Abstract
A major hindrance in gene therapy trials with adeno-associated virus (AAV) vectors is the presence of neutralizing antibodies (NAbs) that inhibit AAV transduction. In this study, we used directed evolution techniques in vitro and in mouse muscle to select novel NAb escape AAV chimeric capsid mutants in the presence of individual patient serum. AAV mutants isolated in vitro escaped broad patient-specific NAb activity but had poor transduction ability in vivo. AAV mutants isolated in vivo had enhanced NAb evasion from cognate serum and had high muscle transduction ability. More importantly, structural modeling identified a 100 amino acid motif from AAV6 in variable region (VR) III that confers this enhanced muscle tropism. In addition, a predominantly AAV8 capsid beta barrel template with a specific preference for AAV1/AAV9 in VR VII located at threefold symmetry axis facilitates NAb escape. Our data strongly support that chimeric AAV capsids composed of modular and nonoverlapping domains from various serotypes are capable of evading patient-specific NAbs and have enhanced muscle transduction.
Introduction
Adeno-associated virus (AAV) vectors have been explored extensively in numerous preclinical studies and a number of these studies are currently translating into encouraging phase 1, 2, and 3 clinical trials.1,2,3,4,5 While AAV gene therapy continues to yield clinical results supportive of the hope for eventual treatment of many diseases, the presence of patient neutralizing antibodies (NAbs) remains a challenge. NAb-mediated elimination of AAV vectors has become a rate-limiting step in advancing the field and a determinant for repeat administration of AAV gene transfer.1,2,6 The fact that more than 90% of the population has been exposed to natural AAV2 infection, and half of those infected carry NAbs in their blood,7,8,9,10,11,12,13,14,15,16,17 highlights the significance of this problem.
Although AAV2 vectors are used in the majority of clinical trials,2,18,19 other serotypes, such as AAV1, 8, or customized vectors such as AAV2.5 are being explored.1,6,20 In an early phase 1 clinical trial with hemophilia B patients using an AAV2 vector encoding the factor IX (F.IX) transgene, ~10% of normal F.IX levels were achieved in one patient who lacked NAbs and no detectable F.IX was observed in a second patient with preexisting AAV2 NAbs.2 More recently, we performed a phase 1 clinical trial in six patients with Duchenne muscular dystrophy (DMD) using the chimeric AAV2.5 vector encoding a mini-dystrophin transgene for muscle delivery. Before intramuscular (i.m.) injection, no AAV2 NAbs were found in three patients, low NAb titers were detected in one patient, and high NAb titers were detected in the remaining two patients. After injection, vector genomes were only detected in muscle biopsies from patients with no or low titers of NAbs.6 This observation suggests that NAbs inhibit AAV transduction following direct i.m. injection.
Several approaches to overcome AAV NAbs include: plasmapheresis,21 reduction of NAb production with B-cell depletion,22 use of empty AAV capsid decoys,23 alternative AAV serotypes that are naturally resistant to NAbs,9,24,25,26,27 selection of NAb escape mutants from an error-prone PCR library,28,29,30 site-directed modification of AAV capsid proteins,31 and AAV capsid-associated polymers that prevent NAb recognition.32,33,34 Based on kinetic analysis of the NAbs in our phase 1 clinical trial with hemophilia patients, NAb cross-reactivity was suggested as a primary mechanism of neutralization of unrelated AAV serotypes.17 This observation was confirmed in results from our DMD phase 1 clinical trial. After i.m. injection of chimeric AAV2.5, all three patients initially lacking NAbs had AAV2 NAb titers at 1:100 (ref. 6). They also displayed differential inhibition to various AAV serotypes. Regarding the AAV capsid modification strategies to develop AAV mutants for NAb evasion, Maheshri et al.30 used an AAV2 library derived from error prone PCR to isolate AAV NAb escape mutants in the presence of rabbit anti-AAV2 sera in vitro. Other studies, including ours, have selected mutants with high tissue tropism using an AAV library from DNA shuffling of different serotypes; however, NAb evasion ability was only tested postselection in cell lines or in the presence of human intravenous immunglobin.35,36,37 In the current study, we isolated AAV NAb escape mutants in the presence of individual patient serum from our DMD clinical trial using AAV directed evolution in vitro. Most importantly, we also performed the selection of AAV mutants with NAb evasion in vivo to determine whether the mutants isolated from one patient's serum are able to escape NAb activity from other sera and retain high tissue transduction. In HEK293 cells, AAV mutants selected from individual patient serum escaped NAb activity in sera from all subjects; however, all in vitro selected mutants were compromised in transducing target tissue in vivo. Importantly, we also selected AAV mutants with NAb evasion in vivo in BALB/c mouse muscle to determine whether the mutants isolated from one patient's serum escaped NAb activity from other patient sera. The in vivo selection resulted in several capsid mutants recovered from skeletal muscle that not only escaped NAbs but also displayed high muscle transduction efficiency compared to most naturally occurring AAV serotypes. To explore the capsid protein amino acid (aa) sequence required for NAb evasion and enhanced tropism, we conducted sequence alignment and structural analysis studies. Phylogenetic analysis and structural modeling of several of the selected mutants helped to identify AAV capsid motifs that can be further engineered to modulate NAb evasion while retaining muscle tropism. This approach of generating personalized, patient-specific NAb AAV escape mutants represents a paradigm shift in steps required when exploiting AAV directed evolution to overcome immune limitations currently impacting gene transfer studies.
Results
Characterization of AAV NAb escaping mutants isolated from HEK293 cells
In our phase 1 DMD clinical trial, six patients were treated via i.m. injection with a chimeric AAV2.5 encoding the mini-dystrophin transgene.6 Prior to vector administration, three patients had no AAV2 NAbs; however, sera collected 100 days posttreatment showed that AAV2 NAb titers increased to 1:1006. To select AAV NAb escape mutants, undiluted sera from these patients were combined with an AAV capsid shuffled library35,38 and applied to HEK293 cells in the presence of adenovirus (Ad). Forty-eight hours posttreatment, cell lysates containing the NAb escape mutants were again used to infect HEK293 cells in the presence of Ad for a second cycle of AAV amplification. After four rounds of amplification, NAb AAV mutants were isolated and sequenced. Nine, 8, and 12 mutants were isolated from patient sera 1, 2, and 3, respectively (Supplementary Figures S1). Most mutants from sera 1 and 2 contained a chimeric VP3 of AAV8 and AAV9 (Table 1 and Supplementary Figure S1A,B). Eleven mutants isolated from serum 3 contained the VP3 C-terminal from AAV8 (Table 1 and Supplementary Figure S1C), and nine of them had the VP3-N-terminal of AAV1. In addition to capsid composition from different serotype of AAV, all mutants contained at least one point mutation. Most mutants produced comparable virus yields to AAV2, but lower titers were produced by mutants AAV001-12 and 15, AAV002-9, and AAV003-19 (Table 1). The AAV001-12 and 15 mutants have proline residues, L636P and N737P, respectively, close to VP interfaces, and the G640E mutation in AAV002-9 is in a buried position inside the capsid likely to cause steric hindrance that may prevent viral capsid assembly. For AAV003-19 the T233Q residue is located in a β-strand inside the capsid. Thus, while the mechanism(s) behind a reduced or lack of virus production is unclear, it is likely that the point mutations in these constructs changed the VP structure, although individual AAV VP subunit can be formed, it is possible that the mutations result in insufficient AAV virion assembly from 60 VP subunits.
Table 1. Characterization of AAV mutants isolated from HEK293 cells.
After identification and virus production in HEK293 cells, recombinant mutants packaging firefly luciferase transgene (luc) were tested for their ability to transduce HEK293 cells. The mutants possessed different transduction efficiency as compared with AAV2 (Table 1). Lower transduction was demonstrated for the mutants with the exception of mutant AAV001-14, which had similar transduction ability to AAV2.
