Abstract
In this study, mitoxantrone and its halogenated derivatives have been designed by density functional theory (DFT) to explore their structural and thermodynamical properties. The performance of these drugs was also evaluated to inhibit DNA topoisomerase type IIα (TOP2A) by molecular docking calculation. Noncovalent interactions play significant role in improving the performance of halogenated drugs. The combined quantum and molecular mechanics calculations revealed that CF3 containing drug shows better preference in inhibiting the TOP2A compared to other modified drugs.
1. Introduction
Cancer is one of the most devastating diseases and causes of million deaths, and it is predicted to continue to be catastrophic in the coming years [1]. Surgery and radiation therapies are limited to treating cancers that are confined to highly precise areas. Chemotherapy is advantageous over such treatments because of its ability to treat widespread or metastatic cancers. Chemotherapy, a key way to treat the malignant tumors, uses various chemical agents to destroy cancer cells [2]. The chemotherapy treatment has earned ample attention and a great deal of recent efforts have been concentrating on the design and development of varied anticancer drugs. Mitoxantrone (1,4-dihydroxy-5,8-bis({2-[(2-hydroxyethyl)amino]ethyl}amino)-9,10-dihydroanthracene-9,10-dione) is a potent synthetic anticancer drug which blocks DNA synthesis by inhibiting the function of DNA topoisomerase II [3–5]. The drug was selected amongst a series of anthracenedione derivatives that have structural similarities to the anthracyclines. Due to the absence of amino-sugar moiety in mitoxantrone, it has less cardio-toxicity [6]. In several cardio-toxicity models, mitoxantrone appeared to have less toxicity than doxorubicin [7]. Currently, mitoxantrone has been used for treating different type of cancers including breast cancer, leukemia, lymphoma, and prostate cancer [8]. Moreover, this drug provides a new therapeutic option for patients with worsening relapsing-remitting and secondary progressive multiple sclerosis and hepatocellular carcinoma [9, 10].
The widely used target of existing anticancer drugs including mitoxantrone is DNA topoisomerase type IIα (TOP2A) and the expression of this enzyme has been used as cancer cell marker because of its role in cell proliferation [11–13]. During DNA replication, TOP2A plays a key role and its main functions are chromosome segregation and chromosome condensation [14]. Humans express another isoform of topoisomerase II which is known as DNA topoisomerase II β (TOP2B) [15, 16]. The two isoforms of topoisomerase II are 68% [17] identical and their catalytic portion share ~78% similarity. ATP dependent type II topoisomerases [18–21] operate by a complex mechanism that involves the organized association and dissociation of subunit dimerization elements [22–25]. For this cleavage reaction, one segment of the DNA duplex (defined as “G-segment”) is bound and cleaved by the enzyme whereas a second double stranded DNA (defined as “T-segment”) is transported through the break. The assembly of topoisomerase and DNA is called the cleavage complex in which a pair of symmetrically related tyrosine residues (Tyr) is responsible for G-segment breakage [26, 27]. Eukaryotic TOP2A contains three regions known as the N-gate, DNA-gate, the C-gate, and the catalytic Tyr805, which is responsible for cleavage present in the DNA-gate [28, 29]. The clinically active anticancer agent, mitoxantrone, inhibits topoisomerase II by increasing its levels in TOP2-DNA complexes [30, 31]. In addition, antitopoisomerase agents that bind within the DNA-gate either impede or stabilize the cleavage and relegation events [32].
Nonbonding interactions between the drug and amino acid residues of the receptor play a crucial role in preventing/obstructing the active/enzymatic site(s), which are responsible for causing certain diseases. Various nonbonding interactions have been identified in drug-receptor complex including hydrogen bonding, halogen bonding, cation-π interactions, anion-pi interactions, pi-alkyl interaction, π-π stacking, and T-shape interactions [33–36]. Molecular level interpretation of these nonbonding interactions appeared as a key factor to design superior drug which can effectively inhibit the receptor protein.
In this study, quantum mechanical calculations were carried out to model and explore the structural, thermodynamical, and molecular orbital properties of 10 halogenated mitoxantrone drugs. Moreover, the binding affinity and nonbonding interactions of these drugs with TOP2A are evaluated by molecular docking study.
2. Computational Methods
2.1. Drug Design by Quantum Mechanical Calculations
All electronic calculations were carried out using Gaussian 09 program package [37]. The initial geometry of 3D structure of mitoxantrone (D) was taken from PubChem Open Chemistry Database [38]. The structure of mitoxantrone was fully optimized by density functional theory employing Becke's (B3) [39, 40] exchange functional combining Lee, Yang, and Parr's (LYP) correlation functional [41]. For all modified drug molecules (D1–D10), Cramer and Truhlar's MidiX basis set was employed [42]. MidiX basis set is originally developed from the Huzinaga MidiX basis and applied to H, C-F, S-Cl, Br, and I atoms. The MidiX basis set is comparatively smaller than the popular 6–31G(d,p) and can provide excellent geometries and charge balances with reasonable computational time and accuracy [43].
After optimization, subsequent vibrational frequency calculation was performed in order to confirm that the stationary points correspond to minima on the potential energy surface. Electronic energies, enthalpies, Gibb's free energies, dipole moments, and partial charge analysis were also explored for all optimized-energy geometries. Molecular orbital calculations were performed at the same level of theory. Hardness and softness of all drugs were also determined from the energies of frontier HOMOs and LUMOs. Considering Parr and Pearson interpretation [43–45] of DFT and Koopmans theorem [46] on the correlation of ionization potential (I) and electron affinities (E) with HOMO and LUMO energies (ε), hardness (η) and softness (S) of the drugs were calculated according to the following equation:
| (1) |
2.2. Preparation of Protein
The mitoxantrone and all modified drugs were subjected to molecular docking against human topoisomerase IIα (TOP2A). The crystal structure of TOP2A was collected from the Protein Data Bank (PDB) database (PDB ID: 4FM9; Chain A) [32]. Since the crystal structure has some issues related to improper bond order, side chains geometry, and missing hydrogen atoms, the structure was checked and an energy minimization was performed with the Swiss-Pdb Viewer software packages (version 4.1.0) [47]. Prior to docking, all the heteroatoms and water molecules were removed from the crystal structure using PyMol (version 1.3) software packages [48]. Addition of nonpolar hydrogen atoms is performed by AutoDock Tools (ADT) of MGL software packages (version 1.5.6). Subsequently the fully optimized structures of the halogenated compounds were opened using ADT to add Gasteiger charges and to set TORSDOF followed by the conversion of all rotatable bonds into nonrotatable (rigid). Finally, both the proteins and ligand structures were saved in .pdbqt format as it is the only one supported file format that required by AutoDock Vina software (version 1.1.2, May 11, 2011) for docking analysis [49].
