Abstract
The central nervous system (CNS) is a highly organised structure. Many signalling systems work in concert to ensure that neural stem cells are appropriately directed to generate progenitor cells, which in turn mature into functional cell types including projection neurons, interneurons, astrocytes, and oligodendrocytes. Herein we explore the role of the low density lipoprotein (LDL) receptor family, in particular family members LRP1 and LRP2, in regulating the behaviour of neural stem and progenitor cells during development and adulthood. The ability of LRP1 and LRP2 to bind a diverse and extensive range of ligands, regulate ligand endocytosis, recruit nonreceptor tyrosine kinases for direct signal transduction and signal in conjunction with other receptors, enables them to modulate many crucial neural cell functions.
1. Low Density Lipoprotein Receptor Related Proteins 1 and 2
The LDL receptor family is a large family of multiligand receptors. Core family members include the LDL receptor; very low density lipoprotein (VLDL) receptor [1]; LDL receptor related protein (LRP)1, also known as CD91 and the α-2-macroglobulin receptor [2–4]; LRP2, also known as GP330 and Megalin [5]; LRP5 [6]; LRP6 [7]; and LRP8, also known as the apolipoprotein receptor-2 [8]. Each family member is a single-pass transmembrane receptor, containing two or more extracellular cysteine-rich complement type repeats, which act as ligand binding domains [9].
At 600 kDa, LRP1 and LRP2 are the largest and most promiscuous members of the LDL receptor family. Transcription of the Lrp1 gene can be activated by a number of transcription factors including sterol regulatory element binding protein 2 [10], hypoxia-induced factor 1α [11], and nitric oxide-dependent transcription factors [12], but is negatively regulated by naturally occurring antisense transcripts that are inversely coded within exons 5 and 6 of the Lrp1 gene [13]. The Lrp1 gene codes for a precursor protein that binds to the receptor associated protein (RAP), a chaperone that occupies the ligand binding domains of the precursor [14] to prevent the binding of other ligands [15], and ensure its correct folding in the endoplasmic reticulum [16, 17] (Figure 1). RAP remains bound to the LRP1 precursor and transports it to the Golgi apparatus. This transport involves the proximal NPXY motif in the intracellular domain of the protein [18]. In the trans-Golgi network, the low pH of the secretory pathway causes protonation of the histidine residues in domain 3 of RAP [19], triggering its dissociation from the LRP1 precursor [14, 20]. The protease Furin then cleaves the LRP1 precursor at the RX(K/R)R consensus sequence, to generate a large α-chain (515 kDa) and a smaller β-chain (85 kDa) [21]. The two fragments remain noncovalently linked on their way to the cell membrane, where they are embedded as one functional unit, comprising mature LRP1 (Figure 1). LRP2 is similarly chaperoned by RAP [22] and also contains an RX(K/R)R consensus sequence, but there is no evidence that LRP2 undergoes intracellular proteolytic processing prior to its insertion into the plasma membrane [5].
Figure 1.
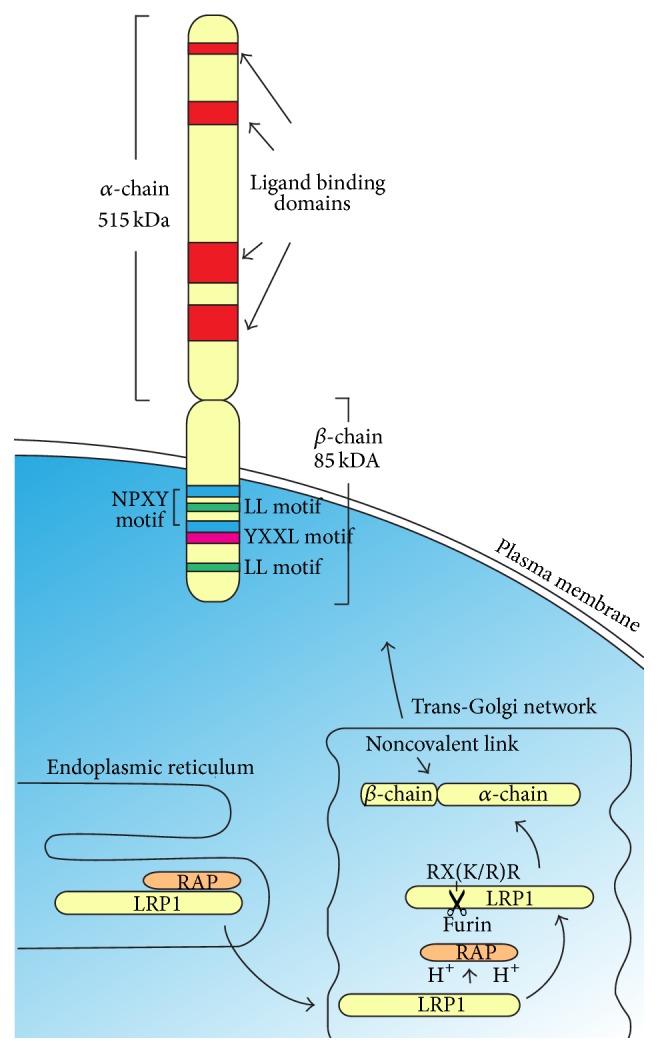
LRP1 maturation and structure. This schematic depicts the LRP1 precursor protein, which is synthesized in the endoplasmic reticulum and is bound to the chaperone protein, receptor associated protein (RAP). The LRP1 precursor is transported to the trans-Golgi network where the low pH causes RAP to dissociate. The protease Furin cleaves the LRP1 precursor at the RX(K/R)R consensus sequence to generate a large α-chain (515 kDa) and a smaller β-chain (85 kDa) which are noncovalently linked and shuttled to the cell membrane, where they are embedded as one functional unit. The α-chain contains four ligand-binding domains (red) that interact with a large number of ligands. The β-chain contains a small extracellular region, a transmembrane region which anchors the LRP1 protein within the plasma membrane, as well as two dileucine (LL, green) motifs and two asparagine-proline-x-tyrosine (NPXY, blue) motifs, where the distal motif is contiguous with a tyrosine-x-x-leucine (YXXL, pink) motif which interact with intracellular adaptor proteins and the endocytotic machinery.
1.1. Soluble LRP1 and LRP2
Once LRP1 is inserted into the plasma membrane, the soluble extracellular domain (sLRP1) can be cleaved from the cell surface by enzymes such as the beta-site APP cleaving enzyme 1 (BACE1) [23] and metalloproteinase [24] (Figure 2). sLRP1 contains the α-chain and a 55 kDa fragment of the β-chain [25] and can be detected in plasma and cerebral spinal fluid [26, 27]. Similarly, soluble fragments of LRP2 have been shown to be released from cultured choroid plexus epithelial cells and can be detected in cerebral spinal fluid [28]. LRP1 and LRP2 can also undergo intramembrane proteolysis mediated by γ-secretase, in either the plasma or endosomal membrane [29], to liberate an intracellular fragment which reportedly enters the nucleus [30, 31] (Figure 2(a)). The physiological function of soluble LRP fragments in normal neural cell development is poorly understood, but they have the potential to bind LRP ligands and prevent them from binding to full-length LRPs or, in the case of the intracellular domain, modulate gene transcription.
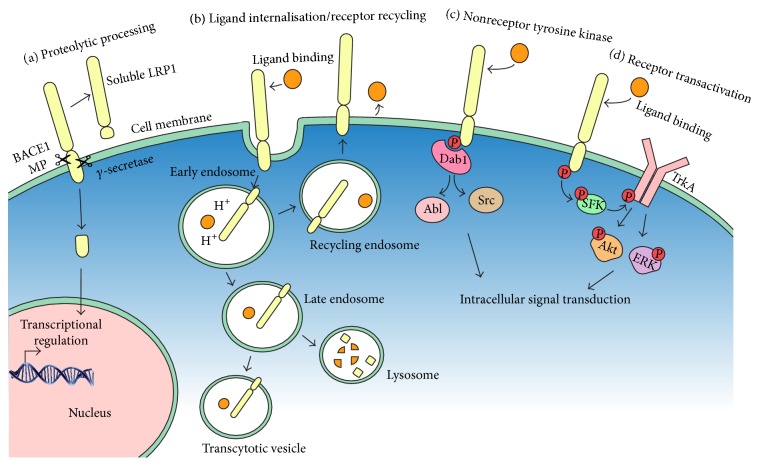
Figure 2.
Signalling mechanisms employed by LRP1. (a) The extracellular domain of LRP1 can be shed following cleavage by beta-site APP cleaving enzyme 1 (BACE1) and metalloproteinases (MP) producing a soluble form of LRP1 (sLRP1). The intracellular domain can be cleaved by γ-secretase and is thought to translocate to the nucleus to influence gene transcription. (b) Ligand binding to LRP1 can result in receptor and ligand internalisation. Once internalised, the ligand/receptor complex can be processed in a multitude of ways, including degradation by lysosomes or resecretion via transcytotic and recycling vesicles. Note that while they are depicted together, ligand and receptor/s are trafficked independently. (c) Specific regions on the intracellular region of LRP1 interact with adaptor proteins such as Disabled-1 (Dab1), which interacts with the NPXY motifs and can recruit nonreceptor tyrosine kinases such as Src and Abl allowing signal transduction. (d) Activation of LRP1 by specific ligands can transactivate other receptors such as tropomyosin receptor kinase A (TrkA), which can then activate downstream signalling pathways to regulate cell function.
1.2. LRP1 and LRP2 as Mediators of Endocytosis
While the proteolytic processing of these receptors is becoming increasingly well understood, LRP1 and LRP2 remain best known for their role in mediating endocytosis (Figure 2(b)). Following ligand binding to mature LRP1 in the plasma membrane, it was originally believed that the two NPXY motifs of the cytoplasmic domain interacted with the endocytotic machinery to mediate rapid clathrin-dependant endocytosis of the receptor-ligand complex, as has been previously shown for other members of this receptor family [32]. However for LRP1, the YXXL motif and the distal dileucine motif independently mediate endocytosis, and the NPXY motifs are not required [33]. The rate of endocytosis is regulated by cAMP-dependent protein kinase A, which constitutively phosphorylates LRP1, predominantly at serine 76 of the cytoplasmic tail [34].
Like LRP1, LRP2 has two intracellular NPXY domains [5]; however unlike LRP1, the distal NPXY motif of LRP2 has been shown to interact with the phosphotyrosine-binding domain of Disabled-2 [35], a clathrin-associated sorting protein, to mediate endocytosis [29, 36, 37]. Interestingly, endocytosis does not occur during mitosis, due to the phosphorylation of Disabled-2, which removes it from the cell surface, so that it no longer colocalizes with clathrin and cannot mediate this process [38]. LRP2-directed endocytosis may still occur via clathrin-independent pathways, instead relying on the small GTPase Arf6 and caveolin 1 [39, 40]. Furthermore, LRP1- and LRP2-mediated endocytosis can be influenced by the expression of miR199a and miR199b family members, which regulate the expression of a number of genes critical for clathrin-dependent and clathrin-independent endocytosis [41]. Following endocytosis, the extracellular beta-propeller regions of LRP1 and LRP2 facilitate ligand dissociation [42], so that the ligands and receptors can be differentially sorted in early endosomes.
The mechanisms regulating the recycling of LRP1 back to the plasma membrane are not fully characterised and may vary between cell types. However, it is known that this process requires binding of the adaptor protein sorting nexin 17 to the first NPXY domain of LRP1 in early endosomes [43, 44], so that LRP1 is recycled back to the cell surface in approximately 30 minutes [45]. In early endosomes, the first NPXY domain of LRP2 instead binds the phosphotyrosine-binding domain of autosomal recessive hypercholesterolemia (ARH) [46], a clathrin-associated sorting protein that couples LRP2 to the dynein motor complex [47] and transports it from the sorting endosomes to the endocytic recycling compartment [29]. The constitutive phosphorylation of LRP2 by GSK3β is also involved in directing LRP2 to the endocytic recycling compartment, from which it is slowly recycled to the plasma membrane [48].