NAb escaping ability of the mutants isolated in HEK293 cells
To investigate the capacity of mutants recovered from HEK293 cells to escape NAbs from individual patient serum, we incubated mutants with undiluted serum at 4 °C for 2 hours. The unbound virus transduction was determined by adding the AAV mutant/serum mixture to HEK293 cells. For escape mutants from serum 1, all tested, with the exception of AAV001-7, exhibited transduction >50% of control without serum, especially mutants AAV001-1, 4, and 5 which showed >80% transduction (Table 1 and Supplementary Figure S2). This may be because the N-terminus of AAV001-7 VP3 was derived from AAV1, and AAV1 transduction was inhibited by serum 1. In contrast, the other eight mutants' VP3 was derived from AAV8 and 9, and AAV8 and 9 transduction was not inhibited to the same extent as was AAV1.
Among the escape mutants from serum 2, a diverse capsid composition was observed. Based on NAb escape analysis, mutants AAV002-9, 14, and 15 were resistant to serum 2, whereas the other five mutants were strongly inhibited (Table 1 and Supplementary Figure S2). Among the inhibited mutants, AAV002-16 was derived entirely from the AAV2 capsid but contained two mutations, S292L and K556E. It is interesting to note that serum 2 also inhibited transduction from mutants AAV002-1 and 3, composed of an AAV1 VP1 N-terminus (VP1u) and AAV8 and 9 VP3, while wild-type (wt) AAV8 and 9 escape the NAb activity of serum 2. This suggests the involvement of the VP1u in antibody recognition. Also, although serum 2 completely suppressed AAV2 transduction and partially inhibited AAV1 transduction, mutant AAV002-14, a chimera of AAV1 and 2, transduced HEK293 cells efficiently in the presence of serum 2. This is likely due to the presence of sequence variation at or close to antigenic regions of AAV1 and AAV2.31,39 In the presence of serum 3, transgene expression was decreased for AAV1, 2, 4, 8, and 9 as well as chimeric AAV2.5. Among the 12 mutants, AAV003-7 and 19 were able to escape the NAb activity to induce transgene expression >50% of control (Table 1 and Supplementary Figure S2). The other isolated mutants could not escape the NAb activity of serum 3.
Cross-reactivity of sera from patients against AAV mutants isolated in HEK293 cells
A number of mutants isolated from patient serum had the ability to escape the NAb activity of their cognate serum as described above. Next, we analyzed the cross-reactivity of patient NAbs with mutants isolated against other patient sera. Some mutants isolated from one serum could also escape the NAb activity of the other two sera, such as AAV001-1, 11, 12, 14, 17, AAV002-9, 14, and AAV003-19 (Table 1 and Supplementary Figure S2). These results indicate that it is possible to isolate AAV mutants that possess a generic ability to evade NAbs from multiple subjects. Other mutants had an ability to escape the NAbs from one but not another alternative patient serum such as AAV001-4, 5, 15, AAV002-15, and AAV003-7. Interestingly, most mutants isolated from serum 3 could only evade NAbs of serum 3 but could not evade NAbs from serum 1 or 2 (Table 1 and Supplementary Figure S2). These data suggest that AAV capsid NAb escape mutants isolated from individual serum may be different from those from a traditional pooled sera approach.
In vivo transduction profile of AAV mutants isolated in HEK293 cells
To determine the tissue tropism of AAV mutants isolated in HEK293 in the presence of patient serum, 1 × 1011 particles of selected AAV vectors encoding the luc transgene were retro-orbitally injected into 6-week-old female BALB/c mice. After 1 week, in vivo images were captured. Consistent with the literature, AAV8 and 2 mainly transduced the liver, AAV4 transduced the heart, AAV1 transduced both liver and heart, and AAV9 transduced almost every tissue (Supplementary Figure S3). Compared to parental AAV serotypes, all mutants except for AAV002-14 transduced the liver, but to a lesser extent than AAV8. Two weeks postinjection, tissue was harvested and luciferase activity was analyzed. AAV003-7, a chimeric of AAV1 and 2, had a similar muscle transduction to serotypes AAV4 and 9, higher than parental serotypes (Supplementary Figure S3). Overall lower transduction was observed across different tissues using chimeric mutants compared to parental serotypes. These results suggest that mutants with the ability to escape NAb should be further analyzed in vivo for maintenance of tissue-specific transduction.
Isolation of AAV mutants from mouse skeletal muscle in the presence of human serum
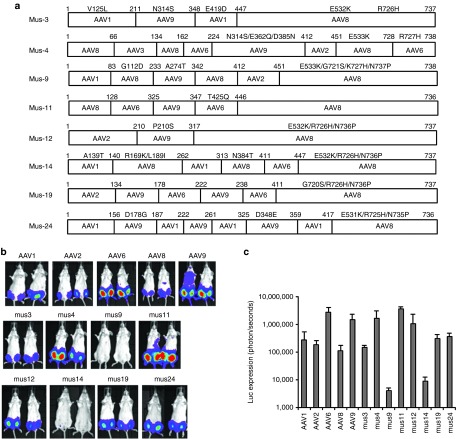
As proof of principle, we successfully obtained chimeric capsid AAV mutants that evade NAb activity of human sera in HEK293 cells. Although most of these mutants transduced cells efficiently in vitro, this may not translate into successful transduction in vivo. To develop tissue-tropic AAV mutants with NAb evasion ability, we directly injected a mixture of the AAV shuffling library and DMD patient 3 serum into the muscle of 6-week-old female BALB/c mice. Three days postinjection, Ad was injected into the same muscle to amplify the AAV genomes. Two days post Ad injection, muscle was collected for total DNA extraction. PCR was used to amplify mutant capsid sequences, and PCR products were subsequently cloned into the pXR2 backbone. Eight colonies were obtained and sequenced; every clone contained the C-terminus of the AAV8 VP3 capsid except for Mus4 for which the C-terminus contained 10 aa residues from AAV6 (Figure 1a).
Figure 1.
Transduction efficiency of AAV mutants recovered from mouse skeletal muscle. 1 × 1010 particles of the AAV shuffled library virus incubated with human serum 3 and administered via i.m. injection to 6-week-old female BALB/c mice. Three days postinjection, Ad dl309 was injected into the same muscle. (a) After 2 days, AAV mutant capsids were recovered from the injected muscle and sequenced. These mutant capsids were used to package the luc transgene. 1 × 109 particles of AAV/luc were administered via i.m. injection in both legs of 6-week-old female BALB/c mice (n = 2 mice). One week post AAV application, (b) imaging was carried out and (c) the photon signal was measured and the average signal from all four legs was calculated. The exposure time for imaging was 10 seconds for AAV6, Mus3, and 12, and 5 minutes for other mutants and AAV serotypes. AAV, adeno-associated virus; i.m., intramuscular.
Transduction efficiencies of AAV mutants recovered from mouse skeletal muscle
To determine the relative muscle tropism of chimeric capsid AAV mutants isolated from mouse muscle, 1 × 109 particles of AAV mutants as well as parental AAV serotypes encoding the luc transgene were delivered via i.m. injection of 6-week-old female BALB/c mice. One week postinjection, imaging was performed. As shown in Figure 1, AAV6 remained superior for muscle transduction, with a transduction efficiency order: AAV6>Mus12>Mus3>Mus11>AAV9=Mus4=Mus19=Mus24= AAV1>AAV2>AAV8>Mus14>Mus9.
To examine muscle transduction after systemic administration, 1 × 1011 particles of AAV mutants as well as parental serotypes were injected via the retro-orbital vein into 6-week-old female BALB/c mice. One week postinjection, imaging was performed. At 2 weeks postinjection, various tissues were collected for in vitro luc activity analysis. Consistent with prior reports, AAV9 was superior in transducing mouse tissue (Supplementary Figure S4). Among six mutants, Mus19 and 24 were able to transduce the liver more efficiently than other mutants, with similar performance compared to parental AAV6, and lower efficiency than AAV8 or 9. Regarding muscle tropism, Mus11 was superior in transducing both heart and skeletal muscles compared to other mutants. However, heart transduction of Mus11 was lower than that of AAV6, 8, and 9. Skeletal muscle transduction by Mus11 was higher than that of AAV6 but lower than that of AAV8 or 9. Lastly, Mus19 induced better skeletal muscle transduction than AAV6. These data indicate that high muscle transduction of mutants delivered via i.m. injection does not translate into high muscle tropism by systemic administration. These results are similar to the observation that AAV6, the serotype superior for muscle transduction following i.m. injection, performs worse than AAV8 or 9 in transducing muscle after systemic application.
