Abstract
Plants interact with a huge variety of soil microbes, ranging from pathogenic to mutualistic. The Fusarium oxysporum (Fo) species complex consists of ubiquitous soil inhabiting fungi that can infect and cause disease in over 120 different plant species including tomato, banana, cotton, and Arabidopsis. However, in many cases Fo colonization remains symptomless or even has beneficial effects on plant growth and/or stress tolerance. Also in pathogenic interactions a lengthy asymptomatic phase usually precedes disease development. All this indicates a sophisticated and fine-tuned interaction between Fo and its host. The molecular mechanisms underlying this balance are poorly understood. Plant hormone signaling networks emerge as key regulators of plant-microbe interactions in general. In this review we summarize the effects of the major phytohormones on the interaction between Fo and its diverse hosts. Generally, Salicylic Acid (SA) signaling reduces plant susceptibility, whereas Jasmonic Acid (JA), Ethylene (ET), Abscisic Acid (ABA), and auxin have complex effects, and are potentially hijacked by Fo for host manipulation. Finally, we discuss how plant hormones and Fo effectors balance the interaction from beneficial to pathogenic and vice versa.
Keywords: plant immunity, root pathogen, vascular wilt disease, effectors, endophyte
Introduction
Fusarium oxysporum (Fo), one of the most relevant plant pathogens in global agriculture, is a widespread soil-borne fungus that invades roots and causes vascular wilt disease through colonization of xylem tissue (Tjamos and Beckman, 1989). Pathogenic Fo strains have been classified in more than 120 formae speciales (ff.spp.), which refers to a specific plant host, as a particular isolate typically produces disease only within a limited range of host species (Armstrong and Armstrong, 1981; Katan and Di Primo, 1999; Table 1). The infection process occurs following attachment to the root surface and subsequent penetration and colonization of the plant root and proliferation within the xylem vessels, leading to both local and systemic induction of a broad spectrum of plant defense responses (Berrocal-Lobo and Molina, 2008). Vascular browning, stunting, progressive wilting, and eventually plant death are typical disease symptoms in infected plants (Pietro et al., 2003; Agrios, 2005). In contrast to the potential of pathogenic Fo isolates to cause destructive plant diseases, many Fo strains are non-pathogenic and survive either saprophytically in the soil, as non-invasive colonizer of the rhizosphere, or as endophyte inside plant tissues (Kuldau and Yates, 2000; Edel-Hermann et al., 2015; Imazaki and Kadota, 2015). There is relatively little known about the lifestyle strategies of these inconspicuous endophytic strains, but some of them have been successfully employed in biocontrol strategies to combat plant diseases (Alabouvette et al., 2009; Vos et al., 2014).
Table 1.
Fo strains and their host described in the manuscript.
| Abbreviations | Formae speciales | Host |
|---|---|---|
| Foal | Fusarium oxysporum f.sp. albedinis | Date palm |
| Focn | Fusarium oxysporum f.sp. conglutinans | Cabbage, Arabidopsis |
| Focb | Fusarium oxysporum f.sp. cubense | Banana |
| Fol | Fusarium oxysporum f.sp. lycopersici | Tomato |
| Fomt | Fusarium oxysporum f.sp. matthioli | Garden stock, Arabidopsis |
| Foph | Fusarium oxysporum f.sp. phaseoli | Common bean |
| Forp | Fusarium oxysporum f.sp. raphani | Radish, Arabidopsis |
| Forl | Fusarium oxysporum f.sp. radicis-lycopersici | Tomato |
Based on their lifestyle plant pathogenic fungi have been classified as biotrophs and necrotrophs. Biotrophic pathogens derive nutrients from living cells and deploy complex manipulation strategies to exploit their hosts while keeping them alive. In contrast, necrotrophic pathogens generally kill host cells and feed on their contents, resulting in extensive necrosis, tissue maceration, and plant rot (Glazebrook, 2005). A third type, termed hemi-biotrophs, displays both forms of nutrient acquisition, shifting from a biotrophic phase early in infection to necrotrophy at later stages. These pathogens typically produce toxins only at later stages of disease development in order to kill the host cells and to complete their life cycle on dead tissues (Horbach et al., 2011). The strategy of different pathogenic Fo strains can vary, but is usually best described by a hemi-biotrophic lifestyle (Michielse and Rep, 2009). Consistently, the Fo genomes show an expansion of genes that encode small, secreted proteins as well as cell-wall degrading enzymes, a feature shared by many hemi-biotrophic fungi (Lo Presti et al., 2015). Analysis of the xylem sap proteome from Fol-infected tomato plants identified numerous fungal proteins, termed Secreted in Xylem (Six) protein. For several Six proteins a contribution to virulence has been demonstrated, designating them as bona fide effectors (Takken and Rep, 2010; de Sain and Rep, 2015). However, their molecular mode of action and putative virulence targets remain unknown.
Phytohormones such as SA, JA, and ET, are known to play major roles in regulating plant defense responses against various pathogens. Generally, SA signaling triggers resistance against biotrophic and hemibiotrophic pathogens, whereas a combination of JA and ET signaling activates resistance against necrotrophic pathogens (Glazebrook, 2005). All these hormones are part of a larger signaling network that integrates environmental inputs and provides robustness against microbial manipulations (Katagiri and Tsuda, 2010; Pieterse et al., 2012). Additional hormones such as auxins, abscisic acid (ABA), gibberellic acids (GAs), and brassinosteroids (BRs) have also been reported to be involved in plant immunity and to fine-tune immunity and growth/development in plants (Table 2; Robert-Seilaniantz et al., 2011). In this review, we summarize the current knowledge on the role of phytohormones in plant disease and resistance triggered by different Fo ff.spp. to uncover how they shape the outcome of this widespread plant-fungal interaction.
Table 2.