But what happens to the internalised ligand? LRP1 and LRP2 have been shown to bind upwards of 40 different ligands, many of which are structurally and functionally unrelated, and the list is always evolving [49]. They both have four LDL receptor homology regions which are the extracellular ligand-binding domains [50, 51] and bind common ligands including tissue-type plasminogen activator [52–55], apolipoprotein E, lactoferrin [17, 52], and metallothioneins I and II [56]; however not all ligands have been shown to bind both receptors. α2-Macroglobulin is a high affinity ligand for LRP1 [57, 58], and like prion protein has only been demonstrated to bind to LRP1 [59], while transthyretin [60] and the complex of vitamin D with the vitamin D binding protein have only been shown to bind LRP2 [61]. Once endocytosed, ligands may be degraded in lysosomes, resecreted from recycling endosomes, or trafficked in transcytotic vesicles from the apical to the basolateral membrane (or vice versa) before being secreted [62] (Figure 2(b)).
1.3. LRP1 and LRP2 Intracellular Signal Transduction
The true complexity of LRP1 and LRP2 signalling lies in the fact that these receptors not only trigger endocytosis but also influence signal transduction. Upon ligand binding, the NPXY motifs can function as a docking sites for intracellular adaptor proteins. LRP1 can bind cytosolic ligands in a phosphorylation-dependent manner, via two dileucine motifs and one YXXL motif in the intracellular domain. For example, the adaptor proteins Disabled-1 and FE65 can bind to the NPXY motifs of LRP1, to recruit and activate nonreceptor tyrosine kinases such as Src and Abl [63] (Figure 2(c)), allowing the receptor to transduce an intracellular signal or form signalling hubs through the binding of coreceptors [49] (Figure 2(d)). A number of coreceptors of LRP1 have been identified, including platelet-derived growth factor receptor (PDGFR) β [64, 65], tropomyosin-related kinase receptor A [66], amyloid precursor protein [67], and insulin-like growth factor 1 receptor [68]. These associations increase the number of intracellular pathways by which distinct LRP ligands may elicit their effects.
2. LRPs as Regulators of Nervous System Development
Despite the large number of common ligands and the structural similarities that exist between LRP1 and LRP2, the two genes are not functionally redundant during development. Both Lrp1 and Lrp2 single knockout mice have severe developmental phenotypes. Lrp1 knockout blastocysts fail to implant and therefore do not develop into embryos [69]. Lrp2 knockout mice are mostly embryonic lethal, presenting with defects including a cleft palate, failure to form an olfactory bulb, and fusion of the forebrain hemispheres, resulting in a single ventricle (holoprosencephaly) [70]. The small number of Lrp2 knockout mice that survive until birth experience severe vitamin D3 deficiency, as the reabsorption of vitamin D and the vitamin D binding protein from the kidney proximal tubule is LRP2-dependant, but die of respiratory failure [61, 70]. Human mutations in Lrp2 are known to cause facio-oculo-acoustico-renal syndrome/Donnai-Barrow syndrome, an autosomal recessive disorder associated with disrupted brain formation, including agenesis of the corpus callosum [71].
The very early developmental defect observed in the Lrp1 knockout mouse, and the gross neural phenotype of the Lrp2 knockout mouse, do not allow us to investigate the importance of these receptors for the functioning of individual neural cell types. However, a variety of expression studies performed alongside knockdown and conditional knockout approaches demonstrate that both receptors mediate ligand endocytosis and intracellular signalling in a number of immature neural cell types. LRP1 is more widely expressed in the CNS than LRP2, being detected in mature neurons, particularly those of the entorhinal cortex, hippocampus [72] and cerebellum [73], and all CNS glia [74]. In contrast, LRP2 expression is restricted to the apical surface of the neural tube and subsequently to the forebrain, optic stalk, and otic vesicle during development [75, 76]. In the CNS of adult mice, LRP2 is predominantly expressed by cells of the choroid plexus [77] and ependymal cells [78] but has also been detected in oligodendrocytes of the spinal cord [79]. The expression patterns of LRP1 and LRP2 are largely spatially and temporally distinct, reflecting their different roles in CNS regulation.
3. LRP1 and LRP2 as Regulators of Neural Stem Cell Function
3.1. Neural Stem Cells in the Developing and Adult CNS
The early neural tube is a pseudostratified epithelium composed of neuroepithelial precursor cells. These early neural stem cells divide symmetrically, expanding their population, before switching to include asymmetric divisions that generate neuroblasts. This switch coincides with a change in gene expression, as the neuroepithelial precursor cells transition into radial glial stem cells, which comprise two molecularly distinct subgroups in the developing human brain, corresponding to those in the outer subventricular zone and those in the ventricular zone [80]. Following neuroblast generation, radial glia switch to glial generation starting with the production of oligodendrocyte progenitor cells (OPCs) and concluding with the production of astrocytic precursors [81]. Towards the end of development a subset of radial glial stem cells adopt a more astrocytic gene expression profile and give rise to the adult neural stem cells [82].
In adulthood neural stem cells reside in two key niches, the subventricular zone of the lateral ventricles and the dentate gyrus of the hippocampus, where they proliferate to generate intermediate progenitor cells and ultimately neuroblasts [83]. Neural stem cells in the subventricular zone also produce a small number of OPCs under normal physiological conditions [84]. The behaviour of neural stem cells (and their intermediate progenitors) is highly controlled by mitogenic and morphogenic signalling. While key ligands and receptors for these pathways are well described, the role of LRP1 and LRP2 in these pathways has only recently been elucidated.
3.2. LRPs as Regulators of Cell Fate Specification
LRP1 and LRP2 have both been shown to facilitate the internalisation of the potent morphogen, sonic hedgehog [85–87], a finding that has provided insight into the significant neurodevelopmental defects observed in patients and mice lacking normal functioning Lrp2 [70, 71, 75]. LRP2 is expressed by neuroepithelial cells, on the apical side of the neural plate, as early as E7.5 in the mouse. After neural tube closure at E9.5, LRP2 expression becomes increasingly restricted to the midline, ultimately being localized to the clathrin-coated pit regions of the apical cell membrane, clustered at the base of the primary cilium (a cellular organelle essential to sonic hedgehog signalling) [88] and in the subapical endosomes of the radial glia [89]. At E8 sonic hedgehog is produced by cells of the axial mesoderm (the notochord and prechordal plate) and by E8.5 its expression expands to include the radial glia at the ventral midline of the rostral diencephalon. This expansion does not occur in Lrp2 knockout embryos, as LRP2 is required for the radial glia to bind and sequester sonic hedgehog as it diffuses, regulating morphogen presentation to the neural stem cells [89].
Once sonic hedgehog is bound to LRP2 it can also bind its receptor patched-1, and the complex undergoes clathrin-mediated endocytosis [89]. All components can then be found within early endosomes and recycling endosomes but do not appear to be targeted to the lysosome for degradation. The internalisation of patched-1 by LRP2 results in activation of the effector smoothened, leading to changes in gene transcription mediated by the Gli transcription factors. Therefore, in the absence of LRP2, radial glia show reduced expression of the sonic hedgehog target genes gli1 and six3 [89]. The loss of sonic hedgehog and Gli3-mediated transcriptional repression has secondary consequences for neural development, including aberrant bone morphogenic protein 4 expression in the dorsal forebrain [75, 89, 90] and disrupted fibroblast growth factor 8 and noggin expression [89]. These data indicate that LRP2 regulates the patched-1-dependent internalisation and trafficking of sonic hedgehog [89], which is necessary for neural stem cell specification and ventral forebrain patterning.
Later in development, the expression of LRP2 by spinal cord radial glial is also necessary for glial cell specification. Lrp2 knockout mice completely lack oligodendrocyte-lineage cells and produce very few astrocytes in the spinal cord [91]. OPC specification from radial glia in the ventral spinal cord is also directed by sonic hedgehog signalling [92–96], and so the lack of spinal cord oligodendrocytes may be explained by a mechanism similar to that detailed above. However OPCs can be generated from cultured neuroepithelial precursors derived from sonic hedgehog and smoothened knockout mice [97, 98], indicating that LRP2 must also interact with other signalling pathways such as basic fibroblast growth factor and insulin-like growth factor I [99], to promote OPC generation from neural stem cells. The decreased number of astrocytes observed in Lrp2 knockout mice is also interesting. LRP2 is expressed by vimentin-positive cells in the E15 ventral spinal cord [79] that most likely correspond to immature astrocytes [84, 100, 101]. While LRP2 may play a role in regulating the behaviour of astrocytic precursors, it is more likely that the observed phenotype is the result of LRP2 being required for astrocyte specification by radial glia, as this immature glial population is not generated in Lrp2 knockout mice. Despite these observations that strongly implicate LRP2 in glial cell specification during neural development, the ligands and signalling mechanisms are unknown.
LRP1 appears to fulfill a similar role in regulating glial generation in the brain. LRP1 is expressed by cells within the embryonic ventricular zone and the early postnatal subventricular zone [102]. While the role of LRP1 in regulating neural stem cell function in vivo is poorly understood, in vitro studies suggest that LRP1 can regulate OPC production. Neural stem cells can be harvested from the cortex of embryonic mice and grown as a suspension culture termed neurospheres. When differentiated, neurospheres generate neurons, astrocytes, and oligodendrocytes. However, neurospheres lacking Lrp1 generate normal numbers of neurons, but significantly fewer O4-positive oligodendrocytes [102]. These data may reflect a requirement of LRP1 signalling in neural stem cells for OPC specification but could equally result if LRP1 is necessary for the proliferation or differentiation of OPCs (see OPC section below).
3.3. LRPs as Regulators of Neural Stem Cell Proliferation
In the subventricular zone of the adult mouse brain, LRP2 is expressed by ependymal cells underlying the neurogenic niche [78, 103]. The importance of LRP2 expression for neural stem and progenitor cell proliferation was examined in Lrp2 267/267 mutant mice, which produce a truncated form of LRP2 [104]. Lrp2 267/267 mice have ~25% fewer proliferating cells in the subventricular zone relative to control mice and a proportional reduction in the number of newborn neurons entering the olfactory bulb [78]. The absence of functional LRP2 from the neurogenic niche was accompanied by increased bone morphogenic proteins 2 and 4, increased phosphorylation of the downstream effectors SMAD1, SMAD5, and SMAD8, and increased activation of the downstream target, inhibitor of DNA binding 3 [78]. It is known that LRP2 can act as an endocytic receptor, sequestering and clearing bone morphogenic protein 4 [75]. However this does not appear to be the mechanism at play here. A ventricular infusion of noggin, the potent bone morphogenic protein 4 antagonist [105], certainly decreases neurogenesis but does so in favour of oligodendrogenesis [106], and this fate-switch is not consistent with the phenotype of the Lrp2 267/267 mouse [78].
The ability of LRPs to regulate proliferation may be more widespread amongst immature neural cell populations, as LRP1 also regulates the proliferation of cerebellar granular neuron precursors. Cerebellar granular neuron precursors are a temporary cell population that proliferate in the external germinal zone of the developing cerebellum, producing granule neurons from birth until ~P15 in the mouse [107]. This cell population is highly responsive to the promitotic effects of sonic hedgehog [108–110]. However, the effect of sonic hedgehog is negatively regulated by an interaction between LRP1 and protease nexin 1, also known as SERPINE2. Protease nexin 1 complexes with its target proteases and binds to LRP1 on the surface of cultured cerebellar granule neuron precursors [111]. Once endocytosed, protease nexin 1 antagonizes sonic hedgehog signalling, reducing the proliferation of cerebellar granule neurons. This regulation is critical for normal cerebellar development, as the absence of protease nexin 1 in vivo delays cerebellar granule neuron precursor differentiation and increases the overall size of the cerebellum [111]. We would predict that conditionally removing Lrp1 from cerebellar granule neuron precursors would have the same effect.
4. LRPs as Regulators of Neuroblast Function
Neuroblast generation and their subsequent migration into the developing cortex has been well characterised [112]. Postmitotic neuroblasts that are generated in the cortical ventricular zone are destined to form cortical projection neurons [113]. They undergo radial migration out of the germinal zone, moving along the apical processes of radial glia. The final laminar position of a newborn neuron is determined by its birth date, with late-born neuroblasts migrating past early-born neurons, to seed progressively more superficial layers of the cortex [114]. In contrast, cortical interneurons are generated from radial glial cells within the ventricular zones of the medial ganglionic eminence, the caudal ganglionic eminence and the preoptic area, and undergo both radial and tangential migration to populate each of the cortical layers [115–117].