NAb escaping ability of muscle-derived AAV mutants in vivo
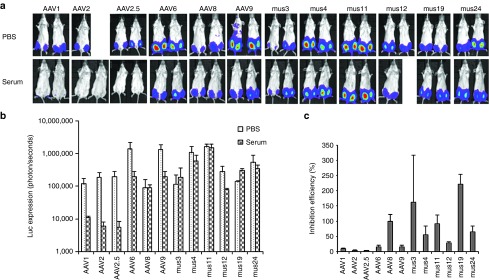
In vitro data showed that undiluted patient serum 3 inhibited transduction by parental AAV serotypes 1, 2, 4, 8, 9, and mutant AAV2.5 (Table 1 and Supplementary Figure S2). To determine whether the chimeric capsid AAV mutants isolated from muscle tissue could evade NAb activity from serum and successfully transduce mouse muscle, we delivered 1 × 109 particles of AAV mutants or parental serotypes encoding the luc transgene mixed with fivefold diluted serum 3 or phosphate-buffered saline (PBS) via i.m. injection to 6-week-old female BALB/c mice. One week postinjection, imaging was performed. Serum 3 inhibited muscle transduction >50% for all parental serotypes and mutant AAV2.5 except for AAV8, and also inhibited Mus12. However, no suppression of luc expression was observed in muscle injected with Mus3, 4, 11, 19, or 24 (Figure 2).
Figure 2.
NAb escape potential for AAV mutants derived from muscle. 1 × 109 particles of AAV/luc from mutants isolated from mouse muscle and wild serotypes were incubated with an equal volume of fivefold diluted patient serum 3 or PBS for 2 hours at 4 °C, then administered via i.m. injection to muscles of both legs in 6-week-old female BALB/c mice (n = 2). Three weeks post AAV application, mice were imaged for (a) luc expression. The photon signal was measured, and (b) the average signal for mice treated with human serum 3 or PBS was calculated for 4 legs. (c) The inhibition of human serum 3 on AAV transduction was analyzed by comparison of transgene expression in serum-treated mice over that of PBS-treated mice. The exposure time for imaging was 10 seconds for AAV6, Mus3, and 12, and 5 minutes for other mutants as well as AAV8 and 9. AAV, adeno-associated virus; i.m., intramuscular; NAb, neutralizing antibody; PBS, phosphate-buffered saline.
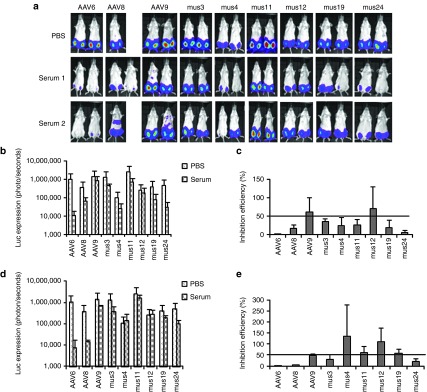
Cross-reactive NAb escaping ability of muscle-derived AAV mutants
In vitro experiments demonstrated that the mutants isolated from serum 3 had a higher capacity to escape NAb activity from serum 3 than from sera 1 and 2. To determine whether muscle-isolated mutants that escaped serum 3 NAbs could also evade NAbs from sera 1 and 2, we delivered 1 × 109 particles of AAV mutants or parental serotypes encoding the luc transgene mixed with fivefold diluted serum 1 or 2 or PBS via i.m. injection to 6-week-old female BALB/c mice. As shown in Figure 3, strong inhibition of AAV6 transduction in the presence of sera 1 or 2 was seen 1 week postinjection. Unlike serum 3, both sera 1 and 2 suppressed AAV8 muscle transduction >50%; AAV9 muscle transduction was not inhibited (>50% of PBS group). Whereas the Mus12 mutant was inhibited by serum 3, sera 1 or 2 did not inhibit muscle transduction. Although serum 1 was able to inhibit muscle transduction from the other 5 Mus mutants >50%, serum 2 only inhibited muscle transduction from Mus3 and 24, but not from Mus4, 11, and 19. These results indicate that some mutants isolated from serum 3 also possess NAb escaping ability toward other sera but that the efficiency of NAb escape varied across sera.
Figure 3.
Cross-reactivity of NAbs on AAV mutants isolated from muscle. 1 × 109 particles of AAV/luc mutants isolated from mouse muscle and wild serotypes were incubated with an equal volume of fivefold diluted patient serum 1, 2, or PBS for 2 hours at 4 °C, then the mixture of AAV/luc and serum was administered via i.m. injection into both legs of 6-week-old BALB/c mice (n = 2). (a) At 1 week post AAV administration, imaging was taken for luc expression. The average signal for legs in mice treated with (b) patient serum 1 or (d) 2 or PBS was calculated (b and d for serum 1 and 2, respectively). The inhibition of human (c) serum 1 or (e) 2 on AAV transduction was analyzed by comparison of transgene expression in serum treated mice over that in PBS mice. The exposure time for imaging was 10 seconds for AAV6, Mus3, and 12, and 5 minutes for other mutants and AAV8 and 9. AAV, adeno-associated virus; i.m., intramuscular; NAb, neutralizing antibody; PBS, phosphate-buffered saline.
Elucidation of the capsid regions for mutants with NAb escape ability and muscle tropism
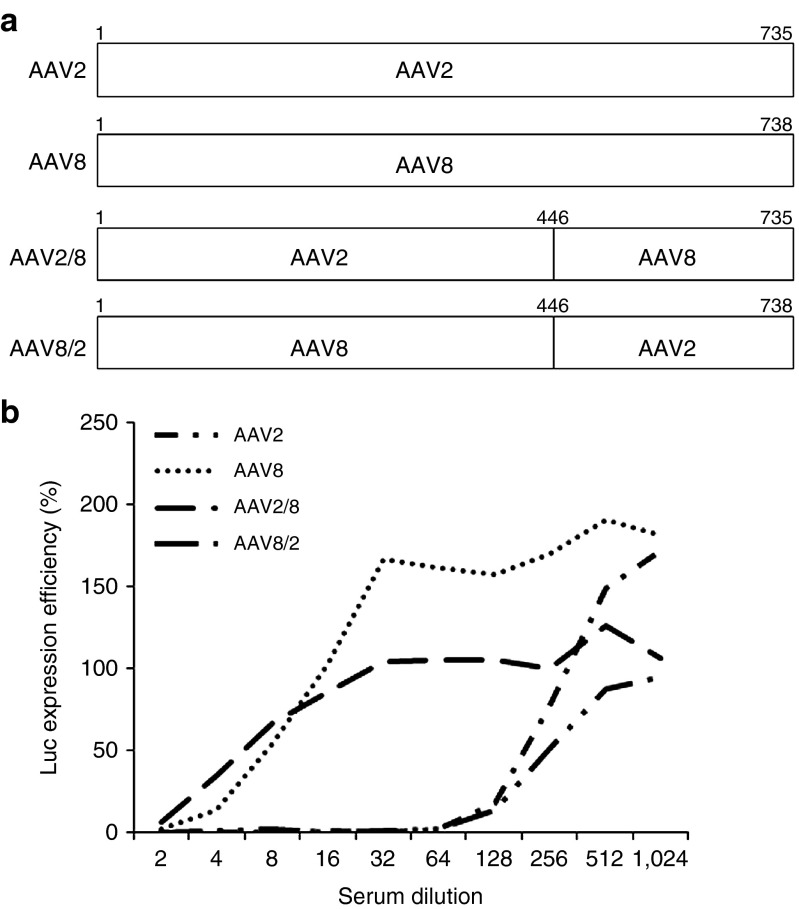
We successfully recovered eight mutants from mouse muscles in the presence of human serum, and the capsid sequence data showed an exclusivity of AAV8 capsid sequence in the C-terminus from aa residues 446 to the end. We wondered whether this VP region might play a role in NAb escape ability for the mutants. We therefore swapped the corresponding domains of AAV2 and AAV8, and subsequently carried out an in vitro NAb titer assay. It was demonstrated that the same dilution of serum 3 inhibited AAV2 and the AAV8/2 chimeric vector transduction by 50% at a 128-fold dilution but inhibited AAV8 and AAV2/8 at a more concentrated 4-fold dilution (Figure 4). These results suggest that it is possible to identify VP regions responsible for NAb binding via selection of AAV mutants in the presence of NAbs.
