Abstract
Staphylococcus aureus is the leading etiologic agent of orthopedic implant infections. Here a ribocluster of 27 S. aureus strains underwent further molecular characterization and subtyping by multilocus sequence typing (MLST) and spa-typing. This cluster had been detected by automated ribotyping (with the EcoRI restriction enzyme) of 200 S. aureus isolates from periprosthetic infections of patients who underwent revision at the Rizzoli Orthopaedic Institute. The ribocluster, consisting of agr type III strains, with a 74% co-occurrence of bone sialoprotein-binding (bbp) and collagen-binding (cna) genes, lacked mecA and IS256, and exhibited a high prevalence of the toxic shock syndrome toxin gene (tst, 85%). Strains' relatedness was analyzed by BURP and eBURST. Two predominant spa types, t012 (32%) and t021 (36%), and one predominant sequence type, ST30 (18/27, 67%) were identified: a S. aureus lineage spread worldwide belonging to MLST CC30. Two new sequence types (ST2954, ST2960) and one new spa type (t13129) were detected for the first time. Interestingly, the 27-strain cluster detected by ribotyping corresponded exactly to MLST CC30, the sole CC identified by eBURST.
Keywords: methicillin-sensitive Staphylococcus aureus, orthopedic implant infections, multilocus sequence typing, spa-typing, virulence factors
Introduction
Staphylococcus aureus is the leading etiologic agent of orthopedic implant-associated infections (Arciola et al., 2005a; Montanaro et al., 2011; Rao et al., 2011; Tande and Patel, 2014). At present, the properties that allow particular S. aureus clones to prevail and become epidemic are not known. Infections related to orthopedic implants occur in ~1.5% of cases (Montanaro et al., 2011; Tsaras et al., 2012; Tande et al., 2014). The high number of primary and revision arthroplasties renders these infections significant in terms of morbidity, mortality, and economic consequences (Montanaro et al., 2011). The potential to colonize host periprosthetic tissues and to cause severe disease differs among clonal lineages, a feature that is attributed to the absence or presence of different virulence factors and to the levels at which they are produced (Li et al., 2009). In orthopedic implant infections, the first microbial adhesion to a biomaterial coated by host extracellular proteins is mediated by Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs; Speziale et al., 2009; Foster et al., 2014). Among these, bone sialoprotein-binding protein (Bbp) and collagen adhesin (Cna) play a crucial role in the onset of device-related infections (Arciola et al., 2005b; Xu et al., 2005; Campoccia et al., 2009; Vazquez et al., 2011; Post et al., 2014). S. aureus can also yield a variety of other virulence factors, such as Panton-Valentine leukocidin (PVL), often associated with necrotizing pneumonia and skin infections (Rasigade et al., 2011), toxic shock syndrome toxin (TSST), a super-antigenic toxin more common in MSSA lineages (He et al., 2013), and the insertion sequence IS256, widespread in genomes of multi-resistant staphylococci (Byrne et al., 1989; Depardieu et al., 2007; Schreiber et al., 2013).
S. aureus has evolved into many clones, some of which are rare and referred to as “sporadic,” and others, with a worldwide prevalence, can be defined “epidemic.” The evolutionary success of different clones of the same microorganism indicates the acquisition of new traits that either boost their virulence or favors their adaptability in the “race for the surface” between bacteria and eukaryotic cells in particular niches of infections, such as the biomaterial/tissue interface (Gristina et al., 1988-1989; Feng et al., 2008; Montanaro et al., 2011).
The clonality of S. aureus was initially revealed by multilocus enzyme electrophoresis (MLEE)-typing and pulsed field gel electrophoresis (PFGE)-typing. Multilocus sequence typing (MLST), based on the profile of alleles at seven loci of housekeeping genes, and spa-typing, based on the variable X-region of the staphylococcal protein A gene, have confirmed the highly clonal structure of S. aureus (Shopsin et al., 1999).
This study was aimed at thoroughly investigating the genetic background of a cluster of 27 S. aureus strains (Campoccia et al., 2009). This cluster was identified when ribotyping 200 S. aureus isolates obtained from patients undergoing revision at the Rizzoli Orthopaedic Institute (IOR) for periprosthetic infections (Campoccia et al., 2009). The ribocluster strains were analyzed by MLST and by spa typing in order to establish whether they belonged to a single clonal complex, and ultimately to ascertain if the most prevalent cluster identified by riboprinting from a collection of orthopedic implant infections corresponded to some known epidemic clonal complex, as the historical CC30 and its lineages (see Table 1S for a biographical sketch of CC30). To this end, the resulting sequences were analyzed by BURP and eBURST algorithms. In the present study, isolates were further characterized by assaying their antibiotic-resistances and checking for the presence of mecA for methicillin-resistance. The search for IS256, Panton-Valentine pvl gene, and TSST gene tst provided additional information for the epidemiological and pathogenetic profiles.
Materials and methods
S. aureus ribocluster
A ribocluster of 27 S. aureus strains was utilized in this study. Three ribogroups form this ribocluster: cra-119-S-8, cra-138-S-2, and cra-53-S-7. These ribogroups had been identified by an automated RiboPrinter® and then recognized as a unique ribocluster when ribotyped among 200 S. aureus isolates from infected prostheses observed at the IOR of Bologna (Campoccia et al., 2009). The automated RiboPrinter® is prone to categorizing the strains that diverge only at the level of bands with molecular weight greater than 50 Kbp as belonging to different ribogroups, and expert supervision is necessary (Brisse et al., 2002). We designated all 27 strains as a single large cluster (Campoccia et al., 2009).
Clinical isolates came from revision of surgical wounds and treatment of infected prostheses of the following categories: external fixation devices (EF), internal fixation devices (IF), knee arthroprostheses (K), and hip arthroprostheses (H).
Staphylococcal species identification was previously performed Api-Staph and/or ID 32 Staph test (BioMérieux, Marcy l'Etoile, France). Following criteria of the Centers for Disease Control and Prevention (CDC) to distinguish community-acquired (CA) and hospital-acquired (HA) infections1, the S. aureus isolates of this study were categorized as hospital acquired (HA). The strains were stored at −80°C. The study was approved and funded by the Scientific Director of the IOR. All microbiological samples were completely de-identified and stripped of all patient identifying information.
Bacterial DNA isolation
The chromosomal DNA used as an amplification template was extracted from the bacterial cultures using QIAmp DNA mini kit (Qiagen, GmbH, Hilden, Germany), according to the manufacturer's instruction.
Detection of mecA, femA, pvl, IS256, and tst genes
PCR conditions and primers used in this study are reported in Table 1.
Table 1.