Phytohormone mutants involved in the defense response against Fo infection.
| Hormones | Mutants and transgenic lines | Process affected | Plant species | Susceptibility | References |
|---|---|---|---|---|---|
| SA | NahG | SA accumulation | Arabidopsis | Increased to Focn and Fol | Berrocal-Lobo and Molina, 2004; Diener and Ausubel, 2005; Thatcher et al., 2009; Trusov et al., 2009 |
| sid2-1 | SA biosynthesis | Arabidopsis | Increased to Focn and Fol | Berrocal-Lobo and Molina, 2004; Diener and Ausubel, 2005 | |
| eds1-1, eds1-22 | SA signaling | Arabidopsis | Unaltered to Focn and Fol | Berrocal-Lobo and Molina, 2004; Trusov et al., 2009 | |
| eds3, eds4, eds10 | SA signaling | Arabidopsis | Increased to Focn | Diener and Ausubel, 2005 | |
| eds5-1 | SA biosynthesis | Arabidopsis | Increased to Focn and Fol | Berrocal-Lobo and Molina, 2004; Diener and Ausubel, 2005; Thatcher et al., 2009; Trusov et al., 2009 | |
| pad4-1 | SA signaling | Arabidopsis | Unaltered to Focn and Fol | Berrocal-Lobo and Molina, 2004 | |
| pad4 | Increased to Focn | Diener and Ausubel, 2005 | |||
| npr1-1 | SA perception | Arabidopsis | Increased to Focn and Fol | Berrocal-Lobo and Molina, 2004 | |
| npr1-1, npr1-2, npr1-3, npr1-4 | SA perception | Arabidopsis | Unaltered to Focn | Diener and Ausubel, 2005; Trusov et al., 2009 | |
| 35S::NPR1 | SA perception | Tomato | Reduced to Fol | Lin et al., 2004 | |
| hpSAMT | SA metabolism | Tomato | Reduced to Fol | Ament et al., 2010 | |
| JA | aos, fad3-2, opr3 fad7-1 fad8 | JA biosynthesis | Arabidopsis | Unaltered to Focn | Thatcher et al., 2009 |
| coi1, coi1-21 | JA perception | Arabidopsis | Reduced to Focn and Fomt, | Thatcher et al., 2009; Trusov et al., 2009; Cole et al., 2014 | |
| jar1-1 | JA-Ile biosynthesis | Arabidopsis | Increased to Focn and Fol | Berrocal-Lobo and Molina, 2004; Trusov et al., 2009 | |
| jar1-1 | Unaltered to Focn | Thatcher et al., 2009 | |||
| jin1-9(atmyc2-3), jin1-9/myc2 | JA signaling | Arabidopsis | Reduced to Focn | Anderson et al., 2004; Trusov et al., 2009 | |
| 35S::AtERF2 | Positive regulator of MeJA response | Arabidopsis | Reduced to Focn | McGrath et al., 2005 | |
| 35S::AtERF4 | Negative regulator of MeJA response | Arabidopsis | Increased to Focn | McGrath et al., 2005 | |
| pft1-1, med8 | JA signaling | Arabidopsis | Reduced to Focn | Kidd et al., 2009 | |
| def1 | JA biosynthesis | Tomato | Increased to Forl and Fol | Thaler et al., 2004; Kavroulakis et al., 2007 | |
| jai1 | JA perception (Coi1 homolog) | Tomato | Unaltered to Fol | Cole et al., 2014 | |
| ET | ein2-1 | ET signaling | Arabidopsis | Reduced to Focn and Forp | Trusov et al., 2009; Cole et al., 2014 |
| ein2, etr1 | ET signaling | Arabidopsis | Unaltered to Focn | Thatcher et al., 2009 | |
| ein2-5 | ET signaling | Arabidopsis | Increased to Focn and Fol | Berrocal-Lobo and Molina, 2004 | |
| etr1-1 | ET perception | Arabidopsis | Reduced to Forp | Pantelides et al., 2013 | |
| 35S::ERF1 | ET signaling | Arabidopsis | Reduced to Focn and Fol | Berrocal-Lobo and Molina, 2004 | |
| Never ripe | ET perception | Tomato | Reduced to Fol | Lund et al., 1998; Francia et al., 2007 | |
| Never ripe, epinastic (epi1) | ET signaling | Tomato | Unaltered to Forl | Kavroulakis et al., 2007 | |
| ABA | aba1-6, aba2-1 | ABA biosynthesis | Arabidopsis | Reduced to Focn | Anderson et al., 2004; Trusov et al., 2009 |
| aba2 | ABA biosynthesis | Arabidopsis | Reduced to Focn | Cole et al., 2014 | |
| Auxin | cyp79b2 cyp79b3, atr4/sur2, myb51/hig1, atr1, atr2d, pad3 | auxin biosynthesis | Arabidopsis | Unaltered to Focn | Kidd et al., 2011 |
| 35S:ATR1/MYB34, atr1d, 35S:ATR2 | |||||
| axr1, axr2, axr3, sgt1b | auxin signaling | Arabidopsis | Reduced to Focn | Kidd et al., 2011 | |
| tir1 | auxin perception | Arabidopsis | Unaltered to Focn | Kidd et al., 2011 | |
| arf1, arf2, arf1arf2 | auxin signaling | Arabidopsis | Reduced to Focn | Lyons et al., 2015 |
SA promotes resistance to Fo
Defense to biotrophic or hemibiotrophic pathogens is frequently mediated via SA signaling (Glazebrook, 2005). Arabidopsis plants with impaired SA accumulation showed increased susceptibility to Fo f.sp. conglutinans (Focn), but not to Fo f.sp. raphani (Forp) pointing to an isolate-specific role of SA-dependent defense responses (Table 2; Berrocal-Lobo and Molina, 2004; Diener and Ausubel, 2005; Trusov et al., 2009; Cole et al., 2014). Interestingly, mutants of the SA master-signaling regulator NPR1 showed wildtype (WT)-like susceptibility to Focn when 2–3-week-old soil grown plants were examined (Diener and Ausubel, 2005; Trusov et al., 2009). In contrast, when seedlings were infected on sterile agar plates npr1 mutants displayed clearly enhanced susceptibility in comparison to WT (Berrocal-Lobo and Molina, 2004). SA accumulation and signaling is also influenced by the nucleo-cytoplasmic proteins PAD4 and EDS1. Soil-grown pad4, but not eds1, Arabidopsis showed increased susceptibility, whereas sterile grown pad4 seedlings behaved like WT (Berrocal-Lobo and Molina, 2004; Diener and Ausubel, 2005; Trusov et al., 2009). Thus, the influence of SA signaling regulators depends on growth and inoculation conditions, plant age and the Fo isolate used. It appears that at least in soil-grown plants, SA enhances immunity to Fo via NPR1- and EDS1-independent pathways.