Figure 4.
The C-terminus of AAV2 VP3 is responsible for NAb binding. (a) A diagram of AAV mutants with swapped domains between the AAV2 and AAV8 capsids. (b) NAb titer analysis of AAV mutants with domain swapping. Patient 3 serum with twofold serial dilutions were incubated with AAV mutants and AAV2 or AAV8 at equal volumes of 1 × 108 particles for 2 hours at 4 °C, then added to RC32 cells in the presence of Ad dl309 (MOI of 5). One day later, luc activity was measured, and transgene expression efficiency was calculated against transduction of AAV mixed with PBS only (control). AAV, adeno-associated virus; MOI, multiplicity of infection; NAb, neutralizing antibody; PBS, phosphate-buffered saline.
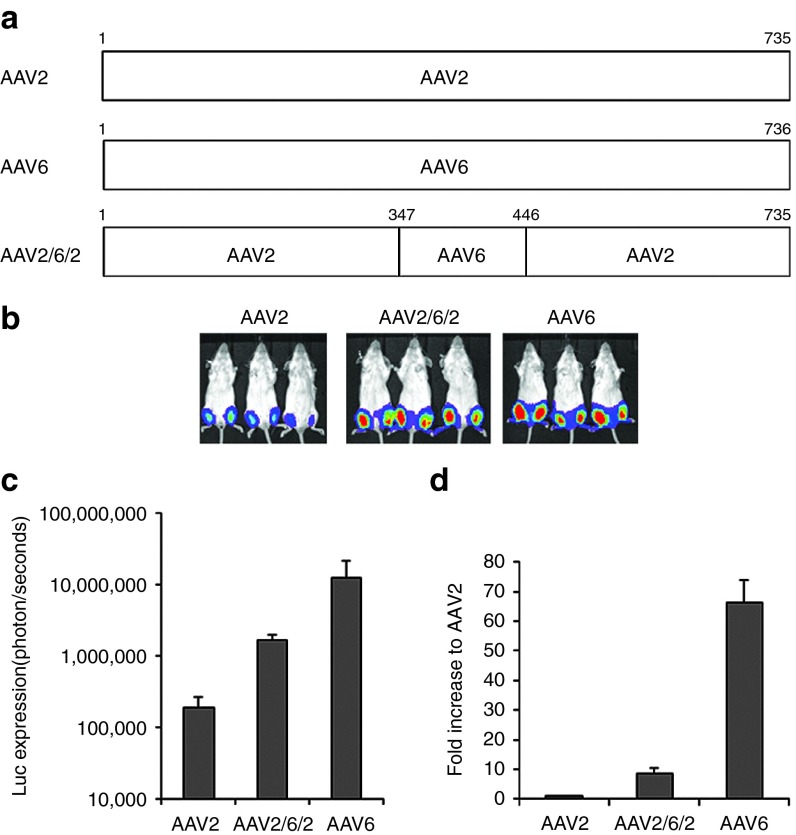
As described above, six out of eight Mus mutants recovered from muscle in the presence of human serum showed higher muscle tropism than parental AAV2, and four mutants (Mus3, 11, 19, and 24) contained a motif from the AAV6 capsid around aa residues 347 to 446 in VP1. To determine whether the motif from AAV6 capsid plays a role in muscle tropism, we made an AAV2 mutant virus (AAV262) by swapping a capsid fragment of AAV6 at aa residues 347 to 446 into the AAV2 capsid, and delivered 1 × 109 particles via i.m. injection to 6-week-old female BALB/c mice. Compared to AAV2 muscle transduction, 3- to 10-fold higher transgene expression was achieved using the AAV262 vector. However, AAV262 muscle transduction remained sevenfold less efficient than AAV6 (Figure 5). This result suggests that aa residues 347 to 446 from AAV6 VP1 contribute to muscle tropism in recovered mutants. Thus, in this study, we are not only able to identify putative capsid motifs responsible for NAb binding, but also determine motifs important for tropism and enhanced muscle transduction by studying in vivo escape mutants.
Figure 5.
A capsid domain of AAV6 contributing to robust muscle tropism. 1 × 109 particles of AAV/luc from mutant AAV262 and wt AAV2 or 6 were administered via i.m. injection to muscles of both legs of 6-week-old female BALB/c mice. One week postinjection, imaging was taken and calculated quantitatively. (a) Diagram of AAV262 mutant. (b) Imaging 1 week post AAV/luc vector i.m. administration. The exposure time was 1 minute for AAV2 and AAV262, and 10 seconds for AAV6. (c) Quantitation of luc imaging. (d) Fold increase of luc expression from AAV262 and AAV6 transduction in muscles over that of the AAV2 vector. AAV, adeno-associated virus; i.m., intramuscular; PBS, phosphate-buffered saline.
Discussion
The objective of this proof-of-principle study was to explore the possibility of isolating AAV mutants that evade NAbs and retain efficient tissue transduction to facilitate repeat administration in patients who were previously treated with AAV. We first isolated AAV mutants in the presence of sera from different human subjects in vitro and demonstrated that these mutants possessed the capacity for evading NAb activity from different sera. Whereas some mutants had the generic ability to escape NAbs across different serum sources, others could escape NAbs from cognate serum far more efficiently than from other sera. Essentially all mutant capsids selected in vitro had reduced transduction in vivo compared to standard serotypes. However, in selecting tissue-specific AAV mutants with NAb evasion ability, we recovered eight mutants that escaped human serum to transduce mouse muscle. Six of the mutants initiated higher muscle transduction than most serotypes. Among the six mutants with high muscle tropism, five were better able to evade NAbs in cognate serum than any parental AAV serotype. Another had the ability to escape NAbs from other sera with efficiency varying by mutant and serum. Finally, based on our NAb escape mutant capsid composition data, we identified the domains responsible for NAb escape activity and muscle tropism.
AAV gene therapy has been successfully used for phase 1 clinical trials in patients with vision and bleeding disorders.1,2,3,4,5 Transgenic coagulation F.IX has been successfully delivered to the bloodstream after AAV vector delivery to the liver in patients with hemophilia B1,2. However, this treatment has only benefited a small set of patients, as more than 50% of people carry NAbs that inhibit AAV transduction. To evade AAV NAb activity, several strategies have been tested to either protect AAV vectors from neutralization or to decrease NAb titers in the patient's blood. Polymer (e.g., polyethylene glycol) has been used to coat the AAV surface and block NAb recognition.32,33,34 This approach is difficult to reproduce with respect to equal coating and has been suggested to change the AAV transduction profile. A second approach has used error-prone PCR or DNA shuffling to generate a library of AAV capsid variants to select for NAb escape mutants in the presence of NAbs in vitro.28,29,30,37 This approach has yielded novel capsids; however, it bears the potential limitation of generating capsids with unknown transduction efficiency in vivo. In the present study, we recovered escape mutants from HEK293 cells in the presence of human NAb sera that had similarly compromised transduction in vivo. In animal models, several studies have found that separate serotypes of AAV show little or no NAb cross-reactivity; thus, it has been suggested to use other serotypes to overcome NAb inhibition.9,13,24,25,26,27 While this strategy is logical, additional studies suggest that concern remains about the possibility of cross-reactivity in most humans which may not be predicted in animals.17 A finally attempted approach to resolve NAb inhibition has been rational mutation of the NAb binding domain on the AAV capsid surface.27 This strategy requires information about monoclonal antibody (mAb) epitopes and the structure of the AAV virion. While successful and informative, this approach is inherently limited due to polyclonal nature of human NAbs, and the difficulty in obtaining mAbs representing all generated NAbs.