Neuroblasts born in the two neurogenic niches of the adult brain also have vastly different migratory requirements. Those born in the hippocampus are destined to be dentate granule neurons, and send axons from the dentate gyrus to CA3 of the hippocampus [118]. After birth these cells move a very short distance as they mature, migrating from the subgranular zone (the inner lip) of the dentate granule neuron layer to their final position within the layer. On the other hand, neuroblasts born in the subventricular zone migrate tangentially, moving as neuroblast chains through the rostral migratory stream into the olfactory bulb [119]. Upon exiting the rostral migratory stream, the neuroblasts turn and migrate radially and differentiate into granule and periglomerular neurons in the olfactory bulb [83]. This type of chain migration is regulated by signals that modify the actin cytoskeleton including contact-mediated signalling between the neuroblasts and the ensheathing glia [120–123] and the chemorepulsion mediated by slit and netrin [124–126]. Recent evidence suggests that, following neural stem cell specification and neuroblast generation, LDL family members, including LRP1 and LRP2, continue to play a significant role in regulating the successful maturation and integration of these new cells in the CNS.
4.1. LRP8 and the VLDL Receptor Are Key Regulators of Neuroblast Migration in Development and Adulthood
While this review focuses on LRP1 and LRP2, it is not possible to discuss the role of LDL family members in regulating neuroblast migration without first detailing the importance of the LRP8 and VLDL receptor in cortical development. Lrp8 and VLDL receptor double knockout mice have abnormalities in the layering of the brain, including the ectopic placement of neurons [127, 128], and also exhibit malformation of the cerebellum and spinal cord [127, 129]. LRP8 and the VLDL receptor are high affinity receptors for reelin [130, 131] a large extracellular matrix protein [127, 129, 130]. Mice that lack reelin largely phenocopy the distinct cortical lamination defects seen in the Lrp8 and VLDL receptor double knockout mice [127, 129, 130]. Oligomeric reelin binds to LRP8 and the VLDL receptor, activates Src family kinases, and induces phosphorylation of Disabled-1. This signalling pathway enables polarisation, adhesion, stabilisation, process outgrowth, and ultimately neuroblast migration [132–134]. During development reelin is first expressed in the cortical marginal zone by Cajal-Retzius cells [135–137] and later by interneurons [138, 139]. Humans with mutations of the VLDL receptor gene have an increased risk of developing schizophrenia, which is thought to result from subtle neuroblast migration defects within the brain [140].
LRP8 and the VLDL receptor can also regulate neuroblast migration when activated by an alternative ligand, thrombospondin-1. Thrombospondin-1 is expressed in the subventricular zone and throughout the rostral migratory stream [141], where it acts on LRP8 and the VLDL receptor to promote neuroblast chain migration. Thrombospondin-1 knockout mice have defective chain migration, with fewer neuroblasts successfully migrating to the olfactory bulb [141]. This phenotype is also observed in mice lacking LRP8 and VLDL receptor, or Disabled-1, but is not observed in reelin knockout mice [142]. However, the successful migration of neurons from the subventricular zone to the olfactory bulb appears to require both ligands. Thrombospondin-1 stabilizes neuroblast chains and increases their length in the subventricular zone and rostral migratory stream, but reelin, produced by mitral cells in the olfactory bulb, is a higher affinity ligand and subsequently directs neuroblast dissociation, allowing them to transition to radial migration [143]. Of the two ligands, only reelin activates the proteasomal degradation of Disabled-1, which is necessary for neuroblast dissociation [141].
There is no evidence that reelin signalling interacts with LRP1 or LRP2. However, thrombospondins are known to interact with membrane proteins such as integrins, CD47, CD36, proteoglycans, and LRP1. Thrombospondin-1 has been shown to interact with LRP1 in combination with calreticulin to promote the focal adhesion of mature oligodendrocytes [144] and microglia [145] but has not been demonstrated to regulate neuroblast migration.
4.2. LRPs, Neuroblast Migration, and Neuronal Development
LRP2 regulates neuroblast migration indirectly. In vitro LRP2 and caveolins are expressed by astrocytes and work together to bind and endocytose albumin [40, 146]. This is significant, as albumin uptake activates the transcription factor sterol regulatory binding element protein 1, inducing expression of stearoyl-coA 9-desaturase-1, the key enzyme required for synthesis of the neurotrophic factor oleic acid [147]. In the lateral periventricular zone of the developing rat brain, oleic acid production regulates neuronal growth, migration, axon generation, and early synaptogenesis [148, 149], with the major neurotrophic effect being mediated by the downstream effectors PAR-α, protein kinase A, and neuro D2 [150]. When stearoyl-coA 9-desaturase-1 is knocked down in lateral periventricular explant cultures, albumin-mediated neuroblast migration is essentially prevented [148].
Once neuroblasts stop migrating, their journey is far from over. The immature neurons extend an axonal process to commence formation of the circuitry of the nervous system. The extending axons are tipped with a growth cone, which navigates the extracellular matrix, guiding the axon to its target cell to ultimately form a synapse [151]. A growth cone comprises membranous, receptor-rich, fan-shaped lamellipodia that extend along finger-like projections known as filopodia. The growth cone cytoskeleton is comprised of closely interacting microtubules and filamentous and globular actin [152–154]. Bundles of filamentous actin give structure to the filopodia, as does the cross-linked filamentous actin along the lamellipodial leading edge [152, 154, 155]. Microtubules are arranged as parallel bundles along the axon and splay outwards within the growth cone, providing structure and transport for proteins and organelles [156].
Growth cones are fitted with an elaborate suite of receptors that allow for the simultaneous integration of a multitude of chemotactic cues [157]. Binding of a chemotactic factor to its specific receptor/s on the growth cone membrane induces an intracellular signalling cascade which manipulates the cytoskeletal elements and dictates whether the response of the growth cone culminates in turning, extension, stasis, retraction, collapse, or bifurcation [154]. Well-defined receptors for chemotactic signals include the Eph family of receptor tyrosine kinases, Neuropilin, Roundabout, Deleted in Colorectal Cancer, L1, and Plexins (reviewed in [158]).
LRP1 and LRP2 are highly expressed on the growth cones of developing neurons in vitro and have been shown to signal in a codependent manner to promote chemotactic axon guidance within developmental neurons in vitro [159]. Together, LRP1 and LRP2 act as chemotactic receptors for a variety of ligands, including metallothioneins and tissue-type plasminogen activator [159]. Metallothioneins are small, highly conserved, inducible heavy metal binding proteins that are avid scavengers of reactive oxygen species [160]. Metallothioneins I and II are widely expressed in the nervous system and elsewhere. They differ by only a few amino acids and appear to have redundant functions. Metallothionein III is highly expressed in the brain, while metallothionein IV appears to be absent from the nervous system [161]. In cultured growth cones from sensory neurons, the activation of LRP1 and LRP2 by metallothionein II stimulated chemoattraction, resulting in growth cones turning towards the source of metallothionein II [159]. Metallothionein III had the opposite effect and induced chemorepulsion. Other LRP1 ligands, such as α2-macroglobulin, and tissue-type plasminogen activator also induced chemorepulsion [159]. The opposing responses induced by different LRP1 ligands are thought to result from differential activation of downstream signaling pathways, with metallothionein II activating Ca2+/calmodulin-dependent protein kinase and other receptors such as the tropomyosin-related kinase A receptor in complex signaling hubs (see Figure 2(d)).
Various LRP ligands have also been shown to alter neurite outgrowth. For example, metallothionein I/II signalling has been shown to transiently activate Akt and ERK, which belong to the mitogen-activated protein kinase and the phosphoinositide-3 kinase/Akt intracellular signalling pathways [162]. Myelin associated glycoprotein, an established chemorepulsive molecule, is known to interact with LRP1 [163] to inhibit axonal outgrowth and induce growth cone collapse [164, 165]. In vitro experiments have demonstrated that myelin associated glycoprotein and LRP1 form a complex with the p75 neurotrophin receptor, to activate RhoA [163], a potent mediator of growth cone collapse and axon retraction [166]. Additionally apolipoprotein E-containing lipoproteins are secreted by astrocytes and have been shown to bind LRP1 on the surface of immature neurons to promote neurite outgrowth generally, without having an effect on directionality [167]. The complexity of LRP signaling interactions in immature neurons remains to be fully deciphered but appear to be context- and ligand-dependent [168].
Mice in which Lrp1 is selectively deleted from neurons exhibit prominent tremor and dystonia, behavioural abnormalities, hyperactivity, motor dysfunction, age-dependent dendritic spine degeneration, synapse loss, neuroinflammation, memory loss, eventual neurodegeneration, and premature death [169–171], clearly demonstrating that LRP1 is crucial to neuronal function. LRP1 is also found postsynaptically, where it can interact with NMDA receptors in vitro, via the intracellular scaffold postsynaptic density protein 95 [169, 172]. LRP1 is able to influence the activity of NMDA receptors and regulate their distribution and internalisation [168, 173, 174], as well as the NMDA-induced internalisation of the AMPA receptor subunit GluR1 [174]. The very nature of this LRP1/NMDA receptor relationship suggests that LRP1 plays an integral role in neurotransmitter-induced calcium signalling, particularly in synaptic plasticity [173, 174].
LRP8 also regulates synaptic plasticity [128, 175]. LRP8 activation, by the addition of reelin to primary mouse cortical neuron cultures, triggers its proteolytic cleavage by γ-secretase. The liberated intracellular domain translocates to the nucleus, along with phosphorylated CREB to enhance the transcription of genes associated with learning and memory [176]. Furthermore, the ability of neurons to produce ATP for synaptic transmission may be tied to LRP1, as cultured neurons lacking Lrp1 have reduced expression of the glutamate transporters GLUT3 and GLUT4 [177].
5. LRP1 and LRP2 as Regulators of Oligodendrocyte Progenitor Cell Function
OPCs, also known as NG2 glia, are a proliferative, immature cell type found in the developing and adult CNS [178, 179]. OPCs can be identified by their expression of specific proteins such as the NG2 proteoglycan [180] and PDGFRα [181]. During the early stages of embryonic development, OPCs are produced from radial glia in the neuroepithelium of the developing brain and spinal cord [182, 183]. In the mouse spinal cord, OPC generation commences from the ventral pMN domain at E12.5 [182, 184]. The pMN domain is named for its role in generating spinal cord motor neurons and is defined by the expression of two transcription factors, OLIG1 and OLIG2 [185], both of which are highly expressed by OPCs and necessary for their generation and subsequent differentiation [186, 187]. Olig1/2 expression by pMN domain neural stem cells is induced by a gradient of ventrally secreted sonic hedgehog, suggesting that specification of this domain would also be LRP1/2-dependant. In the absence of Olig1/2, stem cells in the pMN domain instead form V2 interneurons and astrocytes [188]. Shortly after their birth, OPCs differentiate into myelinating oligodendrocytes in the spinal cord grey and white matter [183, 189]. It is estimated that approximately 85% of all spinal cord oligodendrocytes originate from the pMN domain, but other domains such as the P3 domain [184] and more dorsal domains [190, 191] also produce OPCs, just slightly later in response to different spatiotemporal cues.
Like spinal cord OPCs, forebrain OPCs have multiple origins. They are generated and migrate in three distinct waves [192]. The initial wave commences in the medial ganglionic eminence and the anterior entopeduncular area at E12.5 in mice. The OPCs migrate from their ventral origins to populate all regions of the developing brain, including the developing cortex [96]. The next wave of OPCs is initiated at E15.5 from the lateral- and caudal-ganglionic eminence, followed by the third and final wave from the cortical neuroepithelium [192]. OPCs derived from the initial wave are lost shortly after birth [192] and the function performed by these temporary OPCs and the signals rendering them susceptible to developmental removal are still unknown. By P13, ~80% of oligodendrocyte-lineage cells in the corpus callosum originate from the cortical neuroepithelium, and the remainder originate from the lateral ganglionic eminence [191]. All OPCs that populate the optic nerve arise from the preoptic area [193].
LRP1 may be a critical regulator of OPC behaviour, as recent microarray [194] and RNA sequencing [195] data indicate that Lrp1 mRNA is highly expressed by OPCs in the early postnatal mouse brain. However expression of this gene is rapidly downregulated upon differentiation and is barely detectable in oligodendrocytes. The role of LRP2 in regulating this lineage is more clearly established.
