Abstract
Due to their ability to knock down the expression of any gene, siRNAs have been heralded as ideal candidates for treating a wide variety of diseases, including those involving “undruggable” targets. However, the therapeutic potential of siRNAs remains severely limited by a lack of effective delivery vehicles. Recently, lipid nanoparticles (LNPs) containing ionizable cationic lipids have been developed for hepatic siRNA delivery. However, their suitability for delivery to other cell types has not been determined. We have modified LNPs for preferential targeting to dendritic cells (DCs), central regulators of immune responses. To achieve directed delivery, we coated LNPs with a single-chain antibody (scFv; DEC-LNPs), specific to murine DEC205, which is highly expressed on distinct DC subsets. Here we show that injection of siRNAs encapsulated in DEC-LNPs are preferentially delivered to DEC205+ DCs. Gene knockdown following uptake of DEC-LNPs containing siRNAs specific for the costimulatory molecules CD40, CD80, and CD86 dramatically decreases gene expression levels. We demonstrate the functionality of this knockdown with a mixed lymphocyte response (MLR). Overall, we report that injection of LNPs modified to restrict their uptake to a distinct cell population can confer profound gene knockdown, sufficient to inhibit powerful immune responses like the MLR.
Introduction
The potential of RNAi to silence any gene has made it an attractive therapeutic modality.1 However, the main obstacle to RNAi in the clinic is delivery. To be effective, siRNAs must be transported through the body, bind, and be taken up by target cells where they must traverse the plasma membrane and gain access to the cytosolic compartment, where the RNAi machinery resides. To be useful, ease of formulation and administration, overall cost, and any associated toxicities are essential considerations.
Dendritic cells (DCs) are central regulators in immune responses. DCs are heterogeneous, with subsets defined phenotypically, functionally and by location (reviewed in ref. 2). A delivery vehicle that knocks down expression of specific genes in a distinct DC population(s) would be a valuable tool for targeting diverse diseases including cancers, infectious diseases, autoimmunity and as a vaccine component. Therefore, a platform that delivers siRNAs to specific DC subsets in situ would be useful for inhibiting or activating immune responses. However, gene modulation of primary immune cells in vivo remains a significant challenge.
Lipid nanoparticles (LNPs) are one of the most advanced platforms for siRNA delivery to hepatocytes, and are under clinical evaluation for conditions that require hepatic gene silencing.3,4,5 These LNPs typically contain ionizable cationic lipids (pKa ~6.5) that bind nucleic acids via electrostatic interactions at low pH, but are charge neutral at pH 7.4. As a well-perfused organ the liver is highly amenable to uptake of i.v. injected cargoes. Furthermore, LNP uptake by hepatocytes is mediated by association with serum ApoE leading to efficient uptake via low-density lipoprotein receptors in the liver.6 Following cellular uptake of the LNP, the ionization of the lipid within acidic endosomes is thought to promote siRNA escape to the cytosol.7 Importantly, these LNPs are associated with minimal toxicity, including little induction of proinflammatory cytokines following administration of physiologically relevant doses.8 In contrast to the liver, siRNA delivery to extra-hepatic cells is challenging. In particular, immune cells such as DCs are relatively resistant to siRNA uptake in vitro and in vivo.
Although designed for hepatic gene silencing, the efficacy of these LNPs and their derivatives has been assessed for RNAi-mediated gene silencing in macrophages (MOs) and DCs in vitro and in vivo. LNP uptake and modest gene silencing was achieved at high doses of LNPs (~ 5mg/kg).9 Together with a complementary study,10 this work demonstrates the feasibility of gene silencing in immune cells using LNP technology. Clearly, however, LNP formulations must be modified to achieve optimal gene silencing in immune cells.
Enhanced uptake of LNPs by primary hepatocytes is mediated by apolipoprotein E binding to the neutral LNPs.11 This results in recognition by receptors including the low-density lipoprotein receptor and scavenger receptors largely expressed on hepatocytes.6 The mechanism of uptake strongly suggests that retargeting of these particles to other receptors or other tissues is possible. Indeed, Akinc et al.6 demonstrated that LNPs modified with N-acetylgalactosamine (GalNAc) were retargeted to the asialoglycoprotein receptor (ASGPR) expressed by hepatocytes in vivo. For delivery to nonhepatic cells, we have shown in vitro that anti-transferrin receptor aptamers can be used to redirect similar LNPs.12 More recently, Liang et al.13 described a similar approach using aptamer-coated LNPs to target osteoblasts in vivo. For uptake by immune cells, a recent study coated LNPs with a full-length anti-CD4 antibody for siRNA delivery to primary CD4+ T cells. LNP uptake and gene silencing was observed in CD4+ T cells in various organs including the spleen, lymph nodes, and blood.14 These studies demonstrate the potential for attaching exogenous ligands to LNPs for delivery to hepatic, and importantly nonhepatic primary cells in vivo.