2.3. Binding Site and Docking
The active binding pocket of TOP2A is predicted by CastP [50] having the highest pocket area and volume that are 4390 Å2 and 8674.2 Å3, respectively. The binding pocket and the amino acid residues are presented in Figure S1 (supporting information; see Supplementary Material available online at http://dx.doi.org/10.1155/2016/6817502). The binding site residues predicted by CastP for TOP2A were used for the generation of the grid box.
To dock the mitoxantrone and its halogenated derivatives against TOP2A, the center of the grid box was set at 33.5565, 41.4725, and 15.9145 Å and the box size was set at 25, 25, and 25 Å in x, y, and z directions, respectively. Autodock Vina docking protocol was employed to conduct the docking study. Next, the docked pose of lowest binding free energy conformer with the respective protein was analyzed using PyMOL Molecular Graphics System (version 1.3) [48], Accelrys Discovery Studio 4.1 [51], and LigPlot+ version v1.4.5 [52].
3. Results and Discussions
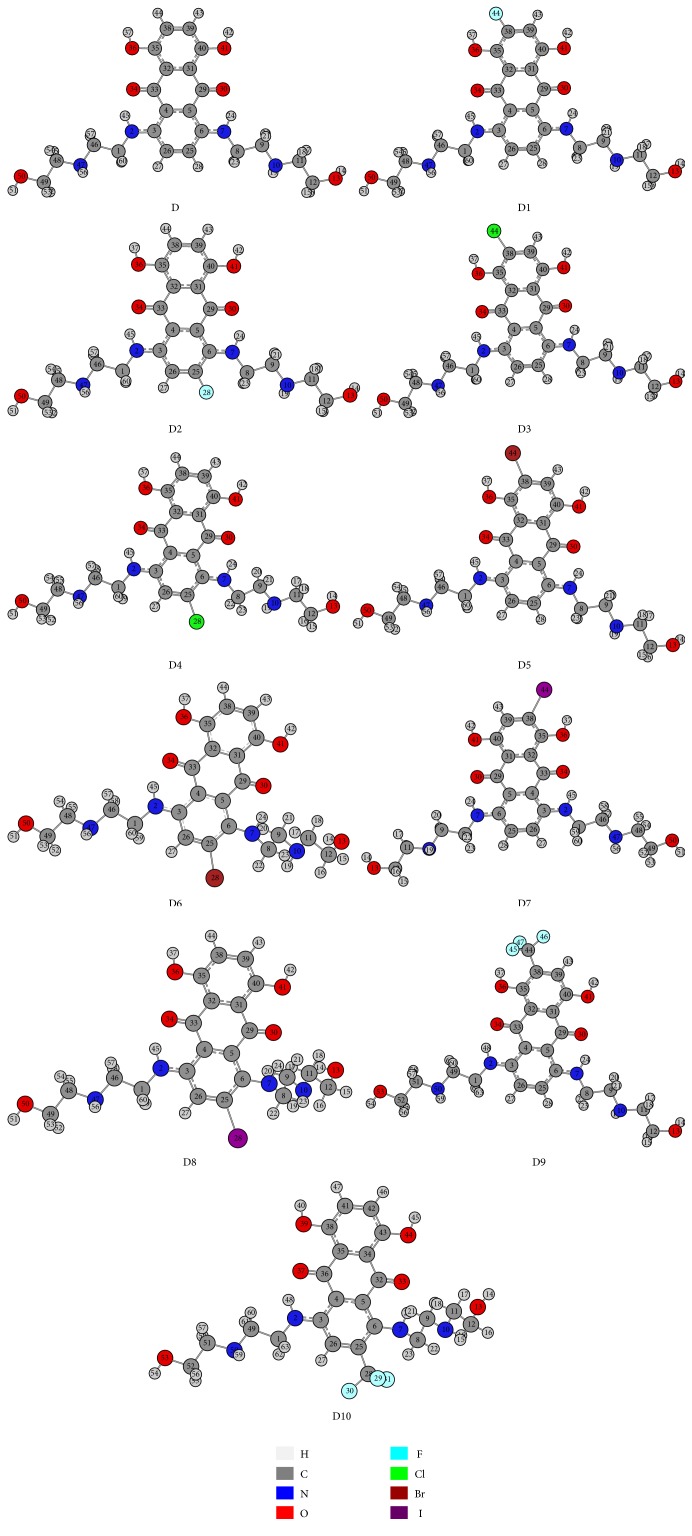
The optimized structures of mitoxantrone (D) and its halogenated derivatives (D1–D10) computed at the B3LYP/MidiX level of theory are presented in Figure 1. Partial charges and direction of the dipole moments of all drugs are illustrated in the supplementary Figure S2. The stoichiometry, electronic energy, enthalpy, Gibbs free energy, and dipole moment of all drugs are reported in Table 1. The HOMO and LUMO energies, HOMO-LUMO gap, hardness, and softness of all drugs are summarized in Table 2. The pictographical presentation of all frontier orbitals is displayed in supplementary Figure S3. The binding affinity and all nonbonding interactions of all drug-receptor complexes are summarized in Table 3 and Figure 2. Aromatic and hydrophobic surface of binding pocket are shown in supplementary information (Figures S4 and S5).
Figure 1.
Optimized structure of mitoxantrone (D) and its halogenated derivatives (D1, D2, D3, D4, D5, D6, D7, D8, D9, and D10) calculated at B3LYP/MidiX level of theory.
Table 1.
The stoichiometry, electronic energy, enthalpy, and Gibbs free energy in Hartree and dipole moment (Debye) of mitoxantrone and its halogenated derivatives.
| Name | Stoichiometry | Electronic energy | Enthalpy | Gibbs free energy | Dipole moment (Debye) |
|---|---|---|---|---|---|
| Mitoxantrone | C22H28N4O6 | −1516.1835 | −1516.1826 | −1516.2838 | 1.5477 |
| D1 | C22H27FN4O6 | −1614.8728 | −1614.8719 | −1614.9746 | 1.3831 |
| D2 | C22H27FN4O6 | −1614.8711 | −1614.8702 | −1614.9731 | 2.2832 |
| D3 | C22H27ClN4O6 | −1973.6278 | −1973.6269 | −1973.7305 | 2.1425 |
| D4 | C22H27ClN4O6 | −1973.6169 | −1973.6159 | −1973.7201 | 5.5153 |
| D5 | C22H27BrN4O6 | −4077.6078 | −4077.6068 | −4077.7124 | 1.3627 |
| D6 | C22H27BrN4O6 | −4077.5965 | −4077.5956 | −4077.6995 | 4.8794 |
| D7 | C22H27IN4O6 | −8405.7319 | −8405.7309 | −8405.8375 | 1.4454 |
| D8 | C22H27IN4O6 | −8405.7210 | −8405.7201 | −8405.8257 | 5.3011 |
| D9 | C23H27F3N4O6 | −1851.3391 | −1851.3382 | −1851.4476 | 1.6814 |
| D10 | C23H27F3N4O6 | −1851.3296 | −1851.3287 | −1851.4370 | 4.5242 |
Table 2.