In addition to vector modification, several clinical approaches have been employed such as plasmapheresis prior to vector delivery.21 Due to the relative inefficiency of each round of plasmapheresis and the fact that even low titers of NAbs (<1:5) can abrogate AAV transduction, this strategy is generally only suitable for patients with lower starting titers of AAV NAbs and requires multiple sessions. Similar to plasmapheresis, the use of balloon catheters in combination with a saline flush have shown efficacy for liver gene transduction in the presence of low to moderate NAb titers.40 More recently, the use of an anti-CD20 antibody (rituximab) to achieve B-cell depletion for 6–9 months has been reported. This approach is not directed at (antibody-producing) plasma cells and is effective in reducing AAV NAb in only a minority of subjects who have a NAb titer less than 1:1,000 (ref. 22). A final clinical approach applies excessive empty AAV capsids as decoys for NAbs.23 Here, a major concern is that the addition of empty particles could increase the AAV capsid load, risking induction of capsid dose-related cytotoxic immune response and perhaps competing with full AAV particles for effective transduction.41 No standardization of processes leading to the generation of “empty particles” exists, and it has recently been reported that partially empty capsids (coproduced with vector) results in greater apparent liver inflammation than when using genome-containing AAV vectors only.42
With this backdrop, we have applied the strategy of directed evolution to develop novel AAV variants in the presence of human NAbs from AAV-treated patients.6,35,38,43 We successfully isolated several AAV mutants in the presence of sera from three patients when selected in vitro. Some of these chimeric capsids could escape the NAb activity from all three patients, while others induced higher transduction in the presence of the cognate serum from which the evolutionary pressure was derived. Such results suggest that it is necessary to develop individual NAb escape mutants in the presence of serum from the specific subject, although it is possible (as we have also found) that some of AAV mutants isolated from one subject serum may confer generic NAb escape ability across other sera. Because the mechanism of varying AAV transduction efficiencies in different cells and tissues is unclear, and it is well known that the AAV transduction profile in vitro does not typically correlate in vivo, we have also determined that it is imperative to develop tissue-tropic AAV vectors with the ability to escape NAbs. Herein, we advanced the in vitro approach to in vivo selection in mouse muscle. Six mutants were isolated from mouse muscle that could evade serum NAbs from patient samples. The data clearly indicate the feasibility of developing NAb evasion mutants with enhanced AAV transduction efficiency but more importantly provide a roadmap for protein domains required for these functions in chimeric capsids. Additionally, the results obtained in this and our studies strongly suggest that selection of NAb escape AAV mutants should be performed in the presence of individual antiserum but not using pooled antisera (e.g., intravenous immunglobin). Pooled sera are collected from different population of human subjects. In this mixed sera, some subjects have no AAV NAb and others have different titers of NAb (as high as 1:1,000).17 When sera from these subjects are pooled together, the titer of AAV NAb actually is decreased due to dilution, and the NAb activity from some subjects may disappear due to low NAb titer. Therefore mutants isolated against pooled antisera may only be applied to subjects with high NAb titer.
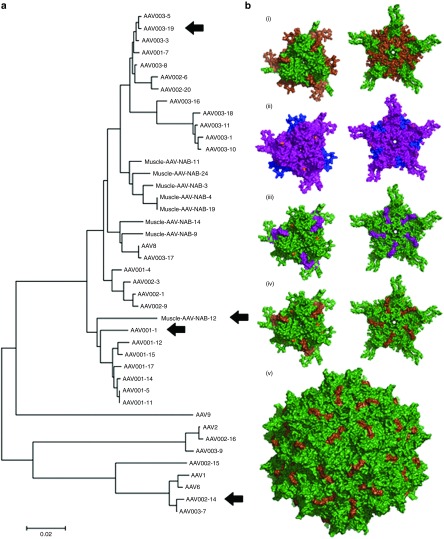
Structural studies have demonstrated that there are nine variable regions (VRs) on the AAV virion surface that are responsible for AAV serotype tropism and transduction as well as NAb recognition.44,45,46,47 For an individual subject, NAb activity will vary in strength to distinct serotypes. The capsid genomes of our AAV shuffled library are a mixture of AAV serotypes 1 through 9. Thus, Nab-binding sites from one but not another serotype may be present on the chimeric virion surface. Therefore, using the directed evolution/shuffling approach we can identify NAb binding domains by analyzing the mutant capsids that possess NAb escape abilities. We first carried out phylogenetic analysis of different isolates obtained from both in vitro and in vivo studies to assess the diversity of mutants obtained by directed evolution (Figure 6a). Consistent with functional data, we observed that the broad NAb evading clones, AAV001-1, AAV002-14, and AAV003-19, are distinct isolates belonging to different clades. It is interesting to note that the broad NAb escaping muscle isolate, Mus12, is phylogenetically similar to AAV001-1, despite being recovered from skeletal muscle tissue in vivo. Furthermore, AAV002-14 is located in the same clade containing AAV1/AAV6. Overall from different muscle isolates, we noted that every escape mutant contained AAV8-derived capsid sequence at the C-terminus. Consistent with this observation, AAV8 had a superior ability to escape NAbs than any other serotype in the presence of patient 3 serum both in vitro and in vivo. Findings based on swapping the C-terminus of AAV2 and AAV8 further supported that NAb binding sites from patient 3 serum were localized to the C-terminus of the AAV2 (but not AAV8) capsid. Therefore, modification of the AAV2 capsid C-terminus can evade NAb inhibition of AAV transduction. It is interesting to note that Mus3 and 12 were most successful at inducing muscle transduction after direct injection among the six mutants with high muscle tropism; however, their NAb evasion abilities varied dramatically: Mus3 was able to escape its cognate serum NAb but not sera from the other two patients, whereas Mus12 could only evade NAb activity of sera from other patient sera but not from patient 3. The sequence alignment between the two mutants revealed AAV8 sequences downstream of residue 448, and that both capsids had AAV9 sequences upstream of residue 317 containing variable point mutations. However, each had aa insertions from a wide variety of parental serotypes located between residues 318–447 (Mus3 has AAV9 and AAV6, Mus12 has AAV8). Other observed differences between the Mus3 and 12 mutants are located in residues 327, 329, and 331, which have previously been shown to be located near the fivefold pore surface, a region that has not been identified as a determinant of antigenic reactivity in previous studies.47
Figure 6.
Phylogeny and structural models of AAV mutants. (a) Neighbor-joining phylogeny of the VP1 capsid sequence of NAb evading AAV mutants. Capsid aa sequences of AAVs were aligned using ClustalW, and the phylogeny was generated using a neighbor-joining algorithm and a Poisson correction to calculate aa distances. HEK293 isolates AAV001-1, AAV002-14, AAV003-19, and the skeletal muscle isolate Mus12 were selected for further structural modeling based on their distinct clades on the phylogenetic tree and functional ability to broadly escape NAbs. (b) Three-dimensional structural models of select AAV NAb escape mutant that broadly escape NAbs. Three-dimensional surface models of capsid subunit trimer/threefold and the pentamer/fivefold symmetry axis regions of AAV NAb escape mutants isolated from 293 cells in vitro or recovered from mouse skeletal muscle in vivo. Amino acid residues derived from different AAV serotypes are highlighted as follows: purple (AAV1), blue (AAV2), hot pink (AAV6), green (AAV8), and brown (AAV9). Point mutations are highlighted in orange. Specific residues and point mutations are listed in Supplementary Figure S1. The surface model of the full capsid (60-mer) of the muscle isolate Mus12 is also shown with aa residues derived from AAV8 highlighted in green and residues derived from AAV9 colored brown. All structural models were visualized using PyMOL (http://www.pymol.org/, The PyMOL Molecular Graphics System, Version 1.5.0.4; Schrödinger). AAV, adeno-associated virus; NAb, neutralizing antibody.
It is well known that AAV vectors from different serotypes display various muscle tropisms: AAV1 or AAV6, which differ by six aa residues, induce the best muscle transduction in mice.48,49 Previous studies have shown that chimeric vectors with aa residues 350 to 430 of AAV2 VP1 induce much higher muscle transduction when substituted with the corresponding sequence from AAV1.50,51 Consistent with the chimeric vector study, 4 mutants (Mus3, 11, 19, and 24) contained aa residues 347 to 446 of VP1 (containing VRIII) derived from AAV1/6, and synthetic AAV2 mutants specifically designed to swap corresponding sequences from AAV6 led to higher muscle transduction. Importantly, the approach used here not only identified chimeric mutants with NAb evasion, but also provided an effective platform to identify motifs responsible for tissue tropism.
