement, or vitamin/mineral supplement as “drug” use. Controls were individuals who did not use any prescription drugs, nonprescription drugs, herbal supplements, and/or vitamin/mineral supplements in the past 30 days.
Each participant submitted a stool sample for bacterial DNA analyses using a home stool specimen kit. PowerSoil®-htp 96 Well Soil DNA Isolation Kits (MO BIO Laboratories, Inc., Carlsbad, CA) were utilized to extract total bacterial DNA with an epMotion® 5075 Liquid Handling Workstation (Eppendorf, Hamburg, Germany). Using the 454 GS FLX pyrosequencing platform, the V35 region of the 16S rRNA gene was amplified and sequenced, followed by curation using mothur software [13,14]. Operational taxonomic unit (OTU) clustering was performed with a 3% distance cutoff and the Ribosomal Database Project naïve Bayesian classifier was used to assign OTUs. Taxonomic assignments were determined by using a naive Bayesian classifier with the Ribosomal Database Project training set (version 9) with an 80% bootstrap confidence threshold. All samples were rarefied to 1,450 sequences per sample [13]. Of the samples with >1,450 sequences, the mean sequence per sample was 6,091 (range, 1450-17120; median, 5986; median absolute deviation, 1296).
For initial microbial analyses, differences in bacterial OTUs from subjects using no medication were compared with individuals taking only one medication. This was followed by an analysis of individuals taking various drug combinations. These included medications previously identified as having gastrointestinal effects: antibiotics (Abx), proton pump inhibitors (PPI), H2 receptor antagonists (H2R), laxatives (Lax), antiemetics, antidepressants (ATD), and NSAIDs. Using previously described methods [13], participants were categorized by ranked differences in relative abundance of bacterial OTUs by drug class. The greatest ranked differences in bacterial relative abundance were used in logit models to assess the discriminatory ability of bacterial OTUs to discriminate categories of medication use. Post-estimation calculations of the area under the curve (AUC) from these logit models were determined for receiver operator characteristic (ROC) curves with 95% confidence intervals (CI). Differences between nested logit models were compared using the test for the equality of ROC areas [15]. Wilcoxon rank-sum test was used to assess relative differences in OTUs by medication class. In addition, two bacterial biodiversity indices were evaluated, the Inverse Simpson Index [16] and the Shannon Index [17]; for both indices, an increased value indicates greater diversity. Principal component analysis was used to generate a biplot of the scores of the OTUs and type of NSAID (ibuprofen, naproxen) for the first two principal components. Data were analyzed using Stata/MP 13.1 (StataCorp LP, College Station, Texas, USA).
RESULTS
There were 155 individuals who participated in the study whose mean age was 52 years (range, 19 to 88). Two-thirds of the participants were women (66%) and 83% were Caucasian. The mean weight was 190 pounds (SD, 38) for men and 165 pounds (SD, 43) for women. Older participants were more likely to use a greater number of medications than younger subjects (p<0.001) but there were no significant associations between the number of medications used in the past 30 days and gender, race, or weight.
The most frequently ingested drugs were NSAIDs; 62 of the 155 participants (40%) used NSAIDs within the past 30 days. The next most commonly used medication class was antibiotics which were taken by 14.8% of the participants, followed by antidepressants taken by 13.5%. Of the 155 participants, only 16.8% did not take any drugs within the past 30 days.
Biodiversity was significantly lower in those who used antibiotics versus those who did not (Shannon Index, p=0.0022; Inverse Simpson Index, p=0.0325). The types of antibiotics used were cephalosporin, penicillin, clindamycin and quinolone. The Inverse Simpson Index was lowest for those individuals taking OTC medications for cold/flu symptoms (mean 5.25), those taking antibiotics (mean 6.01), and those taking antidepressants (mean 6.07). The results were similar for the Shannon Index (mean 2.32, mean 2.20, mean 2.42, respectively).