Energy (atomic unit) of HOMO, LUMO, Gap, hardness, and softness of all drugs.
| Molecules | ε HOMO−1 | ε HOMO | ε LUMO | ε LUMO+1 | Gap | Η (hardness) | S (softness) |
|---|---|---|---|---|---|---|---|
| Mitoxantrone | −0.2027 | −0.1576 | −0.0753 | −0.0092 | 0.0822 | 0.0411 | 24.3161 |
| D1 | −0.2066 | −0.1600 | −0.0774 | −0.0143 | 0.0825 | 0.0412 | 24.2248 |
| D2 | −0.2047 | −0.1608 | −0.0769 | −0.0124 | 0.0838 | 0.0419 | 23.8578 |
| D3 | −0.2117 | −0.1635 | −0.0828 | −0.0179 | 0.0806 | 0.0403 | 24.7892 |
| D4 | −0.2123 | −0.1720 | −0.0872 | −0.0225 | 0.0847 | 0.0423 | 23.5932 |
| D5 | −0.2078 | −0.1612 | −0.0802 | −0.0149 | 0.0810 | 0.0405 | 24.6883 |
| D6 | −0.2138 | −0.1767 | −0.0887 | −0.0259 | 0.0879 | 0.0439 | 22.7298 |
| D7 | −0.2080 | −0.1614 | −0.0806 | −0.0455 | 0.0808 | 0.0404 | 24.7494 |
| D8 | −0.2149 | −0.1791 | −0.0903 | −0.0308 | 0.0888 | 0.0444 | 22.5098 |
| D9 | −0.2109 | −0.1630 | −0.0830 | −0.0217 | 0.0799 | 0.0399 | 25.0250 |
| D10 | −0.2132 | −0.1734 | −0.0887 | −0.0266 | 0.0846 | 0.0423 | 23.6211 |
Table 3.
Binding energy and nonbonding interaction of mitoxantrone and its derivatives.
| Compound | Docking against human topoisomerase II alpha (TOP2A) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Binding energy (kcal mol−1) | H-bond | Halogen bond | Hydrophobic interaction | Electrostatic interaction | |||||
| Amino acid-ligand atom | Distance (Å) | Amino acid-ligand atom | Distance (Å) | Amino acid-ligand atom | Distance (Å) | Amino acid-ligand atom | Distance (Å) | ||
| Mitoxantrone (D) | −9.2 | Lys723 [N-H⋯O] Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ |
2.2693 3.0872 2.9119 3.0135 2.2831 |
Lys723 [alkyl-pi] Lys723 [alkyl-pi] |
4.8912 4.8521 |
||||
|
| |||||||||
| D1 | −9.5 | Glu712 [C-O⋯H] Glu839 [C-O⋯H]∗ His1005 [C-O⋯H]∗ Val1006 [C-O⋯H]∗ Glu839 [C-O⋯H]∗ His1005 [C-O⋯H]∗ Glu712 [C-O⋯H ]∗ |
2.8309 2.4579 2.8559 2.1817 2.7366 2.7720 2.6202 |
Ile715 [C-O⋯F] | 3.6260 | Phe1003 [pi-pi staked] | 4.7087 | Glu839 [anion-pi] | 3.9962 |
|
| |||||||||
| D2 | −9.4 | Lys723 [N-H⋯O] Asp710 [C-H⋯O]∗ Asp710 [C-H⋯O]∗ |
2.2143 2.9100 2.2783 |
Lys723 [alkyl-pi] Lys723 [alkyl-pi] |
4.8686 4.8569 |
||||
|
| |||||||||
| D3 | −9.8 | Ser709 [C-H⋯N]∗
Ser756 [C-H⋯Cl]∗ Asp545 [C-O⋯H]∗ His759 [N⋯H]∗ Glu839 [C-O⋯H]∗ Gln544 [C-O⋯H]∗ |
2.3741 2.8450 2.9559 2.6541 2.1800 2.2268 |
Val836 [alkyl-Cl] Pro724 [alkyl-pi] Val836 [alkyl-pi] His758 [pi-Cl] |
4.5719 4.5000 5.2539 5.3149 |
Lys728 [cation-pi] Asp831 [anion-pi] |
2.2629 3.5500 |
||
|
| |||||||||
| D4 | −10.3 | Asn770 [N-H⋯O] Asn779 [C-O⋯H-O] Ile856 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ |
2.3508 2.4357 2.9606 3.0344 2.5335 2.4966 |
Lys723 [sigma-pi] Lys723 [alkyl-pi] |
2.7069 4.9847 |
Lys723 [cation-pi] | 2.7975 | ||
|
| |||||||||
| D5 | −9.3 | Lys723 [N-H⋯O] Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ |
2.0085 2.9432 2.8721 2.9262 2.3241 |
Lys723 [alkyl-pi] Lys723 [alkyl-pi] Tyr757 [pi-Br] |
4.6518 4.9056 4.8900 |
||||
|
| |||||||||
| D6 | −9.9 | Asn770 [C-O⋯H-N] Asn770 [O⋯H-N] Gly852 [C-O⋯H-O] Ile856 [C-O⋯H]∗ Asn710 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ |
2.3828 2.6450 2.7306 2.8943 2.4152 2.6609 |
Lys723 [sigma-pi] Lys723 [alkyl-pi] |
2.6360 5.1763 |
Lys723 [cation-pi] Lys723 [cation-pi] |
3.5014 2.9085 |
||
|
| |||||||||
| D7 | −9.5 | Glu839 [C-O⋯H-N] Val1006 [C-O⋯H-N] Ser717 [O⋯H] Glu839 [C-O⋯H]∗ Val1006 [C-O⋯H]∗ His1005 [C-O⋯H]∗ |
3.0102 2.9825 2.6782 2.7621 2.2233 3.0479 |
Phe1003 [pi-pi stacked] Pro724 [alkyl-I] Phe1003 [pi-I] |
5.0840 4.6167 4.7419 |
Glu839 [anion-pi] | 3.7952 | ||
|
| |||||||||
| D8 | −9.7 | Asn770 [C-O⋯H-N] Asn770 [O⋯H-N] Gly852 [C-O⋯H] Ile856 [C-O⋯H]∗ Asn710 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ |
2.3318 2.3759 2.8141 2.8758 2.6189 2.4101 |
Lys723 [alkyl-pi] Lys723 [alkyl-pi] |