PCR conditions and primers used in this study.
| Target gene | Primer sequences | Amplicon size (bp) | References |
|---|---|---|---|
| mecA | 5′-TGGCTATCGTGTCACAATCG-3′ 5′-CTGGAACTTGTTGAGCAGAG-3′ |
310 | Vannuffel et al., 1995 |
| femA | 5′-CTTACTTACTGGCTGTACCTG-3′ 5′-ATGTCGCTTGTTATGTGC-3′ |
686 | Vannuffel et al., 1995 |
| Pvl (lukS-PV/lukF-PV) | 5′-ATCATTAGGTAAAATGTCTGGACATGATCCA-3′ 5′-GCATCAASTGTATTGGATAGCAAAAGC-3′ |
433 | Lina et al., 1999 |
| IS256 | 5′-AGTCCTTTTACGGTACAATG-3′ 5′-TGTGCGCATCAGAAATAACG-3′ |
762 | Gu et al., 2005 |
| Tst | 5′-ATGGCAGCATCAGCTTGATA-3′ 5′-TTTCCAATAACCACCCGTTT-3′ |
349 | Jarraud et al., 1999 |
The presence of genes was tested by amplification of the respective gene-fragments using 10 μl RedTaq® ReadyMix™ PCR Reaction Mix (Sigma, St Louis, MO), 1 μl gDNA, and 10 pmol/μl primer. Amplification conditions were as follows: an initial step of 5 min at 95°C, 40 cycles each of 30 s at 95°C, 45 s at 55°C, and 45 s at 72°C, and a final step of 45 s at 72°C.
Antibiotic susceptibility
The agar diffusion (Kirby-Bauer) method was utilized to perform the antibiotic susceptibility tests according to Clinical and Laboratory Standards Institute (CLSI) guidelines (NCCLS, 2002). Antimicrobial susceptibility was tested for a panel of 16 antibiotics: oxacillin (OXA), imipenem (IMP), penicillin (PEN), ampicillin (AMP), cefazolin (CFZ), cefamandole (FAM), gentamicin (GEN), amikacin (AMK), netilmicin (NET), tobramycin (TOB), erythromycin (ERY), clindamycin (CLI), chloramphenicol (CHL), trimethoprim–sulfamethoxazole (SXT), ciprofloxacin (CIP), and vancomycin (VAN).
spa sequencing
The polymorphic X, or short sequence repeat (SSR), region of the S. aureus protein A gene (spa) was amplified by PCR with primers 1113F (5′-TGTAAAACGACGGCCAGT-3′) and 1514R (5′-CAGGAAACAGCTATGACC-3′) according to protocols previously described (Schmid et al., 2013).
Ten microliters of the amplified products were analyzed on 1.5% agarose gels and 5 μl were purified with EXO SAP-IT (GE Health care, Buckinghamshire, GB). Two microliters of the purified amplification products were used for subsequent sequencing using the Big Dye Terminator v3.1 sequencing kit (Applied Biosystems, Carlsbad, CA) and were finally analyzed on ABI Genetic Analyzer 3500Dx (Applied Biosystems).
The chromatograms obtained were analyzed with the Ridom StaphType software (version 1.4; Ridom GmbH, Würzburg, Germany; http://spa.ridom.de/index.shtml) to determine the spa type of each isolate2. The spa types were deduced by the differences in number and sequence of spa repeats. Using the BURP algorithm (Ridom GmbH) and the Ridom SpaServer database (Enright et al., 2000), spa types were clustered into different clonal complexes (spa-CCs) and MLST clonal complexes (CCs) were inferred.
Multilocus sequence typing
MLST genotyping was performed on all 27 S. aureus isolates as described previously by Larsen et al. (2012). The amplification of a portion of seven housekeeping genes (arc, aroE, glp, gmk, pta, tpi, yqiL) was performed and then sequenced. The free cross-platform bioinformatics software package Unipro UGENE 1.133 was used to analyze the sequences. Sequence types (STs) were obtained using the MLST database4. Using the eBURST v3 algorithm5, sequence types (STs) were clustered to assign the clonal complexes (CCs), and assess the population organization and patterns of evolution. S. aureus strains with STs differing by one or two housekeeping genes/loci were considered part of a unique clonal complex.
Results
MLST analysis
The MLST analysis of the 27 strains of the ribocluster identified 5 distinct STs. ST30, the most prevalent, included 18 strains (67%), ST34 consisted of 5 strains (19%), ST2954 of 2 strains (7%), and ST2960 and ST243 were both represented by a single strain. The allelic profile of each identified ST is reported in Table 2. Sequence type attribution, riboprofiles, genotypic characteristics, and clinical origin of each of the 27 strains investigated are summarized in Table 3.
Table 2.
Description of all the STs found in the present study.
| ST | MLST allelic profile* | spa types | spa repeat succession | spa CC |
|---|---|---|---|---|
| ST30 (18) | 2-2-2-2-6-3-2 | t018 (2) | -24-24 | CC021/012 |
| t093 (1) | 12 | |||
| t012 (7) | 24 | |||
| t021 (6) | ||||
| t1382 (1) | 01 | |||
| t11956 (1) | 23 | |||
| ST243 (1) | -5- | t021 (1) | 15-12-----16-02-16-02-25-17-24-------- | |
| ST2960a (1) | -268- | t021 (1) | 15-12-----16-02-16-02-25-17-24-------- | |
| ST2954a (2) | -330 | t298 (2) | 15-12-----16-02-------------17-24-------- | |
| ST34 (5) | 8- | t166 (2) | CC166 | |
| t369 (1) | ||||
| t4437 (1) | ||||
| t13129b (1) | 51 | Singleton |
MLST allelic profile (arc-aroe-glpf-gmk-pta-tpi-yiql); numbers between brackets represent the number of strains;
newly described ST;
newly described spa type; MLST alleles shared with the probable founder of MLST CC30, ST30, are colored in sky blue; the spa sequence repetitions shared with the probable founder of spa CC021/012, t021, appear in blue; the spa sequence repetitions shared with the probable founder of spa CC166, t166, are in red; the spa sequence repetitions of the new spa type t13129 shared with the probable founder of spa CC166 are in green. repeat 23 differs in one base from repeat 16; repeat 01 differs in three bases from repeat 15; repeat 51 differs in three bases from repeat 44.
Table 3.
Detailed genotyping characterization data of the 27 strains.

Genes codifying for: eno, laminin-binding adhesin; ebpS, elastin-binding protein; fib, fibrinogen-binding adhesin; can, collagen-adhesin; fnbB, fibronectin-binding protein B; bbp, bone sialoprotein-binding protein; sdrE, serine-aspartate repeat proteins E. K, H = knee, hip arthroprostheses; EF, IF = external, internal fixation systems. aNew sequence type (ST), bnew spa type found in this study; Singl. = singleton.