Finally, we carried out structural modeling of each representative broad NAb escaping mutant to determine whether structure–function correlation can be established. Specifically, we modeled trimers and pentamers of different VP subunit sequences derived from the clones AAV001-1, AAV002-14, and AAV003-19 as well as the Mus12 isolate (Figure 6b). We noted that the region most varied from the AAV8 capsid sequence is between aa residues 217 and 316. This prominent surface domain contains VR I and has been implicated in earlier antigenicity studies.31 Furthermore, it is interesting to note that AAV001-1, which belongs to the same clade as Mus12 and AAV003-19, also contains the same region, albeit with fewer aa residues (233–315). These results suggest that it is possible to identify single immunodominant epitopes on the AAV capsid that can be engineered to mediate NAb evasion. In particular, AAV8 VR I appears particularly suitable for such applications without adversely affecting transduction efficiency.
The current study has certain limitations. For instance, we only selected AAV mutants in muscle tissue in the presence of human serum and only generated 30 unique clones for sequencing, which may be due to the different muscle fibers transduced from AAV library virus and Ad. Six out of 8 available mutant capsids induced higher muscle transduction than AAV serotypes 1, 2, 8, or 9, but all were slightly reduced compared to the superior muscle transduction of AAV6. Interestingly, most mutants induced higher transgene expression in muscle than AAV6 in the presence of patient NAbs. These results strongly support the feasibility of selection of AAV NAb escape mutants in the presence of NAb. By performing additional rounds of selection in the presence of human serum, perhaps we could have increased the likelihood of isolating a mutant with both NAb evasion and muscle transduction similar to AAV6. Another arguable limitation of this study is that the mutants recovered from muscle had lower muscular transduction than AAV9 upon systemic administration, although most displayed high muscle transduction after i.m. injection. Most likely, these mutants do not have high vascular permeability to transduce muscle as compared to AAV9. Maybe a better strategy to test in future studies would be to isolate AAV mutants from muscles after systemic administration of the AAV library and human serum as our results clearly demonstrate that each parameter in the selection process greatly influences chimeric capsid selection.
In summary, we have successfully recovered AAV mutants in vitro and from mouse muscle after pairing an AAV shuffled library with NAb-containing human serum. Although it is possible to obtain AAV mutants that evade NAb activity from different subjects, we found that mutants isolated from specific patients are better at evading cognate serum-based NAbs than NAbs from others. Most importantly, the isolated mutants provide important information for identifying both NAb binding domains on the AAV virion surface and identifying unique motifs conferring muscle tissue tropism. These results establish a platform to allow us to generate a panel of AAV mutants isolated from different patient sera, and these mutants will be tested for NAb evasion capacity in patients involved in clinical trial. If no mutants are able to escape NAb from a specific patient, it is necessary to develop patient-specific mutants in the presence of cognate serum.
Materials and Methods
Cell lines. HEK293 cells and RC32 cells (HeLa cells with stable AAV2 rep expression) were maintained at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 10% penicillin–streptomycin.
Serum samples. Sera were collected from three DMD patients in our phase 1 clinical trial at day 100 after i.m. injection of AAV2.5/mini-dystrophin vector and stored at −80 °C (ref. 6).
AAV virus production. A three plasmid transfection method was used to produce AAV vector, described previously.52 Briefly, AAV transgene plasmid pTR/CBA-luc, AAV helper plasmid, and Ad helper plasmid pXX6-80 were cotransfected into HEK293 cells. Sixty hours posttransfection, HEK293 cells were collected and lysed. Supernatant was subjected to CsCl gradient ultra-centrifugation. Fractions containing AAV were collected and tittered by dot-blot.
NAb escaping AAV mutants screen in vitro. 2 × 105 HEK293 cells were seeded in a six-well plate at the time of the experiment. 2 × 109 particles of AAV capsid shuffling library in 10 μl PBS was incubated with 10 μl of undiluted patient serum for 2 hours at 4 °C. AAV/serum mixtures were applied to cells with the addition of Ad dl309 at a multiplicity of infection (MOI) of 5. Forty-eight hours posttreatment, cells were harvested and subjected to three freeze/thaw cycles. Lysates were centrifuged at 2,000 rpm for 10 minutes to remove cell debris, and supernatant was used to repeat the process for three more cycles to amplify virus in the absence of patient serum. After the fourth cycle, Hirt DNA was extracted from HEK293 cells, and AAV mutant capsids were amplified by PCR with the following primers: F1 5′-CAACTCCATCACTAGGGGTTC and R1 5′-CATGGGAAAGGTGCCAGA, which are localized at the AAV2 rep and AAV2 ITR, respectively. PCR using PfuUltra High-Fidelity DNA polymerase (Agilent Technologies, Santa Clara, CA) was conducted with the following conditions: 94 °C 30 seconds, 53 °C 30 seconds, 72 °C 3 minutes for 35 cycles. PCR products were digested with SwaI and XbaI and ligated into pXR2 digested with the same endonucleases. Clones were sequenced by the UNC-CH Genome Analysis Facility, and capsid sequence alignment was analyzed with VectorNTI (Invitrogen, Grand Island, NY).
Hirt DNA purification. Low-molecular-weight DNA was extracted from HEK293 cells as described previously.53 Briefly, HEK293 cell pellets were resuspended in 728 μl of Hirt buffer (20 mmol/l Tris–HCl, 20 mmol/l EDTA, pH 8.0) and lysed with the addition of 46 μl of 10% sodium dodecyl sulfate. 218 μl of 5 mol/l NaCl was added to the cell lysate solution and incubated for 1 hour on ice. Lysate was centrifuged at 15K rpm at 4 °C for 30 minutes. Supernatant was collected, and DNA was extracted with phenol/chloroform. DNA was precipitated with isopropranol, and resuspended in 50 μl TE buffer (10 mmol/l Tris–HCl, 1 mmol/l EDTA, pH 8.0) containing 100 μg/ml of DNase-free RNase.
In vitro transduction analysis. 1 × 105 293 cells were seeded in a 48-well plate for 2 hours. 1 × 108 particles of AAV/luc was added. Forty-eight hours postinfection, cells were lysed using Passive Lysis Buffer (Promega, Durham, NC). Cell lysate was transferred to a 96-well plate to measure luciferase activity with a Wallac1420 Victor2 microplate reader. For inhibition assay of transgene expression in the presence of serum, 1 × 108 particles of AAV/luc in 10 μl PBS were incubated with 10 μl serum for 2 hours at 4 °C, then added to HEK293 cells. Luciferase activity was measured 24 hours post-AAV infection.
NAb analyses. NAb analyses were performed as described previously with slight modification.17,27 Sera were serially diluted twofold with PBS. RC32 cells were seeded in 300 μl cell media in a 48-well plate for 3 to 4 hours. 1 × 108 particles of AAV/luc was incubated with each serum dilution in PBS for 2 hours at 4 °C in a total volume of 25 μl. AAV/sera mixtures were added to cells in a final volume of 125 μl, which contained 4 × 106 particles of Ad dl309. Cells with AAV/sera mixtures were incubated for 24 hours at 37 °C. NAb titers were defined as the highest dilution for which luciferase activity was 50% less than controls (no sera).
NAb escaping AAV mutants isolated from mouse muscles. 1 × 1010 particles of the AAV capsid shuffling library virus in 50 μl PBS were mixed with 50 μl undiluted serum from patient 3 for 2 hours at 4 °C. The AAV/serum mixture was delivered via i.m. injection into the hind leg muscle of 6-week-old female BALB/c mice. Three days posttreatment, Ad virus dl309 was delivered via i.m. injection into the same muscle at MOI of 107 to amplify AAV genomes in vivo (Supplementary Figure S5). Muscle was collected 2 days post Ad administration, and total DNA was extracted using Qiagen Kit (Qiagen, Valencia, CA). PCR was carried out for amplification of AAV mutant capsids, as done above, using muscle DNA as template. PCR products were cloned into a pXR2 backbone. Colonies were sequenced and used to generate AAV/luc mutant vectors. Housing and handling of mice was carried out in compliance with the National Institutes of Health guidelines and an Institutional Animal Care and Use Committee approved protocol at UNC-CH.
