Abstract
Oncolytic virotherapy is a novel and emerging treatment modality that uses replication-competent viruses to destroy cancer cells. Although diverse cancer cell types are sensitive to oncolytic viruses, one of the major challenges of oncolytic virotherapy is that the sensitivity to oncolysis ranges among different cancer cell types. Furthermore, the underlying mechanism of action is not fully understood. Here, we report that activation of cyclic adenosine monophosphate (cAMP) signaling significantly sensitizes refractory cancer cells to alphavirus M1 in vitro, in vivo, and ex vivo. We find that activation of the cAMP signaling pathway inhibits M1-induced expression of antiviral factors in refractory cancer cells, leading to prolonged and severe endoplasmic reticulum (ER) stress, and cell apoptosis. We also demonstrate that M1-mediated oncolysis, which is enhanced by cAMP signaling, involves the factor, exchange protein directly activated by cAMP 1 (Epac1), but not the classical cAMP-dependent protein kinase A (PKA). Taken together, cAMP/Epac1 signaling pathway activation inhibits antiviral factors and improves responsiveness of refractory cancer cells to M1-mediated virotherapy.
Introduction
Human cancer remains a global burden and public health problem,1 despite the progresses made in our understanding of tumorigenesis over the past few decades. Oncolytic virotherapy has been reported to be an emerging and promising modality.2 Viruses preferentially target cancer cells as they can exploit the genetic abnormalities of malignant cells to achieve high-replicative capacity, and in turn, contribute to virus-mediated tumor cell killing. Numerous studies have found that a growing number of viruses including adenovirus,3,4 reovirus,5 poxvirus,6,7 vesicular stomatitis virus,8,9 and herpes simplex virus10 can be adapted to cancer therapies for their restricted replication in tumor cells before or after engineering. For example, Talimogene laherparepvec (T-VEC) is the first oncolytic immunotherapy used in a phase 3 clinical trial that demonstrated therapeutic benefit against melanoma.11 The durable response rate was found to be significantly higher with T-VEC (16.3%) than with granulocyte-macrophage colony-stimulatory factor (GM-CSF) (2.1%). Median overall survival was 23.3 months with T-VEC and 18.9 months with GM-CSF.12
Although the evidence for oncolytic viruses inhibiting cancer cell viability in vitro is definitive, the penetrance of oncolytic activity ranges as a consequence of a variety of critical cellular barriers. For example, intratumoral innate immunity can play a crucial role in blocking the therapeutic spread of oncolytic viruses. Pattern-recognition receptors (PRR), both cytoplasmic receptors and Toll-like receptors, recognize invading viruses to induce the Interferon signaling cascade to restrict virus replication.13 IRF3 and IRF7 are crucial transcription factors for Interferon production. Induced Interferon will further lead to the amplified expression of a diverse panel of Interferon stimulated genes, such as MDA5 and IFIT1. MDA5 is a cytoplasmic helicase protein that recognizes viral dsRNA and induces the antiviral response.14 IFIT1 was previously shown to repress viral replication by binding to the eIF3 initiation factor15 and sequestering specific viral RNAs.16
Incidentally, activation of cyclic adenosine monophosphate (cAMP) signal pathway can inhibit the innate immune response of the host,17 lipopolysaccharide (LPS)- or polyinosinic:polycytidylic acid (Poly[I:C])-induced IFNs production in vitro,18 and the production of inflammatory mediators.19 cAMP associates with components related to a variety of cellular activities, including immunosuppression,18 apoptosis,20 generation of reactive oxygen intermediates,21 and phagocytosis.22 In eukaryotic cells, protein kinase A (PKA) is widely known to be the primary effector of cAMP, controlling many cellular mechanisms such as gene transcription, ion transport, and protein phosphorylation.23,24 In addition, the guanine nucleotide exchange factor Epac1 has also been identified as a receptor for cAMP.25 The mediated control of these pathways may be key to overcoming inhibitory antiviral mechanisms.
We have previously identified a naturally occurring alphavirus (M1) as a selective killer that targets tumors deficient in zinc-finger antiviral protein (ZAP) in vitro and in vivo.26 M1 is an alphavirus isolated in the 1960s from the Hainan province of China, and belongs to the Togavirus family of viruses.27,28 The genome of M1 is 11,690 nucleotides (nt) in length and contains two open reading frames, encoding four nonstructural proteins (nsP1-nsP2-nsP3-nsP4), and five structural proteins (C-E3-E2-6K-E1).27 Importantly, we previously observed that M1 causes prolonged and severe endoplasmic reticulum (ER) stress that in turn induced cell apoptosis.
ER stress is primarily caused by endogenous imbalances in the cell, which include increased protein production, loss of Ca2+ homeostasis, inhibition of N-linked glycosylation, and accumulation of misfolded proteins.29 During virus infection, the lumen of the ER rapidly accumulates substantial amounts of viral protein required for virus replication. If the stress is prolonged and unresolvable, three central apoptotic pathways are activated: the C/EBP-homologous protein (CHOP) pathway, the Jun N-terminal kinase (JNK) pathway, and the Caspase-12 pathway.30
In this study, we sought to investigate the anticancer effectiveness of M1/cAMP combination treatment and uncover the mechanisms. cAMP signaling pathway activators, as tumor-specific viral sensitizers, have the potential to dramatically increase the spectrum of malignancies amenable to oncolytic virotherapy.
