Abstract
Candida species can cause severe infections associated with high morbidity and mortality. Therefore, it is essential to gain more insight into the anti-fungal host defense response. The advent of omics technology and development of advanced systems biology tools has permitted to approach this in an unbiased and quantitative manner. This review summarizes the insights gained on anti-Candida immunity from genetic-, transcriptome-, proteome-, metabolome-, microbiome-, mycobiome-, and computational systems biology studies and discusses practical aspects and future perspectives.
Keywords: genome, transcriptome, proteome, metabolome, microbiome, mycobiome, mathematical modeling, antifungal host defense
Introduction
Candida albicans is an opportunistic fungal microorganism which resides in the gastro-intestinal tract in healthy persons (Gouba and Drancourt, 2015). However, in some individuals colonization may lead to invasion and clinical disease: 75% of all woman experience vulvovaginal candidiasis at least once in there lifetime, and up to 5% of woman suffer from recurrent vulvovaginal candidiasis (Sobel, 1988). Furthermore, when the immune system is weakened, as in immunocompromised hosts (Wingard, 1979), Candida species can cause severe skin-, mucosal- and even systemic infections. In this respect, Candida is the fourth most common etiologic agent of sepsis in US hospitals, with 9.0% of all nosocomial blood stream infections being caused by Candida species. Despite current treatment strategies, the crude mortality rate associated with candidiasis remains very high at 39.2% (47.1% in intensive care unit patients and 27.0% in patients from other wards). For some Candida species (C. krusei) the crude mortality rate even reaches 58.7% (Wisplinghoff et al., 2004).
While the current knowledge of the host defense against C. albicans, mostly derived from classical immunological studies, has recently been reviewed (Netea et al., 2015), and the pathogenesis, diagnosis and treatment of invasive candidiasis has been recently described by Kullberg and Arendrup (2015), in the present review we will discuss the novel approaches to understand pathophysiology of Candida infections that make use of the novel omics technologies.
Rationale
Despite current treatment regimens, systems fungal infections are still associated with high morbidity and mortality. In order to develop novel treatment strategies it is crucial to gain more insight into the host response against these infections. In the past, antifungal host defense research was based on hypothesis-driven in vitro or experimental studies. Although these “classical” studies can validate the importance of certain aspects of antifungal defense, they have a slow progress toward providing novel insights, and they are often poor indicators of the relative importance of certain pathways, both of which are crucial when looking for novel therapy targets. In this review we will focus on systems biology research that has been performed on the (human) host defense against fungal infections. Many studies have been performed on the omics of the fungal pathogen itself, but these lie beyond the scope of this review (see other reviews: e.g., Horn et al., 2012).
Genomics
Relatively much research has been performed on the genetic susceptibility to Candida diseases. A recent review summarizes these findings; many of the genetic variants contributing to Candida susceptibility are located in pathogen recognition- or cytokine receptors and downstream signaling molecules (Smeekens et al., 2013c). Recently, Kumar et al. (2014) published the first GWAS on a fungal infection. The authors analyzed 118,989 single-nucleotide polymorphisms (SNPs) across 186 loci known to be associated with immune-mediated diseases in a cohort of 217 systemic candidemia patients and 11,920 controls. There was a significant association between candidemia and SNPs in CD 58 (OR = 4.68), LCE4A (LCE 4A; OR = 4.25) and TAGAP (OR = 2.96) loci. The combination of two or more risk alleles even gave an increased risk for candidemia of 19.4-fold compared to no risk allele (Kumar et al., 2014). CD58, and adhesion molecule on antigen presenting cells, appeared to be involved in the inhibition of Candida germination. TAGAP on the other hand was needed for optimal Candida-induced TNF-α production, as shown by experiments performed in Tagap-/- mice.
Transcriptomics
Barker et al. (2005) published a study on the transcriptional response of THP-1 cells after 3 h of stimulation with live C. albicans SC5314. TNFA, IL8, CD83, MIP1A, and MIP1B were among the genes up-regulated for at least twofold, whereas CCR2 and NCF2 were down-regulated. A novel finding was the up-regulation of RGS1, RGS2, RGS16, down syndrome critical region 1, CXCL2, early growth response 3, FLT4, and TNF-α-induced protein 6, as these genes were not previously known to be responsive to C. albicans (Sobel, 1988; Barker et al., 2005). In 2008, the authors published a similar study on vascular endothelial cells, which responded quite differently on Candida stimulation; genes in chemotaxis, angiogenesis and inhibition of apoptosis were mostly up-regulated in endothelial cells (Wingard, 1979; Barker et al., 2008). Also in human umbilical vein endothelial cell lines C. albicans induced differential expression of genes involved in apoptosis and cell death (Wisplinghoff et al., 2004; Muller et al., 2007; Li et al., 2011). Apparently the cell type under investigation has great influence on the resulting transcriptional profile after Candida stimulation; In human epithelial cells stimulation with C. albicans causes an up-regulation of genes from the NF-κB, mitogen-activated protein kinase and phosphoinositide 3 kinase/protein kinase B immune signaling pathways (Moyes et al., 2014; Netea et al., 2015), and of chemokines and adhesion molecules (Ikuta et al., 2012; Kullberg and Arendrup, 2015). On the other hand, in human granulocytes (neutrophils) C. albicans induces transcription of genes involved in cell-cell signaling, cell signal transduction and cell growth (Fradin et al., 2007; Horn et al., 2012).