Consistent with its defense promoting role in Arabidopsis, exogenous application of SA, or synthetic SA analogs reduced Fo disease symptoms in a broad range of tested plants including tomato, common bean, date palm, and Arabidopsis (Edgar et al., 2006; Mandal et al., 2009; Dihazi et al., 2011; Xue et al., 2014). Furthermore, stable overexpression of Arabidopsis NPR1 in tomato reduced disease symptoms as well as Fo f.sp. lycopersici (Fol) colonization of the stem (Lin et al., 2004). Similarly, preventing SA volatilization by silencing of a Salicylic Acid Methyltransferase reduced tomato susceptibility to Fol, however without significantly changing overall SA levels (Ament et al., 2010). How exactly Methyl-SA levels influence tomato defense to Fol remains to be addressed.
Despite a clear effect of SA on disease severity, global transcriptome profiling of Arabidopsis plants inoculated with Focn revealed relatively mild changes in the expression of known SA marker genes (Kidd et al., 2011; Zhu et al., 2013; Chen et al., 2014; Lyons et al., 2015). In fact, expression of PR1 was even slightly down regulated both in roots and shoots of inoculated plants (Kidd et al., 2011). It is possible that activation of SA signaling occurs at rather late stages, which would be missed by the present studies that focus on the time points 1–6 days-post-inoculation (dpi). Alternatively, SA signaling could activate previously uncharacterized defense mechanisms, in line with the observed NPR1- and EDS1-independency, especially in roots that are still little explored in terms of plant immunity (De Coninck et al., 2015). A third possibility has been suggested by Cole et al.: SA signaling could serve to dampen activation of JA responses that are promoting Arabidopsis infection by Fo (see below; Cole et al., 2014). Similar to Arabidopsis, transcriptome profiling of banana saplings infected with virulent and avirulent Fo f.sp. cubense (Focb) strains also failed to detect activation of typical SA marker genes, at least at the relatively early time points analyzed (Li et al., 2013). This led to the suggestion that in banana defense against Fo is mainly mediated via ET/JA signaling (Swarupa et al., 2014).
Taken together, SA signaling positively regulates defense to Fo in most tested plant species, which is in line with a predominantly hemi-biotrophic lifestyle of this pathogen (Figure 1). However, the exact mechanisms by which SA reduces susceptibility to Fo are not understood and yet unknown SA targets possibly play a role during defense to this root-infecting pathogen.
Figure 1.
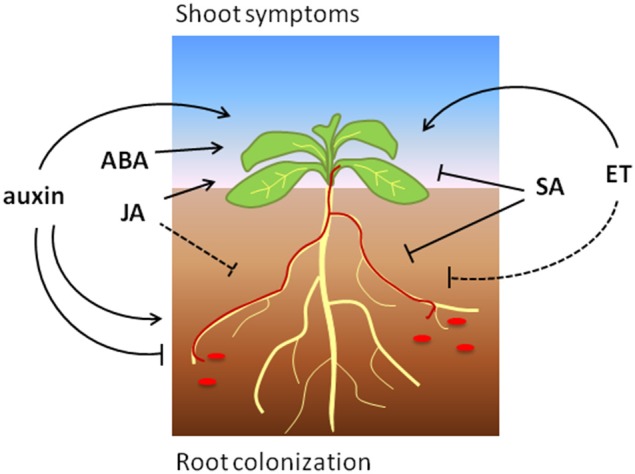
The effects of phytohormone signaling on Arabidopsis root colonization and shoot symptoms induced by Focn (shown in red). Arrows and blunt ends indicate promotion and reduction, respectively. The dashed line indicates a presumed positive role of ET-signaling on (root) defense.
JA signaling can promote either resistance or susceptibility in different host–Fo interactions
JA signaling generally mediates resistance to necrotrophic pathogens and insect herbivores. These two functions are exerted by two separate and often mutually antagonistic branches: the former is regulated by ERF transcription factors and is associated with ET signaling, whereas the latter requires MYC transcription factors and often involves ABA signaling (Pieterse et al., 2012).
In Arabidopsis JA biosynthesis is not critical for defense to Focn as a whole series of mutants with impaired JA accumulation behaved like WT plants (Table 2; Thatcher et al., 2009). Similarly, jar1 mutants that are defective in synthesis of the bioactive JA–Isoleucine conjugate showed WT-like or slightly increased susceptibility (Thatcher et al., 2009; Trusov et al., 2009). Considering these results it was unexpected that mutations in CORONATINE INSENSITIVE1 (COI1), an essential component of JA perception, strongly increased resistance to Focn (Thatcher et al., 2009; Trusov et al., 2009). Importantly, loss-of-function mutations in additional regulators of JA signaling that are involved in pathogen defense, such as the transcriptional regulators MYC2, PFT1, and LBD20, also resulted in increased resistance to Focn (Anderson et al., 2004; Kidd et al., 2009; Thatcher et al., 2012b). These findings indicate that JA signaling-capacity of the host is critical for Fo pathogenesis in Arabidopsis.
Hijacking the host JA signaling machinery is a common strategy employed by many (hemi-)biotrophic pathogens and often involves suppression of SA-dependent defense responses (Pieterse et al., 2012; Kazan and Lyons, 2014). However, COI1 promotes Fo infection in an SA-independent manner as coi1 NahG plants remain as resistant as coi1 plants (Thatcher et al., 2009). This indicates that JA signaling supports Fo pathogenesis by mechanisms other than antagonizing SA responses. Grafting experiments demonstrated that expression of COI1 in roots, but not in shoots, is required for Fo infection (Thatcher et al., 2009). Cole and co-workers observed reduced colonization of the vasculature in coi1 plants pointing to a role of JA signaling at relatively early stages (Cole et al., 2014). Thatcher et al. detected similar levels of fungal biomass in WT and coi1 shoots before the switch to necrotrophic growth, and hence concluded that COI1 is predominantly required for triggering plant decay (Thatcher et al., 2009). These differences might be explained by the inoculation methods used: uprooting of plants before inoculation could have created additional entry sites leading to stronger vascular colonization (as observed in Thatcher et al., 2009). Nevertheless, the studies indicate that COI1 influences the interaction with Fo at several stages. Moreover, the finding that even strongly colonized coi1 plants remain essentially symptomless reveals an uncoupling of colonization and plant disease symptoms (Thatcher et al., 2009).