Drawing from these works, we reasoned that this LNP platform could be a useful foundation for the delivery of siRNAs to DCs to modulate immune responses. To achieve this, we modified LNPs using a single chain antibody (scFv) specific for the DC receptor DEC205, a C-type lectin expressed at high levels on CD8α+ DCs.15 DEC205+ DCs mediate cross-presentation of antigen resulting in modulation of CD8+ T-cell responses.16,17,18,19 Therefore, inhibition of gene expression by DEC205+ DCs is a potentially powerful approach for regulating CD8+ T-cell activation.
Using this approach, we assessed the ability of DEC-LNPs (LNPs coated with scFv specific for murine DEC205, containing siRNAs specific for costimulatory molecules) to inhibit immune responses. Intravenous (i.v.) injection of DEC-LNPs resulted in preferential uptake of the DEC-LNPs by splenic DEC205+ DCs. Furthermore, when coadministered with adjuvant, LNPs containing siRNA targeting CD40, CD80, and CD86 reduced expression of these costimulatory molecules to levels similar to those observed in immature DCs. Most importantly, DCs isolated from mice injected with a low dose (~0.6 mg/kg) of these DEC-LNPs, were able to suppress a robust mixed lymphocyte reaction (MLR), demonstrating the functional efficacy of this approach. Interestingly, when we performed experiments using 2' fluoro (2'F) modified siRNA, a formulation reported to be nonimmunostimulatory and nonimmunogenic,20 significant immune activation of the targeted DCs was detected. This effect was not observed in assays performed with human peripheral blood mononuclear cells (PBMCs) or monocyte-derived DCs (moDCs) and could be mitigated through selective 2′-O-methyl (2′OMe) modification of the siRNA. Overall, our study demonstrates the functionality of LNPs modified for receptor targeting for siRNA delivery to DCs. Additionally our results suggest that specific cell targeting can mediate immunological responses to siRNA formulations. While care needs to be taken in designing and evaluating targeted approaches using LNPs, we found that 2′OMe modification of the siRNA was sufficient to negate immune activation. Our approach enhances the efficacy of siRNA-mediated gene silencing by ~10-fold when compared to nontargeted delivery. Therefore, this targeted strategy should prove effective for regulating multiple immune-mediated diseases.
Results
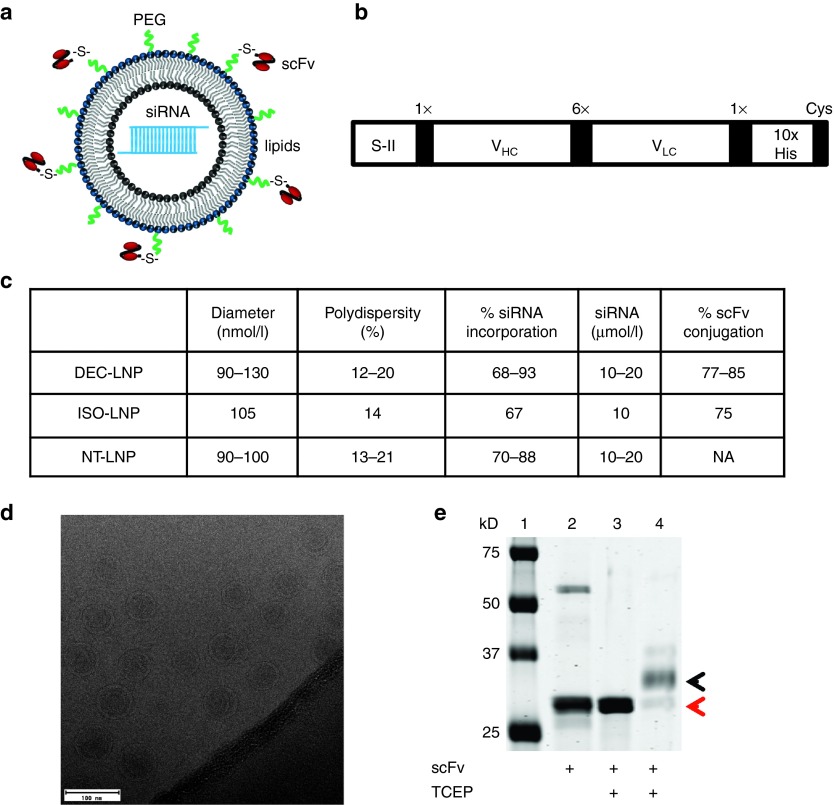
LNPs containing siRNAs specific either for CD80, CD86, CD40, or a control (nontarget) sequence were synthesized by extrusion using a mixture of 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC):DLinDMA:DSPE-PEG:cholesterol at a ratio of 15:40:5:40 (based on refs. 3,7,21). Murine anti-DEC205 scFv with a C-terminal cysteine was attached to the lipid DSPE-PEG by a maleimide group (Figure 1a,b). Following synthesis, we subjected LNPs to quality control to ensure consistency between batches. Dynamic light scattering was used to determine diameter and polydispersity of LNPs, and cryo-electron microscopy confirmed LNP size and their unilamellar structure (Figure 1c,d). The efficiency of siRNA incorporation and scFv binding to DSPE-PEG was also assessed (Figure 1c,e).