In contrast, some stimuli induce such a strong effect that this can be observed in multiple species and cell types. In the murine macrophage cell line J774.2, C. parapsilosis induces transcription of genes from stress-, inflammation-, chemokine-, and cytokine pathways. The highest differential expression was seen with the TNFRSF9 gene, which was 39-fold increased after 8 h of stimulation. Also in peritoneal macrophages from Bagg albino/c mice and in human PBMC-derived macrophages TNFRSF9 was strongly up-regulated after C. parapsilosis stimulation (Kumar et al., 2014; Németh et al., 2014).
Transcriptomic studies on the pathogen side can also throw a light upon the reaction of the host immune system to the fungus. In order to do this, Fradin et al. (2005) exposed C. albicans to different blood fractions. C. albicans thrives in the absence of neutrophils. In contrast, in the presence of neutrophils C. albicans increases responses to overcome nitrogen- and carbohydrate starvation and genes involved in the oxidative stress response are upregulated. These results suggest that neutrophils play an essential role in the defense against C. albicans (Fradin et al., 2005).
Transcriptomics cannot only be used to learn new things about the host immune response, it might also be used as a diagnostic tool; in a murine infection model candidemia could accurately be classified and distinguished from Staphylococcus aureus infection with a sensitivity and specificity of 98 and 96%, respectively. Furthermore, by looking at the transcription profile, also infection progression could be accurately predicted; day one after infection there is increased expression of CXCL2, CXCL13, and IL10R; day two after infection there is increased expression of genes in the TNF-NFκB-B-cell lymphoma 2 pathway; whereas at day three and four, there is down-regulation of apoptotic genes but up-regulation of genes in Toll-like receptor pathways, inflammatory cytokines, of genes involved in NFκB-dependent and mitogen-activated protein kinase-dependent signaling cascades, and of genes involved in T cell activation (Zaas et al., 2010). Also in human blood, the transcriptional profile after bacterial stimulation (S. aureus and Escherichia coli) could be distinguished from that after fungal stimulation (C. albicans and Aspergillus fumigatus) using random forest classification (Dix et al., 2015). This information might be useful in sepsis, where early knowledge on the nature of the causative agent is crucial for selecting the appropriate anti-microbial therapy.
Another transcriptome study indicating the possibility to use transcriptomics as discovery tool was published in 2013. Candida infection could be discriminated from bacterial infection based on a 101-transcript feature set from in vitro experiments with human PBMC. Interestingly, in this set of 101 genes, genes from the type I interferon pathway were significantly overrepresented. This was unexpected as type I interferons had previously been associated with viral and bacterial infections. However, these findings were validated on a functional level in humans (Smeekens et al., 2013b), and also in independent mice studies (Majer et al., 2012; del Fresno et al., 2013). In a separate transcriptome experiment in humans, IFIH1, also part of the type I interferon signaling pathway, was implicated in the host defense against C. albicans hyphae. This finding was also validated on a functional level; patients with CMC express lower levels of IFIH1, there is a strong correlation between genetic variation in IFIH1 and candidemia, and cells from IFIH1 knock out mice and PBMC with different IFIH1 genotypes have altered cytokine production upon C. albicans stimulation (Jaeger et al., 2015).
Tierney et al. (2012) studied the transcriptional profile of C. albicans and mouse dendritic cells simultaneously. Mouse Ptx3 can bind to the C. albicans cell wall, and thereby regulates the transcription of fungal Hap3 target genes, this again alters the immune response to C. albicans, through altered expression of mouse Mta2 target genes like IL-2 and IL-4 (Tierney et al., 2012). Schulze et al. (2015) described a network inference tool (NetGenerator), which constructs gene regulatory networks from time series gene expression data. They re-analyzed the Tierney dataset, taking into account variances in replicated measurement data. Again, the inter-species interactions between Ptx3, Hap3, and Mta2 were considered robust (Schulze et al., 2015). These studies emphasize the need to take into account the fact that there is an interaction between the host and pathogen.
Proteomics
Although studies using transcriptomics are a very insightful tool, they are not able to mirror fully the landscape of active molecules inside an organism; therefore systemic assessment of proteins is an important step for understanding fungal-immune interaction. Since the introduction of proteomics, this can be done in a systematic manner. When the murine macrophage RAW 264.7 cell line was exposed to live C. albicans SC5314, the regulation of several proteins was affected; annexin I, LyGDI, Hspa5, tropomyosin 5, and L-plastin were increased while eukaryotic translation initiation factor 3 subunit 5, Hsp 60, Hspa9a, glucose-regulated protein 58, and Hspa8a were decreased. This indicates that several processes are affected including cytoskeletal organization, oxidative responses and protein biosynthesis and refolding (Martínez-Solano et al., 2006). In contrast, when murine macrophages are stimulated with heat-killed C. albicans the overall response is more anti-inflammatory, although there are some similarities with live C. albicans stimulation as well (Martinez-Solano et al., 2009).
Upon separately analyzing cytosol, organelle/membrane and nucleus enriched fractions from C. albicans-stimulated RAW 264.7 macrophages, 17 new differentially expressed proteins were identified. These proteins are involved in pro-inflammatory and oxidative responses, immune response, unfolded protein response and apoptosis (Reales-Calderón et al., 2012). Also the amount of the membrane receptor Galactin-3, a lectin receptor that recognizes b-mannans (Jouault et al., 2006), was increased upon C. albicans stimulation, especially in phagocytic cups (Reales-Calderón et al., 2012). This receptor had previously been implicated in the pathophysiology of infection by a transcriptome study in mouse macrophages infected with C. albicans, where a 3.4-fold up-regulation of galectin-3 was observed (Shin et al., 2006).