The strains of the studied ribocluster had been previously characterized for their agr type (Montanaro et al., 2010) and for the following panel of MSCRAMM genes (Campoccia et al., 2009): eno, fib, cna, ebpS, fnbB, bbp, and sdrE. Strains were all of the agr type III. All 27 strains turned out negative for mecA, IS256, and sdrE, and positive for eno and ebpS genes.
As shown in Table 4, ST30 was found to consist mainly (86%) of cna- and bbp-positive strains: only one strain out of 18 (6%) was found to be either cna- or bbp-negative. Thus, a remarkably high proportion of the ST30 strains exhibited a typical bbp-cna adhesin co-occurence. The bbp gene encoding the Bbp was observed in 93% of the 27 strains (Campoccia et al., 2009), often in association with the cna gene detected in 78% of the strains.
Table 4.
Genotypic characteristics of the ST, spa-CC, and spa-types.
| ST | spa CC | spa type | bbp-cna tandem | fib | tst | agr | IS256 | mecA |
|---|---|---|---|---|---|---|---|---|
| ST2960 (1) | CC021/012 | t021 (8; 36%) | 100% | 100% | 100% | III | neg | neg |
| ST243 (1) | – | – | ||||||
| ST30 (18) | 33% | 83% | ||||||
| t012 (7; 32%) | 86% | 29% | 100% | |||||
| Other spa types (5; 23%)a | 80% | 20% | 100% | |||||
| ST2954 (2) | t298 (2; 9%) | 100% | 100% | – | ||||
| ST34 (5) | CC166 | t166 (2; 50%) | – | 100% | 100% | |||
| Other spa types (2; 50%)b | – | 50% | 100% | |||||
| singleton | t13129 (1; 100%) | – | 100% | 100% |
Numbers between brackets represent the number of strains and percent frequency.
The remaining spa types are t018 (2), t093, t1382, t11956.
The remaining spa types are t369, t4437.
All ST30 strains except for cra2727 were characterized by the presence of tst gene, which was unusually found to be extremely common in this collection of strains. The ST30 fib-positive strains were 5 (29%) and cra1601 was the only strain carrying the fnbB gene (see Table 4). All ST30 strains were found to be pvl-negative.
All five ST34 strains were cna-negative (100%) in contrast to the remaining strains of the ribocluster, 95% of which were cna-positive.
As far as the other minor STs are concerned, the only ST243 strain, cra2380, and the two ST2954 strains were found to be tst-negative. Apart from these strains belonging to ST243 and ST295, the tst gene was generally present among the other strains of the ribocluster and only one strain (cra2727) out of 24 was found tst-negative. Further, cra2380 was also the only strain of the collection found positive for pvl. All three ST2954 and ST243 strains however exhibited the bbp-cna combination of genes typical of ST30. The single strain of the newly identified ST2960 matched the characteristics of ST30, testing positive to both bbp and cna and presenting the tst gene.
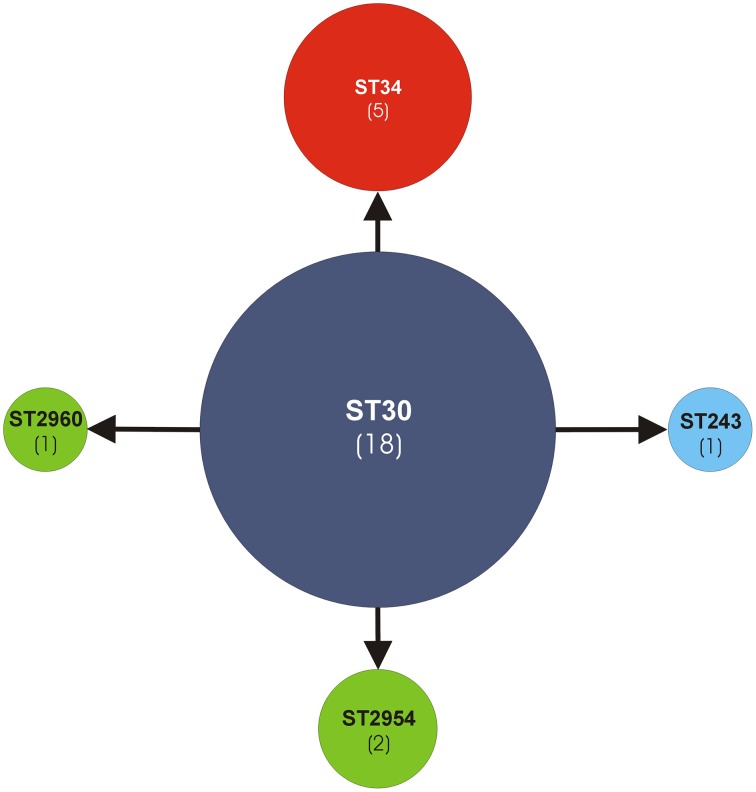
Based on the analyses by the eBURST algorithm, the five STs were clustered within the same MLST clonal complex CC30 (Figure 1). ST30 (18/22, 82%) was recognized as the genotypic founder and ST34 (5/22, 23%) as a subgroup founder. The remaining single locus variant (SLV) STs were ST243 (1/22) and the new alleles first discovered in this work, namely ST2954 (2/22) (strain cra1772 and strain cra1773) and ST2960 (1/22) (strain cra1380). Thus, the 27-strain ribocluster was entirely associated with the CC30, confirming the strict kinship of the 27 strains.
Figure 1.
Analysis of ST allelic profiles of the MLST CC30 by eBURST v3 software. The sequence type ST30, genotypic founder of the MLST CC30, is represented by a blue circle. All other STs are SLV (single locus variant). The new STs (ST2960, ST2954) are represented by green circles. ST34, a sub-group founder, is represented by a red circle. The other ST243 is represented by a sky-blue circle. In parentheses the number of strains.
spa typing
The spa typing analysis revealed 11 distinct spa types within the group of 27 strains. The different spa repeat sequences specific for each identified spa type is reported in Table 2. The spa type including the largest number of strains was t021, enlisting 8 out of 27 strains (30%). It was immediately followed by t012, consisting of 7 strains (26%). All the other 9 spa types included just 1 or at most 2 strains (representing a frequency of 4–7%) as reported in Table 2. Among these less frequent spa types, there was a newly identified spa type, t13129 (strain cra1510), never described before. The spa type attribution of each single clinical strain is reported in Table 3, while the genotypic traits characteristic of the spa types are described in Table 4.