Results
M1 infection inhibits cancer cell viability
To determine the oncolytic efficiency of M1, a panel of commonly used cancer cell lines (Supplementary Table S1) was challenged with M1 for 48 hours and cell viability was subsequently measured (Figure 1a). Sensitivity to M1 treatments was categorized into three groups. We found that 12.3% of cancer cells were inhibited by more than 75% (hypersensitive), 47.1% of cancer cells were inhibited between 25 and 75% (sensitive, S), and in 40.3% of cancer cells, inhibition was lower than 25% (refractory, R) (Figure 1b). Using titer determination, we found that M1 replication was significantly higher in hypersensitive cancer cells and lower in refractory cancer cells (Figure 1c). These results suggest that the replication of M1 highly associates with oncolysis. Furthermore, we chose three additional cell lines representing the three sensitivity groups to validate these results. We observed that M1 rapidly replicates in the hypersensitive bladder cancer cell line T24. M1 replicates slower in the sensitive Huh-6 liver cancer cell line. While in the refractory HCT-116 colorectal cancer cell line, M1 replication lags (Figure 1d,e).
Figure 1.
The oncolytic effects of M1 in diverse cancer lines. (a) Cell viability of 57 cancer cell lines. Cells were infected with or without M1 (10 plaque forming unit (PFU)/cell) and cell viabilities were determined 48 hours postinfection by MTT assay. (b) Sensitivity distribution of cancer cells to M1-mediated inhibition. (c) Virus replication levels in different cancer cells (mean ± SD). All the cells were infected with 0.1 PFU/cell and viral titer were determined 36 hours postinfection. (d) Dose response of M1 in three representative cancer cells. HCT-116 (refractory), Huh-6 (sensitive), and T24 (hypersensitive) were infected with M1 virus at the indicated multiplicity of infection (MOI) (mean ± SD). (e) Time course of viral replication in the three representative cells after infection of M1 (0.01 PFU/cell).
M1 infection induces antiviral factors in refractory cancer cells, while cAMP signal abrogates them
We next probed whether M1 infection affected the expression of antiviral factors related to the Interferon cascade in refractory cancer cells. We discovered that IFNA, IFNB, IRF3, IRF7, MDA-5, and IFIT1 were dramatically upregulated in HCT-116 after M1 treatment (Figure 2a–f). Thus, we speculated that blocking the induced antiviral response may increase M1 replication and enhance M1-induced oncolysis.
Figure 2.
Activation of cAMP signaling inhibits M1 virus-induced antiviral factor expression. (a–f) The colorectal cancer cell line HCT-116 was infected with M1 virus (10 plaque forming unit (PFU)/cell) and interferon cascade genes were quantified. IFNA (a), IFNB (b), IRF3 (c), IRF7 (d), MDA5 (e), and IFIT1 (f) show mRNA fold-expression at 0, 6, 12, and 24 hours after M1 infection (mean ± SD). (g–l) HCT-116 cells were infected with M1 virus (10 PFU/cell) in the presence or absence of db-cAMP (1 mmol/l) or Forskolin (10 μmol/l), and mRNA levels were quantified by reverse transcription-polymerase chain reaction (mean ± SD). IFNA (g), IFNB (h), IRF3 (i), IRF7 (j), MDA5 (k), and IFIT1 (l) show mRNA fold-expression after M1 infection and db-cAMP treatments. Fold-expression of genes was normalized to β-actin. *P < 0.05; **P < 0.01.
cAMP has been reported to block various components of cell activation such as the production of IFN-α and IFN-β, and the production of inflammatory mediators.18 To block the induced antiviral response, we used two cAMP signaling activators to inhibit the M1-induced antiviral response: db-cAMP (an analogue of cAMP, widely used in vitro), and Forskolin (a ubiquitous activator of eukaryotic adenylyl cyclase that increases intracellular cAMP levels). We observed that cAMP signaling activators dramatically abrogated the induced expression of genes related to the Interferon cascade (Figure 2g–l). These results are in accordance with our hypothesis that cAMP activation blocks the induced antiviral response.
Activation of the cAMP signaling pathway cooperates with M1 to selectively kill cancer cells
To examine the oncolytic effects of M1, we infected HCT-116 and Capan-1 cancer cells. L-02 normal human liver cells were also infected as controls. Our results show that M1 alone had little effect on cancer or normal cells, even at 10 plaque forming unit (PFU)/cell (Figure 3a). Interestingly, the cAMP signaling activators db-cAMP, 8-CPT-cAMP (another analogue of cAMP), and Forskolin significantly enhanced oncolytic efficiency of M1 in cancer cells (Figure 3b,c and Supplementary Figure S1). Importantly, we observed that these activators did not inhibit cell viability of L-02 cells and primary human hepatocyte cells in conjunction with M1 infection (Figure 3d,e and Supplementary Figure S1). We next determined the cell viabilities of 11 additional cell lines treated with M1/db-cAMP. We observed that this combination of treatment inhibits the cell viabilities of a diverse range of refractory cancer cells, but not normal cells (Figure 3f). Moreover, treating with db-cAMP significantly increased viral titers in HCT-116 and Capan-1 cells in a time-dependent manner, but had no effect on L-02 and human hepatocyte cell lines (Figure 3g–j). At 24 hours postinfection, viral proteins E1 and NS3, in HCT-116 and Capan-1 cells, were dramatically increased by db-cAMP, but not in L-02 or human hepatocyte cells (Figure 3k). To validate the effect specificities of M1/db-cAMP treatment, we also tested two additional normal human primary cell lines: Human skeletal muscle cells, and Human aortic endothelial cells. Our results show that db-cAMP has no effect on the cell viability of noncancer cells with M1 infection (Supplementary Figure S2). All together, we demonstrate that cAMP signaling activators selectively facilitates M1 oncolysis in cancer cells.
Figure 3.