The first proteomics study in human cells stimulated with Candida was published in 2014. The differences in proteomes between human M1 and M2 macrophages were studied upon stimulation with C. albicans. The biggest differences were found in cytoskeletal rearrangement and metabolic routes; fructose-1,6-biphosphatase 1, a glycogenesis enzyme, is up-regulated in M1 macrophages. Interestingly, stimulation with C. albicans has been suggested to induce a polarization switch from M1 to M2, although one could argue that there is no full correlation between cellular markers and the function of the cell. This switch could be interpreted either as an attempt of the host to reduce damage caused by inflammation, or it could be a Candida pathogenicity mechanism by decreasing the immune response (Reales-Calderón et al., 2014).
Proteomics can also be applied to the in vivo situation; in human cervical-vaginal fluids almost half of the proteins are plasma components. The presence of Alb, transferrin, immunoglobulins, apolipoproteins, alpha-1-acid glycoprotein 1 and calgranulins is positively correlated with the amount of polymorphonuclear leukocytes, but there is no difference in the protein maps of asymptomatic women and women experiencing vulvovaginal candidiasis (Tang et al., 2007). In contrast, in non-neutropenic patients, invasive candidiasis could be predicted by measuring their serum antibody signature against the C. albicans proteins Hsp90 and Eno1 prospectively (Pitarch et al., 2014).
Proteomics can be used to investigate specific proteins. Dectin-1 is an important C-type lectin receptor in the recognition of C. albicans, which recognizes β-1,3- and β-1,6-glucans. In order to investigate early Dectin-1 signaling, the interactome of the intracellular receptor tail was explored using a pull-down proteomics approach in mouse CD11c+ cells. The SHIP-1 colocalizes with Dectin-1 during zymosan phagocytosis, and relocates to C. albicans-containing phagosomes. Further experiments with SHIP-1 deficient granulocyte-macrophage colony-stimulating factor-derived bone marrow cells demonstrated an important role for SHIP-1 in the regulation of reactive oxygen species production (Blanco-Menendez et al., 2015).
Metabolomics
Metabolome studies in individuals with Candida infections are scarce. There is one study in which the urine metabolic profile was compared between a preterm neonate with C. parapsilosis infection and 13 preterm controls. N-glycine, D-serine, L-threonine, D-glucose, and maltose levels are higher in urine during C. parapsilosis infection, whereas citric acid, hexadecanoic acid and octadecanoic acid are decreased. Interestingly, therapy efficacy can be evaluated based on the metabolic profile (Dessì et al., 2014).
Microbiome
Several studies suggest that the microbiome composition has an important impact on health status (Pflughoeft and Versalovic, 2012). The microbiome composition can be influenced by diet. In a mice study, dietary coconut oil reduced C. albicans colonization of the gastro-intestinal tract (Gunsalus et al., 2015). Candida colonization is also influenced by quantitative and qualitative aspects of the microbiome. Kennedy and Volz (1985) described that Lactobacilli inhibit fungal adhesion and growth by producing H2O2 and bacteriocin-like compounds. Also short-chain fatty acids from Lactobacilli can inhibit fungal growth (Noverr and Huffnagle, 2004). Furthermore, Pseudomonas aeruginosa and Enterococcus faecalis can inhibit C. albicans hyphae formation (Hogan et al., 2004). Also in a Caenorhabditis elegans model, E. faecalis inhibits C. albicans hyphal morphogenesis (Cruz et al., 2013). On the other hand, C. albicans can coaggregate with Streptococci, which may facilitate the colonization of oral surfaces by the yeast (Jenkinson et al., 1990).
Not only can the microbiome composition influence the extent of Candida colonization, it can also influence the immune response against Candida species (Kolwijck and van de Veerdonk, 2014; Oever and Netea, 2014; Romani et al., 2014). Mice treated with the short-chain fatty acid propionate, which also increases after the consumption of dietary fermentable fibers, have enhanced generation of macrophage and dendritic cell precursors, more dendritic cells in the lungs and reduced Th2 effector function. This demonstrates that changes in diet and metabolism can alter the immunological environment in the lungs (Trompette et al., 2014).
Also in humans the microbiome composition has been demonstrated to influence the immune response against C. albicans; in patients with hyper IgE syndrome and CMC, there are reduced numbers of regular skin microbiome members (e.g., Corynebacteria), but more Gram-negative bacteria (e.g., Acinetobacter), of which the latter in in vitro stimulation experiments with PBMC suppress the S. aureus and C. albicans-induced cytokine production (Smeekens et al., 2013a). The vaginal microbiome of patients with vulvovaginal candidiasis is highly variable, and could not be described by any single profile (Liu et al., 2013).
How the microbiome influences the immune system is not completely clear yet. One mechanism proposed by Zelante et al. (2013) is that lactobacilli in the gut can use tryptophan as their energy source and produce indole-3-aldehyde in the process. Indole-3-aldehyde stimulates the aryl hydrocarbon receptor, which induces IL-22 production in NKprotein46+NK1.1low cells, that are in turn protective against Candida colonization at mucosal surfaces (Zelante et al., 2013).
Mycobiome
Not only antifungal immunity is affected by microbiome composition; also fungal colonization can be influenced by the host microbiome. The fungal microbiome, also known as mycobiome, is a relatively novel research area. In terms of abundance fungi are relatively rare on human skin (Findley et al., 2013). However, when the normal balance is disturbed, e.g., in immunocompromised hosts, resident fungi can expand (Huffnagle and Noverr, 2013). Indeed, antibiotic treatment in mice causes an altered gut microbiome that coincides with outgrowth of commensal Candida species in the gut (Kim et al., 2014). Also genetic factors contribute to mycobiome composition; in dectin-1 deficient mice there is an altered gut mycobiome, which subsequently increases the susceptibility to experimental colitis (Iliev et al., 2012).