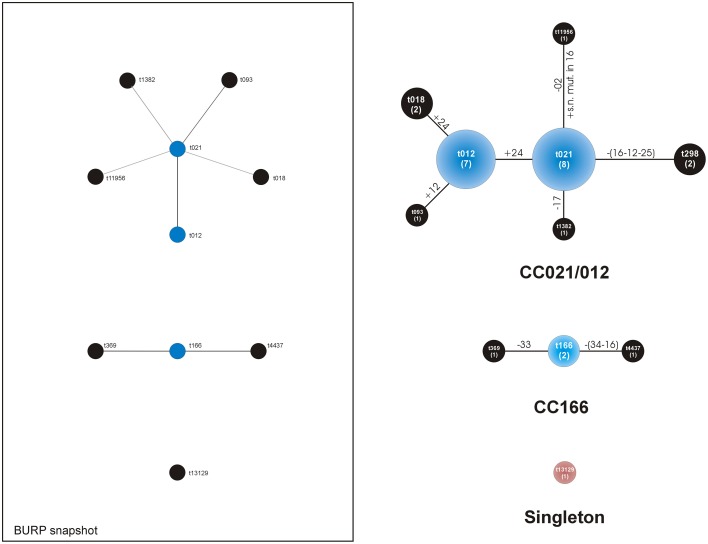
BURP analysis of the spa types yielded two main clonal complexes, spa-CC021/012 (22 strains) and spa-CC166 (4 strains), and a singleton (Table 4). The spa types in the large spa-CC021/012 were: t021 (8 strains out of 22, 36%), t012 (7, 32%), t018 (2, 9%), t298 (2, 9%), t093 (1, 5%), t1382 (1, 5%), and t11956 (1, 5%). Thus, two spa types, t021 and t012 together represented up to 68% of the spa-CC and 59% of the entire collection. These two spa types differed in just one sequence repeat at the end of the repeat sequence succession. The repeat sequence 24 was just in one copy in t021 and double in t012. The two strains of spa-CC021/012 with spa type t298 were belonging to the ST2954.
Apart from the founder t166, spa-CC166 had 2 further spa types: t369 and t4437. Discovered in the present study and identified by BURP analysis as a singleton, spa type t13129 (strain cra1510) exhibited a repeat succession just partially different from that of t166, lacking four repeat sequences (33-31-12-16) and carrying the 51 repeat sequence instead of the repeat 44 (see Table 2).
The population structure analysis is illustrated in Figure 2, which reports the population analysis as per BURP population snapshot. On the right side of Figure 2 an additional drawing shows the type of mutations involved in the transition between spa types.
Figure 2.
Analysis of the spa types clustered by BURP algorithm of the Ridom StaphType Software. On the left, within the frame, the population distribution is illustrated as per BURP population snapshot. The predict founder of a cluster is shown in blue, while the others in black. On the right, a customized representation of the CCs takes into consideration the type of mutations. Near the lines of connection, the mutations involved in the transition from a spa type to the next one are reported in detail. All DNA changes are meant to occur from the founder to the periphery. Legend: numbers along the lines refer to the repeat sequence involved in the mutation; + indicates the acquisition of a repeat sequence; -indicates the loss of a repeat sequence; within circles the numbers of the strains of each CC appear between brackets; extensive losses including more than a repeat sequence are reported along the lines between brackets; s.n. mut.: single nucleotide mutation of a sequence repeat.
As far as the adhesin profile is concerned, differences were observed between the two main spa-CCs. The spa-CC021/012, including the vast majority of the strains of the ribocluster (81%), exhibited a remarkable prevalence of clinical strains endowed with both cna and bbp genes. Indeed, although the co-occurrence of bbp and cna genes was a genetic pattern consistently observed in the ribocluster (74%), its prevalence reached 91% within the spa-CC021/012. The prevalent spa type of CC021/012 was t021, uniquely consisting of bbp- and cna-positive strains. The triple adhesin gene pattern bbp-cna-fib was only observed in 7 strains belonging to spa-CC021/012 (32%).
All four strains of spa-CC166 (15% of the entire ribocluster) were free of bbp gene and, thus, lacked the combination bbp-cna (Tables 3, 4).
The pvl-positive ST243 cra2380 strain belonged to t021 and was tst-negative, positive for the tandem bbp-cna and susceptible to all the antibiotics tested. The overall prevalence of tst gene was 85% (23/27) and the only four strains without tst genes all belonged to the spa-CC021/012.
Antibiotic resistance
Table 2S reports the observed prevalence of antibiotic resistance in the cluster of 27 strains. Apart from the frequent resistance to penicillin observed in 89% of the strains, the clinical strains were found to be nearly all sensitive to the rest of the antibiotics. Indeed, for the other 12 antibiotics, the number of resistant strains varied between 0 and 1 (4%). This profile of low antibiotic resistance was documented by the average multiple antibiotic resistance (MAR) index (Krumperman, 1983; Kaspar et al., 1990), which was as low as 0.24. Three strains were susceptible to all the antibiotics tested.
Discussion
The principal outcome of this work is that the ribocluster initially interpreted as a clone (Campoccia et al., 2009) actually corresponds to a clonal complex, namely the MLST CC30, in which many organisms and diverse sequence types are grouped (Table 2). This finding highlights the higher discriminatory power of MLST and spa-typing with respect to ribotyping. Within CC30, all the strains were methicillin-susceptible and 93% of them belonged to ST30. In contrast to some STs, which are geographically concentrated, ST30 is spread worldwide (Vandenesch et al., 2003; DeLeo et al., 2010). MSSA isolates associated with CC30, particularly the ST30 genetic lineage, are responsible for various infections in different countries of the world (Robinson and Enright, 2003; Vandenesch et al., 2003; Aires de Sousa et al., 2005; Gomes et al., 2006; Vivoni et al., 2006; Hallin et al., 2007; Holtfreter et al., 2007; Fenner et al., 2008; Strommenger et al., 2008; He et al., 2013; Tavares et al., 2014). With regard to the CC30 clone in orthopedic infections, Aamot et al. (2012) reported that CC30 was the most frequent clonal complex in MSSA isolates from surgical site infections in orthopedic patients from Norway. MSSA isolates from orthopedic implant-related infections of Swiss and French hospitals, mainly belonging to the ST30-CC30, were highlighted by Post et al. (2014). Rincón et al. (2013) analyzed numerous isolates obtained from osteomyelitis in South American hospitals and described a high percentage of MSSA belonging to different genetic lineages, including ST30.