Activation of cAMP signaling renders refractory cancer cells, but not normal cells, sensitive to oncolytic virus M1. (a–e) Determination of cell viability by MTT assays. Cancer cells (HCT-116 and Capan-1) and normal cells (L-02 and Human hepatocyte) were infected with M1 virus and treated with or without dbcAMP. Following 72 hours, cell viabilities were determined by MTT assay (mean ± SD). (f) Bar graphs depict the relative differences in AUC (area under the curve) (i.e., gray areas shown in b and c). (g–j) Viral titer determination in different cell lines (mean ± SD). Ctl, control groups. *P < 0.05; **P < 0.01. (k) Western blots showing the expression of viral proteins E1 and NS3 24 hours postinfection. “P” indicates positive controls from M1 infected T24 cancer cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Enhanced oncolysis is mediated by prolonged and severe ER stress
We next sought to determine the mechanism underlying M1-induced oncolysis. We hypothesized that db-cAMP treatment increases viral replication, resulting in an increase in nonfolded or misfolded viral protein accumulating in the lumen of the ER. Such an occurrence could subsequently induce ER stress, and when ER stress levels become insurmountable, this could cause the activation of apoptosis. Of note, viral infections, by HSV-1, rhabdovirus, M1, and others are known to initiate ER stress-mediated apoptosis.31,32,33
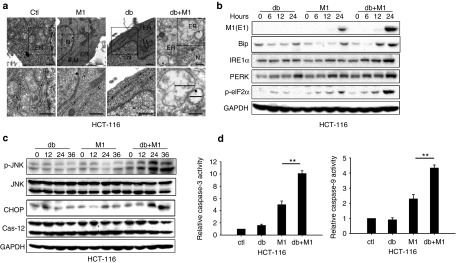
Hence, we reasoned that db-cAMP cooperates with M1 to induce prolonged and severe ER stress, leading to programmed cell death in refractory cancer cells. Using transmission electron microscopy, we observed catastrophic destruction of ER in HCT-116 cells 48 hours after treatment with db-cAMP and M1 (Figure 4a). To confirm the increase of ER stress levels, we probed the expression of markers of the unfolded protein response: the abundant ER chaperone immunoglobulin binding protein, inositol-requiring protein-1α, protein kinase RNA-like ER kinase, and its downstream phosphorylated eukaryotic translational initiation factor 2α (eIF2).34 All of these markers were elevated during M1/db-cAMP treatment (Figure 4b), indicating that ER stress response is activated when cells are exposed to M1 and db-cAMP. We next queried whether specific ER stress-mediated apoptosis pathways were induced by M1/db-cAMP treatments (CHOP pathway, JNK pathway, and caspase-12 pathway).30 Our results showed that CHOP and phosphorylated JNK were elevated in HCT-116 cells after M1/db-cAMP treatment, but cleaved-Caspase-12 was not (Figure 4c). Taken together, M1/db-cAMP treatment induced catastrophic ER stress and cell apoptosis, via CHOP and JNK pathways, but not via Caspase-12 activation.
Figure 4.
M1 infection plus db-cAMP treatment induces severe and prolonged ER stress, leading to cell apoptosis. (a) Transmission electron microscopy images (9,700, up; 13,800, down) of HCT-116 cells. The marker indicates the relative size of the ER. N, nucleus. M, mitochondrion. Scale bars, 500 nm. (b) Expression of ER stress markers by western blot. (c) Western blots of p-JNK, Jun N-terminal kinase (JNK), C/EBP-homologous protein, and Caspase-12 in HCT-116 cells treated with db-cAMP, M1, or M1/db-cAMP. (d) Caspase-3 and Caspase-9 activity assays (mean ± SD). HCT-116 cells were plated on 96-well plates and M1 virus was infected for 72 hours in the presence or absence of db-cAMP. **P < 0.01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine the biological consequences of ER stress as a function of M1/db-cAMP, we next asked whether M1/db-cAMP treatment stimulated the intrinsic apoptosis pathway. We observed marked abnormal nuclear morphology and chromatin condensation within the nucleus during M1/db-cAMP treatments in HCT-116, but not in L-02 cells (Supplementary Figure S3). This observation suggested to us that the apoptosis pathway was activated. Thus, we quantitated the activities of Caspase-9 and its downstream effector Caspase-3 in HCT-116 cells. Notably, these factors were significantly activated during M1/db-cAMP treatment compared with M1 infection alone (Figure 4d). To determine if the mitochondrial apoptotic pathway was activated, we assayed mitochondrial membrane potential using JC-1 staining. JC-1 is a green-fluorescent monomer at low-membrane potential, indicative of apoptotic cells. At higher membrane potentials, JC-1 forms red-fluorescent aggregates, indicative of healthy cells. We found dramatic depolarization of the mitochondrial membrane with M1/db-cAMP treatment (Supplementary Figure S3). These results together demonstrate that the intrinsic apoptosis pathway is triggered in tumor cells.
Epac1 is necessary for cAMP-enhanced viral oncolysis
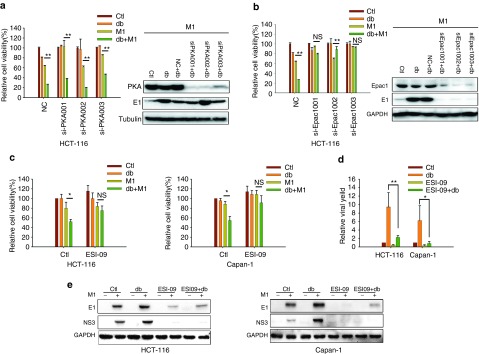
There are a handful of known downstream effectors of cAMP, including PKA/CREB, Epac, and cyclic nucleotide-gated channels.35 We therefore aimed to determine the mechanism by which M1/db-cAMP treatment leads to oncolysis. We first knocked down both PKA and CREB by small interfering RNAs (siRNAs), and observed that neither PKA nor CREB could abrogate the increased oncolytic activity or virus replication by db-cAMP (Figure 5a and Supplementary Figure S4).
Figure 5.