5.1. LRP2 Regulates OPC Proliferation and Migration during Development
One of the signalling molecules regulating OPC proliferation and migration is sonic hedgehog [196, 197], and LRP2 appears to regulate OPC proliferation and migration by modulating sonic hedgehog availability and contributing to the generation of a concentration gradient. In the developing mouse optic nerve, LRP2 is highly expressed by astrocytes [198]. However, LRP2 expression is not homogeneous, being highest in the caudal optic nerve at E14.5, but then changing to be highest in the rostral optic nerve at E16.5. Blocking LRP2 signalling by optic nerve astrocytes leads to a significant reduction in OPC proliferation and migration [198]. In vitro studies suggest that the LRP2-mediated uptake and release of sonic hedgehog by astrocytes promotes OPC proliferation and act as a chemoattractant directing their migration [198]. The temporal regulation of LRP2 expression in the caudal versus rostral regions of the optic nerve would be predicted to “trap” sonic hedgehog in the region being populated by OPCs at that time. The expression pattern of LRP2 in the postnatal optic nerve has not been characterised. However as LRP2 is expressed by mature oligodendrocytes in the postnatal spinal cord [199], it might also be upregulated by optic nerve OPCs upon differentiation.
5.2. How Might LRP1 Influence OPC Behaviour?
When examining LRP1 function in other cell types, there are a number of mechanisms by which LRP1 could feasibly influence OPC behaviour. For example OPC processes share some structural similarities with the growth cones of developing neurons [200, 201]. In particular growth cones comprise specialised cell membrane extensions called lamellipodia and filopodia, which also extend from the cellular processes of OPCs [200]. LRP1 signalling mediates the chemoattraction and chemorepulsion of growth cones in vitro, [159], so perhaps LRP1 could regulate OPC process guidance or even OPC migration. LRP1 is expressed by Schwann cells in vivo and regulates the migration and adhesion of immature Schwann cells in vitro by the activation and repression of two small Rho GTPases, Rac1 and RhoA, respectively [202]. Rac1 activation stimulates the formation of peripheral lamellae by actin remodelling in the leading process [203]. Lrp1 knockdown decreases Rac1 activation and increases RhoA activation, which in turn increases cell adhesion and prevents migration [202]. This is of particular interest, as OPCs take on a bipolar morphology when migrating [201], and their movement has been attributed to the NG2-dependent regulation of small Rho GTPases and polarity complex proteins [204].
LRP1 also has the potential to influence OPC migration by acting as a coreceptor for PDGFRα signalling, in a similar way that it promotes fibroblast migration by cosignalling with PDGFRβ. When PDGFBB binds to PDGFRβ on the surface of cultured mouse embryonic fibroblasts, it induces migration. However this involves the association of LRP1 with PDGFRβ [205, 206]. The two receptors are internalised and colocalize in the endosomal compartment, where the kinase domain of PDGFRβ phosphorylates the distal NPXY motif of LRP1 [65, 205, 207]. Once phosphorylated, LRP1 has an increased affinity for the intracellular domain for SHP-2 [206, 208], outcompeting PDGFRβ for this interaction, and preventing further activation of downstream signalling pathways [206]. While OPCs do not express PDGFRβ, they express high levels of the related receptor, PDGFRα, which is also internalised following ligand binding [209], suggesting an association with an unidentified endocytic receptor which we propose could be LRP1. PDGFAA is known to bind to PDGFRα on the surface of OPCs and activate a phosphorylation cascade involving the Fyn tyrosine kinase and cyclin-dependant kinase 5 [210], a known regulator of the actin cytoskeleton in neurons [211]. By interacting with PDGFRα it is feasible that LRP1 could promote not only OPC migration but also proliferation and cell survival [181, 210, 212, 213]. While the signalling mechanism is likely to be different, a role for LRP1 in regulating cell survival is not unprecedented, as LRP1 has been shown to protect Schwann cells against TNFα-induced cell death in a sciatic nerve crush injury model in vivo and in vitro [214].
LRP1 could equally influence OPC migration by regulating lipid availability within the cell, as the establishment of cell polarity and movement of the leading edge during migration is dependent on the availability of cholesterol [215, 216]. Most lipid-carrying proteins cannot cross the blood brain barrier and therefore must be generated within the CNS. Apolipoprotein E is secreted by astrocytes and functions as an effective lipid transport protein and can bind LRP1 [217, 218]. Lipoproteins form noncovalent aggregates with triglycerides, phospholipids, and cholesterol esters before they bind to specific receptors and are internalised and utilized by the cell [219]. Upon binding of apolipoprotein E to LRP1, the complex is internalised where its lipid content is discharged, making it available to the cell [220], before apolipoprotein E is resecreted [221]. Once internalised, lipoproteins may be utilized by OPCs for a number of functions.
LRP1-mediated lipid uptake may alternatively allow OPCs to sustain their postsynaptic connections with neurons. Forebrain neuron-specific Lrp1 gene knockout mice have severe deficiencies in lipid metabolism and show synapse loss [171]. The presynaptic use of cholesterol by neurons is high, due to the requirements of lipid-rich neurotransmitter vesicles [222]. However, the postsynaptic cell also utilizes cholesterol for receptor recycling in and out of the postsynaptic membrane. Therefore, cholesterol uptake into OPCs may be critical for the formation of axon-OPC synapses and maintenance of the OPC postsynaptic density.
6. Conclusions and Outlook
Our knowledge of LRP1 and LRP2 processing and trafficking has come a long way in the past decade. Without even considering the possibility that cleaved forms of these proteins may regulate gene transcription or perform dominant negative signalling functions, a growing number of studies clearly indicate that LRP1 and LRP2 perform a diverse range of cellular functions in neural stem and progenitor cell populations. The generation of conditional knockout mice has now made it possible to perform the detailed studies that will be necessary to understand the role of LRP1 and LRP2 in each immature cell type, across a variety of developmental stages. This is particularly critical now that we understand that LRP1 and LRP2 can influence the balance of growth factor and morphogen signalling, making them critical spatial and temporal regulators of neural development.
Acknowledgments
The authors would like to thank their colleagues at the University of Tasmania for helpful feedback and comments on this manuscripts. Work in the Young and Foa laboratories was supported by the National Health and Medical Research Council of Australia and the National Stem Cell Foundation of Australia. Mr. Loic Auderset was supported by a Morrell Family Trust Scholarship in Medical Research.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Brown M. S., Goldstein J. L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Binder R. J., Han D. K., Srivastava P. K. CD91: a receptor for heat shock protein gp96. Nature Immunology. 2000;1(2):151–155. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 3.Liu C.-X., Musco S., Lisitsina N. M., Yaklichkin S. Y., Lisitsyn N. A. Genomic organization of a new candidate tumor suppressor gene, LRP1B. Genomics. 2000;69(2):271–274. doi: 10.1006/geno.2000.6331. [DOI] [PubMed] [Google Scholar]
- 4.Marschang P., Brich J., Weeber E. J., et al. Normal development and fertility of knockout mice lacking the tumor suppressor gene LRP1b suggest functional compensation by LRP1. Molecular and Cellular Biology. 2004;24(9):3782–3793. doi: 10.1128/mcb.24.9.3782-3793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito A., Pietromonaco S., Loo A., et al. Complete cloning and sequencing of rat Gp330 megalin, a distinctive member of the low-density-lipoprotein receptor gene family. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(21):9725–9729. doi: 10.1073/pnas.91.21.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hey P. J., Twells R. C. J., Phillips M. S., et al. Cloning of a novel member of the low-density lipoprotein receptor family. Gene. 1998;216(1):103–111. doi: 10.1016/s0378-1119(98)00311-4. [DOI] [PubMed] [Google Scholar]
- 7.Brown S. D., Twells R. C. J., Hey P. J., et al. Isolation and characterization of LRP6, a novel member of the low density lipoprotein receptor gene family. Biochemical and Biophysical Research Communications. 1998;248(3):879–888. doi: 10.1006/bbrc.1998.9061. [DOI] [PubMed] [Google Scholar]
- 8.Riddell D. R., Vinogradov D. V., Stannard A. K., Chadwick N., Owen J. S. Identification and characterization of LRP8 (apoER2) in human blood platelets. Journal of Lipid Research. 1999;40(10):1925–1930. [PubMed] [Google Scholar]
- 9.Daly N. L., Scanlon M. J., Djordjevic J. T., Kroon P. A., Smith R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(14):6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llorente-Cortés V., Costales P., Bernués J. B., Camino-Lopez S., Badimon L. Sterol regulatory element-binding protein-2 negatively regulates low density lipoprotein receptor-related protein transcription. Journal of Molecular Biology. 2006;359(4):950–960. doi: 10.1016/j.jmb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Castellano J., Aledo R., Sendra J., et al. Hypoxia stimulates low-density lipoprotein receptor-related protein-1 expression through hypoxia-inducible factor-1α in human vascular smooth muscle cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31(6):1411–1420. doi: 10.1161/atvbaha.111.225490. [DOI] [PubMed] [Google Scholar]
- 12.Grana T. R., Lamarre J., Kalisch B. E. Nerve growth factor-mediated regulation of low density lipoprotein receptor-related protein promoter activation. Cellular and Molecular Neurobiology. 2013;33(2):269–282. doi: 10.1007/s10571-012-9894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamanaka Y., Faghihi M., Magistri M., Alvarez-Garcia O., Lotz M., Wahlestedt C. Antisense RNA controls LRP1 sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Reports. 2015;11(6):967–976. doi: 10.1016/j.celrep.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudenko G., Henry L., Henderson K., et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298(5602):2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 15.Willnow T. E., Armstrong S. A., Hammer R. E., Herz J. Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(10):4537–4541. doi: 10.1073/pnas.92.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bu G. J., Marzolo M. P. Role of RAP in the biogenesis of lipoprotein receptors. Trends in Cardiovascular Medicine. 2000;10(4):148–155. doi: 10.1016/s1050-1738(00)00045-1. [DOI] [PubMed] [Google Scholar]
- 17.Croy J. E., Shin W. D., Knauer M. F., Knauer D. J., Komives E. A. All three LDL receptor homology regions of the LDL receptor-related protein bind multiple ligands. Biochemistry. 2003;42(44):13049–13057. doi: 10.1021/bi034752s. [DOI] [PubMed] [Google Scholar]
- 18.Reekmans S. M., Pflanzner T., Gordts P. L. S. M., et al. Inactivation of the proximal NPXY motif impairs early steps in LRP1 biosynthesis. Cellular and Molecular Life Sciences. 2010;67(1):135–145. doi: 10.1007/s00018-009-0171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D., Walsh J. D., Mikhailenko I., et al. RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi. Molecular Cell. 2006;22(3):423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Bu G., Geuze H. J., Strous G. J., Schwartz A. L. 39-kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. The EMBO Journal. 1995;14(10):2269–2280. doi: 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willnow T. E., Moehring J. M., Inocencio N. M., Moehring T. J., Herz J. The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochemical Journal. 1996;313(1):71–76. doi: 10.1042/bj3130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czekay R.-P., Orlando R. A., Woodward L., Lundstrom M., Farquhar M. G. Endocytic trafficking of megalin/RAP complexes: dissociation of the complexes in late endosomes. Molecular Biology of the Cell. 1997;8(3):517–532. doi: 10.1091/mbc.8.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Arnim C. A. F., Kinoshita A., Peltan I. D., et al. The low density lipoprotein receptor-related protein (LRP) is a novel beta-secretase (BACE1) substrate. The Journal of Biological Chemistry. 2005;280(18):17777–17785. doi: 10.1074/jbc.m414248200. [DOI] [PubMed] [Google Scholar]