Despite its strong effects on the interaction with Fo, it is not well-understood how the host JA signaling machinery is re-wired by the pathogen to promote disease. Cole and co-workers found that two Arabidopsis-infecting strains, Focn and f.sp. matthioli (Fomt) produce JA-isoleucin and JA-leucin conjugates in culture filtrates that induce senescence-like symptoms on Arabidopsis leaves in a COI1-dependent manner (Cole et al., 2014). However, it is not yet known if these fungal derived hormones are also generated during infection, and to what extent they contribute to virulence. Alternatively, Fo effectors could play a role in JA signaling manipulation as has been suggested for the effector SIX4 (Thatcher et al., 2012a). It will be interesting to explore which JA signaling regulators are targeted by Fo and how their activity is modulated.
The dependency on host JA signaling for successful colonization however, is not conserved among all Fo ff.spp. For instance, Forp colonizes WT and coi1 Arabidopsis plants to a similar extent and this isolate does not produce bioactive JA-conjugates in vitro. Similarly, Fol seems to infect tomato without JA-signaling manipulation as this f.sp. does not produce JAs and is not dependent on the tomato COI1 homolog to cause disease (Cole et al., 2014). Thus, different Fo isolates have developed distinct infection strategies that either strongly rely on host JA signaling manipulation or involve alternative virulence mechanisms that are JA-independent.
In addition, several lines of evidence point to a role of JA in promoting resistance rather than susceptibility in plant species other than Arabidopsis. Tomato def1 mutants that are defective in JA synthesis show enhanced susceptibility to Fol, which can be suppressed by exogenous JA treatment (Thaler et al., 2004). Similarly, def1 tomato plants were more susceptible to root rot caused by Fo f.sp. radicis-lycopersici (Forl; Kavroulakis et al., 2007). Consistently, Sun and co-workers found that spraying of banana plants with Methyl-JA reduced disease incidence and severity caused by Focb Tropical Race 4 (Sun et al., 2013). In addition, resistant cultivars of strawberry and watermelon showed strong induction of the JA biosynthesis enzyme AOC upon inoculation with Fo f.sp. fragariae (Lu et al., 2011; Fang et al., 2013). A positive role of JA for defense activation was also found in date palms inoculated with Fo f.sp. albenidis (Jaiti et al., 2009).
In summary, JA signaling promotes defense to Fo in different plant species, but can also be hijacked to induce pathogenicity in at least Arabidopsis (Figure 1). Further research will be necessary to reveal via which mechanisms JA contributes to disease reduction or induction, and which function is predominant among different plant species.
Dual role of ET in activation of both defense responses and disease symptoms
Generally, ET together with JA mediates the resistance response to necrotrophic pathogens. However, ET can also positively influence defense responses to hemi-biotrophs and induced systemic resistance, which is triggered by beneficial root-associated microbes (Pieterse et al., 2014; Broekgaarden et al., 2015).
Pre-treatment of Arabidopsis seedlings with either MeJA or the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC) resulted in enhanced disease symptoms upon Focn inoculation, indicating that both these hormones promote disease development (Trusov et al., 2009). Accordingly, the ET-insensitive Arabidopsis ein2-1 and etr1-1 mutants showed a reduction of disease symptoms compared to WT Col plants when inoculated with Focn or Forp, respectively (Table 2; Trusov et al., 2009; Pantelides et al., 2013). It is worth mentioning that enhanced disease upon MeJA treatment, as well as reduced disease in ET-insensitive plants was not observed in similar studies, indicating that these effects are either weak or depend on yet unknown factors (Edgar et al., 2006; Thatcher et al., 2009). Moreover, a different ein2 allele (ein2-5) even showed markedly enhanced susceptibility to Focn under sterile conditions (Berrocal-Lobo and Molina, 2004). These findings point to an age- and/or condition-dependent role of ET in Arabidopsis interaction with Fo.
Global transcriptome profiling in Arabidopsis and banana plants inoculated with virulent Fo strains revealed a massive induction of ET/JA-responsive genes such as Plant Defensins (PDFs) and Pathogenesis-Related (PR) genes as well as genes encoding ethylene biosynthesis enzymes (McGrath et al., 2005; Kidd et al., 2011; Li et al., 2013; Zhu et al., 2013; Lyons et al., 2015). Furthermore, the transcriptome profiles indicated that initial activation of ET-dependent genes precedes the activation of JA, SA and ABA signaling (Li et al., 2013; Zhu et al., 2013). Altogether, these findings suggest a model in which initial ET/JA-associated defenses are mounted in response to Fo infection, but these are typically insufficient to stop the pathogen. At later stages however, ET signaling can rather enhance disease symptoms and possibly also pathogen proliferation. This hypothesis is further supported by the observation that Arabidopsis plants overexpressing certain ERF transcription factors, thereby constitutively activating ET/JA-dependent defense responses, become less susceptible to Focn (Berrocal-Lobo and Molina, 2004; McGrath et al., 2005). However, whether ET signaling is actively suppressed and/or at later stages co-opted by Fo to promote pathogenesis remains to be addressed.
The exact role of ET in other plant species is relatively little understood. The tomato Never ripe (Nr) mutant is impaired in ethylene perception and shows reduced disease symptoms upon Fol inoculation (Lund et al., 1998; Francia et al., 2007). Interestingly, previous work revealed a role for ET in mediating xylem occlusion through formation of gels in castor bean (Vandermolen et al., 1983). Xylem occlusion is thought to limit pathogen spread, but also to contribute to the typical wilting symptoms (Yadeta and Thomma, 2013). Thus, it is an interesting question whether Nr tomato plants allow systemic fungal spread and how this would correspond to the observed reduction in disease symptoms. Resistance to Forl, a pathogen which adopts a necrotroph-like lifestyle, was largely unaffected in two tested ET-insensitive tomato lines, Nr and epinastic (epi; Kavroulakis et al., 2007). However, protection mediated by an endophytic Fusarium solani strain was greatly reduced in Nr and epi tomato plants and hence required intact ET signaling (Kavroulakis et al., 2007).