Epac1 is the downstream effector of cAMP. (a and b) The effects of PKA and Epac1. HCT-116 cells were transfected with small interfering RNAs (siRNAs) against PKA or Epac1 and infected with or without M1 (10 plaque forming unit (PFU)/cell) in the presence or absence of db-cAMP. Cell viabilities were determined by MTT assay (left) (mean ± SD). PKAcα (a, right) and Epac1 (b, right) expression levels were determined. (c) The effect of Epac1 inhibitor treatment in cancer cells (mean ± SD). HCT-116 and Capan-1 cells were pretreated with ESI-09 (10 μmol/l) for 1 hour and then infected with M1 (10 PFU/cell) in the presence or absence of 400 μmol/l db-cAMP for 72 hours. (d and e) Viral production after Epac1 inhibitor treatment (mean ± SD). All four groups were infected with M1 (0.1 PFU/cell) (d). NC, negative control (scrambled siRNA). *P < 0.05; **P < 0.01; NS, not significant; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We next performed siRNA knock-down of Epac1, a guanine nucleotide exchange factor for the Ras-like small GTPase Rap1. We observed that both db-cAMP-induced oncolytic activity and M1 viral production were abrogated by siRNAs against Epac1 (Figure 5b). Furthermore, we used an Epac1-specific inhibitor ESI-09 (ref. 36) to pharmacologically block the activity of Epac1. We found that ESI-09 also reversed the decrease in cell viability caused by M1/db-cAMP treatment in both HCT-116 and Capan-1 cells (Figure 5c). Additionally, ESI-09 significantly abrogated db-cAMP-induced viral protein production after 0.1 PFU/cell of M1 infection (Figure 5d). Similarly, we observed that ESI-09 decreased the expression of the viral structural protein E1, and the non-structural protein NS3 in independent of db-cAMP (Figure 5e). These results indicate that the increased oncolytic activities of db-cAMP are mediated by Epac1, but not by PKA.
8-CPT-cAMP inhibits tumor growth in combination with M1 and promotes virus replication in tumors
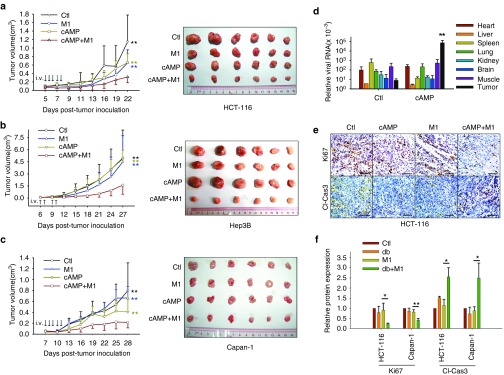
Given the significant oncolytic efficacy of a combined M1 and cAMP activator in vitro treatment, we next sought to evaluate the antitumor efficacy of this combination in a subcutaneous xenograft model. We therefore developed an HCT-116 subcutaneous xenograft model in nude mice. The in vivo use of 8-CPT-cAMP for acute promyelocytic leukemia therapy has been previously reported.37 Thus, we performed intravenous injections of M1, 8-CPT-cAMP, or in combination. Interestingly, M1 infection and 8-CPT-cAMP treatments together significantly restricted tumor growth compared with vehicle, M1, or 8-CPT-cAMP treatment groups (Figure 6a). To validate the combinatorial effects of M1 and 8-CPT-cAMP, we generated hepatocellular carcinoma Hep-3B and pancreatic cancer Capan-1 subcutaneous xenograft models. In the combined group, we also observed significant inhibition of tumor growth in Hep-3B and Capan-1 subcutaneous xenografts by both tumor growth curves and tumor mass indices (Figure 6b,c).
Figure 6.
8-CPT-cAMP plus M1 virus significantly reduces tumor size. (a–c) Measurement of the oncolytic effects of M1 in vivo. Nude mice (NU/NU) bearing subcutaneous HCT-116 (a), Hep-3B (b), and Capan-1 (c) tumors were treated with vehicle, 8-CPT-cAMP (20 mg/kg/day), M1 virus (Hep3B, 5 × 106 plaque forming unit (PFU)/day; HCT-116 and Capan-1, 3 × 107 PFU/day), M1 virus and 8-CPT-cAMP (n ≥ 7). Tumor growth was assessed by tumor volume measurement over time (mean ± SD). At experimental endpoints, mice were anesthetized and sacrificed. Tumors were subsequently dissected and photographed. i.v., intravenously injection (tail vein). **P < 0.01, compared with the combination group. (d–f) In vivo distribution of M1 (d) and intratumoral expression of Ki-67 and cleaved-Caspase-3 (e and f). Immunohistochemistry was performed to analyze the expression of Ki-67 and Cleaved-Caspase-3. Relative protein expressions were quantified with Image-Pro Plus 6.0 (IPP 6.0, MediaCybernetics, Rockville, MD). **P < 0.01, compared with tumor in control group. Scale bars, 50 μmol/l.
We have now demonstrated that M1 infection induces an antiviral response in cancer cells. In addition, we have shown that cAMP signaling activators can inhibit the induced antiviral response and M1 replication. We therefore probed the in vivo distribution of M1 and demonstrated that 8-CPT-cAMP specifically and dramatically increases viral RNA expression in subcutaneous tumor tissues (Figure 6d). We also analyzed Ki-67 and cleaved-Caspase-3 expression in subcutaneous xenograft tumor sections by Immunohistochemistry staining across treatment groups (Figure 6e,f). We observed that Ki-67 was significantly downregulated, while cleaved-Caspase-3 was elevated in the M1/8-CPT-cAMP combined group compared with M1 treatment alone group. Collectively, these results indicate that 8-CPT-cAMP and M1 can inhibit tumor growth and can induce tumor apoptosis in vivo.
8-CPT-cAMP enhances oncolytic efficiency of M1 in primary human tumor specimens
To evaluate the clinical relevance of this therapeutic strategy, we finally examined whether cAMP treatment could sensitize freshly derived patient tumor samples to M1-mediated oncolysis. Dissociated cultures isolated from four patients with primary human hepatocellular carcinoma were treated with or without 8-CPT-cAMP in the presence or absence of M1. Cell viabilities were then determined 96 hours following treatment. Representative images of ex vivo tumor specimens are shown in Figure 7a and quantification of the images are shown in Figure 7b. The clinical drug, 5-fluorouracil, for hepatocellular carcinoma was also applied. In three of the four specimens, the combination of 8-CPT-cAMP and M1 dramatically killed tumor cells compared with M1 and 8-CPT-cAMP treatments alone (Figure 7c). Taken together, these results demonstrate the ability of cAMP to specifically enhance the oncolytic effects of M1 in primary tumor tissues, but not in normal human cells (Figure 3 and Supplementary Figure S2b,c).