- 24.Selvais C., D'Auria L., Tyteca D., et al. Cell cholesterol modulates metalloproteinase-dependent shedding of low-density lipoprotein receptor-related protein-1 (LRP-1) and clearance function. The FASEB Journal. 2011;25(8):2770–2781. doi: 10.1096/fj.10-169508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn K. A., Pye V. J., Dai Y.-P., Chesterman C. N., Owensby D. A. Characterization of the soluble form of the low density lipoprotein receptor-related protein (LRP) Experimental Cell Research. 1999;251(2):433–441. doi: 10.1006/excr.1999.4590. [DOI] [PubMed] [Google Scholar]
- 26.Quinn K. A., Grimsley P. G., Dai Y.-P., Tapner M., Chesterman C. N., Owensby D. A. Soluble low density lipoprotein receptor-related protein (LRP) circulates in human plasma. The Journal of Biological Chemistry. 1997;272(38):23946–23951. doi: 10.1074/jbc.272.38.23946. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q., Zhang J., Tran H., et al. LRP1 shedding in human brain: roles of ADAM10 and ADAM17. Molecular Neurodegeneration. 2009;4(1, article 17) doi: 10.1186/1750-1326-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spuch C., Antequera D., Pascual C., et al. Soluble megalin is reduced in cerebrospinal fluid samples of Alzheimer’s disease patients. Frontiers in Cellular Neuroscience. 2015;9, article 134 doi: 10.3389/fncel.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah M., Baterina O. Y., Jr., Taupin V., Farquhar M. ARH directs megalin to the endocytic recycling compartment to regulate its proteolysis and gene expression. The Journal of Cell Biology. 2013;202(1):113–127. doi: 10.1083/jcb.201211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biemesderfer D. Regulated intramembrane proteolysis of megalin: linking urinary protein and gene regulation in proximal tubule? Kidney International. 2006;69(10):1717–1721. doi: 10.1038/sj.ki.5000298. [DOI] [PubMed] [Google Scholar]
- 31.May P., Krishna Reddy Y., Herz J. Proteolytic processing of low density lipoprotein receptor-related protein mediates regulated release of its intracellular domain. The Journal of Biological Chemistry. 2002;277(21):18736–18743. doi: 10.1074/jbc.m201979200. [DOI] [PubMed] [Google Scholar]
- 32.Chen W.-J., Goldstein J. L., Brown M. S. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. The Journal of Biological Chemistry. 1990;265(6):3116–3123. [PubMed] [Google Scholar]
- 33.Li Y., Marzolo M. P., van Kerkhof P., Strous G. J., Bu G. The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal for low density lipoprotein receptor-related protein. Journal of Biological Chemistry. 2000;275(22):17187–17194. doi: 10.1074/jbc.m000490200. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., van Kerkhof P., Marzolo M. P., Strous G. J., Bu G. Identification of a major cyclic AMP-dependent protein kinase A phosphorylation site within the cytoplasmic tail of the low-density lipoprotein receptor-related protein: implication for receptor-mediated endocytosis. Molecular and Cellular Biology. 2001;21(4):1185–1195. doi: 10.1128/mcb.21.4.1185-1195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oleinikov A. V., Zhao J., Makker S. P. Cytosolic adaptor protein Dab2 is an intracellular ligand of endocytic receptor gp600/megalin. Biochemical Journal. 2000;347(3):613–621. doi: 10.1042/0264-6021:3470613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai J., Christensen E. I., Morris S. M., Willnow T. E., Cooper J. A., Nielsen R. Mutually dependent localization of megalin and Dab2 in the renal proximal tubule. American Journal of Physiology—Renal Physiology. 2005;289(3):F569–F576. doi: 10.1152/ajprenal.00292.2004. [DOI] [PubMed] [Google Scholar]
- 37.Traub L. M. Tickets to ride: Selecting cargo for clathrin-regulated internalization. Nature Reviews Molecular Cell Biology. 2009;10(9):583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- 38.Chetrit D., Barzilay L., Horn G., Bielik T., Smorodinsky N. I., Ehrlich M. Negative regulation of the endocytic adaptor disabled-2 (Dab2) in mitosis. The Journal of Biological Chemistry. 2011;286(7):5392–5403. doi: 10.1074/jbc.m110.161851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff N. A., Lee W.-K., Abouhamed M., Thévenod F. Role of ARF6 in internalization of metal-binding proteins, metallothionein and transferrin, and cadmium-metallothionein toxicity in kidney proximal tubule cells. Toxicology and Applied Pharmacology. 2008;230(1):78–85. doi: 10.1016/j.taap.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Bento-Abreu A., Velasco A., Polo-Hernández E., et al. Albumin endocytosis via megalin in astrocytes is caveola- and Dab-1 dependent and is required for the synthesis of the neurotrophic factor oleic acid. Journal of Neurochemistry. 2009;111(1):49–60. doi: 10.1111/j.1471-4159.2009.06304.x. [DOI] [PubMed] [Google Scholar]
- 41.Aranda J. F., Canfran-Duque A., Goedeke L., Suarez Y., Fernandez-Hernando C. The miR-199-dynamin regulatory axis controls receptor-mediated endocytosis. Journal of Cell Science. 2015;128(17):3197–3209. doi: 10.1242/jcs.165233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon H., Meng W., Takagi J., Eck M. J., Springer T. A., Blacklow S. C. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nature Structural & Molecular Biology. 2001;8(6):499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- 43.Donoso M., Cancino J., Lee J., et al. Polarized traffic of LRP1 involves AP1B and SNX17 operating on Y-dependent sorting motifs in different pathways. Molecular Biology of the Cell. 2009;20(1):481–497. doi: 10.1091/mbc.e08-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farfán P., Lee J., Larios J., Sotelo P., Bu G., Marzolo M.-P. A sorting nexin 17-binding domain within the LRP1 cytoplasmic tail mediates receptor recycling through the basolateral sorting endosome. Traffic. 2013;14(7):823–838. doi: 10.1111/tra.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko K. W. S., McLeod R. S., Avramoglu R. K., et al. Mutation at the processing site of chicken low density lipoprotein receptor-related protein impairs efficient endoplasmic reticulum exit, but proteolytic cleavage is not essential for its endocytic functions. The Journal of Biological Chemistry. 1998;273(43):27779–27785. doi: 10.1074/jbc.273.43.27779. [DOI] [PubMed] [Google Scholar]
- 46.Nagai M., Meerloo T., Takeda T., Farquhar M. G. The adaptor protein ARH escorts megalin to and through endosomes. Molecular Biology of the Cell. 2003;14(12):4984–4996. doi: 10.1091/mbc.e03-06-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehtonen S., Shah M., Nielsen R., et al. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Molecular Biology of the Cell. 2008;19(7):2949–2961. doi: 10.1091/mbc.E07-05-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuseff M. I., Farfán P., Bu G., Marzolo M.-P. A cytoplasmic PPPSP motif determines megalin's phosphorylation and regulates receptor's recycling and surface expression. Traffic. 2007;8(9):1215–1230. doi: 10.1111/j.1600-0854.2007.00601.x. [DOI] [PubMed] [Google Scholar]
- 49.Spuch C., Ortolano S., Navarro C. LRP-1 and LRP-2 receptors function in the membrane neuron. Trafficking mechanisms and proteolytic processing in alzheimer's disease. Frontiers in Physiology. 2012;3, article 269 doi: 10.3389/fphys.2012.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herz J., Strickland D. K. LRP: a multifunctional scavenger and signaling receptor. Journal of Clinical Investigation. 2001;108(6):779–784. doi: 10.1172/jci200113992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marzolo M.-P., Farfán P. New insights into the roles of megalin/LRP2 and the regulation of its functional expression. Biological Research. 2011;44(1):89–105. doi: 10.4067/s0716-97602011000100012. [DOI] [PubMed] [Google Scholar]
- 52.Willnow T. E., Goldstein J. L., Orth K., Brown M. S., Herz J. Low density lipoprotein receptor-related protein and gp330 bind similar ligands, including plasminogen activator-inhibitor complexes and lactoferrin, an inhibitor of chylomicron remnant clearance. The Journal of Biological Chemistry. 1992;267(36):26172–26180. [PubMed] [Google Scholar]
- 53.Bu G., Maksymovitch E. A., Schwartz A. L. Receptor-mediated endocytosis of tissue-type plasminogen activator by low density lipoprotein receptor-related protein on human hepatoma HepG2 cells. The Journal of Biological Chemistry. 1993;268(17):13002–13009. [PubMed] [Google Scholar]
- 54.Grobmyer S. R., Kuo A., Orishimo M., Okada S. S., Cines D. B., Barnathan E. S. Determinants of binding and internalization of tissue-type plasminogen activator by human vascular smooth muscle and endothelial cells. The Journal of Biological Chemistry. 1993;268(18):13291–13300. [PubMed] [Google Scholar]
- 55.Lin S., Racz J., Tai M. F., et al. A role for low density lipoprotein receptor-related protein 1 in the cellular uptake of tissue plasminogen activator in the lungs. Pharmaceutical Research. 2016;33(1):72–82. doi: 10.1007/s11095-015-1763-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ambjørn M., Asmussen J. W., Lindstam M., et al. Metallothionein and a peptide modeled after metallothionein, EmtinB, induce neuronal differentiation and survival through binding to receptors of the low-density lipoprotein receptor family. Journal of Neurochemistry. 2008;104(1):21–37. doi: 10.1111/j.1471-4159.2007.05036.x. [DOI] [PubMed] [Google Scholar]
- 57.Hanover J. A., Willingham M. C., Pastan I. Receptor-mediated endocytosis of α 2-macroglobulin: solubilization and partial purification of the fibroblast α 2-macroglobulin receptor. Annals of the New York Academy of Sciences. 1983;421:410–423. doi: 10.1111/j.1749-6632.1983.tb18135.x. [DOI] [PubMed] [Google Scholar]
- 58.Marynen P., Van Leuven F., Cassiman J.-J., Van den Berghe H. Solubilization and affinity purification of the α2-macroglobulin receptor from human fibroblasts. The Journal of Biological Chemistry. 1984;259(11):7075–7079. [PubMed] [Google Scholar]
- 59.Parkyn C. J., Vermeulen E. G. M., Mootoosamy R. C., et al. LRP1 controls biosynthetic and endocytic trafficking of neuronal prion protein. Journal of Cell Science. 2008;121(6):773–783. doi: 10.1242/jcs.021816. [DOI] [PubMed] [Google Scholar]
- 60.Sousa M. M., Berglund L., Saraiva M. J. Transthyretin in high density lipoproteins: association with apolipoprotein A-I. Journal of Lipid Research. 2000;41(1):58–65. [PubMed] [Google Scholar]
- 61.Nykjaer A., Dragun D., Walther D., et al. An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3 . Cell. 1999;96(4):507–515. doi: 10.1016/s0092-8674(00)80655-8. [DOI] [PubMed] [Google Scholar]
- 62.Willnow T. E., Christ A., Hammes A. Endocytic receptor-mediated control of morphogen signaling. Development. 2012;139(23):4311–4319. doi: 10.1242/dev.084467. [DOI] [PubMed] [Google Scholar]
- 63.Trommsdorff M., Borg J.-P., Margolis B., Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. The Journal of Biological Chemistry. 1998;273(50):33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 64.Boucher P., Liu P., Gotthardt M., Hiesberger T., Anderson R. G. W., Herz J. Platelet-derived growth factor mediates tyrosine phosphorylation of the cytoplasmic domain of the low density lipoprotein receptor-related protein in caveolae. The Journal of Biological Chemistry. 2002;277(18):15507–15513. doi: 10.1074/jbc.m200428200. [DOI] [PubMed] [Google Scholar]
- 65.Loukinova E., Ranganathan S., Kuznetsov S., et al. Platelet-derived growth factor (PDGF)-induced tyrosine phosphorylation of the low density lipoprotein receptor-related protein (LRP). Evidence for integrated co-receptor function between LRP and the PDGF. The Journal of Biological Chemistry. 2002;277(18):15499–15506. doi: 10.1074/jbc.m200427200. [DOI] [PubMed] [Google Scholar]
- 66.Shi Y., Mantuano E., Inoue G., Campana W. M., Gonias S. L. Ligand binding to LRP1 transactivates trk receptors by a src family kinase-dependent pathway. Science Signaling. 2009;2(68, article ra18) doi: 10.1126/scisignal.2000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pietrzik C. U., Yoon I.-S., Jaeger S., Busse T., Weggen S., Koo E. H. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. Journal of Neuroscience. 2004;24(17):4259–4265. doi: 10.1523/jneurosci.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woldt E., Matz R. L., Terrand J., et al. Differential signaling by adaptor molecules LRP1 and ShcA regulates adipogenesis by the insulin-like growth factor-1 receptor. The Journal of Biological Chemistry. 2011;286(19):16775–16782. doi: 10.1074/jbc.m110.212878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herz J., Clouthier D. E., Hammer R. E. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71(3):411–421. doi: 10.1016/0092-8674(92)90511-A. [DOI] [PubMed] [Google Scholar]