In conclusion, the present studies underline a multifaceted role of ET signaling that strongly depends on the interaction stage, the host plant and environmental conditions (Figure 1).
ABA promotes shoot disease symptoms but not root colonization in arabidopsis
Besides its well-described role in development and abiotic stress responses, ABA has been increasingly recognized as a critical regulator of biotic interactions. ABA can either positively or negatively influence resistance largely depending on the encountered pathogen (Ton et al., 2009; Robert-Seilaniantz et al., 2011).
Similarly to Methyl-JA and ACC, exogenous treatment with ABA increased Arabidopsis susceptibility to Focn (Trusov et al., 2009). Consistently, Arabidopsis mutants in which either ABA biosynthesis or signaling is disrupted showed fewer symptoms (Table 2; Anderson et al., 2004; Trusov et al., 2009). The reduced susceptibility of ABA mutants was associated with hyper-activation of ET/JA-dependent defense genes, likely due to antagonistic interactions between ABA and ET signaling (Anderson et al., 2004). ABA could also antagonize SA-dependent responses (Yasuda et al., 2008), but currently it is unknown if the interaction with other hormones explains reduced Fo symptoms in ABA-deficient mutants.
Interestingly, Fo successfully colonized the roots of ABA-deficient mutants to a similar extent as those from WT plants (Cole et al., 2014). This would point to a role of ABA during the switch to the necrotrophic phase. However, transcriptome profiling revealed activation of numerous ABA responsive genes in the roots of Fo-inoculated plants (Lyons et al., 2015). Previous studies indicated that ABA mediates root-to-shoot defense signaling in plants (Balmer et al., 2013). This raises the possibility that Fo co-opts systemic ABA signaling to manipulate root-shoot signaling. Moreover, it has been shown that, for example during defense against herbivorous insects, ABA signaling can serve to activate or enhance the MYC2-regulated branch of JA signaling (Kazan and Manners, 2013; Vos et al., 2013). However, if and how exactly Fo manipulates ABA signaling, is currently unknown.
Auxins affects both root colonization and shoot symptom development
Auxins are major regulators of plant growth and development, but have also profound effects on interactions with both pathogenic and mutualistic microbes (Robert-Seilaniantz et al., 2011; Zamioudis et al., 2013).
Exogenous application of auxin or auxin biosynthesis inhibitors did not affect disease development in Focn-inoculated Arabidopsis (Kidd et al., 2011). Similarly, mutants with either reduced or increased auxin levels behaved like WT plants. In contrast, Focn-inoculated auxin-signaling mutants showed markedly reduced symptoms relative to WT plants (Table 2). Additionally, alteration of polar auxin transport, either by chemical inhibitors or in mutants, resulted in increased resistance to Focn (Kidd et al., 2011). These data indicate that local changes of auxin levels and/or distribution are important for disease susceptibility. Indeed, histological visualization of DR5 expression, a well-known auxin reporter gene, revealed activation of auxin signaling at root tips and lateral root initials, two preferred Fo entry sites in Arabidopsis (Czymmek et al., 2007; Kidd et al., 2011; Diener, 2012). Additionally, Diener revealed that fewer root tips are colonized in plants mutated in the auxin efflux carrier PIN2/EIR1 (Diener, 2012). In contrast, tir3 mutants which are defective in polar auxin transport show 2–3 fold higher Fo biomass in roots, but disease symptoms of the shoot remained strongly reduced (Kidd et al., 2011; Diener, 2012). These findings suggest that auxin signaling and transport affect several stages of the Fo–Arabidopsis interaction from initial root tip colonization to disease symptom expression in the shoot. However, the mechanisms by which auxin promotes colonization and symptom development are still enigmatic. Previous studies describe an antagonistic relationship between auxin and SA signaling, however the Fo disease phenotypes of auxin signaling mutants were not SA-dependent (Kidd et al., 2011).
Auxin accumulation, transport, and signaling are modulated by numerous different symbiotic and pathogenic organisms including bacteria, fungi, nematodes, and even parasitic plants during their interaction with roots (Grunewald et al., 2009; Kazan and Manners, 2009; Zamioudis and Pieterse, 2012). This suggests that manipulation of the host auxin signaling pathway represents a common strategy employed by diverse root colonizers resulting in either detrimental or beneficial effects for plants.
Are phytohormones determinants of Fo lifestyle?
Interestingly, changes in the phytohormone network can uncouple colonization by Fo from plant disease development. For instance, specific mutants with impaired JA, ABA, and auxin signaling still allow extensive root (and sometimes shoot) colonization but have greatly reduced disease symptoms (Thatcher et al., 2009; Diener, 2012; Cole et al., 2014). Similarly, resistant tomato plants that are completely free of symptoms can have their shoots and stems extensively colonized by Fol (Mes et al., 1999). Furthermore, a Fol knockout strain lacking the Six6 effector triggered vascular browning in a susceptible tomato cultivar, indicative of successful xylem colonization, but exerted almost no negative effects on plant growth and development (Gawehns et al., 2014). Altogether, these findings suggest that manipulation of plant hormone signaling rather than colonization triggers disease symptom development.
How does Fo manipulate the plant hormone network? One mechanism could be the production by the fungus of hormone-like secondary metabolites, including JAs, auxins, gibberellic acids, and ethylene (Hasan, 2002; Cole et al., 2014; Bitas et al., 2015). Fo also secretes numerous small proteins during plant infection, which might be another means to manipulate the host. For several of these proteins a virulence-promoting function has been shown designating them as effectors sensu strictu (Takken and Rep, 2010; de Sain and Rep, 2015). Among these, SIX4 was found to enhance JA signaling during infection of Arabidopsis (Thatcher et al., 2012a). Infection of tomato plants with Fol knockout strains lacking specific effectors revealed common and unique effects on the xylem proteome composition raising the possibility that each effector targets a distinct hormone signaling pathway (Gawehns et al., 2015). However, for the vast majority of Fo effectors their working mechanism remains unknown. It will be interesting to explore if plant hormone-synthesis or -signaling represents a recurrent virulence target of Fo strains on various hosts. Furthermore, it is tempting to speculate that at least a subset of the proteinacious effectors mediate immune suppression enabling (endophytic) colonization during the biotrophic phase of infection, whereas secondary metabolites with hormonal- or toxic-activity trigger plant damage/death during necrotrophic growth.