Oral microbiome analysis revealed that C. albicans was the most common fungal microorganism in healthy controls and HIV-infected participants. Candida colonization is negatively correlated with Pichia abundance. Pichia conditioned medium inhibits Candida growth and biofilms. Interestingly, in a murine oral candidiasis model Pichia conditioned medium lowers infection score, fungal burden, and tongue epithelial damage. This study demonstrates that findings from mycobiome data can lead to novel interesting antifungals (Zelante et al., 2014).
Computational Systems Biology
Technical advances in the last years have increased the availability of ‘omics’ data, thereby augmenting the demands for mathematical modeling. As mentioned before, it is important to consider there is an ongoing interaction between cells, proteins and metabolites of pathogens and the host, which influences the outcome of infection. Dühring et al. (2015) recently gave an overview of host defense strategies and the corresponding fungal evasion mechanisms, and the computational systems biology approaches to investigate these interactions. Durmuş et al. (2015) also reviewed the available literature on the computational analysis of PHI networks, and in addition the use of web-based databases and text-mining tools in order to optimize use of currently existing data. Last year, PHI networks for C. albicans and A. fumigatus were constructed, based on protein orthology and gene function. These networks predicted Eno1, phospholipase B, Hsp70, pyruvate kinase (CDC19) as Candida virulence factors, which interact with the host factors CD4, Alb and amyloid beta (A4) precursor protein, Toll-like receptor 2, and epidermal growth factor receptor, respectively. These networks can serve as a framework in the analysis of host-pathogen transcriptome and proteome data (Remmele et al., 2015). It is also possible to incorporate the migration of interacting immune and Candida cells in three-dimensional space in a PHI network (Lehnert et al., 2015). Building a more detailed and accurate PHI model may aid timely stratification of sepsis patients based on their inflammatory status in the future.
Discussion
Several studies have demonstrated that Candida infection can be discriminated from bacterial infection based on the induced transcriptome profile (Zaas et al., 2010; Smeekens et al., 2013b), suggesting that in the future transcriptomics might be used in diagnostics. However, these results should be replicated first in independent studies. This will be challenging because it is difficult to recruit large cohorts of candidemia patients. Furthermore, transcriptome studies are fairly costly and time-consuming, while rapid diagnostics are desirable. Moreover, analysis of transcriptome data is not straightforward and should be standardized between the studies (Jafari and Azuaje, 2006). Still, functional genomics studies are very useful, because they can lead to the discovery of novel therapy options. Microbiome studies have indicated several (products of) microorganisms that can inhibit Candida growth, like Pichia (Zelante et al., 2014), Lactobacilli (Kennedy and Volz, 1985; Noverr and Huffnagle, 2004; Zelante et al., 2013), P. aeruginosa and E. faecalis (Hogan et al., 2004). More research is necessary to utilize this knowledge in the development of novel treatment options.
There are some things to take into consideration when conducting or analyzing functional genomics studies. First of all, many studies do not demonstrate sample size calculations, and cohorts are usually small, introducing the possibility of false-positive findings and over-fitting of data to the specific cohort under investigation. Therefore, validation is an import issue. Many findings have not been validated in independent cohorts, making it difficult to assess their value. Of course, for in vivo studies it is difficult to gather large cohorts of patients, as invasive Candida infections are relatively scarce, especially from an individual medical center point of view. In the urine metabolome study there was one premature baby (32 weeks gestational age) with a fungal chest infection, who was compared to 13 premature healthy babies (mean gestational age 34.6). With only one patient it is difficult to discern the true effect on the metabolome of fungal infection from other (external) factors like in this case respiratory distress and gestational age (Dessì et al., 2014).
Next to adequate samples size and validation in independent cohorts it is also important to keep in mind the functional validation of novel findings. Employing systems biology will always lead to a top list of genes, proteins, metabolites, or microorganisms associated with a certain phenotype like Candida infection. However, this does not mean that these associations are also relevant. Furthermore, it is difficult to interpret novel, unexpected findings, without knowing their functional consequence(s). Therefore results need to be validated on a functional level. For example, Zelante et al. (2014) demonstrated that Candida colonization was negatively correlated with Pichia abundance in HIV patients, which is a purely statistical finding. They strengthened this finding by demonstrating that Pichia conditioned medium inhibits Candida growth and biofilm formation. More importantly, Pichia conditioned medium ameliorated a murine oral candidiasis model (Zelante et al., 2014). Furthermore, Kumar et al. (2014) demonstrated in a GWAS study that SNPs in CD58, LCE4A, and TAGAP were significantly associated with candidemia. Two of the genes were validated on a functional level; in siRNA experiments CD58 appeared to be involved in the inhibition of Candida germination. Tagap-/- mice had reduced Candida-induced TNF-α production (Kumar et al., 2014).
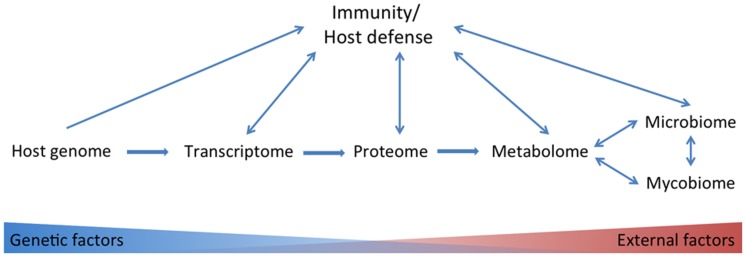
There is no golden standard in systems biology approaches. The correlation between different systems biology approaches is not 100%; in mice it has been reported that for only about half of the genes tested there was a correlation between transcripts and proteins, with an average correlation of 0.27 (Ghazalpour et al., 2011). Compared to the proteome the metabolome is even less subject to genetic influences but more to environmental factors (Figure 1). In a study with healthy male volunteers, up to 12% of the observed variance in metabolite concentrations could be explained by genetic variation in common SNPs (Gieger et al., 2008). Although the transcriptome, proteome and metabolome are less subject to genetic influences, they are often more closely related to phenotypic traits compared to genome data (Homuth et al., 2012). In contrast, clinical traits, like adiposity, were more strongly correlated to transcript levels than to protein levels in a study with mice (Ghazalpour et al., 2011).