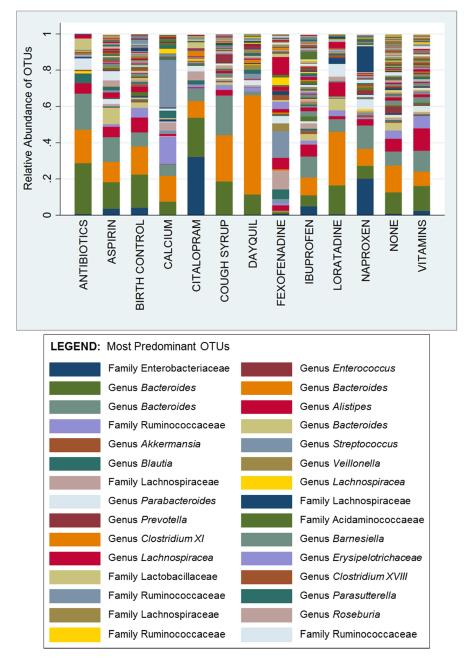
Of the 155 people, 14.8% used only 1 medication. The gut bacterial composition for participants taking only 1 medication versus those taking none is shown in Figure 1. The relative abundance of a specific OTU in the Enterobacteriaceae family was significantly higher (p<0.0001) in persons taking the antidepressant citalopram (32%) and persons taking the NSAID naproxen (20%) than in individuals not taking any medications (0.7%). There was an enrichment of a specific Bacteroides spp. in persons taking DayQuil® and those taking cough syrup.
FIGURE 1.
Gut Bacterial Operational Taxonomic Units (OTUs) for Subjects using only one Medication versus Subjects using no Medication
To assess discriminatory ability of the gut bacteria for aspirin use versus no medication, the AUC for the ROC curve was 0.96 (95% CI 0.84, 1.00) when 4 bacteria were entered into the model (Prevotella spp., Bacteroides spp., an OTU from family Ruminococaceae, Barnesiella spp.). This group of bacteria was more discriminatory than individual OTUs, although a specific Bacteroides spp. did yield an AUC of 0.83 (95% CI: 0.61, 1.00) when modeled alone. In comparison, the discriminatory ability of age and gender in determining aspirin use was lower, with an AUC of 0.77 (95% CI: 0.51, 1.00).
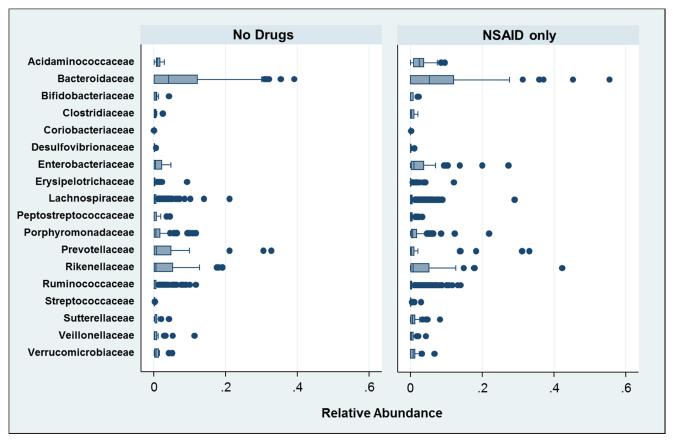
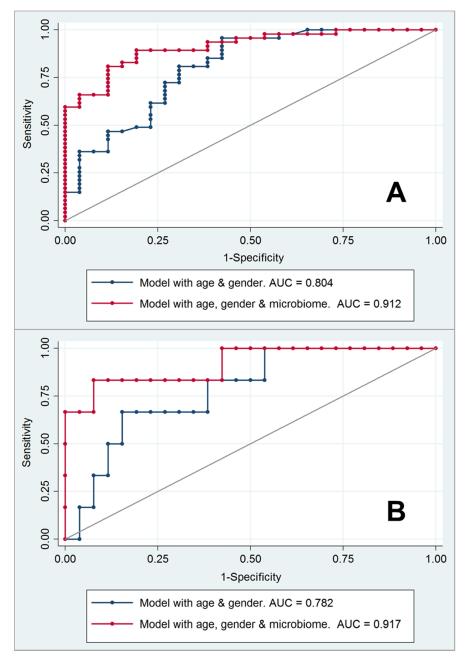
Microbial differences in relative abundance for NSAID users (without other medications) and controls are shown in Figure 2. Bacteria of the family Acidaminococcaceae were significantly more prevalent in those taking NSAIDs compared to those not taking any medications (p=0.0246). In a logit model, there were 4 specific bacterial OTUs which significantly discriminated between individuals taking NSAIDs versus those taking no medication. These OTUs were: (b) 2 distinct organisms of Bacteroides spp.; (b) bacterium in the family Enterobacteriaceae; and (c) bacterium in the family Acidaminococcaceae. Of the 2 OTUs of Bacteroides spp., 1 