A growing body of evidence suggests that the majority of Fo strains survive in soil, in the rhizosphere or within plant tissues without causing disease symptoms, and some strains even confer extensive beneficial effects (Alabouvette et al., 2009; Edel-Hermann et al., 2015; Imazaki and Kadota, 2015). The existence of such a widespread and intimate co-habitation points to a finely balanced interaction between plant and fungus. The finding that also pathogenic strains can reside inside plant tissues without damaging the host indicate that one Fo isolate can employ diverse interaction/colonization strategies whose outcome possibly depends on the “compatibility” of a putative host plant. Thus, the “infection tools” of a Fo strain, likely comprised of a combination of effectors, enzymes, and secondary metabolites, determine the outcome of an interaction: either endophytic with potential beneficial effects for the host or pathogenic with various levels of disease and in extreme cases plant death. This idea is supported by the observation that transfer of one specific Fol chromosome, which contains most of its effector genes plus a secondary metabolite cluster, conferred pathogenicity to an endophytic strain (Ma et al., 2010).
Clearly more research in the areas of genomics and effector biology is required to understand how Fo manages to trick the hormonal network of its hosts, and how these interactions can have opposite outcomes ranging from pathogenesis to mutualism.
Conclusions and outlook
Phytohormones determine colonization and disease symptom development during interactions with pathogenic Fo strains. However, their roles vary depending on the host plant and the fungal strain involved, suggesting that the manipulation of the host hormonal network differs between individual Fo strains. Genetic interference with hormone regulators mostly reduced disease symptoms, as seen for JA, ABA, and auxin, indicating that the ability to hijack plant hormone pathways is a requirement for pathogenesis. This scenario implies a strong adaptation to a particular host plant, potentially leading to the narrow host range observed in the Fo species complex. How exactly the manipulation of phytohormone signaling differs between Fo strains—and if this is indeed the key difference between pathogenic and endophytic interactions—remains an intriguing question for future research. Comparison of the respective effector repertoires as well as a better understanding of their mode of action will help answering these questions and may furthermore reveal novel approaches for plant protection, either by breeding or by optimizing Fo strains for biocontrol.
Author contributions
XD and NT wrote the manuscript together with input from FT. The scope and the topic were developed by XD, NT, and FT.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to our colleagues whose work is not cited due to space limitation. The authors wish to thank Prof. Ben Cornelissen and Dr. Martijn Rep for critically reading and commenting on the manuscript. XD was supported by funding from the program of China Scholarship Council. FT received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skodowska-Curie grant agreement No. 676480 (Bestpass) and from the NWO-Earth and Life Sciences funded VICI project No. 865.14.003. NT was financed from a NWO VICI grant (project No. 865.10.002) granted to Dr. Martijn Rep.
References
- Agrios G. N. (2005). Plant Pathology. St. Louis, MO: Academic Press. [Google Scholar]
- Alabouvette C., Olivain C., Migheli Q., Steinberg C. (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New phytol. 184, 529–544. 10.1111/j.1469-8137.2009.03014.x [DOI] [PubMed] [Google Scholar]
- Ament K., Krasikov V., Allmann S., Rep M., Takken F. L., Schuurink R. C. (2010). Methyl salicylate production in tomato affects biotic interactions. Plant J. 62, 124–134. 10.1111/j.1365-313X.2010.04132.x [DOI] [PubMed] [Google Scholar]
- Anderson J. P., Badruzsaufari E., Schenk P. M., Manners J. M., Desmond O. J., Ehlert C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. 10.1105/tpc.104.025833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong G. M., Armstrong J. K. (1981). Formae speciales and races of Fusarium oxysporum causing wilt diseases, in Fusarium, Diseases, Biology and Taxonomy, eds Nelson P. E., Toussoun T. A., Cook R. J. (University park, TX: Pennsylvania State University Press; ), 391–399. [Google Scholar]
- Balmer D., De Papajewski D. V., Planchamp C., Glauser G., Mauch-Mani B. (2013). Induced resistance in maize is based on organ-specific defence responses. Plant J. 74, 213–225. 10.1111/tpj.12114 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A. (2004). Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microb. Interact. 17, 763–770. 10.1094/MPMI.2004.17.7.763 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A. (2008). Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci. 13, 145–150. 10.1016/j.tplants.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Bitas V., McCartney N., Li N., Demers J., Kim J. E., Kim H. S., et al. (2015). Volatiles enhance plant growth via affecting Auxin transport and signaling. Front. Microbiol. 6:1248. 10.3389/fmicb.2015.01248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekgaarden C., Caarls L., Vos I. A., Pieterse C. M., Van Wees S. C. (2015). Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 169, 2371–2379. 10.1104/pp.15.01020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C., Wong C. L., Muzzi F., Vlaardingerbroek I., Kidd B. N., Schenk P. M. (2014). Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci. Rep. 4:5584. 10.1038/srep05584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. J., Yoon A. J., Faull K. F., Diener A. C. (2014). Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol. Plant Pathol. 15, 589–600. 10.1111/mpp.12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czymmek K. J., Fogg M., Powell D. H., Sweigard J., Park S. Y., Kang S. (2007). In vivo time-lapse documentation using confocal and multi-photon microscopy reveals the mechanisms of invasion into the Arabidopsis root vascular system by Fusarium oxysporum. Fungal Genet. Biol. 44, 1011–1023. 10.1016/j.fgb.2007.01.012 [DOI] [PubMed] [Google Scholar]