Figure 7.
8-CPT-cAMP enhances M1 oncolysis in primary tumor specimens. Surgical liver cancer specimens 50–100 mg were cut into small pieces (1 mm3) and cultured in Dulbecco's modified Eagle's medium containing 15% fetal bovine serum at 37 °C. Tissues were then treated with vehicle (OPTI-SFM), 5-fluorouracil (5-Fu, 100 μl/ml), M1 (5 × 107 plaque forming unit (PFU)), 8-CPT-cAMP (1 mmol/l), M1 plus 8-CPT-cAMP, or HgCl2. Tissue viability was assessed by tissue culture-end point staining-computer image analysis (TECIA) after MTT staining. (a) Representative images of ex vivo tumor specimens. The left panel indicates the area of the tissue and the right panel indicates living cells after drug treatment (MTT staining) captured by the TECIA system. (b) Inhibition percentages corresponding to the calculated areas displayed in a. (c) Cytotoxicity after treatment of four specimens (*, not detectable).
Discussion
Our large-scale cell line screen revealed that the sensitivities to M1 are quite variable. We evaluated viral replication in these cancer cells, and we found that the replication of M1 is highest in hypersensitive cancer cells, intermediate in sensitive cancer cells and lowest in refractory cancer cells. These results prompted us to seek strategies to enhance the oncolytic efficiency of M1 in refractory cancer.
We have demonstrated that the activation of cAMP signal pathway can only enhance the oncolytic effects of M1 in refractory cancer cells. Oncolytic viruses exploit the immunodeficiency of cancer cells to achieve replication. This may be the reason that most cancer cell lines are sensitive to the oncolytic virus M1. Nevertheless, M1 infection induces the activation of antiviral factors in refractory cancer cells, restricting M1 replication and oncolysis. We therefore have established that the targeted enhancement of M1-mediated oncolysis by immunosuppressive cAMP in refractory cancer cells, but not in sensitive cells is intrinsically important. Furthermore, we do not find that the intracellular levels of cAMP correlate with the sensitivity of cell lines (data not shown). Therefore, cAMP signaling pathway activation can only improve the oncolytic effects of M1 in refractory cancer cells, but not in sensitive cells.
Increased replication of M1 induces prolonged and severe ER stress. It has been reported that oncolytic Maraba virus induces cancer cell death through ER stress and inhibiton of unfolded protein response, which leads to ER preload, facilitates the oncolytic effects of Maraba virus.38 We found that M1 infection alone induces prolonged and severe ER stress in sensitive cancer cells but not in refractory cancer cells. Thus, cAMP signaling activators sensitize refractory cancer cells to M1 replication, which further induces ER stress-mediated apoptosis. It would be worth testing whether the use of unfolded protein response inhibitors might further lead to enhanced oncolysis by M1.
The immunosuppressive effect of cAMP improves M1 replication and oncolysis in vitro, but its effects of inducing an antitumor immune response during oncolytic virus treatment are still unknown. Our combinatorial use of M1 infection and cAMP activation still requires further exploration in immunocompetent mice and patient tumor samples. It is important that further systematic and detailed investigation regarding the effects of cAMP on the M1-mediated immune response be pursued.
An emerging strategy to safely cope with the rapid clearance of virus is to reactivate oncolytic viruses within tumors.39 Interestingly, based on the induced antiviral response in refractory cancer cells, we have revealed that cAMP activators can transiently diminish induced antiviral response and can increase M1 enrichment in refractory tumors. Most importantly, cAMP activators do not increase viral replication and do not inhibit cell viability in normal cells. Additionally, we did not observe any toxicity in vivo. Thus, cAMP activators can efficiently enrich oncolytic virus within malignant tumors, without changing the tropism of M1. Our studies highlight a novel and safe strategy that enhances the potency of oncolytic viruses.
In summary, our study shows that cAMP, together with M1, dramatically inhibits cancer cell growth in vitro, in vivo, and ex vivo. With systemic administration of cAMP analogues to maintain the robust oncolytic virus replication within tumors, this combinatorial treatment can reduce cancer malignancy and induce cancer cell apoptosis through prolonged and severe ER stress. At the same time, this approach does not induce death in normal cells and visible toxicity in mice. Notably, the ability of cAMP to increase virus replication may identify a novel and potentially targetable strategy to therapeutically kill cancer cells.
Materials and Methods
Reagents, viruses, and cell lines. Production of Alphavirus M1 in this study was described previously.27,28 All viruses were propagated in Vero cells (OPTI-SFM, 12309-019, Thermo Fisher, Waltham, MA) and virus titers were determined by TCID50 in the BHK-21 cell line. All cell lines were maintained at 37 °C with 5% CO2, in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum, penicillin/streptomycin. Cell lines were purchased from American Type Culture Collection and Shanghai Institute of Cell Biology. Reagents used in this study are listed as follows: dbcAMP (100 mm, dissolved in ddH2O, D0627-1G, Sigma, St. Louis, MO), Forskolin (20 mmol/l, dissolved in dimethylsulfoxide, F6886-10MG, Sigma), 8-cpt-cAMP (50 mmol/l, dissolved in ddH2O, C 010-500, Biolog, Germany), ESI-09 (10 mmol/l, dissolved in dimethylsulfoxide, B 133, Biolog).