was significantly more common in the NSAIDs group (p=0.040) while the other was more common in controls (p=0.022). Discrimination in a logit model produced an AUC of 0.86 with inclusion of these 4 OTUs (95% CI: 0.78, 0.94). In comparison, the AUC for age and gender was 0.80 (95% 69, 0.92). The addition of microbiome data significantly added to the predictive abilities of the model with age and gender (AUC=0.91; 95% CI: 0.85, 0.97; p=0017 for the difference between models) and is shown in Figure 3.
FIGURE 2.
Prevalence of Gut Bacterial Families for Subjects using only NSAIDs versus Subjects not using Medications
FIGURE 3.
ROC Curves for Individuals Taking (A) NSAIDs or (B) Naproxen showing Discrimination by Age, Gender and Microbiome Data
Bacterial relative abundance also distinguished persons taking naproxen versus those taking no medication; the AUC was 0.92 (95% CI: 0.77, 1.00) as shown in Figure 3. The OTUs entered into this model were: a bacterium of family Enterobacteriaceae, 3 bacteria of Bacteroides spp., and one bacterium of Alistipes spp. In comparison, the AUC for age and gender was 0.78 (95% CI: 0.59, 0.97). For ketorolac, one OTU of Alistipes spp. was highly predictive of this drug use (relative abundance 42% with ketorolac, relative abundance 7% with no medication, p<0.0001). The microbiome profile of celecoxib users was more similar to ibuprofen users, with both showing an enrichment of Acidaminococcaceae and Enterobacteriaceae.
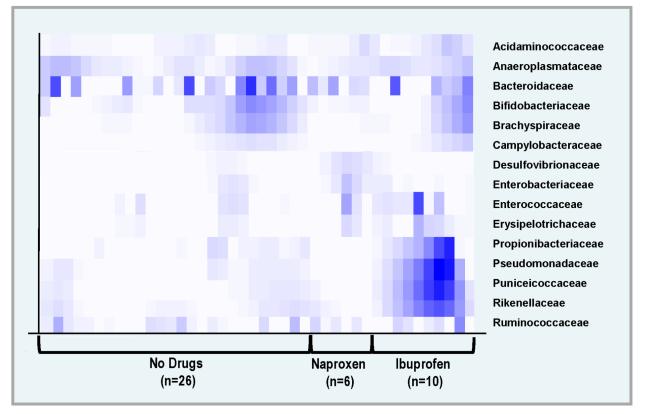
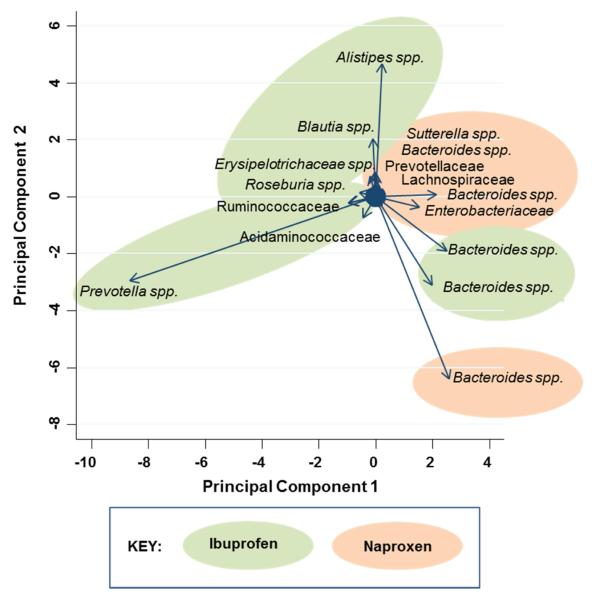
Figure 4 shows a heat map of bacterial families in naproxen users, ibuprofen users, and controls (for families with greatest differences in relative abundance). Bacteria from families Propionibacteriaceae, Pseudomonadaceae, Puniceicoccaceae, and Rikenellaceae were more abundant in individuals who were taking ibuprofen than in controls or in individuals who were taking naproxen. Bacteria of families Acidaminococcaceae, Desulfovibrionaceae, Enterococcaceae, and Erysipelotrichaceae