- De Coninck B., Timmermans P., Vos C., Cammue B. P. A., Kazan K. (2015). What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 20, 91–101. 10.1016/j.tplants.2014.09.007 [DOI] [PubMed] [Google Scholar]
- de Sain M., Rep M. (2015). The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 16, 23970–23993. 10.3390/ijms161023970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener A. (2012). Visualizing and quantifying Fusarium oxysporum in the plant host. Mol. Plant Microbe Interact. 25, 1531–1541. 10.1094/MPMI-02-12-0042-TA [DOI] [PubMed] [Google Scholar]
- Diener A. C., Ausubel F. M. (2005). Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321. 10.1534/genetics.105.042218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihazi A., Serghini M. A., Jaiti F., Daayf F., Driouich A., Dihazi H., et al. (2011). Structural and biochemical changes in salicylic-acid-treated date palm roots challenged with Fusarium oxysporum f. sp. albedinis. J. Pathog. 2011:280481. 10.4061/2011/280481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel-Hermann V., Gautheron N., Mounier A., Steinberg C. (2015). Fusarium diversity in soil using a specific molecular approach and a cultural approach. J. Microbiol. Methods 111, 64–71. 10.1016/j.mimet.2015.01.026 [DOI] [PubMed] [Google Scholar]
- Edgar C. I., McGrath K. C., Dombrecht B., Manners J. M., Maclean D. C., Schenk P. M., et al. (2006). Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Australas. Plant Pathol. 35, 581–591. 10.1071/AP06060 [DOI] [Google Scholar]
- Fang X. L., Jost R., Finnegan P. M., Barbetti M. J. (2013). Comparative proteome analysis of the strawberry-Fusarium oxysporum f. sp fragariae pathosystem reveals early activation of defense responses as a crucial determinant of host resistance. J. Proteome Res. 12, 1772–1788. 10.1021/pr301117a [DOI] [PubMed] [Google Scholar]
- Francia D., Demaria D., Calderini O., Ferraris L., Valentino D., Arcioni S., et al. (2007). Wounding induces resistance to pathogens with different lifestyles in tomato: role of ethylene in cross-protection. Plant Cell Environ. 30, 1357–1365. 10.1111/j.1365-3040.2007.01709.x [DOI] [PubMed] [Google Scholar]
- Gawehns F., Houterman P. M., Ichou F. A., Michielse C. B., Hijdra M., Cornelissen B. J. C., et al. (2014). The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microb. Interact. 27, 336–348. 10.1094/MPMI-11-13-0330-R [DOI] [PubMed] [Google Scholar]
- Gawehns F., Ma L. S., Bruning O., Houterman P. M., Boeren S., Cornelissen B. J. C., et al. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 6:967. 10.3389/fpls.2015.00967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Grunewald W., Van Noorden G., Van Isterdael G., Beeckman T., Gheysen G., Mathesius U. (2009). Manipulation of Auxin transport in plant roots during rhizobium symbiosis and nematode parasitism. Plant Cell 21, 2553–2562. 10.1105/tpc.109.069617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan H. A. H. (2002). Gibberellin and auxin production by plant root-fungi and their biosynthesis under salinity-calcium interaction. Rost. Vyroba 48, 101–106. 10.1556/amicr.49.2002.1.11 [DOI] [PubMed] [Google Scholar]
- Horbach R., Navarro-Quesada A. R., Knogge W., Deising H. B. (2011). When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J. Plant Physiol. 168, 51–62. 10.1016/j.jplph.2010.06.014 [DOI] [PubMed] [Google Scholar]
- Imazaki I., Kadota I. (2015). Molecular phylogeny and diversity of Fusarium endophytes isolated from tomato stems. FEMS Microbiol. Ecol. 91:fiv098. 10.1093/femsec/fiv098 [DOI] [PubMed] [Google Scholar]
- Jaiti F., Verdeil J. L., El Hadrami I. (2009). Effect of jasmonic acid on the induction of polyphenoloxidase and peroxidase activities in relation to date palm resistance against Fusarium oxysporum f. sp albedinis. Physiol. Mol. Plant Pathol. 74, 84–90. 10.1016/j.pmpp.2009.09.005 [DOI] [Google Scholar]
- Katagiri F., Tsuda K. (2010). Understanding the plant immune system. Mol. Plant Microb. Interact. 23, 1531–1536. 10.1094/MPMI-04-10-0099 [DOI] [PubMed] [Google Scholar]
- Katan T., Di Primo P. (1999). Current status of vegetative compatibility groups in Fusarium oxysporum. Phytoparasitica 27(Suppl.), 273–277. 10.1007/BF02981483 [DOI] [Google Scholar]
- Kavroulakis N., Ntougias S., Zervakis G. I., Ehaliotis C., Haralampidis K., Papadopoulou K. K. (2007). Role of ethylene in the protection of tomato plants against soil-borne fungal pathogens conferred by an endophytic Fusarium solani strain. J. Exp. Bot. 58, 3853–3864. 10.1093/jxb/erm230 [DOI] [PubMed] [Google Scholar]
- Kazan K., Lyons R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26, 2285–2309. 10.1105/tpc.114.125419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14, 373–382. 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2013). Myc2: the master in action. Mol. Plant 6, 686–703. 10.1093/mp/sss128 [DOI] [PubMed] [Google Scholar]
- Kidd B. N., Edgar C. I., Kumar K. K., Aitken E. A., Schenk P. M., Manners J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. 10.1105/tpc.109.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B. N., Kadoo N. Y., Dombrecht B., Tekeoglu M., Gardiner D. M., Thatcher L. F., et al. (2011). Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol. Plant Microb. Interact. 24, 733–748. 10.1094/MPMI-08-10-0194 [DOI] [PubMed] [Google Scholar]
- Kuldau G., Yates I. (2000). Evidence for Fusarium endophytes in cultivated and wild plants, in Microbial Endophytes, eds Bacon C. W., White J. F. (New York, NY: Marcel Dekker; ), 85–117. [Google Scholar]
- Li C. Q., Shao J. F., Wang Y. J., Li W. B., Guo D. J., Yan B., et al. (2013). Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp cubense. BMC Genomics 14:851. 10.1186/1471-2164-14-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. C., Lu C. F., Wu J. W., Cheng M. L., Lin Y. M., Yang N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13, 567–581. 10.1007/s11248-004-2375-9 [DOI] [PubMed] [Google Scholar]
- Lo Presti L., Lanver D., Schweizer G., Tanaka S., Liang L., Tollot M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant. Biol. 66, 513–545. 10.1146/annurev-arplant-043014-114623 [DOI] [PubMed] [Google Scholar]