Figure 1.
Synthesis of scFv-targeted lipid nanoparticles (LNPs). Schematic of (a) LNP structure and (b) scFv construct. S-II = Strep-tag II (IBA, Germany), 1× and 6× = G4S linker repeats, VHC, variable heavy chain domain; VLC, variable light chain domain, 10× His = 10 histidine repeats, Cys = free cysteine. (c) Quality control assays to determine LNP size (dynamic light scattering), siRNA incorporation, and scFv conjugation. For DEC-LNPs and NT-LNPs, a representative range for these parameters is shown for LNPs generated from multiple batches (n ≥ 3). The ISO-LNP was only generated once for our studies. (d) CryoEM analysis of LNPs. The scale bar = 100 nm. (e) Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis showing scFv conjugation to the PEG lipid. scFv were pretreated with Tris-(2-carboxyethyl) phosphine hydrochloride to ensure efficient conjugation to PEG-maleimide (Lanes 3 and 4). Conjugation of scFv to DSPE-PEG results in retardation of the scFv by ~3 kD (corresponds to the MW of DSPE-PEG; Lane 4; black arrowhead). Unreacted scFv is indicated by a red arrowhead (Lane 4). Lane 1 = protein marker; Lane 2 = nonreduced scFv.
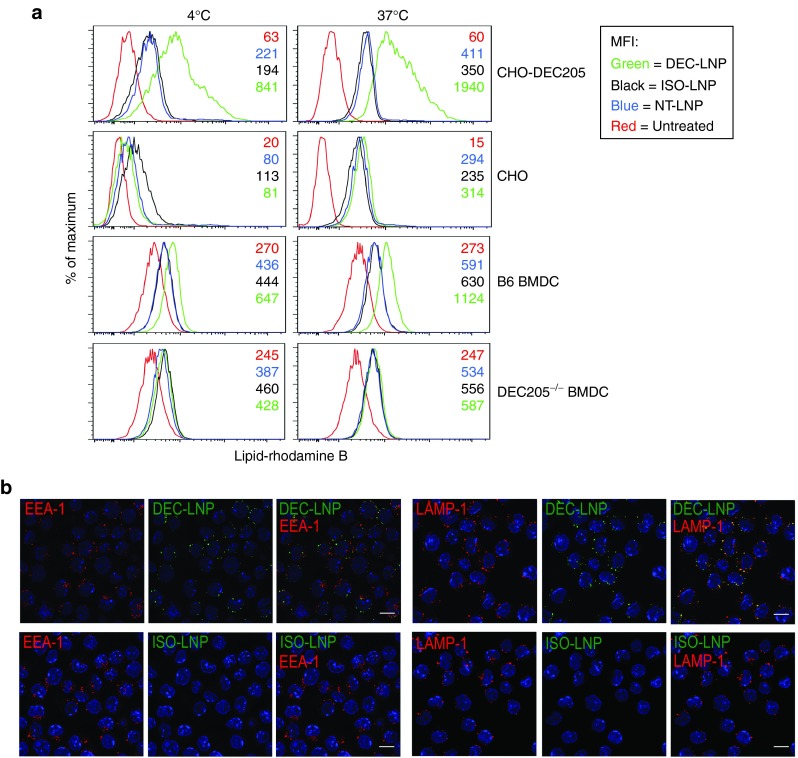
To show specificity of binding to DEC205, scFv-LNPs containing Dy547-labeled siRNA were cultured with either parental (DEC205–) or DEC205+ CHO cells. As seen in Figure 2a, at 4 °C DEC205+ cells bind DEC-LNPs approximately fourfold better compared with nontargeted LNPs (NT-LNPs: LNPs not coated with any scFv), or LNPs coated with an isotype scFv (ISO-LNPs; CHO-DEC205 panel). When similar assays were performed at 37 °C to allow cell uptake, cell staining increased by at least fivefold. Little uptake was observed when LNPs (targeted or nontargeted) were incubated with parental CHO cells (CHO panel). Similar results were obtained when bone marrow-derived DCs (BMDCs), derived from wild-type mice (B6) were incubated with DEC-LNPs. We note that as DEC205 is expressed at lower levels on BMDCs compared with CHO-DEC205, less DEC-LNP binding and uptake was observed in BMDCs (~2-fold enhancement compared with NT- or ISO-LNPs; B6 BMDC panel). To further confirm target-specific uptake we performed experiments using BMDCs derived from DEC205–/– mice, and no specific cell binding or uptake was observed with DEC-LNPs (DEC205–/– panel). Following receptor binding, the DEC205 antibody is internalized via receptor-mediated endocytosis, and is targeted to late endosomes and lysosomes.22 Confocal microscopy confirmed that similar to DEC205 antibody, DEC-LNPs were targeted to LAMP-1+ late endosomes or lysosomes (Figure 2b).