- 70.Willnow T. E., Hilpert J., Armstrong S. A., et al. Defective forebrain development in mice lacking gp330/megalin. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(16):8460–8464. doi: 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kantarci S., Al-Gazali L., Hill R., et al. Mutations in LRP2, coding for the multi-ligand receptor megalin, cause Donnai-Barrow and Faciooculoacousticorenal (FOAR) syndromes. European Journal of Human Genetics. 2007;15(supplement 1):p. 3. [Google Scholar]
- 72.Wolf B. B., Lopes M. B. S., VandenBerg S. R., Gonias S. L. Characterization and immunohistochemical localization of α2-macroglobulin receptor (low-density lipoprotein receptor-related protein) in human brain. American Journal of Pathology. 1992;141(1):37–42. [PMC free article] [PubMed] [Google Scholar]
- 73.Bu G., Maksymovitch E. A., Nerbonne J. M., Schwartz A. L. Expression and function of the low-density-lipoprotein receptor-related protein (LRP) in mammalian central neurons. The Journal of Biological Chemistry. 1994;269(28):18521–18528. [PubMed] [Google Scholar]
- 74.Gaultier A., Wu X., Le Moan N., et al. Low-density lipoprotein receptor-related protein 1 is an essential receptor for myelin phagocytosis. Journal of Cell Science. 2009;122(part 8):1155–1162. doi: 10.1242/jcs.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spoelgen R., Hammes A., Anzenberger U., et al. LRP2/megalin is required for patterning of the ventral telencephalon. Development. 2005;132(2):405–414. doi: 10.1242/dev.01580. [DOI] [PubMed] [Google Scholar]
- 76.Assémat E., Châtelet F., Chandellier J., et al. Overlapping expression patterns of the multiligand endocytic receptors cubilin and megalin in the CNS, sensory organs and developing epithelia of the rodent embryo. Gene Expression Patterns. 2005;6(1):69–78. doi: 10.1016/j.modgep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 77.Chun J. T., Wang L., Pasinetti G. M., Finch C. E., Zlokovic B. V. Glycoprotein 330/megalin (LRP-2) has low prevalence as mRNA and protein in brain microvessels and choroid plexus. Experimental Neurology. 1999;157(1):194–201. doi: 10.1006/exnr.1999.7052. [DOI] [PubMed] [Google Scholar]
- 78.Gajera C. R., Emich H., Lioubinski O., et al. LRP2 in ependymal cells regulates BMP signaling in the adult neurogenic niche. Journal of Cell Science. 2010;123(11):1922–1930. doi: 10.1242/jcs.065912. [DOI] [PubMed] [Google Scholar]
- 79.Wicher G., Larsson M., Rask L., Aldskogius H. Low-density lipoprotein receptor-related protein (LRP)-2/megalin is transiently expressed in a subpopulation of neural progenitors in the embryonic mouse spinal cord. Journal of Comparative Neurology. 2005;492(2):123–131. doi: 10.1002/cne.20673. [DOI] [PubMed] [Google Scholar]
- 80.Pollen A. A., Nowakowski T. J., Chen J., et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163(1):55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kessaris N., Pringle N., Richardson W. D. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363(1489):71–85. doi: 10.1098/rstb.2006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Young K. M., Fogarty M., Kessaris N., Richardson W. D. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. The Journal of Neuroscience. 2007;27(31):8286–8296. doi: 10.1523/jneurosci.0476-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Rourke M., Gasperini R., Young K. M. Adult myelination: wrapping up neuronal plasticity. Neural Regeneration Research. 2014;9(13):1261–1264. doi: 10.4103/1673-5374.137571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Young K. M., Mitsumori T., Pringle N., Grist M., Kessaris N., Richardson W. D. An Fgfr3-iCreER T2 transgenic mouse line for studies of neural stem cells and astrocytes. Glia. 2010;58(8):943–953. doi: 10.1002/glia.20976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Capurro M. I., Shi W., Filmus J. LRP1 mediates the Shh-induced endocytosis of the GPC3-Shh complex. Journal of Cell Science. 2012;125(part 14):3380–3389. doi: 10.1242/jcs.098889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCarthy R. A., Barth J. L., Chintalapudi M. R., et al. Megalin functions as an endocytic sonic hedgehog receptor. The Journal of Biological Chemistry. 2002;277(28):25660–25667. doi: 10.1074/jbc.m201933200. [DOI] [PubMed] [Google Scholar]
- 87.Morales C. R., Zeng J., El Alfy M., et al. Epithelial trafficking of Sonic hedgehog by megalin. Journal of Histochemistry and Cytochemistry. 2006;54(10):1115–1127. doi: 10.1369/jhc.5A6899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choudhry Z., Rikani A. A., Choudhry A. M., et al. Sonic hedgehog signalling pathway: a complex network. Annals of Neurosciences. 2014;21(1):1–4. doi: 10.5214/ans.0972.7531.210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christ A., Christa A., Kur E., et al. LRP2 is an auxiliary SHH receptor required to condition the forebrain ventral midline for inductive signals. Developmental Cell. 2012;22(2):268–278. doi: 10.1016/j.devcel.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 90.Fuccillo M., Joyner A. L., Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nature Reviews Neuroscience. 2006;7(10):772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 91.Wicher G., Aldskogius H. Megalin deficiency induces critical changes in mouse spinal cord development. NeuroReport. 2008;19(5):559–563. doi: 10.1097/WNR.0b013e3282f94267. [DOI] [PubMed] [Google Scholar]
- 92.Poncet C., Soula C., Trousse F., et al. Induction of oligodendrocyte progenitors in the trunk neural tube by ventralizing signals: effects of notochord and floor plate grafts, and of sonic hedgehog. Mechanisms of Development. 1996;60(1):13–32. doi: 10.1016/s0925-4773(96)00595-3. [DOI] [PubMed] [Google Scholar]
- 93.Pringle N. P., Yu W.-P., Guthrie S., et al. Determination of neuroepithelial cell fate: induction of the oligodendrocyte lineage by ventral midline cells and sonic hedgehog. Developmental Biology. 1996;177(1):30–42. doi: 10.1006/dbio.1996.0142. [DOI] [PubMed] [Google Scholar]
- 94.Orentas D. M., Hayes J. E., Dyer K. L., Miller R. H. Sonic hedgehog signaling is required during the appearance of spinal cord oligodendrocyte precursors. Development. 1999;126(11):2419–2429. doi: 10.1242/dev.126.11.2419. [DOI] [PubMed] [Google Scholar]
- 95.Nery S., Wichterle H., Fishell G. Sonic hedgehog contributes to oligodendrocyte specification in the mammalian forebrain. Development. 2001;128(4):527–540. doi: 10.1242/dev.128.4.527. [DOI] [PubMed] [Google Scholar]
- 96.Tekki-Kessaris N., Woodruff R., Hall A. C., et al. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128(13):2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- 97.Cai J., Qi Y., Hu X., et al. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45(1):41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 98.Chandran S., Kato H., Gerreli D., Compston A., Svendsen C. N., Allen N. D. FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development. 2003;130(26):6599–6609. doi: 10.1242/dev.00871. [DOI] [PubMed] [Google Scholar]
- 99.Bolós M., Fernandez S., Torres-Aleman I. Oral administration of a GSK3 inhibitor increases brain insulin-like growth factor I levels. The Journal of Biological Chemistry. 2010;285(23):17693–17700. doi: 10.1074/jbc.m109.096594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pringle N. P., Yu W.-P., Howell M., Colvin J. S., Ornitz D. M., Richardson W. D. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003;130(1):93–102. doi: 10.1242/dev.00184. [DOI] [PubMed] [Google Scholar]
- 101.Wu Y. Y., Liu Y., Levine E. M., Rao M. S. Hes1 but not Hes5 regulates an astrocyte versus oligodendrocyte fate choice in glial restricted precursors. Developmental Dynamics. 2003;226(4):675–689. doi: 10.1002/dvdy.10278. [DOI] [PubMed] [Google Scholar]
- 102.Hennen E., Safina D., Haussmann U., et al. A LewisX glycoprotein screen identifies the low density lipoprotein receptor-related protein 1 (LRP1) as a modulator of oligodendrogenesis in mice. The Journal of Biological Chemistry. 2013;288(23):16538–16545. doi: 10.1074/jbc.m112.419812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zheng G., Bachinsky D. R., Stamenkovic I., et al. Organ distribution in rats of two members of the low-density lipoprotein receptor gene family, gp330 and LRP/alpha 2MR, and the receptor-associated protein (RAP) Journal of Histochemistry & Cytochemistry. 1994;42(4):531–542. doi: 10.1177/42.4.7510321. [DOI] [PubMed] [Google Scholar]
- 104.Zarbalis K., May S. R., Shen Y., Ekker M., Rubenstein J. L. R., Peterson A. S. A focused and efficient genetic screening strategy in the mouse: identification of mutations that disrupt cortical development. PLoS Biology. 2004;2(8):1177–1187. doi: 10.1371/journal.pbio.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lim D. A., Tramontin A. D., Trevejo J. M., Herrera D. G., García-Verdugo J. M., Alvarez-Buylla A. Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron. 2000;28(3):713–726. doi: 10.1016/S0896-6273(00)00148-3. [DOI] [PubMed] [Google Scholar]
- 106.Sabo J. K., Kilpatrick T. J., Cate H. S. Effects of bone morphogenic proteins on neural precursor cells and regulation during central nervous system injury. NeuroSignals. 2009;17(4):255–264. doi: 10.1159/000231892. [DOI] [PubMed] [Google Scholar]
- 107.Marzban H., Del Bigio M. R., Alizadeh J., Ghavami S., Zachariah R. M., Rastegar M. Cellular commitment in the developing cerebellum. Frontiers in Cellular Neuroscience. 2015;8, article 450 doi: 10.3389/fncel.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dahmane N., Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126(14):3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 109.Wechsler-Reya R. J., Scott M. P. Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron. 1999;22(1):103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 110.Haldipur P., Bharti U., Govindan S., et al. Expression of sonic hedgehog during cell proliferation in the human cerebellum. Stem Cells and Development. 2012;21(7):1059–1068. doi: 10.1089/scd.2011.0206. [DOI] [PubMed] [Google Scholar]
- 111.Vaillant C., Michos O., Orlicki S., et al. Protease nexin 1 and its receptor LRP modulate SHH signalling during cerebellar development. Development. 2007;134(9):1745–1754. doi: 10.1242/dev.02840. [DOI] [PubMed] [Google Scholar]
- 112.Stiles J., Jernigan T. L. The basics of brain development. Neuropsychology Review. 2010;20(4):327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parnavelas J. G. The origin and migration of cortical neurones: new vistas. Trends in Neurosciences. 2000;23(3):126–131. doi: 10.1016/s0166-2236(00)01553-8. [DOI] [PubMed] [Google Scholar]
- 114.Kriegstein A., Noctor S., Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nature Reviews Neuroscience. 2006;7(11):883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 115.Lavdas A. A., Grigoriou M., Pachnis V., Parnavelas J. G. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. Journal of Neuroscience. 1999;19(18):7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Letinic K., Zoncu R., Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417(6889):645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 117.Nery S., Fishell G., Corbin J. G. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nature Neuroscience. 2002;5(12):1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 118.Hastings N. B., Gould E. Rapid extension of axons into the CA3 region by adult-generated granule cells (vol 413, pg 146, 1999) Journal of Comparative Neurology. 1999;415(1):p. 144. doi: 10.1002/(sici)1096-9861(19991206)415:1<144::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 119.Doetsch F., Caillé I., Lim D. A., Garcia-Verdugo J. M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 120.Alberti S., Krause S. M., Kretz O., et al. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(17):6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tomasiewicz H., Ono K., Yee D. L., et al. Genetic deletion of a neural cell-adhesion molecule variant (N-CAM-180) produces distinct defects in the central-nervous-system. Neuron. 1993;11(6):1163–1174. doi: 10.1016/0896-6273(93)90228-j. [DOI] [PubMed] [Google Scholar]