4.2645 5.3858 |
Lys723 [cation-pi] Lys723 [cation-pi] |
3.5624 2.7612 |
||
|
| |||||||||
| D9 | −10.3 | Lys723 [N-H⋯O-C] Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ Ile856 [C-O⋯H]∗ Asp710 [C-O⋯H]∗ |
2.0587 2.9275 2.9210 2.8580 2.3996 |
Gly725 [N-H⋯F] Gly725 [N-H⋯F] Gly725 [C-H⋯F] |
2.9348 2.6339 2.2421 |
Tyr757 [pi-alkyl] Lys723 [alkyl-pi] Lys723 [alkyl-pi] |
5.1734 4.7419 4.9257 |
||
|
| |||||||||
| D10 | −10.0 | Arg713 [N-H⋯O-C] Arg713 [N-H⋯O] Lys723 [N-H⋯F] Ser763 [C-O⋯N] Arg929 [N-H⋯O] Arg929 [N-H⋯O] His759 [C-O⋯H]∗ |
2.4985 2.3481 2.2497 2.4413 2.9278 2.8935 2.8837 |
Arg713 [C-O⋯F] | 3.3749 | Lys723 [alkyl-alkyl] Lys723 [alky-pi] Ille856 [alkyl-pi] |
4.2984 5.4568 5.0991 |
||
∗Nonconventional hydrogen bond.
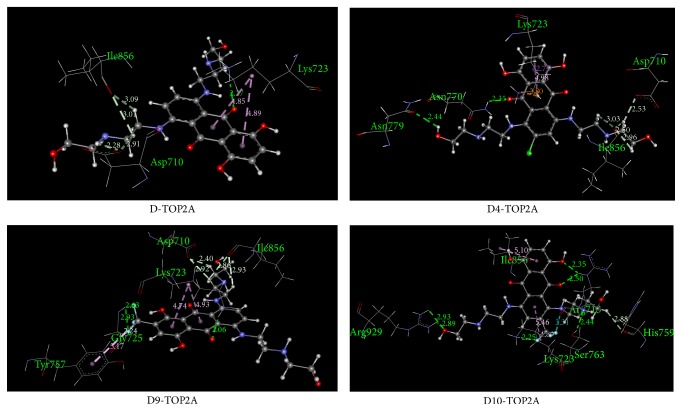
Figure 2.
Nonbonding interaction of D, D4, D9, and D10 with TOP2A.
3.1. The Electronic Structure of Mitoxantrone and Its Halogenated Derivatives
Installation of F, Cl, Br, I, and CF3 on mitoxantrone at positions 44 and 28 significantly influences the structural properties of these drugs in terms of energy, partial charge distribution, and dipole moment. The electronic energy, enthalpy, and Gibbs free energy appear to be more negative following halogenation which indicates that structures become more stable after modification (Table 1). The highest Gibbs free energy is observed for D5. In D5, bromination at position 44, replacing the hydrogen atom, changes the free energy to −4077.7124 Hartree from −1516.2828 Hartree. The highest electronic energy and enthalpy are also detected for D5.
In general, the polar nature of the molecule provides a higher value of the dipole moment. This parameter is a good indicator for studying the drug-receptor interaction [53] and plays a significant role on the formation of hydrogen bonds in biological systems. Incorporation of F, Br, I, and CF3 at position 44, replacing the hydrogen atom, decreases the dipole moment. To the contrary, increased values of the dipole moment are observed when F, Br, I, and CF3 groups are installed at position 25 (Table 1). Moreover, Table 1 shows that the dipole moment of D9 is 1.6814 Debye which is closer to that of mitoxantrone (1.5477 Debye).
The two global chemical descriptors known as hardness and softness are also calculated for all drugs and presented in Table 2. It is observed that D9 has the highest softness and lowest hardness in comparison to mitoxantrone. Among all the modified drugs, the lowest HOMO-LUMO gap has been observed for D9. The lowest HOMO-LUMO gap indicates that the molecule is more chemically reactive [54]. Pearson found that the HOMO-LUMO gap has a relation to the chemical hardness and softness of a molecule [55, 56].
The partial charges on the halogen atom of the modified drugs are changed due to their position. In modified drugs D1, D3, D5, and D7 the partial charges of F, Cl, Br, and I in the 44 position are −0.324 (a.u), −0.218 (a.u), −0.066 (a.u), and +0.125 (a.u), respectively. It is interesting to note that iodine bears partial positive charge. On the other hand, in D2, D4, D6, and D8 drugs, the partial charges are changed to −0.319 (a.u), −0.208 (a.u), −0.048 (a.u), and +0.133 (a.u), respectively. Several recent studies showed that halogen (particularly Br and I) atoms can form a halogen bond similar to a hydrogen bond and these noncovalent interactions can play remarkable roles in biological and chemical systems [57–62]. In halogen bonding, the X atom can act as an electron deficient Lewis acid and this acid is attracted by Lewis bases that are electron rich (such as the carbonyl oxygen and amine nitrogen). In our study, we notice that the I atom of modified drugs D7 and D8 shows positive charge of +0.125 (a.u) and +0.133 (a.u), respectively.