The absence of mecA and IS256 (Tables 3, 4) reaffirms that antibiotic-resistance and hypervirulence (Benson et al., 2014) are not strictly necessary for the success of S. aureus lineages within CC30 and that other determinants might play a more relevant role, conferring greater fitness, and efficiency in host colonization and invasion (Ziebuhr et al., 1999; Arciola et al., 2002, 2004, 2015; Kiem et al., 2004; Valle et al., 2007; McAdam et al., 2012; Cheung et al., 2014; Lin et al., 2015).
In the Bayesian phylogenetic reconstruction of the CC30 lineage, presented in a recent work of McAdam et al. (2012), many of the CC30 related to hospital acquired EMRSA-16 exhibited the same genetic properties of the isolates of the present study. McAdam et al. (2012) showed that tst gene is absent in the isolates of the pandemic phage type 80/81 and SWP lineages, while present in 83% of isolates from the HA-EMRSA-16 lineage (ST36). Moreover, while phage type 80/81 and SWP clones were pvl-positive with a prevalence of 90%, EMRSA-16 and the other epidemic CC30 were all pvl-negative.
In our results, we observed the absence of pvl in all strains except one and 85% prevalence for tst. It is likely that MSSA-CC30 strains, especially those harboring tst, were strictly associated with the contemporary epidemic CC30 and related to the hospital acquired EMRSA-16 clone. In contrast to phage type 80/81 and SWP clones, the other epidemic CC30 related to EMRSA-16 appeared to be restricted to hospital settings and had reduced virulence, due in part to the low levels of expression of toxins such as PVL and LukE/LukD (Campoccia et al., 2008). McGavin et al. (2012) suggested that clade 3, comprising the contemporary hospital-associated MSSA-CC30 clone, brought a large burden of diseases due to its ability to persist in human hosts at the expense of an attenuated virulence. With regard to the strain cra2380, it turned out pvl-positive and tst-negative; this strain belongs to exactly the same MLST allelic profile (ST243) and spa type (t021) of the ATCC25923 reference strain. As reported by Chen et al. (2013), the ATCC25923 strain, isolated in USA (WA) in 1945, is a PVL-positive (PVL haplotype: H1a) CA-MSSA clinical strain belonging to the phage type 80/81 lineage.
The analysis of spa sequences by BURP algorithm allowed identification of two spa clonal complexes (CC021/012, CC166) and a singleton. As in the work of Wiśniewska et al. (2014), spa-CC021/012, with the main type t021 related to CC30, was the most frequent genetic lineage among our strains. CC166 strains and the singleton strain were all found to belong to the sequence type ST34 and exhibited some characteristic traits such as the absence of cna, a gene otherwise frequent within the predominant spa-CC021/012. S. aureus ST34 genetic background has been suggested to be of hybrid origin, being derived from recombination of large, contiguous portions of the chromosomes of genetically distinct parent backgrounds, with only part of the genome coming from CC30 (Robinson and Enright, 2004; Thomas et al., 2012). This notion may explain why ST34 lineage lacks the bbp-cna gene combination and may also provide an explanation why these strains belong to a different spa type. With the exclusion of the strains of the ST34 lineage, 91% of the remaining 22 strains possessed the bbp-cna gene combination, encoding for a couple of adhesins able to bind the most abundant bone proteins and crucial in the pathogenesis of orthopedic implant infections (Campoccia et al., 2009).
Moreover, all the strains shared the agr type III polymorphism. It should be remarked that strains with the same agr tend to be characterized by a well-defined pattern of virulence genes generated by intraspecific cross-inhibitions (Goerke et al., 2003).
In Tables 2–4, the MSSA ST30-t012 and ST30-t021 genetic patterns emerged as the two most prevalent spa repeat successions t021, differing from t012 in only one repeat sequence, indicating that they are likely to be close relatives. Holtfreter et al. (2007) showed that in MSSA-CC30 isolates, the spa type t012 was the most prevalent among nasal isolates, while the spa type t021 was most prevalent among blood culture isolates. In Denmark, in a study of MSSA clinical isolates from blood cultures, the authors presented ST30-t021 as the most frequent genetic lineage (Gomes et al., 2006). Fenner et al. also observed t021 and t012 in invasive MSSA (Fenner et al., 2008) in a Swiss University Hospital, as did Nulens et al. (2008) in bloodstream isolates collected from a Dutch university hospital. Post et al. (2014) indicated the presence of spa type t012 (ST30) as the most frequent among the MSSA isolates.
The most prevalent clonal type ST30-t012 of our collection has also been recurrently observed in Portugal, Spain, Belgium, the Netherlands and in the USA, by analyzing MSSA isolates from different time periods (Aires de Sousa et al., 2005; Rijnders et al., 2009; Argudín et al., 2013; Miko et al., 2013; Tavares et al., 2014). In other countries, such as China, Taiwan, the African countries of Cameroon, Madagascar, Senegal, Niger and Morocco, MSSA genetic lineages from community infections were dissimilar (Rijnders et al., 2009; Breurec et al., 2011; He et al., 2013). The reason for the differences in MSSA lineages among various countries still remains not well understood. Also the discovery of a new spa type (t13129) and of two new sequence types (ST2954, ST2960) suggests that there is still much to be revealed about the evolution of CC30. The success of CC30 could be multifactorial and its panel of adhesins represents a successful strategy for renewing and continuing its adaptation to different niches.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge a financial contribution by “5 per mille” grants for Health Research to the Rizzoli Orthopedic Institute of Bologna.