FIGURE 1.
Overview of omics tools used in studies on the anti-Candida host defense. Host genetic-, transcriptome-, proteome-, metabolome-, microbiome-, and mycobiome data have been linked to immunity/host defense. Host genetic factors play to a lesser extent a role in consecutively transcriptome, proteome, metabolome, and microbiome and mycobiome data.
Also the models and experimental setups that are chosen have great influence on systems biology approaches. As discussed above, C. albicans induces the transcription of different principal pathways in human monocyte-like cells (Barker et al., 2005), vascular endothelial cells (Muller et al., 2007; Barker et al., 2008; Li et al., 2011), human epithelial cells (Ikuta et al., 2012; Moyes et al., 2014) and human granulocytes (neutrophils; Fradin et al., 2007). The fungal species is of importance as well, and TNFRSF9 transcription has so far only been shown to be up-regulated after C. parapsilosis stimulation (Németh et al., 2014). Most studies have been performed with C. albicans, while other fungal species, like C. tropicalis, C. glabrata, and C. krusei, are also medically relevant (Kullberg and Arendrup, 2015). These should also be the focus of future studies. Another aspect is fungal strain and morphology; many studies use heat-killed fungi, while these are fundamentally different from live fungi. In PBMC cultures live C. albicans can actively suppress IL-17 production (Cheng et al., 2010), and important cytokine in the anti-Candida host defense (van de Veerdonk et al., 2009). So although using live fungal pathogens more closely resembles an in vivo infection, it is more difficult to delineate host defense responses from fungal influences.
In the past few years, functional genomics studies have led to interesting novel discoveries regarding the anti-Candida host defense (Table 1), making them a valuable addition to classical immunological studies.
Table 1.
Research highlights from functional genomics studies on the host immune response against Candida albicans
| Tool | Model | Main findings | Reference |
|---|---|---|---|
| Genomics | GWAS with human candidemia patients | SNPs in CD58, LCE4A, TAGAP increase risk for candidemia | Kumar et al., 2014 |
| Transcriptomics | In vitro studies with human monocyte cell line or mouse dendritic cells | Differential expression of specific genes: RGS1, RGS2, RGS16, down syndrome critical region 1, CXCL2, early growth response 3, FLT4, TNF-α-induced protein 6, Ptx3, and Mta2 | Barker et al., 2005; Tierney et al., 2012; Schulze et al., 2015 |
| Endothelial cells stimulated with C. albicans | Differential regulation of certain pathways: Type I interferons; chemotaxis, angiogenesis, apoptosis, cell death, NF-κB- MAPK- and PI3K/Akt signaling; cell–cell signaling, cell signal transduction and cell growth, chemokines and adhesion molecules | Wisplinghoff et al., 2004; Fradin et al., 2007; Muller et al., 2007; Barker et al., 2008; Li et al., 2011; Horn et al., 2012; Ikuta et al., 2012; Smeekens et al., 2013b; Moyes et al., 2014; Kullberg and Arendrup, 2015; Netea et al., 2015 | |
| Murine candidemia model, in vitro studies with human blood | Transcriptomics can be used as a diagnostic tool to discriminate fungal from bacterial infection | Zaas et al., 2010; Dix et al., 2015 | |
| Proteomics | Serum from non-neutropenic patients | Proteomics can be useful in prognosis: the serum antibody signature against the C. albicans proteins Hsp90 and Eno1 is predictive of invasive candidiasis | Pitarch et al., 2014 |
| In vitro studies with human and murine macrophages | Differential regulation of certain processes: cytoskeletal organization, oxidative responses, protein biosynthesis and refolding, pro-inflammatory responses, immune response, unfolded protein response and apoptosis and metabolism | Martínez-Solano et al., 2006; Reales-Calderón et al., 2012, 2014; Blanco-Menendez et al., 2015 | |
| Metabolomics | Urine metabolome in C. parapsilosis neonate | Increased levels of N-glycine, D-serine, L-threonine, D-glucose and maltose levels. Decreased levels of citric acid, hexadecanoic acid and octadecanoic acid | Dessì et al., 2014 |
| Microbiome | In vitro experiments | Microbiome composition can influence Candida colonization | Hogan et al., 2004; Noverr and Huffnagle, 2004 |
| Microbiome studies in patients with skin/mucosal C. albicans infections, mice studies | Microbiome composition influences the host immune response against C. albicans | Zelante et al., 2013; Smeekens et al., 2013a; Trompette et al., 2014 | |
| Mycobiome | Oral microbiome analysis in HIV patients, in vitro studies and a murine oral candidiasis model | Pichia inhibits C. albicans | Zelante et al., 2014 |
| Computational systems biology | Construction of PHI network | The host factors CD4, Alb and amyloid beta (A4) precursor protein, Toll-like receptor 2, epidermal growth factor receptor are predicted to interact with C. albicans virulence factors | Remmele et al., 2015 |
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MN was supported by an ERC Consolidator Grant (#310372). FvdV was supported by the ERA-Net for Research Programs on Rare Diseases “EURO-CMC”.