were more common in the NSAID users than in the controls. In secondary analyses, we conducted a principal component analysis of the relative abundance of OTUs distinguishing ibuprofen from naproxen. The biplot with vectors is shown in Figure 5. The most influential bacteria distinguishing ibuprofen from naproxen were Prevotella spp. and Alistipes spp. in ibuprofen users and a specific Bacteroides spp. in naproxen users.
FIGURE 4.
Heat Map for Bacterial Families showing differences in Relative Abundance by Medication Use
FIGURE 5.
Principal Component Analysis Biplot of Bacteria to distinguish Ibuprofen Users from Naproxen Users
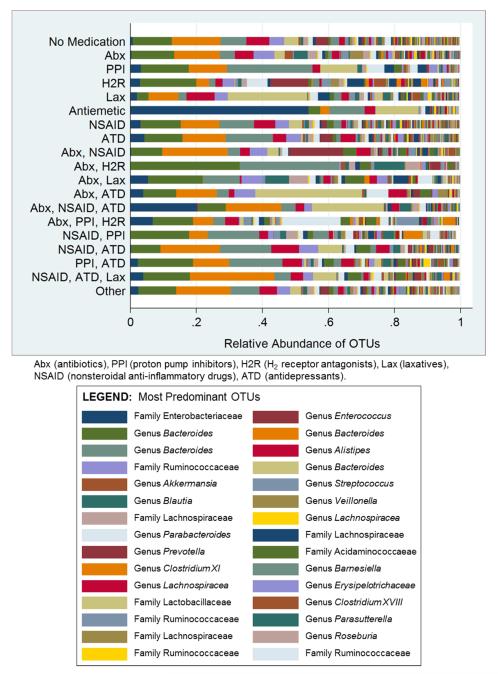
The gut microbiome of participants who used multiple medications was also examined (Figure 6). The most predominant bacterium in the antiemetic group was from the family Enterobacteriaceae (53.9% relative abundance). For participants taking both antibiotics and H2 receptor antagonists, two bacteria of the species Bacteroides were most abundant (33% and 30%). There were differences in the gut microbiome for individuals taking NSAIDs with PPIs versus those taking NSAIDs without PPIs. In a logit model comparing persons taking NSAIDs with PPIs versus those taking only NSAIDs, the AUC was 0.96 (95% CI: 0.87, 1.00) when 2 specific OTUs were included (Bacteroides spp., Erysipelotrichaceae spp.). Logit models comparing participants taking NSAIDs with antidepressants versus those taking NSAIDs (without antidepressants) also were predictive, with an AUC of 0.85 (95% CI: 0.55, 1.00) for 4 OTUs (3 Bacteroides spp. and 1 Lachnospiracea spp.). When the next organism was added (family Acidaminococcaceae), the AUC was 0.96 (95% CI: 0.87, 1.00). Three specific OTUs (2 Bacteroides spp., 1 family Ruminococcaceae) significantly discriminated those individuals who were taking NSAIDS in combination with antidepressants and laxatives versus individuals who were taking NSAIDs (without antidepressants or laxatives). The AUC was 0.98 (95% CI: 0.93, 1.00).