Footnotes
1Center for Disease Control and Prevention (CDC). Available online at http://www.cdc.gov/
2Ridom SpaServer database. Available online at http://www.spaserver.ridom.de
3MLST website. Available online at http://saureus.mlst.net/sql/multiplelocus.asp
4MLST database. Available online at http://www.mlst.net
5eBURST algorithm. Available online http://eburst.mlst.net/v3/mlst_datasets/
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00008
References
- Aamot H. V., Blomfeldt A., Skråmm I., Müller F., Monecke S. (2012). Molecular characterisation of methicillin-sensitive Staphylococcus aureus from deep surgical site infections in orthopaedic patients. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1999–2004. 10.1007/s10096-011-1532-3 [DOI] [PubMed] [Google Scholar]
- Aires de Sousa M., Conceição T., Simas C., de Lencastre H. (2005). Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 43, 5150–5157. 10.1128/JCM.43.10.5150-5157.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciola C. R., An Y. H., Campoccia D., Donati M. E., Montanaro L. (2005a). Etiology of implant orthopedic infections: a survey on 1027 clinical isolates. Int. J. Artif. Organs 28, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Arciola C. R., Baldassarri L., Montanaro L. (2002). In catheter infections by Staphylococcus epidermidis the intercellular adhesion (ica) locus is a molecular marker of the virulent slime-producing strains. J. Biomed. Mater. Res. 59, 557–562. 10.1002/jbm.10006 [DOI] [PubMed] [Google Scholar]
- Arciola C. R., Campoccia D., Gamberini S., Baldassarri L., Montanaro L. (2005b). Prevalence of cna, fnbA and fnbB adhesin genes among Staphylococcus aureus isolates from orthopedic infections associated to different types of implant. FEMS Microbiol. Lett. 246, 81–86. 10.1016/j.femsle.2005.03.035 [DOI] [PubMed] [Google Scholar]
- Arciola C. R., Campoccia D., Gamberini S., Rizzi S., Donati M. E., Baldassarri L., et al. (2004). Search for the insertion element IS256 within the ica locus of Staphylococcus epidermidis clinical isolates collected from biomaterial-associated infections. Biomaterials 25, 4117–4125. 10.1016/j.biomaterials.2003.11.027 [DOI] [PubMed] [Google Scholar]
- Arciola C. R., Campoccia D., Ravaioli S., Montanaro L. (2015). Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front. Cell. Infect. Microbiol. 5:7. 10.3389/fcimb.2015.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argudín M. A., Argumosa V., Mendoza M. C., Guerra B., Rodicio M. R. (2013). Population structure and exotoxin gene content of methicillin-susceptible Staphylococcus aureus from Spanish healthy carriers. Microb. Pathog. 54, 26–33. 10.1016/j.micpath.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Benson M. A., Ohneck E. A., Ryan C., Alonzo F., III., Smith H., Narechania A., et al. (2014). Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol. Microbiol. 93, 664–681. 10.1111/mmi.12682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J. M., Stemper M. E., Weigel A., Chyou P. H., Reed K. D., Shukla S. K. (2007). Sporadic “transitional” community-associated methicillin-resistant Staphylococcus aureus strains from health care facilities in the United States. J. Clin. Microbiol. 45, 2654–2661. 10.1128/JCM.02579-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breurec S., Fall C., Pouillot R., Boisier P., Brisse S., Diene-Sarr F., et al. (2011). Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 17, 633–639. 10.1111/j.1469-0691.2010.03320.x [DOI] [PubMed] [Google Scholar]
- Brisse S., Fussing V., Ridwan B., Verhoef J., Willems R. J. (2002). Automated ribotyping of vancomycin-resistant Enterococcus faecium isolates. J. Clin. Microbiol. 40, 1977–1984. 10.1128/JCM.40.6.1977-1984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E., Rouch D. A., Skurray R. A. (1989). Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin- resistance transposon Tn4001. Gene 81, 361–367. 10.1016/0378-1119(89)90197-2 [DOI] [PubMed] [Google Scholar]
- Campanile F., Bongiorno D., Falcone M., Vailati F., Pasticci M. B., Perez M., et al. (2012). Changing Italian nosocomial-community trends and heteroresistance in Staphylococcus aureus from bacteremia and endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 31, 739–745. 10.1007/s10096-011-1367-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoccia D., Baldassarri L., Pirini V., Ravaioli S., Montanaro L., Arciola C. R. (2008). Molecular epidemiology of Staphylococcus aureus from implant orthopaedic infections: ribotypes, agr polymorphism, leukocidal toxins and antibiotic resistance. Biomaterials 29, 4108–4116. 10.1016/j.biomaterials.2008.07.006 [DOI] [PubMed] [Google Scholar]
- Campoccia D., Speziale P., Ravaioli S., Cangini I., Rindi S., Pirini V., et al. (2009). The presence of both bone sialoprotein-binding protein gene and collagen adhesin gene as a typical virulence trait of the major epidemic cluster in isolates from orthopedic implant infections. Biomaterials 30, 6621–6628. 10.1016/j.biomaterials.2009.08.032 [DOI] [PubMed] [Google Scholar]
- Chambers H. F., Deleo F. R. (2007). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. J., Siu L. K., Lin J. C., Wang C. H., Lu P. L. (2012). Molecular typing and characterization of nasal carriage and community-onset infection methicillin-susceptible Staphylococcus aureus isolates in two Taiwan medical centers. BMC Infect. Dis. 12:343. 10.1186/1471-2334-12-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Solanki M., Mediavilla J. R., Mathema B., Schlievert P. M., et al. (2013). Genetic variation among Panton-Valentine leukocidin-encoding bacteriophages in Staphylococcus aureus clonal complex 30 strains. J. Clin. Microbiol. 51, 914–919. 10.1128/JCM.03015-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Kretschmer D., Duong A. C., Yeh A. J., Ho T. V., Chen Y., et al. (2014). Production of an attenuated phenol-soluble modulin variant unique to the MRSA clonal complex 30 increases severity of bloodstream infection. PLoS Pathog. 10:e1004298. 10.1371/journal.ppat.1004298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs G. W., Nimmo G. R., Bell J. M., Huygens F., O'Brien F. G., Malkowski M. J., et al. (2004). Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42, 4735–4743. 10.1128/JCM.42.10.4735-4743.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M. Z., Daum R. S. (2010). Community-associated methicillin resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23, 616–687. 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo F. R., Kennedy A. D., Chen L., Bubeck Wardenburg J., Kobayashi S. D., Mathema B., et al. (2011). Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 108, 18091–18096. 10.1073/pnas.1111084108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo F. R., Otto M., Kreiswirth B. N., Chambers H. F. (2010). Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depardieu F., Podglajen I., Leclercq R., Collatz E., Courvalin P. (2007). Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20, 79–114. 10.1128/CMR.00015-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg R. H., Stobberingh E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. 10.1016/j.meegid.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Diep B. A., Carleton H. A., Chang R. F., Sensabaugh G. F., Perdreau-Remington F. (2006). Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 193:1495–1503. 10.1086/503777 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015. 10.1128/IAI.69.4.2416-2427.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Chen C. J., Su L. H., Hu S., Yu J., Chiu C. H. (2008). Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 32, 23–37. 10.1111/j.1574-6976.2007.00086.x [DOI] [PubMed] [Google Scholar]