Characterization of AAV mutants in mice. Six-week-old female BALB/c mice received 1 × 1011 particles of AAV/luc via retro-orbital injection. Luciferase expression was imaged 1 week postinjection using a Xenogen IVIS Lumina (Caliper Lifesciences, Waltham, MA) following i.p. injection of d-luciferin substrate at 120 mg/kg (Nanolight Pinetop, AZ). Bioluminescent images were analyzed using Living Image (PerkinElmer, Waltham, MA). For muscle transduction, 1 × 109 particles of AAV/luc were injected into the gastrocnemius of 6-week-old female BALB/c mice. Mice were imaged at the indicated time points.
Quantitation of luciferase expression in tissues. Animals utilized for imaging studies were sacrificed 2 weeks post AAV injection, and the following organs were collected: liver, spleen, kidney, heart, lung, skeletal muscle (gastrocnemius), and brain. Tissue was minced and homogenized in passive lysis buffer. Tissue lysates were centrifuged at 10K rpm for 5 minutes to remove cellular debris. Supernatant was transferred to 96-well plates for luciferase activity analysis as described above. Total protein concentration in tissue lysates were measured using the Bradford assay (BioRad Laboratories, Philadelphia, PA).
AAV mutant capsid cloning. AAV capsid swap cassettes were generated by PCR with primers listed in Supplementary Table S1. PCR products were ligated into the pXR2 backbone. To generate the pXR-262 clone, we first made the construct pXR-62. To generate pXR-62, we obtained a PCR product with primers F/R2-262 using pXR6 as a template and a PCR product with primers F3-262/R using pXR2 as a template. Both PCR products were digested with SalI and NotI and ligated into pXR2 digested with the same endonucleases. Next, to generate pXR-262, we obtained a PCR product with primers F2-262/R using pXR-62 as a template and a PCR product with primers F/R1-262 using pXR2 as a template. Both PCR products were digested with SalI and NotI and ligated into pXR2 digested with the same endonucleases. To generate clone pXR-2/8, we obtained a PCR product with primers F/R1-2/8 using pXR2 as a template and a PCR product with primers F2-2/8/R using pXR8 as a template. Both PCR products were digested with SalI and NotI and ligated into pXR2 digested with the same endonucleases. To generate clone pXR-8/2, we obtained PCR products with primers F3-262/R using pXR2 as a template and a PCR product using F/R2-2/8 using pXR8 as template. Both PCR products were digested with SalI and NotI and ligated into pXR2 digested with the same endonucleases. Colonies were verified by DNA sequencing.
Molecular modeling. Structural homology models of the AAV capsid mutants were obtained using the SWISS-MODEL online server (http://swissmodel.expasy.org/),54 with the crystal structure of AAV8 VP3 (PDB ID 2QA0) serving as a template. Structure-based alignment was generated using the secondary structure matching application in the WinCoot software,55,56,57 with the AAV4 VP3 (PDB ID 3NG9) monomer being used as a template. The threefold and fivefold symmetry axes/VP3 trimers and pentamers and the whole capsid were generated using the oligomer generator utility in VIPERdb (http://viperdb.scripps.edu/oligomer_multi.php)58 and visualized using the PyMOL Molecular Graphics System (Schrödinger).
Phylogenetic analysis. The entire capsid aa sequences of different AAV isolates were aligned using ClustalW,59 and phylogenetic trees were generated with the MEGAv5.05 software package.60 Computational algorithms including maximum likelihood, neighbor joining, minimum evolution, and maximum parsimony were used. The phylogeny was produced using the neighbor-joining algorithm, and aa distances were calculated using a Poisson correction. Statistical testing was done by bootstrapping with 1,000 replicates to test the confidence of the phylogenetic analysis.
SUPPLEMENTARY MATERIAL Figure S1. Mutants selected from patient sera. Figure S2. NAb evasion of AAV mutants isolated from HEK293 cells. Figure S3. Transduction profile in vivo for NAb escape mutants isolated from HEK293 cells. Figure S4. Transduction profile in vivo for NAb escape mutants isolated from muscles with systemic administration. Figure S5. AAV genome copy number in mouse muscle. Table S1. List of primers
Acknowledgments
We thank Xiaojing Chen, Karen Hogan, and Violeta Zaric for their excellent technical assistance, and Eric Hastie, Marc Weinberg, and Ashley Crosby for their critical reading of the manuscript. We also thank the Ohio State University College of Medicine, Department of Pediatrics, for providing sera. The authors acknowledge the UNC Biomedical Research Imaging Center (BRIC) Small Animal Imaging (SAI) facility for assistance of mouse imaging. This work was supported by NIH grants R01DK084033 (to C.L. and R.J.S.), P01HL112761 (to R.J.S. and A.A.), R01AI072176, R01AR064369 (to M.H. and R.J.S.), U54AR056953 (to R.J.S), R01GM082946 (to M.A.M), P30-CA016086-35–37 and U54-CA151652-01-04 (to the BRIC SAI facility). R.J.S. is the founder and a shareholder at Asklepios Biopharmaceutical. He holds patents that have been licensed by UNC to Asklepios Biopharmaceutical , for which he receives royalties.
Supplementary Material
References
- Nathwani, AC, Tuddenham, EG, Rangarajan, S, Rosales, C, McIntosh, J, Linch, DC et al. (2011). Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 365: 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno, CS, Pierce, GF, Arruda, VR, Glader, B, Ragni, M, Rasko, JJ et al. (2006). Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med 12: 342–347. [DOI] [PubMed] [Google Scholar]
- Maguire, AM, Simonelli, F, Pierce, EA, Pugh, EN Jr, Mingozzi, F, Bennicelli, J et al. (2008). Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med 358: 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, AM, High, KA, Auricchio, A, Wright, JF, Pierce, EA, Testa, F et al. (2009). Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374: 1597–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelli, F, Maguire, AM, Testa, F, Pierce, EA, Mingozzi, F, Bennicelli, JL et al. (2010). Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther 18: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles, DE, McPhee, SW, Li, C, Gray, SJ, Samulski, JJ, Camp, AS et al. (2012). Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol Ther 20: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow, NR, Hoggan, MD, Kapikian, AZ, Austin, JB and Rowe, WP (1968). Epidemiology of adenovirus-associated virus infection in a nursery population. Am J Epidemiol 88: 368–378. [DOI] [PubMed] [Google Scholar]
- Georg-Fries, B, Biederlack, S, Wolf, J and zur Hausen, H (1984). Analysis of proteins, helper dependence, and seroepidemiology of a new human parvovirus. Virology 134: 64–71. [DOI] [PubMed] [Google Scholar]
- Xiao, W, Chirmule, N, Berta, SC, McCullough, B, Gao, G and Wilson, JM (1999). Gene therapy vectors based on adeno-associated virus type 1. J Virol 73: 3994–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko, M, Chen, L, van Roey, M, Donahue, BA, Snyder, RO, McArthur, JG et al. (2000). Epitope mapping of human anti-adeno-associated virus type 2 neutralizing antibodies: implications for gene therapy and virus structure. J Virol 74: 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles, K, Sebökovà, P and Schlehofer, JR (1999). Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV). J Med Virol 59: 406–411. [DOI] [PubMed] [Google Scholar]
- Chirmule, N, Propert, K, Magosin, S, Qian, Y, Qian, R and Wilson, J (1999). Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther 6: 1574–1583. [DOI] [PubMed] [Google Scholar]
- Halbert, CL, Miller, AD, McNamara, S, Emerson, J, Gibson, RL, Ramsey, B et al. (2006). Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5, and 6 in cystic fibrosis and normal populations: Implications for gene therapy using AAV vectors. Hum Gene Ther 17: 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan, CD, Jiang, H, Liu, T, Patarroyo-White, S, Sommer, JM, Zhou, S et al. (2006). Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood 107: 1810–1817. [DOI] [PubMed] [Google Scholar]
- Calcedo, R, Vandenberghe, LH, Gao, G, Lin, J and Wilson, JM (2009). Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin, S, Monteilhet, V, Veron, P, Leborgne, C, Benveniste, O, Montus, MF et al. (2010). Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21: 704–712. [DOI] [PubMed] [Google Scholar]
- Li, C, Narkbunnam, N, Samulski, RJ, Asokan, A, Hu, G, Jacobson, LJ et al.; Joint Outcome Study Investigators. (2012). Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther 19: 288–294. [DOI] [PubMed] [Google Scholar]