FIGURE 6.
Relative Abundance of Bacterial Operational Taxonomic Units (OTUs) by Medication Type
The total number of medications used in the past 30 days was calculated for each subject (mean=4; range, 0 to 20). The number of drugs used was not significantly associated with either the Shannon Index (coefficient 0.01, p=0.448) or the Inverse Simpson Index (coefficient 0.11, p=0.340).
DISCUSSION
Bacteria in the human gastrointestinal tract reflect the types of medications that people ingest. There were differences in the relative abundance of specific bacteria for individuals who took a single drug, people who used drug combinations, and those who did not use any medications. The number of drugs did not significantly impact gut biodiversity but the specific types of medications did. In particular, NSAID users exhibited a different gut microbiome profile than nonusers, as did users of specific types of NSAID (e.g., ibuprofen, naproxen).
There is evidence that the adverse effects of NSAIDs vary with the particular type of NSAID ingested [6]. Ketoprofen, naproxen and ketorolac tend to exhibit more aggressive gastrointestinal effects than ibuprofen or celecoxib [18-20]. We found that individuals taking ibuprofen or celecoxib had similar gut microbiome profiles, while those taking naproxen or ketorolac had distinctly different profiles. There has also been recent attention regarding the vascular effects of NSAIDs; the Food and Drug Administration strengthened its warnings on NSAID use because of the increased risks of myocardial infarction and stroke [21]. In a meta-analysis of 280 randomized trials of NSAIDs published in The Lancet, ibuprofen increased the risk of coronary events 2-fold, yet naproxen did not increase the risk of vascular events [6]. Wang and colleagues demonstrated that the gut microbiota can promote cardiovascular disease through phosphatidylcholine metabolism [22]; their results suggested that dietary choline (generally from high consumption of animal-derived foods) is used by the gut microbiota to produce trimethyl amine (TMA) and trimethylamine N-oxide (TMAO), thus raising cardiovascular risk. Naproxen use has been shown to significantly decrease urinary choline concentrations [23]. Cracium and Balskus identified the gene cluster responsible for the conversion of choline to TMA by anaerobic bacteria [24]. Their data indicate that specific types of anaerobes such as Desulfovibrio, Clostridia, Streptococcus, Klebsiella, Proteus and others, are choline degraders – while bacteria in the phylum Bacteroidetes generally do not. Our results, taken with this existing evidence, could suggest that naproxen influences TMA and TMAO production (thereby lowering vascular risk) through alteration of the types of choline-utilizing anaerobes that are present.
Although NSAIDs are commonly used for the treatment of pain, they are often prescribed with proton pump inhibitors or H2 receptor antagonists which alleviate upper gastrointestinal effects such as heartburn. However, such combinations have been shown to exacerbate enteropathy in the more distal small intestine [4,5]. Our findings demonstrate that the gut microbiome is different in individuals who take such combinations of medications (e.g., NSAIDs with proton pump inhibitors) than in those using NSAIDs alone. Moreover, the addition of antidepressants and laxatives further changed the bacterial composition of the gastrointestinal tract. While the microbial-induced effects of NSAIDs are of considerable concern, there are some preliminary studies of prostaglandin-inducers showing enhancement of the healing of duodenal ulcers [25]. Studies of prostaglandin-inducers may provide additional insights into the microbial interplay between mucosal integrity and ulcerative injury in the gut.
Medication use in our study was similar to what was found in the Mayo Clinic study using data from the Rochester Epidemiology Project [26]. In both of these investigations, the most common prescription medication was antibiotics followed by antidepressants. However, in our study, we also measured OTC drug use and found that such medications (particularly, NSAIDs) were frequently used (40% in the past 30 days). In the Mayo Clinic study, 20% of individuals received prescriptions for 5 or more medications in a 1-year period, while we found that 35% of community residents took 5 or more medications (OTC and prescription combined) in the past 30 days.