- Fenner L., Widmer A. F., Frei R. (2008). Molecular epidemiology of invasive methicillin-susceptible Staphylococcus aureus strains circulating at a Swiss University Hospital. Eur. J. Clin. Microbiol. Infect. Dis. 27, 623–626. 10.1007/s10096-008-0463-0 [DOI] [PubMed] [Google Scholar]
- Foster T. J., Geoghegan J. A., Ganesh V. K., Höök M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V., Jr., Nelson C. L., McIntyre L. M., Kreiswirth B. N., Monk A., et al. (2007). Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J. Infect. Dis. 196, 738–747. 10.1086/520088 [DOI] [PubMed] [Google Scholar]
- Goerke C., Kümmel M., Dietz K., Wolz C. (2003). Evaluation of intraspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J. Infect. Dis. 188, 250–256. 10.1086/376450 [DOI] [PubMed] [Google Scholar]
- Gomes A. R., Westh H., de Lencastre H. (2006). Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 50, 3237–3244. 10.1128/AAC.00521-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristina A. G., Naylor P., Myrvik Q. (1988-1989). Infections from biomaterials implants: a race for the surface. Med. Prog. Technol. 14, 205–224. [PubMed] [Google Scholar]
- Gu J., Li H., Li M., Vuong C., Otto M., Wen Y., et al. (2005). Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61, 342–348. 10.1016/j.jhin.2005.04.017 [DOI] [PubMed] [Google Scholar]
- Hallin M., Denis O., Deplano A., De Mendonça R., De Ryck R., Rottiers S., et al. (2007). Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey. J. Antimicrob. Chemother. 59, 465–472. 10.1093/jac/dkl535 [DOI] [PubMed] [Google Scholar]
- Hassall J. E., Rountree P. M. (1959). Staphylococcal septicaemia. Lancet 1, 213–217. 10.1016/S0140-6736(59)90047-9 [DOI] [PubMed] [Google Scholar]
- He W., Chen H., Zhao C., Zhang F., Li H., Wang Q., et al. (2013). Population structure and characterisation of Staphylococcus aureus from bacteraemia at multiple hospitals in China: association between antimicrobial resistance, toxin genes and genotypes. Int. J. Antimicrob. Agents 42, 211–219. 10.1016/j.ijantimicag.2013.04.031 [DOI] [PubMed] [Google Scholar]
- Hennig S., Ziebuhr W. (2010). Characterization of the transposase encoded by 17, the prototype of a major family of bacterial insertion sequence elements. J. Bacteriol. 192, 4153–4163. 10.1128/JB.00226-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtfreter S., Grumann D., Schmudde M., Nguyen H. T., Eichler P., Strommenger B., et al. (2007). Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 45, 2669–2680. 10.1128/JCM.00204-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarraud S., Cozon G., Vandenesch F., Bes M., Etienne J., Lina G. (1999). Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 37, 2446–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar C. W., Burgess J. L., Knight I. T., Colwell R. R. (1990). Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can. J. Microbiol. 36, 891–894. 10.1139/m90-154 [DOI] [PubMed] [Google Scholar]
- Kiem S., Oh W. S., Peck K. R., Lee N. Y., Lee J. Y., Song J. H., et al. (2004). Phase variation of biofilm formation in Staphylococcus aureus by IS 256 insertion and its impact on the capacity adhering to polyurethane surface. J. Korean Med. Sci. 19, 779–782. 10.3346/jkms.2004.19.6.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumperman P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J., Enright M. C., Godoy D., Spratt B. G., Larsen A. R., Skov R. L. (2012). Multilocus sequence typing scheme for Staphylococcus aureus: revision of the gmk locus. J. Clin. Microbiol. 50, 2538–2539. 10.1128/JCM.00290-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Diep B. A., Villaruz A. E., Braughton K. R., Jiang X., DeLeo F. R., et al. (2009). Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 5883–5888. 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. H., Shu J. C., Lin L. P., Chong K. Y., Cheng Y. W., Du J. F., et al. (2015). Elucidating the crucial role of poly N-acetylglucosamine from Staphylococcus aureus in cellular adhesion and pathogenesis. PLoS ONE 10:e0124216. 10.1371/journal.pone.0124216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G., Piémont Y., Godail-Gamot F., Bes M., Peter M. O., Gauduchon V., et al. (1999). Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132. 10.1086/313461 [DOI] [PubMed] [Google Scholar]
- McAdam P. R., Templeton K. E., Edwards G. F., Holden M. T., Feil E. J., Aanensen D. M., et al. (2012). Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 109, 9107–9112. 10.1073/pnas.1202869109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin M. J., Arsic B., Nickerson N. N. (2012). Evolutionary blueprint for host- and niche-adaptation in Staphylococcus aureus clonal complex CC30. Front. Cell. Infect. Microbiol. 2:48. 10.3389/fcimb.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miko B. A., Hafer C. A., Lee C. J., Sullivan S. B., Hackel M. A., Johnson B. M., et al. (2013). Molecular characterization of methicillin-susceptible Staphylococcus aureus clinical isolates in the United States, 2004 to 2010. J. Clin. Microbiol. 51, 874–879. 10.1128/JCM.00923-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L., Speziale P., Campoccia D., Pirini V., Ravaioli S., Cangini I., et al. (2010). Polymorphisms of agr locus correspond to distinct genetic patterns of virulence in Staphylococcus aureus clinical isolates from orthopedic implant infections. J. Biomed. Mater. Res. A 94, 825–832. 10.1002/jbm.a.32764 [DOI] [PubMed] [Google Scholar]
- Montanaro L., Speziale P., Campoccia D., Ravaioli S., Cangini I., Pietrocola G., et al. (2011). Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 6, 1329–1349. 10.2217/fmb.11.117 [DOI] [PubMed] [Google Scholar]
- Munckhof W. J., Schooneveldt J., Coombs G. W., Hoare J., Nimmo G. R. (2003). Emergence of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infection in Queensland, Australia. Int. J. Infect. Dis. 7, 259–264. 10.1016/S1201-9712(03)90104-4 [DOI] [PubMed] [Google Scholar]
- NCCLS (2002). Performance Standards for Antimicrobial Susceptibility Testing: Twelfth Informational Supplement NCCLS Document M100-S12 NCCLS. Wayne, PA: NCCLS. [Google Scholar]