- 122.Chazal G., Durbec P., Jankovski A., Rougon G., Cremer H. Consequences of neural cell adhesion molecule deficiency on cell migration in the rostral migratory stream of the mouse. The Journal of Neuroscience. 2000;20(4):1446–1457. doi: 10.1523/JNEUROSCI.20-04-01446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu H. Y., Tomasiewicz H., Magnuson T., Rutishauser U. The role of polysialic acid in migration of olfactory bulb interneuron precursors in the subventricular zone. Neuron. 1996;16(4):735–743. doi: 10.1016/s0896-6273(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 124.Nguyen-Ba-Charvet K. T., Picard-Riera N., Tessier-Lavigne M., Evercooren A. B.-V., Sotelo C., Chédotal A. Multiple roles for slits in the control of cell migration in the rostral migratory stream. The Journal of Neuroscience. 2004;24(6):1497–1506. doi: 10.1523/jneurosci.4729-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu W., Wong K., Chen J.-H., et al. Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature. 1999;400(6742):331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murase S.-I., Horwitz A. F. Deleted in colorectal carcinoma and differentially expressed integrins mediate the directional migration of neural precursors in the rostral migratory stream. Journal of Neuroscience. 2002;22(9):3568–3579. doi: 10.1523/JNEUROSCI.22-09-03568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Trommsdorff M., Gotthardt M., Hiesberger T., et al. Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97(6):689–701. doi: 10.1016/S0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 128.Weeber E. J., Beffert U., Jones C., et al. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. The Journal of Biological Chemistry. 2002;277(42):39944–39952. doi: 10.1074/jbc.m205147200. [DOI] [PubMed] [Google Scholar]
- 129.Su J., Klemm M. A., Josephson A. M., Fox M. A. Contributions of VLDLR and LRP8 in the establishment of retinogeniculate projections. Neural Development. 2013;8(1, article 11) doi: 10.1186/1749-8104-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.D'Arcangelo G., Nakajima K., Miyata T., Ogawa M., Mikoshiba K., Curran T. Reelin is a secreted glycoprotein recognized by the CR-50 monoclonal antibody. The Journal of Neuroscience. 1997;17(1):23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hiesberger T., Trommsdorff M., Howell B. W., et al. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24(2):481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 132.Niu S., Renfro A., Quattrocchi C. C., Sheldon M., D'Arcangelo G. Reelin promotes hippocampal dendrite development through the VLDLR/ApoER2-Dab1 pathway. Neuron. 2004;41(1):71–84. doi: 10.1016/s0896-6273(03)00819-5. [DOI] [PubMed] [Google Scholar]
- 133.Nichols A. J., Olson E. C. Reelin promotes neuronal orientation and dendritogenesis during preplate splitting. Cerebral Cortex. 2010;20(9):2213–2223. doi: 10.1093/cercor/bhp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sekine K., Kawauchi T., Kubo K.-I., et al. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron. 2012;76(2):353–369. doi: 10.1016/j.neuron.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Frotscher M. Cajal-Retzius cells, Reelin, and the formation of layers. Current Opinion in Neurobiology. 1998;8(5):570–575. doi: 10.1016/S0959-4388(98)80082-2. [DOI] [PubMed] [Google Scholar]
- 136.Marín-Padilla M. Cajal-Retzius cells and the development of the neocortex. Trends in Neurosciences. 1998;21(2):64–71. doi: 10.1016/s0166-2236(97)01164-8. [DOI] [PubMed] [Google Scholar]
- 137.Soriano E., del Río J. A. The cells of cajal-retzius: still a mystery one century after. Neuron. 2005;46(3):389–394. doi: 10.1016/j.neuron.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 138.Alcántara S., Ruiz M., D'Arcangelo G., et al. Regional and cellular patterns of reelin mRNA expression in the forebrain of the developing and adult mouse. Journal of Neuroscience. 1998;18(19):7779–7799. doi: 10.1523/JNEUROSCI.18-19-07779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Drakew A., Frotscher M., Deller T., Ogawa M., Heimrich B. Developmental distribution of a reeler gene-related antigen in the rat hippocampal formation visualized by CR-50 immunocytochemistry. Neuroscience. 1997;82(4):1079–1086. doi: 10.1016/s0306-4522(97)00326-6. [DOI] [PubMed] [Google Scholar]
- 140.Deutsch S. I., Burket J. A., Katz E. Does subtle disturbance of neuronal migration contribute to schizophrenia and other neurodevelopmental disorders? Potential genetic mechanisms with possible treatment implications. European Neuropsychopharmacology. 2010;20(5):281–287. doi: 10.1016/j.euroneuro.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 141.Blake S. M., Strasser V., Andrade N., et al. Thrombospondin-1 binds to ApoER2 and VLDL receptor and functions in postnatal neuronal migration. The EMBO Journal. 2008;27(22):3069–3080. doi: 10.1038/emboj.2008.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Andrade N., Komnenovic V., Blake S. M., et al. ApoER2/VLDL receptor and Dab1 in the rostral migratory stream function in postnatal neuronal migration independently of Reelin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(20):8508–8513. doi: 10.1073/pnas.0611391104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hack I., Bancila M., Loulier K., Carroll P., Cremer H. Reelin is a detachment signal in tangential chain-migration during postnatal neurogenesis. Nature Neuroscience. 2002;5(10):939–945. doi: 10.1038/nn923. [DOI] [PubMed] [Google Scholar]
- 144.Gudz T. I., Schneider T. E., Haas T. A., Macklin W. B. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. Journal of Neuroscience. 2002;22(17):7398–7407. doi: 10.1523/JNEUROSCI.22-17-07398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Le Marrec-Croq F., Bocquet-Garcon A., Vizioli J., et al. Calreticulin contributes to C1q-dependent recruitment of microglia in the leech Hirudo medicinalis following a CNS injury. Medical Science Monitor. 2014;20:644–653. doi: 10.12659/msm.890091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bento-Abreu A., Velasco A., Polo-Hernández E., Pérez-Reyes P. L., Tabernero A., Medina J. M. Megalin is a receptor for albumin in astrocytes and is required for the synthesis of the neurotrophic factor oleic acid. Journal of Neurochemistry. 2008;106(3):1149–1159. doi: 10.1111/j.1471-4159.2008.05462.x. [DOI] [PubMed] [Google Scholar]
- 147.Tabernero A., Velasco A., Granda B., Lavado E. M., Medina J. M. Transcytosis of albumin in astrocytes activates the sterol regulatory element-binding protein-1, which promotes the synthesis of the neurotrophic factor oleic acid. The Journal of Biological Chemistry. 2002;277(6):4240–4246. doi: 10.1074/jbc.m108760200. [DOI] [PubMed] [Google Scholar]
- 148.Polo-Hernández E., Tello V., Arroyo Á. A., et al. Oleic acid synthesized by stearoyl-CoA desaturase (SCD-1) in the lateral periventricular zone of the developing rat brain mediates neuronal growth, migration and the arrangement of prospective synapses. Brain Research C. 2014;1570:13–25. doi: 10.1016/j.brainres.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 149.Polo-Hernández E., de Castro F., García-García A. G., Tabernero A., Medina J. M. Oleic acid synthesized in the periventricular zone promotes axonogenesis in the striatum during brain development. Journal of Neurochemistry. 2010;114(6):1756–1766. doi: 10.1111/j.1471-4159.2010.06891.x. [DOI] [PubMed] [Google Scholar]
- 150.Granda B., Tabernero A., Tello V., Medina J. M. Oleic acid induces GAP-43 expression through a protein kinase C-mediated mechanism that is independent of NGF but synergistic with NT-3 and NT-4/5. Brain Research. 2003;988(1-2):1–8. doi: 10.1016/S0006-8993(03)03253-0. [DOI] [PubMed] [Google Scholar]
- 151.Caudy M., Bentley D. Pioneer growth cone morphologies reveal proximal increases in substrate affinity within leg segments of grasshopper embryos. The Journal of Neuroscience. 1986;6(2):364–379. doi: 10.1523/JNEUROSCI.06-02-00364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lewis A. K., Bridgman P. C. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. The Journal of Cell Biology. 1992;119(5):1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bentley D., O'Connor T. P. Cytoskeletal events in growth cone steering. Current Opinion in Neurobiology. 1994;4(1):43–48. doi: 10.1016/0959-4388(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 154.Suter D. M., Forscher P. Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. Journal of Neurobiology. 2000;44(2):97–113. doi: 10.1002/1097-4695(200008)44:2<97::aid-neu2>3.3.co;2-l. [DOI] [PubMed] [Google Scholar]
- 155.Korey C. A., Van Vactor D. From the growth cone surface to the cytoskeleton: one journey, many paths. Journal of Neurobiology. 2000;44(2):184–193. doi: 10.1002/1097-4695(200008)44:2<184::aid-neu8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 156.Dailey M. E., Bridgman P. C. Dynamics of the endoplasmic reticulum and other membranous organelles in growth cones of cultured neurons. Journal of Neuroscience. 1989;9(6):1897–1909. doi: 10.1523/JNEUROSCI.09-06-01897.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tessier-Lavigne M., Goodman C. S. The molecular biology of axon guidance. Science. 1996;274(5290):1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 158.Dickson B. J. Molecular mechanisms of axon guidance. Science. 2002;298(5600):1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 159.Landowski L. M., Pavez M., Brown L. S., et al. Low-density lipoprotein receptor-related proteins in a novel mechanism of axon guidance and peripheral nerve regeneration. The Journal of Biological Chemistry. 2016;291(3):1092–1102. doi: 10.1074/jbc.M115.668996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Thornalley P. J., Vašák M. Possible role for metallothionein in protection against radiation-induced oxidative stress. Kinetics and mechanism of its reaction with superoxide and hydroxyl radicals. Biochimica et Biophysica Acta (BBA)—Protein Structure and Molecular Enzymology. 1985;827(1):36–44. doi: 10.1016/0167-4838(85)90098-6. [DOI] [PubMed] [Google Scholar]
- 161.Hidalgo J., Aschner M., Zatta P., Vašák M. Roles of the metallothionein family of proteins in the central nervous system. Brain Research Bulletin. 2001;55(2):133–145. doi: 10.1016/S0361-9230(01)00452-X. [DOI] [PubMed] [Google Scholar]
- 162.Leung J. Y. K., Bennett W. R., Herbert R. P., et al. Metallothionein promotes regenerative axonal sprouting of dorsal root ganglion neurons after physical axotomy. Cellular and Molecular Life Sciences. 2012;69(5):809–817. doi: 10.1007/s00018-011-0790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stiles T. L., Dickendesher T. L., Gaultier A., et al. LDL receptor-related protein-1 is a sialic-acid-independent receptor for myelin-associated glycoprotein that functions in neurite outgrowth inhibition by MAG and CNS myelin. Journal of Cell Science. 2013;126(part 1):209–220. doi: 10.1242/jcs.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Henley J. R., Huang K.-H., Wang D., Poo M.-M. Calcium mediates bidirectional growth cone turning induced by myelin-associated glycoprotein. Neuron. 2004;44(6):909–916. doi: 10.1016/j.neuron.2004.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hines J. H., Abu-Rub M., Henley J. R. Asymmetric endocytosis and remodeling of β 1-integrin adhesions during growth cone chemorepulsion by MAG. Nature Neuroscience. 2010;13(7):829–837. doi: 10.1038/nn.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu K. Y., Hengst U., Cox L. J., et al. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nakato M., Matsuo M., Kono N., et al. Neurite outgrowth stimulation by n-3 and n-6 PUFAs of phospholipids in apoE-containing lipoproteins secreted from glial cells. Journal of Lipid Research. 2015;56(10):1880–1890. doi: 10.1194/jlr.M058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mantuano E., Lam M. S., Gonias S. L. LRP1 assembles unique co-receptor systems to initiate cell signaling in response to tissue-type plasminogen activator and Myelin-associated glycoprotein. The Journal of Biological Chemistry. 2013;288(47):34009–34018. doi: 10.1074/jbc.m113.509133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.May P., Rohlmann A., Bock H. H., et al. Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Molecular and Cellular Biology. 2004;24(20):8872–8883. doi: 10.1128/MCB.24.20.8872-8883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mulder M., Jansen P. J., Janssen B. J. A., et al. Low-density lipoprotein receptor-knockout mice display impaired spatial memory associated with a decreased synaptic density in the hippocampus. Neurobiology of Disease. 2004;16(1):212–219. doi: 10.1016/j.nbd.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 171.Liu Q., Trotter J., Zhang J., et al. Neuronal LRP1 knockout in adult mice leads to impaired brain lipid metabolism and progressive, age-dependent synapse loss and neurodegeneration. Journal of Neuroscience. 2010;30(50):17068–17078. doi: 10.1523/JNEUROSCI.4067-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gotthardt M., Trommsdorff M., Nevitt M. F., et al. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. The Journal of Biological Chemistry. 2000;275(33):25616–25624. doi: 10.1074/jbc.m000955200. [DOI] [PubMed] [Google Scholar]