Abbreviations
- Alb
albumin
- CD
cluster of differentiation
- CMC
chronic mucocutaneous candidiasis
- CXCL
chemokine (C-X-C motif) ligand
- Eno
enolase
- GWAS
genome wide association study
- Hap
hook-associated protein
- Hsp
heat shock protease protein
- IFIH
interferon induced with helicase c domain
- IL
interleukin
- LCE
late cornified envelope
- Mta
metastasis-associated protein
- NF
nuclear factor
- OR
odds ratio
- PHI
pathogen-host interaction
- Ptx
pentraxin
- MIP
macrophage inflammatory protein
- PBMC
peripheral blood mononuclear Cells
- RGS
regulator of G-protein signaling
- SHIP
SH2 domain containing inositol phosphatase
- SNP
single nucleotide polymorphism
- TAGAP
T cell activation RHO-GTPase-activating protein
- TNF
tumor necrosis factor
- TNFRSF
tumor necrosis factor super family
References
- Barker K. S., Liu T., Rogers P. D. (2005). Coculture of THP-1 human mononuclear cells with Candida albicans results in pronounced changes in host gene expression. J. Infect. Dis. 192 901–912. 10.1086/432487 [DOI] [PubMed] [Google Scholar]
- Barker K. S., Park H., Phan Q. T., Xu L., Homayouni R., Rogers P. D., et al. (2008). Transcriptome profile of the vascular endothelial cell response to Candida albicans. J. Infect. Dis. 198 193–202. 10.1086/589516 [DOI] [PubMed] [Google Scholar]
- Blanco-Menendez N., del Fresno C., Fernandes S., Calvo E., Conde-Garrosa R., Kerr W. G., et al. (2015). SHIP-1 couples to the dectin-1 hemITAM and selectively modulates reactive oxygen species production in dendritic cells in response to Candida albicans. J. Immunol. 195 4466–4478. 10.4049/jimmunol.1402874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.-C., van de Veerdonk F., Smeekens S., Joosten L. A. B., van der Meer J. W. M., Kullberg B. J., et al. (2010). Candida albicans dampens host defense by downregulating IL-17 production. J. Immunol. 185 2450–2457. 10.4049/jimmunol.1000756 [DOI] [PubMed] [Google Scholar]
- Cruz M. R., Graham C. E., Gagliano B. C., Lorenz M. C., Garsin D. A. (2013). Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 81 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Fresno C., Soulat D., Roth S., Blazek K., Udalova I., Sancho D., et al. (2013). Interferon-β production via dectin-1-Syk-IRF5 signaling in dendritic cells is crucial for immunity to C. albicans. Immunity 38 1176–1186. 10.1016/j.immuni.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Dessì A., Liori B., Caboni P., Corsello G., Giuffrè M., Noto A., et al. (2014). Monitoring neonatal fungal infection with metabolomics. J. Matern. Fetal. Neonatal. Med. 27 34–38. 10.3109/14767058.2014.954787 [DOI] [PubMed] [Google Scholar]
- Dix A., Hunniger K., Weber M., Guthke R., Kurzai O., Linde J. (2015). Biomarker-based classification of bacterial and fungal whole-blood infections in a genome-wide expression study. Front. Microbiol. 6:171 10.3389/fmicb.2015.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühring S., Germerodt S., Skerka C., Zipfel P. F., Dandekar T., Schuster S. (2015). Host-pathogen interactions between the human innate immune system and Candida albicans—understanding and modeling defense and evasion strategies. Front. Microbiol. 6:625 10.3389/fmicb.2015.00625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmuş S., Cakir T., Ozgur A., Guthke R. (2015). A review on computational systems biology of pathogen–host interactions. Front. Microbiol. 6:235 10.3389/fmicb.2015.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J. A., et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498 367–370. 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., et al. (2005). Granulocytes govern the transcriptional response, morphology, and proliferation of Candida albicans in human blood. Mol. Microbiol. 56 397–415. 10.1111/j.1365-2958.2005.04557.x [DOI] [PubMed] [Google Scholar]
- Fradin C., Mavor A. L., Weindl G., Schaller M., Hanke K., Kaufmann S. H. E., et al. (2007). The early transcriptional response of human granulocytes to infection with Candida albicans is not essential for killing but reflects cellular communications. Infect. Immun. 75 1493–1501. 10.1128/IAI.01651-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A., Bennett B., Petyuk V. A., Orozco L., Hagopian R., Mungrue I. N., et al. (2011). Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 7:e1001393 10.1371/journal.pgen.1001393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieger C., Geistlinger L., Altmaier E., de Angelis M. H., Kronenberg F., Meitinger T., et al. (2008). Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4:e1000282 10.1371/journal.pgen.1000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouba N., Drancourt M. (2015). Digestive tract mycobiota: a source of infection. Méd. Malad. Infect. 45 9–16. 10.1016/j.medmal.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Gunsalus K., Tornberg-Belanger S. N., Matthan N. R., Lichtenstein A. H., Kumamoto C. A. (2015). Manipulation of host diet to reduce gastrointestinal colonization by the opportunistic pathogen Candida albicans. mSphere 1:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. A., Vik Å, Kolter R. (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54 1212–1223. 10.1111/j.1365-2958.2004.04349.x [DOI] [PubMed] [Google Scholar]
- Homuth G., Teumer A., Völker U., Nauck M. (2012). A description of large-scale metabolomics studies: increasing value by combining metabolomics with genome-wide SNP genotyping and transcriptional profiling. J. Endocrinol. 215 17–28. 10.1530/JOE-12-0144 [DOI] [PubMed] [Google Scholar]