Figure 2.
DEC-lipid nanoparticles (LNPs) are taken up by DEC205+ cells and colocalize to lysosomes. (a) CHO-DEC205 (top panel) and parental CHO (second panel), B6 bone marrow-derived dendritic cell (BMDC) (third panel) and DEC205–/– BMDC (fourth panel) were incubated with DEC-LNPs (green), ISO-LNP (black), NT-LNP (blue) or were left untreated (red). Cells were incubated at 4 °C (left panel) or 37 °C (right panel) for 30 minutes, washed and analyzed by flow cytometry. Numbers in panels denote mean fluorescence intensity. (b) DEC-LNPs and ISO-LNPs containing fluorescently labeled siRNAs were incubated for 1 hour, 37 °C with A20 cells (DEC205+). Following fixation and permeabilization, cells were stained with EEA-1 and LAMP-1 and cellular localization of LNPs determined by confocal microscopy. DEC-LNP, ISO-LNP = green; EEA-1, LAMP-1 = red; DAPI = blue. Images acquired with 63× objective. 10 μm scale bars are shown in the merged images.
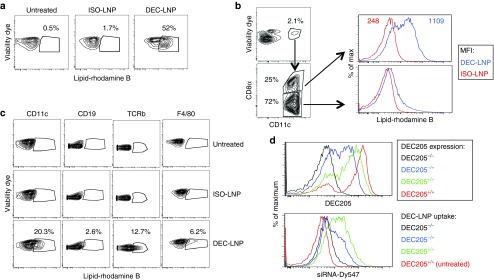
Having demonstrated DEC205-mediated uptake of DEC-LNPs in vitro, we investigated their localization in vivo. B6 mice were injected systemically (intravenous, i.v.) with fluorescently labeled scFv-LNPs. One day later LNP uptake and distribution was determined by flow cytometry. As the DEC205 receptor is endocytosed when it binds its ligand, we could not use this molecule to identify DEC205+ DCs.22 Therefore, we used CD8α, a protein that is coexpressed on DEC205+ DCs, as a surrogate marker for tracking these cells.16,23 More than 50% of CD11c+CD8α+ cells took up DEC-LNPs, and uptake was largely restricted to the CD8α+ DC population, with little uptake observed by CD8α– DCs (Figure 3a,b). Specificity was demonstrated by comparison of uptake of DEC-LNPs and ISO-LNPs. Significant uptake was only observed in DEC205+ DCs following DEC-LNP injection, and other splenic immune cells took up little DEC- or ISO-LNPs (Figure 3a,c). When DEC-LNPs were injected into DEC205–/– mice, no uptake was detected (Figure 3d).
Figure 3.