- 173.Maier W., Bednorz M., Meister S., et al. LRP1 is critical for the surface distribution and internalization of the NR2B NMDA receptor subtype. Molecular Neurodegeneration. 2013;8(1, article 25) doi: 10.1186/1750-1326-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nakajima C., Kulik A., Frotscher M., et al. Low density lipoprotein receptor-related protein 1 (LRP1) modulates N-methyl-D-aspartate (NMDA) receptor-dependent intracellular signaling and NMDA-induced regulation of postsynaptic protein complexes. The Journal of Biological Chemistry. 2013;288(30):21909–21923. doi: 10.1074/jbc.m112.444364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Qiu S., Zhao L. F., Korwek K. M., Weeber E. J. Differential reelin-induced enhancement of NMDA and AMPA receptor activity in the adult hippocampus. Journal of Neuroscience. 2006;26(50):12943–12955. doi: 10.1523/jneurosci.2561-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Telese F., Ma Q., Perez P. M., et al. LRP8-Reelin-regulated neuronal enhancer signature underlying learning and memory formation. Neuron. 2015;86(3):696–710. doi: 10.1016/j.neuron.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu C., Hu J., Tsai C., et al. Neuronal LRP1 regulates glucose metabolism and insulin signaling in the brain. The Journal of Neuroscience. 2015;35(14):5851–5859. doi: 10.1523/jneurosci.5180-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dimou L., Simon C., Kirchhoff F., Takebayashi H., Götz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. Journal of Neuroscience. 2008;28(41):10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang S., Young K. M. Review: white matter plasticity in adulthood. Neuroscience. 2014;276:148–160. doi: 10.1016/j.neuroscience.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 180.Nishiyama A., Lin X.-H., Giese N., Heldin C.-H., Stallcup W. B. Co-localization of NG2 proteoglycan and PDGF α-receptor on O2A progenitor cells in the developing rat brain. Journal of Neuroscience Research. 1996;43(3):299–314. doi: 10.1002/(sici)1097-4547(19960201)43:3<299::aid-jnr5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 181.Richardson W. D., Pringle N., Mosley M. J., Westermark B., Dubois-Dalcg M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53(2):309–319. doi: 10.1016/0092-8674(88)90392-3. [DOI] [PubMed] [Google Scholar]
- 182.Noll E., Miller R. H. Oligodendrocyte precursors originate at the ventral ventricular zone dorsal to the ventral midline region in the embryonic rat spinal cord. Development. 1993;118(2):563–573. doi: 10.1242/dev.118.2.563. [DOI] [PubMed] [Google Scholar]
- 183.Pringle N. P., Richardson W. D. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117(2):525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- 184.Richardson W. D., Kessaris N., Pringle N. Oligodendrocyte wars. Nature Reviews Neuroscience. 2006;7(1):11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou Q., Wang S., Anderson D. J. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25(2):331–343. doi: 10.1016/S0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 186.Lu Q. R., Yuk D.-I., Alberta J. A., et al. Sonic hedgehog—regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25(2):317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 187.Dai J., Bercury K. K., Ahrendsen J. T., Macklin W. B. Olig1 function is required for oligodendrocyte differentiation in the mouse brain. Journal of Neuroscience. 2015;35(10):4386–4402. doi: 10.1523/jneurosci.4962-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhou Q., Anderson D. J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 189.Fok-Seang J., Miller R. H. Distribution and differentiation of A2B5+ glial precursors in the developing rat spinal cord. Journal of Neuroscience Research. 1994;37(2):219–235. doi: 10.1002/jnr.490370208. [DOI] [PubMed] [Google Scholar]
- 190.Fogarty M., Richardson W. D., Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132(8):1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- 191.Tripathi R. B., Clarke L. E., Burzomato V., et al. Dorsally and ventrally derived oligodendrocytes have similar electrical properties but myelinate preferred tracts. The Journal of Neuroscience. 2011;31(18):6809–6819. doi: 10.1523/jneurosci.6474-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kessaris N., Fogarty M., Iannarelli P., Grist M., Wegner M., Richardson W. D. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nature Neuroscience. 2006;9(2):173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Small R. K., Riddle P., Noble M. Evidence for migration of oligodendrocyte-type-2 astrocyte progenitor cells into the developing rat optic nerve. Nature. 1987;328(6126):155–157. doi: 10.1038/328155a0. [DOI] [PubMed] [Google Scholar]
- 194.Cahoy J. D., Emery B., Kaushal A., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of Neuroscience. 2008;28(1):264–278. doi: 10.1523/jneurosci.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang Y., Chen K., Sloan S. A., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gao L., Miller R. H. Specification of optic nerve oligodendrocyte precursors by retinal ganglion cell axons. The Journal of Neuroscience. 2006;26(29):7619–7628. doi: 10.1523/jneurosci.0855-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Murray K., Calaora V., Rottkamp C., Guicherit O., Dubois-Dalcq M. Sonic Hedgehog is a potent inducer of rat oligodendrocyte development from cortical precursors in vitro. Molecular and Cellular Neuroscience. 2002;19(3):320–332. doi: 10.1006/mcne.2001.1079. [DOI] [PubMed] [Google Scholar]
- 198.Ortega M. C., Cases O., Merchán P., Kozyraki R., Clemente D., De Castro F. Megalin mediates the influence of sonic hedgehog on oligodendrocyte precursor cell migration and proliferation during development. Glia. 2012;60(6):851–866. doi: 10.1002/glia.22316. [DOI] [PubMed] [Google Scholar]
- 199.Wicher G., Larsson M., Svenningsen Å. F., Gyllencreutz E., Rask L., Aldskogius H. Low density lipoprotein receptor-related protein-2/megalin is expressed in oligodendrocytes in the mouse spinal cord white matter. Journal of Neuroscience Research. 2006;83(5):864–873. doi: 10.1002/jnr.20774. [DOI] [PubMed] [Google Scholar]
- 200.Schmidt C., Ohlemeyer C., Labrakakis C., Walter T., Kettenmann H., Schnitzer J. Analysis of motile oligodendrocyte precursor cells in vitro and in brain slices. Glia. 1997;20(4):284–298. doi: 10.1002/(sici)1098-1136(199708)20:4<284::aid-glia2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 201.Simpson P. B., Armstrong R. C. Intracellular signals and cytoskeletal elements involved in oligodendrocyte progenitor migration. Glia. 1999;26(1):22–35. doi: 10.1002/(sici)1098-1136(199903)26:1<22::aid-glia3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 202.Mantuano E., Jo M., Gonias S. L., Campana W. M. Low density Lipoprotein Receptor-related Protein (LRP1) regulates Rac1 and RhoA reciprocally to control Schwann cell adhesion and migration. The Journal of Biological Chemistry. 2010;285(19):14259–14266. doi: 10.1074/jbc.m109.085126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pankov R., Endo Y., Even-Ram S., et al. A Rac switch regulates random versus directionally persistent cell migration. Journal of Cell Biology. 2005;170(5):793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Binamé F., Sakry D., Dimou L., Jolivel V., Trotter J. NG2 regulates directional migration of oligodendrocyte precursor cells via Rho GTPases and polarity complex proteins. The Journal of Neuroscience. 2013;33(26):10858–10874. doi: 10.1523/jneurosci.5010-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Muratoglu S. C., Mikhailenko I., Newton C., Migliorini M., Strickland D. K. Low density lipoprotein receptor-related protein 1 (LRP1) forms a signaling complex with platelet-derived growth factor receptor-β in endosomes and regulates activation of the MAPK pathway. The Journal of Biological Chemistry. 2010;285(19):14308–14317. doi: 10.1074/jbc.m109.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Craig J., Mikhailenko I., Noyes N., Migliorini M., Strickland D. K. The LDL receptor-related protein 1 (LRP1) regulates the PDGF signaling pathway by binding the protein phosphatase SHP-2 and modulating SHP-2- mediated PDGF signaling events. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0070432.e70432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Newton C. S., Loukinova E., Mikhailenko I., et al. Platelet-derived growth factor receptor-β (PDGFR-β) activation promotes its association with the low density lipoprotein receptor-related protein (LRP): evidence for co-receptor function. The Journal of Biological Chemistry. 2005;280(30):27872–27878. doi: 10.1074/jbc.m505410200. [DOI] [PubMed] [Google Scholar]
- 208.Rönnstrand L., Arvidsson A.-K., Kallin A., et al. SHP-2 binds to Tyr763 and Tyr1009 in the PDGF β-receptor and mediates PDGF-induced activation of the Ras/MAP kinase pathway and chemotaxis. Oncogene. 1999;18(25):3696–3702. doi: 10.1038/sj.onc.1202705. [DOI] [PubMed] [Google Scholar]
- 209.Avrov K., Kazlauskas A. The role of c-Src in platelet-derived growth factor alpha receptor internalization. Experimental Cell Research. 2003;291(2):426–434. doi: 10.1016/j.yexcr.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 210.Miyamoto Y., Yamauchi J., Tanoue A. Cdk5 phosphorylation of WAVE2 regulates oligodendrocyte precursor cell migration through nonreceptor tyrosine kinase fyn. The Journal of Neuroscience. 2008;28(33):8326–8337. doi: 10.1523/jneurosci.1482-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chae T., Kwon Y. T., Bronson R., Dikkes P., En L., Tsai L.-H. Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron. 1997;18(1):29–42. doi: 10.1016/S0896-6273(01)80044-1. [DOI] [PubMed] [Google Scholar]
- 212.Rosenkranz S., DeMali K. A., Gelderloos J. A., Bazenet C., Kazlauskas A. Identification of the receptor-associated signaling enzymes that are required for platelet-derived growth factor-AA-dependent chemotaxis and DNA synthesis. The Journal of Biological Chemistry. 1999;274(40):28335–28343. doi: 10.1074/jbc.274.40.28335. [DOI] [PubMed] [Google Scholar]
- 213.McKinnon R. D., Waldron S., Kiel M. E. PDGF α-receptor signal strength controls an RTK rheostat that integrates phosphoinositol 3′-kinase and phospholipase Cγ pathways during oligodendrocyte maturation. The Journal of Neuroscience. 2005;25(14):3499–3508. doi: 10.1523/jneurosci.5049-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Campana W. M., Li X., Dragojlovic N., Janes J., Gaultier A., Gonias S. L. The low-density lipoprotein receptor-related protein is a pro-survival receptor in Schwann cells: possible implications in peripheral nerve injury. The Journal of Neuroscience. 2006;26(43):11197–11207. doi: 10.1523/jneurosci.2709-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mañes S., Martínez-A C. Cholesterol domains regulate the actin cytoskeleton at the leading edge of moving cells. Trends in Cell Biology. 2004;14(6):275–278. doi: 10.1016/j.tcb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 216.Mañes S., Mira E., Gómez-Moutón C., et al. Membrane raft microdomains mediate front-rear polarity in migrating cells. The EMBO Journal. 1999;18(22):6211–6220. doi: 10.1093/emboj/18.22.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Boyles J. K., Pitas R. E., Wilson E., Mahley R. W., Taylor J. M. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. The Journal of Clinical Investigation. 1985;76(4):1501–1513. doi: 10.1172/jci112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Beisiegel U., Weber W., Ihrke G., Herz J., Stanley K. K. The LDL-receptor-related protein, LRP, is an apolipoprotein E-binding protein. Nature. 1989;341(6238):162–164. doi: 10.1038/341162a0. [DOI] [PubMed] [Google Scholar]
- 219.Morrisett J. D., Jackson R. L., Gotto A. M., Jr. Lipoproteins: structure and function. Annual Review of Biochemistry. 1975;44:183–207. doi: 10.1146/annurev.bi.44.070175.001151. [DOI] [PubMed] [Google Scholar]
- 220.Willnow T. E., Hammes A., Eaton S. Lipoproteins and their receptors in embryonic development: more than cholesterol clearance. Development. 2007;134(18):3239–3249. doi: 10.1242/dev.004408. [DOI] [PubMed] [Google Scholar]
- 221.Laatsch A., Panteli M., Sornsakrin M., Hoffzimmer B., Grewal T., Heeren J. Low density lipoprotein receptor-related protein 1 dependent endosomal trapping and recycling of apolipoprotein. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0029385.e29385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pfrieger F. W. Role of cholesterol in synapse formation and function. Biochimica et Biophysica Acta (BBA)—Biomembranes. 2003;1610(2):271–280. doi: 10.1016/s0005-2736(03)00024-5. [DOI] [PubMed] [Google Scholar]