Because our study was not experimental in design, we cannot ascertain whether the gut microbiome is a reflection of the medications per se or of the underlying disease that the medications were intended to treat. There is ample evidence that antibiotics will reduce bacterial abundance [27,28] but for some medications (NSAIDs, antidepressants), it is possible that the bacterial composition could partly reflect the underlying condition. For example, in those participants who were taking medications for symptoms of a cold or flu, the biodiversity index was rather low which could be a reflection of the medication itself or, alternatively, may be a marker for microbial shifts during an infection. On the other hand, drugs are developed specifically for their ability to alter metabolic pathways and expectations would include possible bacterial response and alterations in end products.
In conclusion, both OTC and prescription drug use are common in adults and the gut microbiome reflects this use. For NSAIDs, bacterial composition varied for specific drugs, with ibuprofen and celecoxib having similar microbiome profiles while naproxen and ketorolac had different profiles. Bacterial composition was altered when NSAIDs were used with other drugs such as proton pump inhibitors, antidepressants and laxatives. We found that it was the types of medications used, rather than the number of medications, which yielded the greatest differences in the microbiome. Additional investigations evaluating microbial transience with drug dosages or with intermittent drug use may be informative, as would metabolic studies linked to functional outcomes in individuals. Hopefully, such studies will eventually help people better tolerate multiple medications or, alternatively, assist decision-making regarding the types of medications that would provide greatest benefit.
ACKNOWLEDGMENTS
This work was supported by NIH grants: 1R01GM099514, R01HG005975, U19AI090871, and P30DK034933.
We thank Alyxandria Schubert, Cathrin Ring and Jill Mogle for their assistance with clinical samples. We thank Patrick Schloss and Vincent Young for securing funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST: None.
REFERENCES
- 1.National Center for Health Statistics . Health, United States, 2013: with special feature on prescription drugs. Hyattsville, Maryland: 2014. [PubMed] [Google Scholar]
- 2.Consumer Healthcare Products Association . Understanding trust in OTC medicines: consumer and healthcare provider perspectives. Nielsen and IMS; 2013. [Google Scholar]
- 3.Zhou Y, Boudreau DM, Freedman AN. Trends in the use of aspirin and nonsteroidal anti-inflammatory drugs in the general U.S. population. Pharmacoepidemiol Drug Saf. 2014;23:43–50. doi: 10.1002/pds.3463. [DOI] [PubMed] [Google Scholar]
- 4.Wallace JL. NSAID gastropathy and enteropathy: distinct pathogenesis likely necessitates distinct prevention strategies. Br J Pharmacol. 2012;165:67–74. doi: 10.1111/j.1476-5381.2011.01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marlicz W, Loniewski I, Grimes DS, Quigley EM. Nonsteroidal anti-inflammatory drugs, proton pump inhibitors, and gastrointestinal injury: contrasting interactions in the stomach and small intestine. Mayo Clin Proc. 2014;89(12):1699–1709. doi: 10.1016/j.mayocp.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Coxib and traditional NSAID Trialists' (CNT) Collaboration. Bhala N, Emberson J, Merhi A, et al. Vascular and upper gastrointestinal effects of non-steroidal anti-inflammatory drugs: meta-analyses of individual participant data from randomised trials. Lancet. 2013;382:769–779. doi: 10.1016/S0140-6736(13)60900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwo PY, Tremaine WJ. Nonsteroidal anti-inflammatory drug-induced enteropathy: case discussion and review of the literature. Mayo Clin Proc. 1995;70:55–61. doi: 10.1016/S0025-6196(11)64666-1. [DOI] [PubMed] [Google Scholar]