- Manno, CS, Chew, AJ, Hutchison, S, Larson, PJ, Herzog, RW, Arruda, VR et al. (2003). AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood 101: 2963–2972. [DOI] [PubMed] [Google Scholar]
- Brantly, ML, Spencer, LT, Humphries, M, Conlon, TJ, Spencer, CT, Poirier, A et al. (2006). Phase I trial of intramuscular injection of a recombinant adeno-associated virus serotype 2 alphal-antitrypsin (AAT) vector in AAT-deficient adults. Hum Gene Ther 17: 1177–1186. [DOI] [PubMed] [Google Scholar]
- Brantly, ML, Chulay, JD, Wang, L, Mueller, C, Humphries, M, Spencer, LT et al. (2009). Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci USA 106: 16363–16368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteilhet, V, Saheb, S, Boutin, S, Leborgne, C, Veron, P, Montus, MF et al. (2011). A 10 patient case report on the impact of plasmapheresis upon neutralizing factors against adeno-associated virus (AAV) types 1, 2, 6, and 8. Mol Ther 19: 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi, F, Chen, Y, Edmonson, SC, Zhou, S, Thurlings, RM, Tak, PP et al. (2013). Prevalence and pharmacological modulation of humoral immunity to AAV vectors in gene transfer to synovial tissue. Gene Ther 20: 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi, F, Anguela, XM, Pavani, G, Chen, Y, Davidson, RJ, Hui, DJ et al. (2013). Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci Transl Med 5: 194ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert, CL, Rutledge, EA, Allen, JM, Russell, DW and Miller, AD (2000). Repeat transduction in the mouse lung by using adeno-associated virus vectors with different serotypes. J Virol 74: 1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildinger, M, Auricchio, A, Gao, G, Wang, L, Chirmule, N and Wilson, JM (2001). Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J Virol 75: 6199–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière, C, Danos, O and Douar, AM (2006). Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Ther 13: 1300–1308. [DOI] [PubMed] [Google Scholar]
- Li, C, Diprimio, N, Bowles, DE, Hirsch, ML, Monahan, PE, Asokan, A et al. (2012). Single amino acid modification of adeno-associated virus capsid changes transduction and humoral immune profiles. J Virol 86: 7752–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner, NA, Girod, A, Perabo, L, Edbauer, D, Kleinschmidt, JA, Büning, H et al. (2003). Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther 10: 2139–2147. [DOI] [PubMed] [Google Scholar]
- Perabo, L, Endell, J, King, S, Lux, K, Goldnau, D, Hallek, M et al. (2006). Combinatorial engineering of a gene therapy vector: directed evolution of adeno-associated virus. J Gene Med 8: 155–162. [DOI] [PubMed] [Google Scholar]
- Maheshri, N, Koerber, JT, Kaspar, BK and Schaffer, DV (2006). Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol 24: 198–204. [DOI] [PubMed] [Google Scholar]
- Lochrie, MA, Tatsuno, GP, Christie, B, McDonnell, JW, Zhou, S, Surosky, R et al. (2006). Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol 80: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, GK, Maheshri, N, Kaspar, B and Schaffer, DV (2005). PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnol Bioeng 92: 24–34. [DOI] [PubMed] [Google Scholar]
- Carlisle, RC, Benjamin, R, Briggs, SS, Sumner-Jones, S, McIntosh, J, Gill, D et al. (2008). Coating of adeno-associated virus with reactive polymers can ablate virus tropism, enable retargeting and provide resistance to neutralising antisera. J Gene Med 10: 400–411. [DOI] [PubMed] [Google Scholar]
- Le, HT, Yu, QC, Wilson, JM and Croyle, MA (2005). Utility of PEGylated recombinant adeno-associated viruses for gene transfer. J Control Release 108: 161–177. [DOI] [PubMed] [Google Scholar]
- Li, W, Asokan, A, Wu, Z, Van Dyke, T, DiPrimio, N, Johnson, JS et al. (2008). Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther 16: 1252–1260. [DOI] [PubMed] [Google Scholar]
- Lisowski, L, Dane, AP, Chu, K, Zhang, Y, Cunningham, SC, Wilson, EM et al. (2014). Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 506: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, D, Lee, JS, Wang, L, Desai, T, Akache, B, Storm, TA et al. (2008). In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 82: 5887–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W, Zhang, L, Johnson, JS, Zhijian, W, Grieger, JC, Ping-Jie, X et al. (2009). Generation of novel AAV variants by directed evolution for improved CFTR delivery to human ciliated airway epithelium. Mol Ther 17: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraw, DM, O'Donnell, JK, Taylor, KA, Stagg, SM and Chapman, MS (2012). Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology 431: 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimuro, J, Mizukami, H, Hishikawa, S, Ikemoto, T, Ishiwata, A, Sakata, A et al. (2013). Minimizing the inhibitory effect of neutralizing antibody for efficient gene expression in the liver with adeno-associated virus 8 vectors. Mol Ther 21: 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y, Weinberg, MS, Hirsch, M, Johnson, MC, Tisch, R, Samulski, RJ et al. (2013). Kinetics of adeno-associated virus serotype 2 (AAV2) and AAV8 capsid antigen presentation in vivo are identical. Hum Gene Ther 24: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, K, Li, M, Zhong, L, Su, Q, Li, J, Li, S et al. (2014). Empty virions in AAV8 vector preparations reduce transduction efficiency and may cause total viral particle dose-limiting side-effects. Mol Ther Methods Clin Dev 1: 20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, SJ, Blake, BL, Criswell, HE, Nicolson, SC, Samulski, RJ, McCown, TJ et al. (2010). Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB). Mol Ther 18: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q, Bu, W, Bhatia, S, Hare, J, Somasundaram, T, Azzi, A et al. (2002). The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci USA 99: 10405–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindasamy, L, Padron, E, McKenna, R, Muzyczka, N, Kaludov, N, Chiorini, JA et al. (2006). Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol 80: 11556–11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbandje-McKenna, M and Kleinschmidt, J (2011). AAV capsid structure and cell interactions. Methods Mol Biol 807: 47–92. [DOI] [PubMed] [Google Scholar]
- Gurda, BL, DiMattia, MA, Miller, EB, Bennett, A, McKenna, R, Weichert, WS et al. (2013). Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol 87: 9111–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, H, Liu, Y, Rabinowitz, J, Li, C, Samulski, RJ and Walsh, CE (2000). Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol Ther 2: 619–623. [DOI] [PubMed] [Google Scholar]
- Blankinship, MJ, Gregorevic, P, Allen, JM, Harper, SQ, Harper, H, Halbert, CL et al. (2004). Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther 10: 671–678. [DOI] [PubMed] [Google Scholar]
- Hauck, B and Xiao, W (2003). Characterization of tissue tropism determinants of adeno-associated virus type 1. J Virol 77: 2768–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, B, Xu, RR, Xie, J, Wu, W, Ding, Q, Sipler, M et al. (2006). Efficient AAV1-AAV2 hybrid vector for gene therapy of hemophilia. Hum Gene Ther 17: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X, Li, J and Samulski, RJ (1998). Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 72: 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C and Samulski, RJ (2005). Serotype-specific replicating AAV helper constructs increase recombinant AAV type 2 vector production. Virology 335: 10–21. [DOI] [PubMed] [Google Scholar]
- Arnold, K, Bordoli, L, Kopp, J and Schwede, T (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Krissinel, E and Henrick, K (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2256–2268. [DOI] [PubMed] [Google Scholar]
- Emsley, P, Lohkamp, B, Scott, WG and Cowtan, K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4): 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P and Cowtan, K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2126–2132. [DOI] [PubMed] [Google Scholar]
- Carrillo-Tripp, M, Shepherd, CM, Borelli, IA, Venkataraman, S, Lander, G, Natarajan, P et al. (2009). VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res 37(Database issue): D436–D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, JD, Higgins, DG and Gibson, TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K, Peterson, D, Peterson, N, Stecher, G, Nei, M and Kumar, S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.