Cell viability assays. Cell lines were purchased from American Type Culture Collection, Shanghai Institute of Cell Biology. All primary cell lines were purchased from Sciencell Research Laboratories. Cells were seeded in 96-well plates at 4,000 cells per well, and were infected with M1 virus (10 PFU/cell) and various drugs were added as described in the figure legends where applicable. Seventy-two hours later, cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Cells were stained with MTT at a concentration of 1 mg/ml and plates were further cultured at 37 °C for another 3 hours. Media was removed and precipitates were dissolved in 100 μl dimethylsulfoxide. The optical absorbance was determined at 570 nm using an iMark microplate reader (Bio-Rad).
RNA interference. Specific and nontargeting siRNAs, were synthesized by Ribobio (Guangzhou, China). Cells were replaced with 10% fetal bovine serum DMEM (without penicillin/streptomycin). siRNAs were transfected using Lipofectamine RNAiMAX (13778-150, Thermo Fisher) with OPTI-MEM (31985070, Thermo Fisher).
Quantitative reverse transcription-polymerase chain reaction. Total RNA was extracted using TRIzol (Life Technologies) reagent and reverse transcribed to cDNA with oligo (dT). Specific gene expression was quantified using SuperReal PreMix SYBR Green (FP204-02, TIANGEN, Beijing, China) on an Applied Biosystems 7500 Fast Real-Time PCR system (Life Technologies). All genes were normalized to β-actin. Amplification primers (Thermo Fisher) are listed as follows: M1 NS1 sense (GTTCCAACAGGCGTCACCATC), M1 NS1 antisense (ACACATTCTTGTCTAGCACAGTCC); ACTB sense (GATCATTGCTCCTCCTGAGC), ACTB antisense (ACTCCTGCTTGCTGATCCAC); DDX58 sense (ATCCCAGTGTATGAACAGCAG), DDX58 antisense (GCCTGTAACTCTATACCCATGTC). MDA5 sense (TCACAAGTTGATGGTCCTCAAGT), MDA5 antisense (CTGATGAGTTATTCTCCATGCCC). IRF3 sense (AGAGGCTCGTGATGGTCAAG), IRF3 antisense (AGGTCCACAGTATTCTCCAGG). IRF7 sense (CCCACGCTATACCATCTACCT), IRF7 antisense (GATGTCGTCATAGAGGCTGTTG). IFNB sense (GCTTGGATTCCTACAAAGAAGCA), IFNB antisense (ATAGATGGTCAATGCGGCGTC).
Transmission electron microscopy. HCT-116 cells were infected with M1 (10 PFU/cell) in the presence or absence of db-cAMP for 48 hours. In brief, cells were harvested and pelleted at 1,000×g for 5 minutes at room temperature. Cell pellets were then resuspended, washed once with phosphate-buffered saline, pelleted at 1,500×g for 5 minutes, and fixed on ice for 4 hours in 0.1 M phosphate-buffered saline (pH 7.4) containing 2.5% glutaraldehyde and 2% paraformaldehyde. Samples were then submitted to the Zhongshan School of Medicine (Sun Yat-sen University) Electron Microscopy Facility for standard transmission electron microscopy ultrastructural analysis.
Antibodies and western blot analyses. Cells were lysed using M-PER Mammalian Protein Extraction Reagent (Thermo Scientific) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed. Antibodies used in this study are listed as follows: Human immunoglobulin binding protein (3177, Cell Signaling Technology, Danvers, MA), eIF-2α (5324, Cell Signaling Technology), phosphorylated eIF-2α (3398, Cell Signaling Technology), JNK (9252, Cell Signaling Technology), phosphorylated JNK(9255, Cell Signaling Technology), Caspase-12 (2202, Cell Signaling Technology), glyceraldehyde-3-phosphate dehydrogenase (AP0060, Bioworld, St. Louis Park, MN), β-actin (AP0063, Bioworld), M1 E1 and NS3 (produced by Beijing Protein Innovation, Beijing, China) PKAcα (4782s, Cell Signaling Technology), Epac1 (4155s, Cell Signaling Technology, Danvers, MA), IRF7 (4920s, Cell Signaling Technology), Ki-67 (9449s, Cell Signaling Technology), and Cleaved-Caspase-3 (9664s, Cell Signaling Technology).
Caspase activity analyses. Cells were cultured in 96-well plates and infected with M1 virus (10 PFU/cell) in the presence or absence of db-cAMP. Caspase-3/7 and Caspase-9 activities were determined by Caspase-Glo Assay Systems (Promega, Madison, WI) according to manufacturer's protocols. The results were normalized to cellular viability (MTT assay).
Animal models. Mouse studies were approved by the Animal Ethical and Welfare Committee of Sun Yat-sen University. HCT-116 (5 × 106 cells/mouse), Hep3B (5 × 106 cells/mouse), and Capan-1 (1x107 cells/mouse) cancer cells were inoculated subcutaneously into the hind-flank of 4-week-old female BALB/c-nu/nu mice. After 5–7 days, palpable tumors developed (50 mm3), and mice were divided into four groups by random. The four groups were intravenously injected with vehicle (OPTI-SFM), M1, 8-CPT-cAMP, and in combination (M1/8-CPT-cAMP) in a total volume of 200 μl. Tumor lengths and widths were measured every other day and the volume was calculated according to the formula (length × width2)/2. Measurements were performed blinded to group allocations.
Immunohistochemistry assay. The expressions of Cleaved-Caspase 3 and Ki-67 in tumors were characterized by immunohistochemistry using specific antibodies. Briefly, tumor sections (4 μm) were dewaxed in xylene, hydrated in descending concentrations of ethanol, immersed in 0.3% H2O2-methanol for 30 minutes, washed with phosphate-buffered saline, and probed with monoclonal anti-Cleaved-Caspase 3 (1:100) or Ki-67 antibodies (1:100) or isotype control at 4 °C overnight. After washing, the sections were incubated with biotinylated goat anti-rabbit or anti-mouse IgG at room temperature for 2 hours. Immunostaining was visualized with streptavidin/peroxidase complex and diaminobenzidine, and sections were then counterstained with hematoxylin. We quantified the relative protein expression with software Image-Pro Plus 6.0 (MediaCybernetics).