- Nienaber J. J., Sharma Kuinkel B. K., Clarke-Pearson M., Lamlertthon S., Park L., Rude T. H., et al. (2011). International-collaboration on endocarditis-microbiology investigators. Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J. Infect. Dis. 204, 704–713. 10.1093/infdis/jir389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo G. R., Schooneveldt J., O'Kane G., McCall B., Vickery A. (2000). Community acquisition of gentamicin-sensitive methicillin-resistant Staphylococcus aureus in southeast Queensland, Australia. J. Clin. Microbiol. 38, 3926–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nulens E., Stobberingh E. E., van Dessel H., Sebastian S., van Tiel F. H., Beisser P. S., et al. (2008). Molecular characterization of Staphylococcus aureus bloodstream isolates collected in a Dutch University Hospital between 1999 and 2006. J. Clin. Microbiol. 46, 2438–2441. 10.1128/JCM.00808-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post V., Wahl P., Uçkay I., Ochsner P., Zimmerli W., Corvec S., et al. (2014). Phenotypic and genotypic characterisation of Staphylococcus aureus causing musculoskeletal infections. Int. J. Med. Microbiol. 304, 565–576. 10.1016/j.ijmm.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Rao N., Cannella B. A., Crossett L. S., Yates A., Jr., McGough R., 3rd., et al. (2011). Preoperative screening/decolonization for Staphylococcus aureus to prevent orthopedic surgical site infection: prospective cohort study with 2-year follow-up. J. Arthroplasty 26, 1501–1507. 10.1016/j.arth.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Rasigade J. P., Sicot N., Laurent F., Lina G., Vandenesch F., Etienne J. (2011). A history of Panton-Valentine leukocidin (PVL)-associated infection protects against death in PVL-associated pneumonia. Vaccine 29, 4185–4186. 10.1016/j.vaccine.2011.04.033 [DOI] [PubMed] [Google Scholar]
- Rijnders M. I., Deurenberg R. H., Boumans M. L., Hoogkamp-Korstanje J. A., Beisser P. S., Antibiotic Resistance Surveillance Group et al. (2009). Population structure of Staphylococcus aureus strains isolated from intensive care unit patients in the Netherlands over an 11-year period (1996 to 2006). J. Clin. Microbiol. 47, 4090–4095. 10.1128/JCM.00820-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón S., Reyes J., Carvajal L. P., Rojas N., Cortés F., Panesso D., et al. (2013). Cefazolin high-inoculum effect in methicillin-susceptible Staphylococcus aureus from South American hospitals. J. Antimicrob. Chemother. 68, 2773–2778. 10.1093/jac/dkt254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. A., Enright M. C. (2003). Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47, 3926–3934. 10.1128/AAC.47.12.3926-3934.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. A., Enright M. C. (2004). Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186, 1060–1064. 10.1128/JB.186.4.1060-1064.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. A., Kearns A. M., Holmes A., Morrison D., Grundmann H., Edwards G., et al. (2005). Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet 365, 1256–1258. 10.1016/S0140-6736(05)74814-5 [DOI] [PubMed] [Google Scholar]
- Rountree P. M., Beard M. A. (1958). Further observations on infection with phage type 80 Staphylococci in Australia. Med. J. Aust. 45, 789–795. [PubMed] [Google Scholar]
- Schmid D., Simons E., Ruppitsch W., Hrivniaková L., Stoeger A., Wechsler-Fördös A., et al. (2013). Limited value of routine spa typing: a cross-sectional study of methicillin-resistant Staphylococcus aureus-positive patients in an Austrian hospital. Am. J. Infect. Control 41, 617–624. 10.1016/j.ajic.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Schreiber F., Szekat C., Josten M., Sahl H. G., Bierbaum G. (2013). Antibiotic-induced autoactivation of IS256 in Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 6381–6384. 10.1128/AAC.01585-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B., Gomez M., Montgomery S. O., Smith D. H., Waddington M., Dodge D. E., et al. (1999). Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37, 3556–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speziale P., Pietrocola G., Rindi S., Provenzano M., Provenza G., Di Poto A., et al. (2009). Structural and functional role of Staphylococcus aureus surface components recognizing adhesive matrix molecules of the host. Future Microbiol. 4, 1337–1352. 10.2217/fmb.09.102 [DOI] [PubMed] [Google Scholar]
- Strommenger B., Braulke C., Heuck D., Schmidt C., Pasemann B., Nübel U., et al. (2008). spa Typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 46, 574–581. 10.1128/JCM.01599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tande A. J., Osmon D. R., Greenwood-Quaintance K. E., Mabry T. M., Hanssen A. D., Patel R. (2014). Clinical characteristics and outcomes of prosthetic joint infection caused by small colony variant staphylococci. MBio 5, e01910–14. 10.1128/mBio.01910-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tande A. J., Patel R. (2014). Prosthetic joint infection. Clin. Microbiol. Rev. 27, 302–345. 10.1128/CMR.00111-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A., Faria N. A., de Lencastre H., Miragaia M. (2014). Population structure of methicillin-susceptible Staphylococcus aureus (MSSA) in Portugal over a 19-year period (1992-2011). Eur. J. Clin. Microbiol. Infect. Dis. 33, 423–432. 10.1007/s10096-013-1972-z [DOI] [PubMed] [Google Scholar]
- Thomas J. C., Godfrey P. A., Feldgarden M., Robinson D. A. (2012). Draft genome sequences of Staphylococcus aureus sequence type 34 (ST34) and ST42 hybrids. J. Bacteriol. 194, 2740–2741. 10.1128/JB.00248-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaras G., Osmon D. R., Mabry T., Lahr B., St Sauveur J., Yawn B., et al. (2012). Incidence, secular trends, and outcomes of prosthetic joint infection: a population-based study, Olmsted county, Minnesota, Z. (1969-2007). Infect. Control Hosp. Epidemiol. 33, 1207–1212. 10.1086/668421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J., Vergara-Irigaray M., Merino N., Penadés J. R., Lasa I. (2007). sigmaB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J. Bacteriol. 189, 2886–2896. 10.1128/JB.01767-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F., Naimi T., Enright M. C., Lina G., Nimmo G. R., Heffernan H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9, 978–984. 10.3201/eid0908.030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannuffel P., Gigi J., Ezzedine H., Vandercam B., Delmee M., Wauters G., et al. (1995). Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J. Clin. Microbiol. 33, 2864–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez V., Liang X., Horndahl J. K., Ganesh V. K., Smeds E., Foster T. J., et al. (2011). Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J. Biol. Chem. 286, 29797–29805. 10.1074/jbc.M110.214981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivoni A. M., Diep B. A., de Gouveia Magalhães A. C., Santos K. R., Riley L. W., Sensabaugh G. F., et al. (2006). Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J. Clin. Microbiol. 44, 1686–1691. 10.1128/JCM.44.5.1686-1691.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska K., Piórkowska A., Kasprzyk J., Bronk M., Swieć K. (2014). Clonal distribution of bone sialoprotein-binding protein gene among Staphylococcus aureus isolates associated with bloodstream infections. Folia Microbiol. (Praha). 59, 465–471. 10.1007/s12223-014-0321-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Rivas J. M., Brown E. L., Liang X., Höök M. (2005). Virulence potential of the staphylococcal adhesin CNA in experimental arthritis is determined by its affinity for collagen. J. Infect. Dis. 189, 2323–2333. 10.1086/420851 [DOI] [PubMed] [Google Scholar]
- Ziebuhr W., Krimmer V., Rachid S., Lössner I., Götz F., Hacker J. (1999). A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32, 345–356. 10.1046/j.1365-2958.1999.01353.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.