3.2. Interaction and Binding Affinity of Mitoxantrone (D) and Modified Drug (D4) against TOP2A
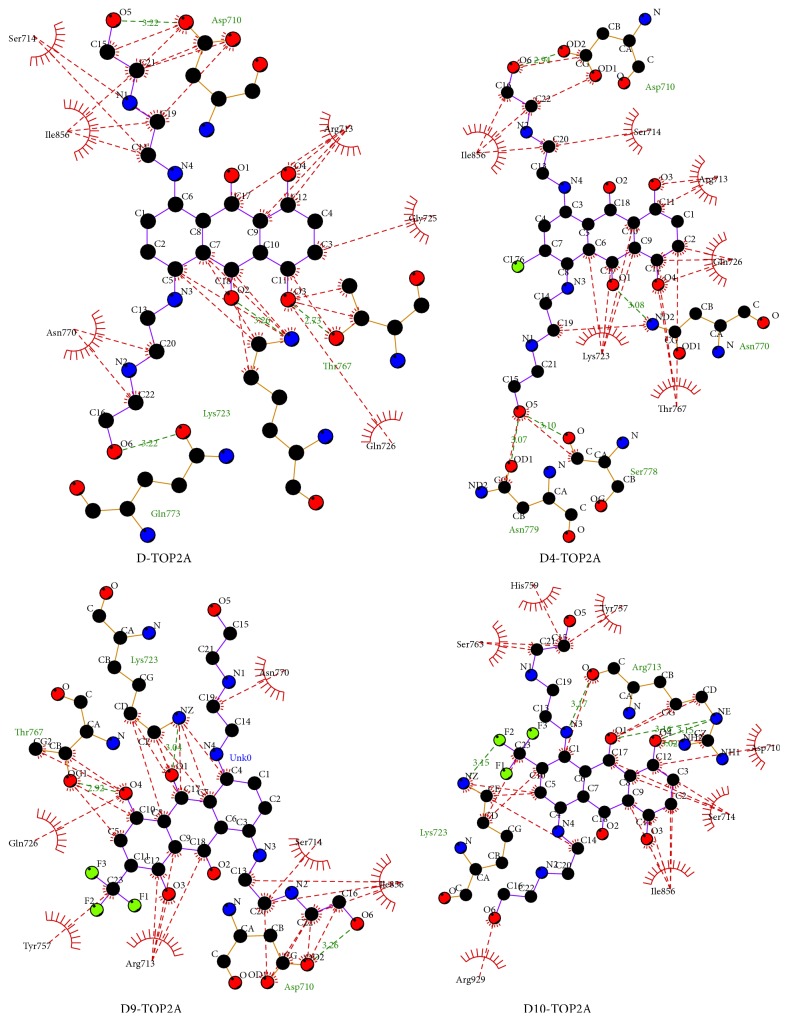
The binding affinity of D and D4 against TOP2A is −9.2 and −10.3 kcal mol−1, respectively. The surrounding residues (generated by LigPlot program) of TOP2A which interact with D and D4 are demonstrated in Figure 3. Both drugs have significant interaction with amino acid residues such as Ile, Lys, and Gly. Details of nonbonding interactions are examined by Discovery Studios Software. In the D-TOP2A complex, no halogen bond is detected. There are two pi-alkyl interactions (4.89 Å and 4.85 Å) observed between the aromatic ring of D and Lys723. Frontier molecular orbital calculations revealed that the LUMO orbital of D contributes the second pi-alkyl interaction. Both conventional and nonconventional hydrogen bonds are observed in D-TOP2A complex. The C-H⋯O interaction, known as nonconventional hydrogen bond, slightly weaker than its classical O-H⋯O hydrogen bonding, is believed to be critical in a large number of biomacromolecules' crystal structures [63, 64]. Five hydrogen bonds are observed in which four hydrogen bonds are nonconventional (C-H⋯O) and these bonds are formed with Ile856 (3.09 Å), Asp710 (2.91 Å), Ile856 (3.01 Å), and Asp710 (2.28 Å) (Figure 2). This nonconventional hydrogen bond plays crucial role in biological systems.
Figure 3.
D, D4, D9, and D10 interactions with surrounding residues of TOP2A generated by LigPlus.
The D4-TOP2A complex is stabilized by one electrostatic, two hydrophobic, and six hydrogen bonds. Strong pi-cation interaction is detected between N-H of Lys723 (2.80 Å) and benzene ring of D4. The Lys723 residue also participates in the hydrophobic interaction in which one is sigma-pi (2.71 Å) and another one is alkyl-pi (4.95 Å) interaction. In the D4-TOP2A complex, two strong C-H⋯O (distance of 2.96 Å and 2.50 Å) interactions and one relatively weak interaction with a distance of 3.03 Å has been detected with Ile856.
3.3. Interaction and Binding Affinity of Modified Drugs D1, D2, D3, D5, D6, D7, and D8 against TOP2A
The binding affinities of D1, D2, and D3 are −9.5, −9.4, and −9.8 kcal mol−1, respectively (Table 3). The surrounding residues (generated by LigPlot program) of TOP2A which interact with D1–D3 and D5–D8 are demonstrated in Figure S6. In D1 and D2, incorporation of fluorine at position 44 and 28 by replacing the hydrogen atom does not significantly change the binding affinity. An electrostatic interaction (anion-pi interaction) with a distance of 3.99 Å is found in the D1-TOP2A complex between CO of Glu839 and aromatic ring of D1. In the D1-TOP2A complex, Glu839, Phe1003, Ile715, Glu712, Glu839, His1005, and Val1006 are actively involved in the noncovalent interaction (Figure S7). Pi-pi stacked interaction between aromatic ring of Phe1003 and D1 plays a crucial role in the D1-TOP2A complex.
In the D2-TOP2A complex, only two amino acids (Lys723 and Asp710) participate in noncovalent interactions and no other interactions are detected (Figure S8). In D3-TOP2A complex, two unusual hydrophobic interactions are identified between Cl of D3 and CH2 of Val836 and another is formed by the imidazole ring of His758 with Cl of D3 (Figure S9). Hydrogen bonding plays a significant role on the binding affinity in which Ser709, Ser756, Asp545, His759, Glu839, and Gln544 residues are involved. Electrostatic interactions are also formed by Lys728 and Asp831.
The binding affinities of D5, D6, D7, and D8 against TOP2A are −9.3, −9.9, −9.5, and −9.7 kcal mol−1, respectively. These binding energies are slightly higher than that of mitoxantrone. No halogen bond is detected for modified drugs D5-D8 (Figures S10–S13). A strong electrostatic interaction is observed in both modified drugs D6 (2.90 Å) and D8 (2.76 Å) which are formed by Lys723. Pi-alkyl interactions are common for drugs D5, D8, and D9 due to Lys723. In the D5-TOP2A complex, Lys723, Ile856, and Asp710 are responsible for forming hydrogen bonds. However, in the D6-TOP2A complex, Asn770, Asn710, Gly852, and Ile856 residues are involved in hydrogen bonding (Figure S11). Hydrogen boning plays a noticeable role on the binding affinity of D7 and D8 with TOP2A (Figures S12 and S13).