- Horn F., Heinekamp T., Kniemeyer O., Pollmacher J., Valiante V., Brakhage A. A. (2012). Systems biology of fungal infection. Front. Microbiol. 3:108 10.3389/fmicb.2012.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G. B., Noverr M. C. (2013). The emerging world of the fungal microbiome. Trends Microbiol. 21 334–341. 10.1016/j.tim.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T., Bhawal U. K., Tsushima K., Aoki A., Kuboyama N., Abiko Y. (2012). Identification by DNA microarray of genes involved in Candida albicans-treated gingival epithelial cells. J. Oral Pathol. Med. 41 769–778. 10.1111/j.1600-0714.2012.01149.x [DOI] [PubMed] [Google Scholar]
- Iliev I. D., Funari V. A., Taylor K. D., Nguyen Q., Reyes C. N., Strom S. P., et al. (2012). Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science 336 1314–1317. 10.1126/science.1221789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger M., van der Lee R., Cheng S. C., Johnson M. D., Kumar V., Ng A., et al. (2015). The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. Eur. J. Clin. Microbiol. Infect. Dis. 34 963–974. 10.1007/s10096-014-2309-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari P., Azuaje F. (2006). An assessment of recently published gene expression data analyses: reporting experimental design and statistical factors. BMC Med. Informat. Decis. Mark. 6:27 10.1186/1472-6947-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H. F., Lala H. C., Shepherd M. G. (1990). Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouault T., Abed-El Behi M., Martínez-Esparza M., Breuilh L., Trinel P.-A., Chamaillard M., et al. (2006). Specific recognition of Candida albicans by macrophages requires galectin-3 to Discriminate Saccharomyces cerevisiae and needs association with tlr2 for signaling. J. Immunol. 177 4679–4687. 10.4049/jimmunol.177.7.4679 [DOI] [PubMed] [Google Scholar]
- Kennedy M. J., Volz P. A. (1985). Ecology of Candida albicans gut colonization: inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect. Immun. 49:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-G., Udayanga K. G. S., Totsuka N., Weinberg J. B., Nuñez G., Shibuya A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 15 95–102. 10.1016/j.chom.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolwijck E., van de Veerdonk F. L. (2014). The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur. J. Immunol. 44 3156–3165. 10.1002/eji.201344404 [DOI] [PubMed] [Google Scholar]
- Kullberg B. J., Arendrup M. C. (2015). Invasive Candidiasis. N. Engl. J. Med. 373 1445–1456. 10.1056/NEJMra1315399 [DOI] [PubMed] [Google Scholar]
- Kumar V., Cheng S.-C., Johnson M. D., Smeekens S. P., Wojtowicz A., Giamarellos-Bourboulis E., et al. (2014). Immunochip SNP array identifies novel genetic variants conferring susceptibility to candidaemia. Nat. Commun. 5:4675 10.1038/ncomms5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert T., Timme S., Pollmacher J., Hunniger K., Kurzai O., Figge M. T. (2015). Bottom-up modeling approach for the quantitative estimation of parameters in pathogen-host interactions. Front. Microbiol. 6:608 10.3389/fmicb.2015.00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. S. Y., Rosli R., Seow H. F., Chong P. P. (2011). Transcriptome profiling of endothelial cells during infections with high and low densities of C. albicans cells. Int. J. Med. Microbiol. 301 536–546. 10.1016/j.ijmm.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Liu M.-B., Xu S.-R., He Y., Deng G.-H., Sheng H.-F., Huang X.-M., et al. (2013). Diverse vaginal microbiomes in reproductive-age women with vulvovaginal candidiasis. PLoS ONE 8:e79812 10.1371/journal.pone.0079812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer O., Bourgeois C., Zwolanek F., Lassnig C., Kerjaschki D., Mack M., et al. (2012). Type i interferons promote fatal immunopathology by regulating inflammatory monocytes and neutrophils during Candida Infections. PLoS Pathog 8:e1002811 10.1371/journal.ppat.1002811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Solano L., Nombela C., Molero G., Gil C. (2006). Differential protein expression of murine macrophages upon interaction with Candida albicans. Proteomics 6 S133–S144. 10.1002/pmic.200500581 [DOI] [PubMed] [Google Scholar]
- Martinez-Solano L., Reales-Caldero J. A., Nombela C., Molero G., Gil C. (2009). Proteomics of RAW 264.7 macrophages upon interaction with heat-inactivated Candida albicans cells unravel an anti-inflammatory response. Proteomics 9 2995–3010. 10.1002/pmic.200800016 [DOI] [PubMed] [Google Scholar]
- Moyes D. L., Shen C., Murciano C., Runglall M., Richardson J. P., Arno M., et al. (2014). Protection against epithelial damage during Candida albicans infection is mediated by pi3k/akt and mammalian target of rapamycin signaling. J. Infect. Dis. 209 1816–1826. 10.1093/infdis/jit824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller V., Viemann D., Schmidt M., Endres N., Ludwig S., Leverkus M., et al. (2007). Candida albicans triggers activation of distinct signaling pathways to establish a proinflammatory gene expression program in primary human endothelial cells. J. Immunol. 179 8435–8445. 10.4049/jimmunol.179.12.8435 [DOI] [PubMed] [Google Scholar]
- Németh T., Tóth A., Hamari Z., Falus A., Éder K., Vágvölgyi C., et al. (2014). Transcriptome profile of the murine macrophage cell response to Candida parapsilosis. Fungal Genet. Biol. 65 48–56. 10.1016/j.fgb.2014.01.006 [DOI] [PubMed] [Google Scholar]