Mitochondrial membrane potential assays. 5 μmol/l of the fluorescent probe, 5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine iodide (JC-1, Sigma-Aldrich, St. Louis, MO) was treated on cultured cells seeded onto 35-mm dishes, and incubated for 20 minutes at 37 °C. Cells were then washed twice with DMEM and imaged by fluorescence microscopy (Olympus, Tokyo, Japan) using a “dual-bandpass” filter.
Ex vivo. Tissue culture-end point staining-computer image analysis (TECIA) was used to evaluate the ex vivo anticancer activity. TECIA is an improved histoculture drug response assay, and is described elsewhere.40 Primary tumor tissue specimens were obtained from consenting patients who underwent tumor resection. Informed consent was obtained from the patients before tissue collection. The work was approved by an ethics review committee at Sun Yat-sen University (Guangzhou, China). The institutional review board of Sun Yat-sen University Cancer Center has approved all human studies. Tumor samples were received in cell culture medium and processed within 2–6 hours. Samples were manually divided using a scalpel blade into approximately 1 mm3 blocks using sterile techniques. The explants were placed on moist but not submerged filter-paper inserted into single wells of 24-well plates with 1 ml DMEM containing 15% fetal bovine serum, and cultured at 37 °C with 5% CO2 for 24 hours. The A-score was recorded by volumetric integral of samples using the Image Analysis System. Samples were then exposed to saline (negative control), 5-fluorouracil, M1, cAMP, cAMP plus M1, and HgCl2 (positive control) for 4 days. After treatment, 100 μl of MTT (5 mg/ml) was added and cultured for 4 hours. The B-score was determined by the area and intensity of staining using the Image Analysis System. Every treatment on each sample was tested in quadruplicate. The efficacy of different treatments was presented as percentage inhibition that was calculated using the following formula: Inhibition (%) = (1-(mean of B-scores of treated sample/mean of A-scores of treated sample)/(mean of B-scores of control)/(mean of A-scores of control)) × 100%. Neither negative control samples with low MTT staining nor positive control samples with low inhibition (<80%) were accepted for analysis.
Statistical analysis. All statistical analyses were performed using SPSS 13.0 software (SPSS, IBM, Armonk, NY). Most of the data were subjected to Student's t-test or one-way analysis of variance followed by Dunnett's multiple post-hoc tests. Values of tumor volume were analyzed by repeated measures one-way analysis of variance. Unless otherwise indicated, all error bars indicate SD. Significance was defined as P < 0.05.
Study approval. All animal studies were approved by the Sun Yat-sen University Institutional Animal Care and Use Committee. Primary cancer tissue specimens were approved by an ethics review committee at Sun Yat-sen University.
SUPPLEMENTARY MATERIAL Figure S1. 8-CPT-cAMP and Foskolin also selectively enhance oncolytic effects of M1 in refractory cancer cells. Figure S2. Combination of M1 with db-cAMP does not affect the cell viability of normal cells. Figure S3. Combination of M1 with db-cAMP induces nuclear concentration and mitochondrial potential loss in HCT-116 cancer cells. Figure S4. Knockdown of CREB did not affect viral protein expression level during db-cAMP treatment. Table S1. Cancer cell line screening for M1 anti-cancer efficacy.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81273531, 81373428, 81202555 and 81573447) and the Guangdong Provincial Natural Science Foundation Grant (2015A030313081). The authors have declared that no conflict of interest exists.
Supplementary Material
Cancer cell line screening for M1 anti-cancer efficacy.
References
- Jemal, A, Bray, F, Center, MM, Ferlay, J, Ward, E and Forman, D (2011). Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Russell, SJ, Peng, KW and Bell, JC (2012). Oncolytic virotherapy. Nat Biotechnol 30: 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise, C and Kirn, DH (2000). Replication-selective adenoviruses as oncolytic agents. J Clin Invest 105: 847–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell, CM, Shibata, A, Tookman, LA, Archibald, KM, Flak, MB, Pirlo, KJ et al. (2011). Genomic DNA damage and ATR-Chk1 signaling determine oncolytic adenoviral efficacy in human ovarian cancer cells. J Clin Invest 121: 1283–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, KL and Lee, PW (2000). Reovirus as a novel oncolytic agent. J Clin Invest 105: 1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne, SH, Hwang, TH, O'Gorman, WE, Bartlett, DL, Sei, S, Kanji, F et al. (2007). Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J Clin Invest 117: 3350–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parato, KA, Breitbach, CJ, Le Boeuf, F, Wang, J, Storbeck, C, Ilkow, C et al. (2012). The oncolytic poxvirus JX-594 selectively replicates in and destroys cancer cells driven by genetic pathways commonly activated in cancers. Mol Ther 20: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl, DF, Lichty, BD, tenOever, BR, Paterson, JM, Power, AT, Knowles, S et al. (2003). VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4: 263–275. [DOI] [PubMed] [Google Scholar]
- Wongthida, P, Diaz, RM, Galivo, F, Kottke, T, Thompson, J, Melcher, A et al. (2011). VSV oncolytic virotherapy in the B16 model depends upon intact MyD88 signaling. Mol Ther 19: 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todo, T (2012). Active immunotherapy: oncolytic virus therapy using HSV-1. Adv Exp Med Biol 746: 178–186. [DOI] [PubMed] [Google Scholar]
- Liu, TC, Galanis, E and Kirn, D (2007). Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat Clin Pract Oncol 4: 101–117. [DOI] [PubMed] [Google Scholar]