3.4. Interaction and Binding Affinity of Modified Drugs D9 and D10 against TOP2A
In modified drugs D9 and D10, the trifluoromethyl group (CF3) has been added to carbon (position 38 and 25, resp.) by replacing hydrogen atom (Figure 1). Addition of CF3 to organic molecules has wide application in polymers, pharmaceuticals products, material science, and agriculture [65–68]. Due to enhanced binding selectivity, increased lipophilicity, elevated electronegativity, and improved metabolic stability, this group has earned great attention in drug design [69].
The binding affinities of D9 and D10 against TOP2A are −10.3 and −10.0 kcal mol−1, respectively (Table 3). The D9-TOP2A complex is stabilized by five hydrogen bonds, three hydrophobic interaction, and three halogen bonds. Three fluorine bonds are formed between fluorine atoms of CF3 and Gly725 (Table 3). Moreover, CF3 formed one pi-alkyl interaction (5.17 Å) with the aromatic ring of Tyr757. In the D9-TOP2A complex, there is only one conventional hydrogen bond in which the CO group of D9 acts as hydrogen bond acceptor and the NH of Lys723 acts as hydrogen bond donor. Moreover, the HOMO orbital of D1 on CO plays a remarkable role on the formation of this hydrogen bond (Figure S3). The CH2 of Lys723 contributes two pi-alkyl interactions (Figure S4) with a distance of 4.74 Å and 4.93 Å. Moreover, Asp710 and Ile856 amino acids form two nonconventional hydrogen bonds (C-O⋯H-C) separately with D9 (Figure 2).
Noncovalent interactions such as hydrogen bonding, halogen bonding, and hydrophobic interactions contributed more towards the binding affinity of TOP2A with modified drug D10. The LUMO region of 38O plays a significant role in the formation of the conventional hydrogen bond (2.50 Å) between the CO of D10 and NH of Arg713. In addition, another hydrogen bond is formed between Arg713 and D10 with a distance of 2.35 Å. In the D10-TOP2A complex, three pi-alkyl interactions are detected in which two are contributed by Lys723 (Table 3). One halogen bond is also observed with a distance of 3.37 Å between CO of Arg713 and F of D10. Among all modified drugs, more conventional hydrogen bonds are found in the D10-TOP2A complex.
4. Conclusion
This study reveals that halogenated mitoxantrone drugs also interact with TOP2A. Density functional theory calculation demonstrated some interesting features related to charge distribution, dipole moment, enthalpy, free energy, and molecular orbital of the drug molecules. Halogenation made the modified drugs thermodynamically more stable, evident from the enthalpy and Gibbs free energies. The HOMO-LUMO gap of the modified drugs D3, D5, D7, and D9 is reasonably lower than that of mitoxantrone which indicates that these compounds are more chemically reactive than the unmodified one. The dipole moment of D9 is slightly increased but close to mitoxantrone and the HOMO-LUMO gap is significantly lower than mitoxantrone. Modified drug D9 has the greatest softness amongst all drugs considered. The strongest binding affinity (−10.3 kcal mol−1) has been found for D4-TOP2A and D9-TOP2A which is higher than the parent drug. DFT and molecular docking results demonstrated that D4 and D9 show the better performance on inhibiting human topoisomerase 2 alpha. The details of nonbonding interactions detected between the modified drugs and TOP2A may help to develop new anticancer drug which can effectively target the TOP2A receptor.
Supplementary Material
The binding pocket of the protein; dipole moment, partial charge, HOMO, and LUMO orbitals of all drugs; hydrophobic and aromatic surface, noncovalent interactions in the drug-receptor complexes are included in the supplementary Figures S1-S13.
Acknowledgments
The authors are grateful to their donors who helped build a computational platform in Bangladesh; http://computchembiochem.com/1_8_Donate.html. The authors are also thankful to Professor Raymond Poirier, Department of Chemistry, Memorial University, Canada, and the Atlantic Computational Excellence Network (ACENET) for allocating computational resource for some calculations. The authors like to thank Dr. Steven Daly, Institut Lumière Matière, CNRS et Université Lyon 1, Villeurbanne, France, for reading this paper.
Conflict of Interests
Authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Gibbs J. B. Mechanism-based target identification and drug discovery in cancer research. Science. 2000;287(5460):1969–1973. doi: 10.1126/science.287.5460.1969. [DOI] [PubMed] [Google Scholar]
- 2.Yadav D. K., Khan F. QSAR, docking and ADMET studies of camptothecin derivatives as inhibitors of DNA topoisomerase-I. Journal of Chemometrics. 2013;27(1-2):21–33. doi: 10.1002/cem.2488. [DOI] [Google Scholar]
- 3.Braña M. F., Cacho M., Gradillas A., De Pascual-Teresa B., Ramos A. Intercalators as anticancer drugs. Current Pharmaceutical Design. 2001;7(17):1745–1780. doi: 10.2174/1381612013397113. [DOI] [PubMed] [Google Scholar]
- 4.Burns C. P., Haugstad B. N., North J. A. Membrane transport of mitoxantrone by L1210 leukemia cells. Biochemical Pharmacology. 1987;36(6):857–860. doi: 10.1016/0006-2952(87)90176-6. [DOI] [PubMed] [Google Scholar]
- 5.Christmann-Franck S., Bertrand H.-O., Goupil-Lamy A., et al. Structure-based virtual screening: an application to human topoisomerase II α . Journal of Medicinal Chemistry. 2004;47(27):6840–6853. doi: 10.1021/jm049745w. [DOI] [PubMed] [Google Scholar]
- 6.Posner L. E., Dukart G., Goldberg J., Bernstein T., Cartwright K. Mitoxantrone: an overview of safety and toxicity. Investigational New Drugs. 1985;3(2):123–132. doi: 10.1007/BF00174159. [DOI] [PubMed] [Google Scholar]
- 7.Henderson B. M., Dougherty W. J., James V. C. Safety assessment of a new anticancer compound, mitoxantrone, in beagle dogs: comparison with doxorubicin. I. Clinical observations. Cancer Treatment Reports. 1982;66(5):1129–1143. [PubMed] [Google Scholar]
- 8.Seiter K. Toxicity of the topoisomerase II inhibitors. Expert Opinion on Drug Safety. 2005;4(2):219–234. doi: 10.1517/14740338.4.2.219. [DOI] [PubMed] [Google Scholar]
- 9.Dunk A. A., Scott S. C., Johnson P. J., Melia W. Mitozantrone as single agent therapy in hepatocellular carcinoma: a phase II study. Journal of Hepatology. 1985;1(4):395–404. doi: 10.1016/s0168-8278(85)80777-7. [DOI] [PubMed] [Google Scholar]