- Netea M. G., Joosten L. A. B., van der Meer J. W. M., Kullberg B. J., van de Veerdonk F. L. (2015). Immune defence against Candida fungal infections. Nat. Rev. Immunol. 15 630–642. 10.1038/nri3897 [DOI] [PubMed] [Google Scholar]
- Noverr M. C., Huffnagle G. B. (2004). Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72 6206–6210. 10.1128/IAI.72.11.6206-6210.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oever J. T., Netea M. G. (2014). The bacteriome-mycobiome interaction and antifungal host defense. Eur. J. Immunol. 44 3182–3191. 10.1002/eji.201344405 [DOI] [PubMed] [Google Scholar]
- Pflughoeft K. J., Versalovic J. (2012). Human microbiome in health and disease. Annu. Rev. Pathol. Mech. Dis. 7 99–122. 10.1146/annurev-pathol-011811-132421 [DOI] [PubMed] [Google Scholar]
- Pitarch A., Gil C., Nombela C. (2014). Serum antibody signature directed against Candida albicans hsp90 and enolase detects invasive candidiasis in non-neutropenic patients. J. Proteome Res. 13 5165–5184. 10.1021/pr500681x [DOI] [PubMed] [Google Scholar]
- Reales-Calderón J. A., Aguilera-Montilla N., Corbí Á. L., Molero G., Gil C. (2014). Proteomic characterization of human proinflammatory M1 and anti-inflammatory M2 macrophages and their response to Candida albicans. Proteomics 14 1503–1518. 10.1002/pmic.201300508 [DOI] [PubMed] [Google Scholar]
- Reales-Calderón J. A., Martínez-Solano L., Martinez-Gomariz M., Nombela C., Moleroa G., Gil C. (2012). Sub-proteomic study on macrophage response to Candida albicans unravels new proteins involved in the host defense against the fungus. J. Proteomics 75 4734–4746. 10.1016/j.jprot.2012.01.037 [DOI] [PubMed] [Google Scholar]
- Remmele C. W., Luther C. H., Balkenhol J., Dandekar T., Müller T., Dittrich M. T. (2015). Integrated inference and evaluation of host–fungi interaction networks. Front. Microbiol. 6:764 10.3389/fmicb.2015.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L., Zelante T., Palmieri M., Napolioni V., Picciolini M., Velardi A., et al. (2014). The cross-talk between opportunistic fungi and the mammalian host via microbiota’s metabolism. Semin. Immunopathol. 37 163–171. 10.1007/s00281-014-0464-2 [DOI] [PubMed] [Google Scholar]
- Schulze S., Henkel S. G., Driesch D., Guthke R., Linde J. R. (2015). Computational prediction of molecular pathogen-host interactions based on dual transcriptome data. Front. Microbiol. 6:65 10.3389/fmicb.2015.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.-K., Lee H.-J., Lee J. S., Paik Y.-K. (2006). Proteomic analysis of mammalian basic proteins by liquid-based two-dimensional column chromatography. Proteomics 6 1143–1150. 10.1002/pmic.200500433 [DOI] [PubMed] [Google Scholar]
- Smeekens S. P., Huttenhower C., Riza A., van de Veerdonk F. L., Zeeuwen P. L. J. M., Schalkwijk J., et al. (2013a). Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J. Innate Immun. 6 253–262. 10.1159/000351912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. P., Ng A., Kumar V., Johnson M. D., Plantinga T. S., van Diemen C., et al. (2013b). Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat. Commun. 4:1342 10.1038/ncomms2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. P., van de Veerdonk F. L., Kullberg B. J., Netea M. G. (2013c). Genetic susceptibility to Candida infections. EMBO Mol. Med. 5 805–813. 10.1002/emmm.201201678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel J. D. (1988). Pathogenesis and epidemiology of vulvovaginal candidiasis. Ann. N. Y. Acad. Sci. 544 547–557. 10.1111/j.1749-6632.1988.tb40450.x [DOI] [PubMed] [Google Scholar]
- Tang L.-J., De Seta F., Odreman F., Venge P., Piva C., Guaschino S., et al. (2007). Proteomic analysis of human cervical-vaginal fluids. J. Proteome Res. 6 2874–2883. 10.1021/pr0700899 [DOI] [PubMed] [Google Scholar]
- Tierney L., Linde J., Muller S., Brunke S., Molina J. C., Hube B., et al. (2012). An interspecies regulatory network inferred from simultaneous RNA-seq of Candida albicans invading innate immune cells. Front. Microbiol. 3:85 10.3389/fmicb.2012.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E. S., Yadava K., Sichelstiel A. K., Sprenger N., Ngom-Bru C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20 159–166. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- van de Veerdonk F. L., Marijnissen R. J., Kullberg B. J., Koenen H. J. P. M., Cheng S.-C., Joosten I., et al. (2009). The macrophage mannose receptor induces IL-17 in response to Candida albicans. Cell Host Microbe 5 329–340. 10.1016/j.chom.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Wingard J. R. (1979). Candida tropicalis: a major pathogen in immunocompromised patients. Ann. Intern. Med. 91 539–543. 10.7326/0003-4819-91-4-539 [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- Zaas A. K., Aziz H., Lucas J., Perfect J. R., Ginsburg G. S. (2010). Blood gene expression signatures predict invasive candidiasis. Sci. Transl. Med. 2:ra17–ra21. 10.1126/scitranslmed.3000715 [DOI] [PubMed] [Google Scholar]
- Zelante T., Iannitti R. G., Cunha C., De Luca A., Giovannini G., Pieraccini G., et al. (2013). Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39 372–385. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Zelante T., Mukherjee P. K., Iannitti R. G., Chandra J., Cunha C., Retuerto M., et al. (2014). Oral mycobiome analysis of HIV-infected patients: identification of pichia as an antagonist of opportunistic fungi. PLoS Pathog. 10:e1003996 10.1371/journal.ppat.1003996 [DOI] [PMC free article] [PubMed] [Google Scholar]