- Andtbacka, RH, Kaufman, HL, Collichio, F, Amatruda, T, Senzer, N, Chesney, J et al. (2015). Talimogene Laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol 33: 2780–2788. [DOI] [PubMed] [Google Scholar]
- Akira, S, Uematsu, S and Takeuchi, O (2006). Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- Yoneyama, M, Kikuchi, M, Matsumoto, K, Imaizumi, T, Miyagishi, M, Taira, K et al. (2005). Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175: 2851–2858. [DOI] [PubMed] [Google Scholar]
- Guo, J, Hui, DJ, Merrick, WC and Sen, GC (2000). A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J 19: 6891–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas, YM, Pichlmair, A, Górna, MW, Superti-Furga, G and Nagar, B (2013). Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins. Nature 494: 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon, D, Vanwalleghem, G, Morias, Y, Denoeud, J, Krumbholz, C, Lhommé, F et al. (2012). Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science 337: 463–466. [DOI] [PubMed] [Google Scholar]
- Xu, XJ, Reichner, JS, Mastrofrancesco, B, Henry, WL Jr and Albina, JE (2008). Prostaglandin E2 suppresses lipopolysaccharide-stimulated IFN-beta production. J Immunol 180: 2125–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, K, Takaesu, G, Yoshida, R, Nakaya, M, Kobayashi, T, Kinjyo, I et al. (2009). Cyclic adenosine monophosphate suppresses the transcription of proinflammatory cytokines via the phosphorylated c-Fos protein. Immunity 30: 372–383. [DOI] [PubMed] [Google Scholar]
- Lizcano, JM, Morrice, N and Cohen, P (2000). Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem J 349(Pt 2): 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, G, Giembycz, MA, Rabe, KF, Wolf, B, Barnes, PJ and Magnussen, H (1994). Theophylline suppresses human alveolar macrophage respiratory burst through phosphodiesterase inhibition. Am J Respir Cell Mol Biol 10: 565–572. [DOI] [PubMed] [Google Scholar]
- Aronoff, DM, Canetti, C and Peters-Golden, M (2004). Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol 173: 559–565. [DOI] [PubMed] [Google Scholar]
- Walsh, DA, Perkins, JP and Krebs, EG (1968). An adenosine 3',5'-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem 243: 3763–3765. [PubMed] [Google Scholar]
- Dumaz, N and Marais, R (2003). Protein kinase A blocks Raf-1 activity by stimulating 14-3-3 binding and blocking Raf-1 interaction with Ras. J Biol Chem 278: 29819–29823. [DOI] [PubMed] [Google Scholar]
- Kraemer, A, Rehmann, HR, Cool, RH, Theiss, C, de Rooij, J, Bos, JL et al. (2001). Dynamic interaction of cAMP with the Rap guanine-nucleotide exchange factor Epac1. J Mol Biol 306: 1167–1177. [DOI] [PubMed] [Google Scholar]
- Lin, Y, Zhang, H, Liang, J, Li, K, Zhu, W, Fu, L et al. (2014). Identification and characterization of alphavirus M1 as a selective oncolytic virus targeting ZAP-defective human cancers. Proc Natl Acad Sci USA 111: E4504–E4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, JS, Zhao, WZ, Liu, JW, Zhou, H, Tao, JP, Yan, HJ et al. (2007). Genomic analysis of a Chinese isolate of Getah-like virus and its phylogenetic relationship with other Alphaviruses. Virus Genes 35: 597–603. [DOI] [PubMed] [Google Scholar]
- Hu, J, Cai, XF and Yan, G (2009). Alphavirus M1 induces apoptosis of malignant glioma cells via downregulation and nucleolar translocation of p21WAF1/CIP1 protein. Cell Cycle 8: 3328–3339. [DOI] [PubMed] [Google Scholar]
- Kaufman, RJ (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13: 1211–1233. [DOI] [PubMed] [Google Scholar]
- Szegezdi, E, Logue, SE, Gorman, AM and Samali, A (2006). Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7: 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, HL, Liao, CL and Lin, YL (2002). Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol 76: 4162–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif, KD, Mori, K and Siddiqui, A (2002). Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol 76: 7453–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, JY, Hurwitz, BS, Bolyard, C, Yu, JG, Zhang, J, Selvendiran, K et al. (2014). Bortezomib-induced unfolded protein response increases oncolytic HSV-1 replication resulting in synergistic antitumor effects. Clin Cancer Res 20: 3787–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, D and Walter, P (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529. [DOI] [PubMed] [Google Scholar]
- Beavo, JA and Brunton, LL (2002). Cyclic nucleotide research – still expanding after half a century. Nat Rev Mol Cell Biol 3: 710–718. [DOI] [PubMed] [Google Scholar]
- Almahariq, M, Tsalkova, T, Mei, FC, Chen, H, Zhou, J, Sastry, SK et al. (2013). A novel EPAC-specific inhibitor suppresses pancreatic cancer cell migration and invasion. Mol Pharmacol 83: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, B, Ren, ZH, Liu, P, Chen, LJ, Shi, JY, Dong, Y et al. (2013). 8-CPT-cAMP/all-trans retinoic acid targets t(11;17) acute promyelocytic leukemia through enhanced cell differentiation and PLZF/RARα degradation. Proc Natl Acad Sci USA 110: 3495–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, DJ, Lefebvre, C, Allan, K, Brun, J, Sanaei, CA, Baird, S et al. (2011). Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell 20: 443–456. [DOI] [PubMed] [Google Scholar]
- Nguyên, TL, Abdelbary, H, Arguello, M, Breitbach, C, Leveille, S, Diallo, JS et al. (2008). Chemical targeting of the innate antiviral response by histone deacetylase inhibitors renders refractory cancers sensitive to viral oncolysis. Proc Natl Acad Sci USA 105: 14981–14986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, SY, Jeon, DG, Cho, WH, Song, WS, Kim, MB and Park, JH (2006). Preliminary study of chemosenstivity tests in osteosarcoma using a histoculture drug response assay. Anticancer Res 26(4B): 2929–2932. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cancer cell line screening for M1 anti-cancer efficacy.