- 8.Syer SD, Wallace JL. Environmental and NSAID-enteropathy: dysbiosis as a common factor. Curr Gastroenterol Rep. 2014;16:377. doi: 10.1007/s11894-014-0377-1. [DOI] [PubMed] [Google Scholar]
- 9.Montalto M, Gallo A, Gasbarrini A, Landolfi R. NSAID enteropathy: could probiotics prevent it? J Gastroenterol. 2013;48:689–697. doi: 10.1007/s00535-012-0648-2. [DOI] [PubMed] [Google Scholar]
- 10.Robert A, Asano T. Resistance of germfree rats to indomethacin-induced intestinal lesions. Prostaglandins. 1977;14:333–341. doi: 10.1016/0090-6980(77)90178-2. [DOI] [PubMed] [Google Scholar]
- 11.Uejima M, Kinouchi T, Kataoka K, Hiraokam I, Ohnishi Y. Role of intestinal bacteria in ileal ulcer formation in rats treated with a nonsteroidal anti-inflammatory drug. Microbiol Immunol. 1996;40:553–560. doi: 10.1111/j.1348-0421.1996.tb01108.x. [DOI] [PubMed] [Google Scholar]
- 12.Wallace JL, Syer S, Denou E, et al. Proton pump inhibitors exacerbate NSAID-induced small intestinal injury by inducing dysbiosis. Gastroenterology. 2011;141:1314–1322. doi: 10.1053/j.gastro.2011.06.075. [DOI] [PubMed] [Google Scholar]
- 13.Schubert AM, Rogers MA, Ring C, et al. 2014 Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio. 2014;5:e01021–14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 16.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 17.Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- 18.Castellsague J, Riera-Guardia N, Calingaert B, et al. Individual NSAIDs and upper gastrointestinal complications: a systematic review and meta-analysis of observational studies (the SOS project) Drug Saf. 2012;35:1127–1146. doi: 10.1007/BF03261999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis SC, Langman MJ, Laporte JR, Matthews JN, Rawlins MD, Wiholm BE. Dose-response relationships between individual nonaspirin nonsteroidal anti-inflammatory drugs (NANSAIDs) and serious upper gastrointestinal bleeding: a meta-analysis based on individual patient data. Br J Clin Pharmacol. 2002;54:320–326. doi: 10.1046/j.1365-2125.2002.01636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapeyre-Mestre M, Grolleau S, Montastruc JL, Association Française des Centres Régionaux de Pharmacovigilance (CRPV) Adverse drug reactions associated with the use of NSAIDs: a case/noncase analysis of spontaneous reports from the French pharmacovigilance database 2002-2006. Fundam Clin Pharmacol. 2013;27:223–230. doi: 10.1111/j.1472-8206.2011.00991.x. [DOI] [PubMed] [Google Scholar]
- 21.United States Food and Drug Administration FDA Drug Safety Communication: FDA strengthens warning that non-aspirin nonsteroidal anti-inflammatory drugs (NSAIDs) can cause heart attacks or strokes. Safety Announcement. 2015 Jul 9; [Google Scholar]
- 22.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung J, Park M, Park HJ, Shim SB, et al. 1H NMR-based metabolic profiling of naproxen-induced toxicity in rats. Toxicol Lett. 2011;200:1–7. doi: 10.1016/j.toxlet.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 24.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa S, Katsuki S, Fujita T, Saitoh Y, Ohta H, Nishikawa K, Sato Y, Sato Y, Ohira K, Yamada M, Kato M. A randomized, double-blinded, placebo-controlled, multicenter trial, healing effect of rebamipide in patients with low-dose aspirin and/or non-steroidal anti-inflammatory drug induced small bowel injury. J Gastroenterol. 2014;49(2):239–44. doi: 10.1007/s00535-013-0805-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhong W, Maradit-Kremers H, St Sauver JL, et al. Age and sex patterns of drug prescribing in a defined American population. Mayo Clin Proc. 2013;88:697–707. doi: 10.1016/j.mayocp.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology. 2014;146:1547–1553. doi: 10.1053/j.gastro.2014.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]