- 10.Hartung H.-P., Gonsette R., König N., et al. Mitoxantrone in progressive multiple sclerosis: a placebo-controlled, double-blind, randomised, multicentre trial. The Lancet. 2002;360(9350):2018–2025. doi: 10.1016/s0140-6736(02)12023-x. [DOI] [PubMed] [Google Scholar]
- 11.Dingemans A.-M. C., Pinedo H. M., Giaccone G. Clinical resistance to topoisomerase-targeted drugs. Biochimica et Biophysica Acta (BBA)—Gene Structure and Expression. 1998;1400(1–3):275–288. doi: 10.1016/s0167-4781(98)00141-9. [DOI] [PubMed] [Google Scholar]
- 12.Fry A. M., Chresta C. M., Davies S. M., et al. Relationship between topoisomerase II level and chemosensitivity in human tumor cell lines. Cancer Research. 1991;51(24):6592–6595. [PubMed] [Google Scholar]
- 13.Boege F., Andersen A., Jensen S., Zeidler R., Kreipe H. Proliferation-associated nuclear antigen Ki-S1 is identical with topoisomerase IIα: delineation of a carboxy-terminal epitope with peptide antibodies. American Journal of Pathology. 1995;146(6):1302–1308. [PMC free article] [PubMed] [Google Scholar]
- 14.Watt P. M., Hickson I. D. Structure and function of type II DNA topoisomerases. Biochemical Journal. 1994;303(3):681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung T. D. Y., Drake F. H., Tan K. B., Per S. R., Crooke S. T., Mirabelli C. K. Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(23):9431–9435. doi: 10.1073/pnas.86.23.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Austin C. A., Fisher L. M. Isolation and characterization of a human cDNA clone encoding a novel DNA topoisomerase II homologue from HeLa cells. FEBS Letters. 1990;266(1-2):115–117. doi: 10.1016/0014-5793(90)81520-X. [DOI] [PubMed] [Google Scholar]
- 17.Austin C. A., Sng J.-H., Patel S., Fisher L. M. Novel HeLa topoisomerase II is the IIβ isoform: complete coding sequence and homology with other type II topoisomerases. Biochimica et Biophysica Acta (BBA)—Gene Structure and Expression. 1993;1172(3):283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 18.Goto T., Wang J. C. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. The Journal of Biological Chemistry. 1982;257(10):5866–5872. [PubMed] [Google Scholar]
- 19.Baldi M. I., Benedetti P., Mattoccia E., Tocchini-Valentini G. P. In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase. Cell. 1980;20(2):461–467. doi: 10.1016/0092-8674(80)90632-7. [DOI] [PubMed] [Google Scholar]
- 20.Liu L. F., Liu C. C., Alberts B. M. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979;281(5731):456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- 21.Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roca J., Wang J. C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71(5):833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 23.Roca J., Wang J. C. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77(4):609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 24.Roca J., Berger J. M., Harrison S. C., Wang J. C. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(9):4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wigley D. B., Davies G. J., Dodson E. J., Maxwell A., Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351(6328):624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 26.Morrison A., Cozzarelli N. R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979;17(1):175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- 27.Tse Y. C., Kirkegaard K., Wang J. C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. Journal of Biological Chemistry. 1980;255(12):5560–5565. [PubMed] [Google Scholar]
- 28.Schoeffler A. J., Berger J. M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Quarterly Reviews of Biophysics. 2008;41(1):41–101. doi: 10.1017/s003358350800468x. [DOI] [PubMed] [Google Scholar]
- 29.Bates A. D., Berger J. M., Maxwell A. The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Research. 2011;39(15):6327–6339. doi: 10.1093/nar/gkr258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker J. V., Nitiss J. L. DNA topoisomerase II as a target for cancer chemotherapy. Cancer Investigation. 2002;20(4):570–589. doi: 10.1081/CNV-120002156. [DOI] [PubMed] [Google Scholar]
- 31.Nitiss J. L., Beck W. T. Antitopoisomerase drug action and resistance. European Journal of Cancer. 1996;32(6):958–966. doi: 10.1016/0959-8049(96)00056-1. [DOI] [PubMed] [Google Scholar]
- 32.Wendorff T. J., Schmidt B. H., Heslop P., Austin C. A., Berger J. M. The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. Journal of Molecular Biology. 2012;424(3-4):109–124. doi: 10.1016/j.jmb.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Hoog P., Gamez P., Mutikainen I., Turpeinen U., Reedijk J. An aromatic anion receptor: anion-π interactions do exist. Angewandte Chemie—International Edition. 2004;43(43):5815–5817. doi: 10.1002/anie.200460486. [DOI] [PubMed] [Google Scholar]
- 34.Metrangolo P., Neukirch H., Pilati T., Resnati G. Halogen bonding based recognition processes: a world parallel to hydrogen bonding. Accounts of Chemical Research. 2005;38(5):386–395. doi: 10.1021/ar0400995. [DOI] [PubMed] [Google Scholar]
- 35.Slutsky M. M., Marsh E. N. G. Cation-pi interactions studied in a model coiled-coil peptide. Protein Science. 2004;13(8):2244–2251. doi: 10.1110/ps.04702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umezawa Y., Nishio M. Ch/π interactions as demonstrated in the crystal structure of guanine- nucleotide binding proteins, src homology-2 domains and human growth hormone in complex with their specific ligands. Bioorganic and Medicinal Chemistry. 1998;6(4):493–504. doi: 10.1016/s0968-0896(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 37.Frisch M., Trucks G., Schlegel H. Gaussian 09, Revision D. 01. Wallingford, Conn, USA: Gaussian; 2009. [Google Scholar]
- 38.Wang Y., Xiao J., Suzek T. O., Zhang J., Wang J., Bryant S. H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Research. 2009;37(2):W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becke A. D. Density-functional exchange-energy approximation with correct asymptotic behavior. Physical Review A. 1988;38(6):3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 40.Becke A. D. A new mixing of Hartree-Fock and local density-functional theories. The Journal of Chemical Physics. 1993;98(2):1372–1377. doi: 10.1063/1.464304. [DOI] [Google Scholar]