- Lu G. Y., Guo S. G., Zhang H. Y., Geng L. H., Song F. M., Fei Z. J., et al. (2011). Transcriptional profiling of watermelon during its incompatible interaction with Fusarium oxysporum f. sp niveum. Eur. J. Plant Pathol. 131, 585–601. 10.1007/s10658-011-9833-z [DOI] [Google Scholar]
- Lund S. T., Stall R. E., Klee H. J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10, 371–382. 10.1105/tpc.10.3.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R., Stiller J., Powell J., Rusu A., Manners J. M., Kazan K. (2015). Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS ONE 10:e0121902. 10.1371/journal.pone.0121902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. J., Van Der Does H. C., Borkovich K. A., Coleman J. J., Daboussi M. J., Di Pietro A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. 10.1038/nature08850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Mallick N., Mitra A. (2009). Salicylic acid-induced resistance to Fusarium oxysporum f. sp lycopersici in tomato. Plant Physiol. Biochem. 47, 642–649. 10.1016/j.plaphy.2009.03.001 [DOI] [PubMed] [Google Scholar]
- McGrath K. C., Dombrecht B., Manners J. M., Schenk P. M., Edgar C. I., Maclean D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. 10.1104/pp.105.068544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes J. J., Weststeijn E. A., Herlaar F., Lambalk J. J., Wijbrandi J., Haring M. A., et al. (1999). Biological and molecular characterization of Fusarium oxysporum f. sp lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 89, 156–160. 10.1094/PHYTO.1999.89.2.156 [DOI] [PubMed] [Google Scholar]
- Michielse C. B., Rep M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10, 311–324. 10.1111/j.1364-3703.2009.00538.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelides I. S., Tjamos S. E., Pappa S., Kargakis M., Paplomatas E. J. (2013). The ethylene receptor ETR1 is required for Fusarium oxysporum pathogenicity. Plant Pathol. 62, 1302–1309. 10.1111/ppa.12042 [DOI] [Google Scholar]
- Pieterse C. M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C., Bakker P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- Pietro A. D., Madrid M. P., Caracuel Z., Delgado-Jarana J., Roncero M. I. (2003). Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. 10.1046/j.1364-3703.2003.00180.x [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Sun D. Q., Lu X. H., Hu Y. L., Li W. M., Hong K. Q., Mo Y. W., et al. (2013). Methyl jasmonate induced defense responses increase resistance to Fusarium oxysporum f. sp cubense race 4 in banana. Sci. Hortic. 164, 484–491. 10.1016/j.scienta.2013.10.011 [DOI] [Google Scholar]
- Swarupa V., Ravishankar K. V., Rekha A. (2014). Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta 239, 735–751. 10.1007/s00425-013-2024-8 [DOI] [PubMed] [Google Scholar]
- Takken F., Rep M. (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 11, 309–314. 10.1111/j.1364-3703.2009.00605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J. S., Owen B., Higgins V. J. (2004). The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. 10.1104/pp.104.041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher L. F., Gardiner D. M., Kazan K., Manners J. M. (2012a). A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant Microb. Interact. 25, 180–190. 10.1094/MPMI-08-11-0212 [DOI] [PubMed] [Google Scholar]
- Thatcher L. F., Manners J. M., Kazan K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58, 927–939. 10.1111/j.1365-313X.2009.03831.x [DOI] [PubMed] [Google Scholar]
- Thatcher L. F., Powell J. J., Aitken E. A., Kazan K., Manners J. M. (2012b). The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 160, 407–418. 10.1104/pp.112.199067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjamos E. C., Beckman C. H. (1989). Vascular Wilt Diseases of Plants. Berlin: Springer-Verlag. [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. 10.1016/j.tplants.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Trusov Y., Sewelam N., Rookes J. E., Kunkel M., Nowak E., Schenk P. M., et al. (2009). Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J. Cell Mol. Biol. 58, 69–81. 10.1111/j.1365-313X.2008.03755.x [DOI] [PubMed] [Google Scholar]
- Vandermolen G. E., Labavitch J. M., Strand L. L., Devay J. E. (1983). Pathogen-induced vascular gels - ethylene as a host intermediate. Physiol. Plantarum 59, 573–580. 10.1111/j.1399-3054.1983.tb06282.x [DOI] [Google Scholar]
- Vos C. M., Yang Y., De Coninck B., Cammue B. P. A. (2014). Fungal (-like) biocontrol organisms in tomato disease control. Biol. Control 74, 65–81. 10.1016/j.biocontrol.2014.04.004 [DOI] [Google Scholar]
- Vos I. A., Verhage A., Schuurink R. C., Watt L. G., Pieterse C. M., Van Wees S. C. (2013). Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 4:539. 10.3389/fpls.2013.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R. F., Wu J., Wang L. F., Blair M. W., Wang X. M., Ge W. D., et al. (2014). Salicylic acid enhances resistance to Fusarium oxysporum f. sp. phaseoli in common beans (Phaseolus vulgaris L.). J. Plant Growth Regul. 33, 470–476. 10.1007/s00344-013-9376-y [DOI] [Google Scholar]
- Yadeta K., Thomma B. P. (2013). The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4:97. 10.3389/fpls.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M., Ishikawa A., Jikumaru Y., Seki M., Umezawa T., Asami T., et al. (2008). Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20, 1678–1692. 10.1105/tpc.107.054296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C., Mastranesti P., Dhonukshe P., Blilou I., Pieterse C. M. (2013). Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 162, 304–318. 10.1104/pp.112.212597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C., Pieterse C. M. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microb. Interact. 25, 139–150. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]
- Zhu Q. H., Stephen S., Kazan K., Jin G. L., Fan L. J., Taylor J., et al. (2013). Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene 512, 259–266. 10.1016/j.gene.2012.10.036 [DOI] [PubMed] [Google Scholar]