- 41.Lee C., Yang W., Parr R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Physical Review B. 1988;37(2):785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 42.Easton R. E., Giesen D. J., Welch A., Cramer C. J., Truhlar D. G. The MIDI! basis set for quantum mechanical calculations of molecular geometries and partial charges. Theoretical Chemistry Accounts. 1996;93(5):281–301. [Google Scholar]
- 43.Li J., Cramer C. J., Truhlar D. G. MIDI! basis set for silicon, bromine, and iodine. Theoretical Chemistry Accounts. 1998;99(3):192–196. doi: 10.1007/s002140050323. [DOI] [Google Scholar]
- 44.Parr R., Yang W. Density-Functional Theory of Atoms and Molecules. Oxford University Press; 1989. [Google Scholar]
- 45.Pearson R. G. The HSAB principle—more quantitative aspects. Inorganica Chimica Acta. 1995;240(1-2):93–98. doi: 10.1016/0020-1693(95)04648-8. [DOI] [Google Scholar]
- 46.Pearson R. G. Absolute electronegativity and hardness correlated with molecular orbital theory. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(22):8440–8441. doi: 10.1073/pnas.83.22.8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 48.DeLano W. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- 49.Trott O., Olson A. J. Software news and update: autodock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Research. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Accelrys Software Inc. Discovery Studio Modeling Environment, Release 4.0. San Diego, Calif, USA: Accelrys Software Inc; 2013. [Google Scholar]
- 52.Laskowski R. A., Swindells M. B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 53.Lien E. J., Guo Z. R., Li R., Su C. T. Use of dipole moment as a parameter in drug-receptor interaction and quantitative structure-activity relationship studies. Journal of Pharmaceutical Sciences. 1982;71(6):641–655. doi: 10.1002/jps.2600710611. [DOI] [PubMed] [Google Scholar]
- 54.Hoque M. M., Halim M. A., Sarwar M. G., Khan M. W. Palladium-catalyzed cyclization of 2-alkynyl-N-ethanoyl anilines to indoles: synthesis, structural, spectroscopic, and mechanistic study. Journal of Physical Organic Chemistry. 2015;28(12):732–742. doi: 10.1002/poc.3477. [DOI] [Google Scholar]
- 55.Parr R. G., Zhou Z. Absolute hardness: unifying concept for identifying shells and subshells in nuclei, atoms, molecules, and metallic clusters. Accounts of Chemical Research. 1993;26(5):256–258. doi: 10.1021/ar00029a005. [DOI] [Google Scholar]
- 56.Ayers P. W., Parr R. G., Pearson R. G. Elucidating the hard/soft acid/base principle: a perspective based on half-reactions. Journal of Chemical Physics. 2006;124(19) doi: 10.1063/1.2196882.194107 [DOI] [PubMed] [Google Scholar]
- 57.Sarwar M. G., Ajami D., Theodorakopoulos G., Petsalakis I. D., Rebek J., Jr. Amplified halogen bonding in a small space. Journal of the American Chemical Society. 2013;135(37):13672–13675. doi: 10.1021/ja407815t. [DOI] [PubMed] [Google Scholar]
- 58.Sarwar M. G., Dragisic B., Salsberg L. J., Gouliaras C., Taylor M. S. Thermodynamics of halogen bonding in solution: Substituent, structural, and solvent effects. Journal of the American Chemical Society. 2010;132(5):1646–1653. doi: 10.1021/ja9086352. [DOI] [PubMed] [Google Scholar]
- 59.Sarwar M. G., Dragisic B., Sagoo S., Taylor M. S. A tridentate halogen-bonding receptor for tight binding of halide anions. Angewandte Chemie—International Edition. 2010;49(9):1674–1677. doi: 10.1002/anie.200906488. [DOI] [PubMed] [Google Scholar]
- 60.Hoque M. M., Halim M. A., Rahman M. M., Hossain M. I., Khan M. W. Synthesis and structural insights of substituted 2-iodoacetanilides and 2-iodoanilines. Journal of Molecular Structure. 2013;1054-1055:367–374. doi: 10.1016/j.molstruc.2013.10.011. [DOI] [Google Scholar]
- 61.Beale T. M., Chudzinski M. G., Sarwar M. G., Taylor M. S. Halogen bonding in solution: thermodynamics and applications. Chemical Society Reviews. 2013;42(4):1667–1680. doi: 10.1039/c2cs35213c. [DOI] [PubMed] [Google Scholar]
- 62.Sarwar M. G., Dragisić B., Dimitrijević E., Taylor M. S. Halogen bonding between anions and iodoperfluoroorganics: solution-phase thermodynamics and multidentate-receptor design. Chemistry—A European Journal. 2013;19(6):2050–2058. doi: 10.1002/chem.201202689. [DOI] [PubMed] [Google Scholar]
- 63.Kuduva S., Craig D. Cubanecarboxylic acids. Crystal engineering considerations and the role of C − H ⋯ O hydrogen bonds in determining O − H ⋯ O networks. Journal of the American Chemical Society. 1999;121(9):1936–1944. doi: 10.1021/ja981967u. [DOI] [Google Scholar]
- 64.Meadows E. S., De Wall S. L., Barbour L. J., Fronczek F. R., Kim M.-S., Gokel G. W. Structural and dynamic evidence for C–H···O hydrogen bonding in lariat ethers: implications for protein structure. Journal of the American Chemical Society. 2000;122(14):3325–3335. doi: 10.1021/ja9940672. [DOI] [Google Scholar]
- 65.Ma J.-A., Cahard D. Strategies for nucleophilic, electrophilic, and radical trifluoromethylations. Journal of Fluorine Chemistry. 2007;128(9):975–996. doi: 10.1016/j.jfluchem.2007.04.026. [DOI] [Google Scholar]
- 66.McClinton M. A., McClinton D. A. Trifluoromethylations and related reactions in organic chemistry. Tetrahedron. 1992;48(32):6555–6666. doi: 10.1016/S0040-4020(01)80011-9. [DOI] [Google Scholar]
- 67.Furuya T., Kamlet A. S., Ritter T. Catalysis for fluorination and trifluoromethylation. Nature. 2011;473(7348):470–477. doi: 10.1038/nature10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji Y., Brueckl T., Baxter R. D., et al. Innate C-H trifluoromethylation of heterocycles. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(35):14411–14415. doi: 10.1073/pnas.1109059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lishchynskyi A., Novikov M. A., Martin E., Escudero-Adán E. C., Novák P., Grushin V. V. Trifluoromethylation of aryl and heteroaryl halides with fluoroform-derived CuCF3: scope, limitations, and mechanistic features. Journal of Organic Chemistry. 2013;78(22):11126–11146. doi: 10.1021/jo401423h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The binding pocket of the protein; dipole moment, partial charge, HOMO, and LUMO orbitals of all drugs; hydrophobic and aromatic surface, noncovalent interactions in the drug-receptor complexes are included in the supplementary Figures S1-S13.