Injection of DEC-lipid nanoparticles (LNPs) results in uptake by receptor-positive cells. B6 mice were injected (i.v.) with 0.6 mg/kg DEC-LNPs or ISO-LNPs (LNPs synthesized with 0.1% DOPE-rhodamine B) or nothing (Untreated). After 24 hours, spleens were isolated and uptake by various cell types was determined by flow cytometry. (a) LNP uptake by CD11c+CD8α+ cells. (b) CD11c+ splenocytes, isolated from mice injected with DEC-LNPs or ISO-LNPs, were analyzed based on CD8α expression (left and center panels depict gating strategy). Uptake of DEC-LNPs was observed predominantly in the CD8α+ (DEC205+) DCs (top panel), little uptake is observed in CD8α– (DEC205–) DCs (lower panel). Blue lines = DCs from DEC-LNP-injected mice; red lines = DCs from ISO-LNP injected mice. Top panel shows mean fluorescence intensity values. (c) Splenocytes were stained with CD11c, CD19, TCRβ, or F4/80 to determine LNP uptake by various cell subsets. (d) DEC205–/– and B6 mice were injected with DEC-LNPs or nothing. DEC-LNP injected mice: Black = DEC205–/–, Blue = DEC205–/+, green = B6; untreated B6 = red. Top panel shows DEC205 expression, bottom panel LNP uptake (gated on CD11c+CD8α+ cells). Data shown is representative of two to six experiments, containing groups of three to five mice.
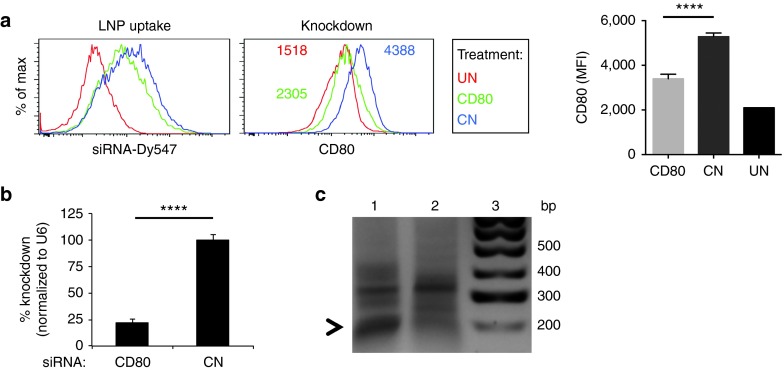
To show that DEC-LNPs were competent for specific gene silencing, we generated DEC-LNPs containing a siRNA specific for CD80 (having identified the most effective siRNA sequence; see Supplementary Figure S1) and injected into B6 mice. CD11c+DEC205+ cells took up similar amounts of LNPs following injection with DEC-LNP containing either CD80- or control-siRNA (LNP uptake panel, mean fluorescence intensity (MFI): DEC-LNP-control = 443 versus DEC-LNP-siCD80 = 464). In mice injected with DEC-LNP encapsulating CD80-specific siRNA, CD80 protein expression approached basal levels, and was reduced by ~2-fold when compared with DEC-LNP containing control-siRNA (Figure 4a). A 75% reduction in CD80 mRNA was observed in DEC205+ DCs that had taken up DEC-LNPs containing CD80-specific siRNA (compared with DEC-LNPs containing control siRNAs; Figure 4b). We confirmed that gene knockdown was RNAi-mediated using 5′ RACE to detect siRNA-directed mRNA cleavage products (Figure 4c). Sequencing of the polymerase chain reaction (PCR) fragment verified that it was derived from CD80 mRNA and that the cut site corresponded with nucleotide positions 10–11 of the siRNA guide strand (not shown).
Figure 4.
In situ uptake of DEC-lipid nanoparticles (LNPs) results in gene-specific knockdown that is mediated via the RNAi pathway. B6 mice were injected with 0.6 mg/kg DEC-LNPs, containing either CD80 or control (CN) siRNAs. After 24 hours, spleens were harvested. LNPs contained Dy547-labeled siRNA to monitor cell uptake. (a) DEC-LNP uptake by CD11c+CD8α+ cells (left panel) and CD80 expression in CD11c+CD8α+ DCs (right panel) was determined by flow cytometry. Red = untreated, green = CD80 siRNA, blue = control (CN) siRNA. Data from all mice are quantified in the bar graphs. Significance was determined by one-way ANOVA. ****P < 0.0001. (b) and (c) CD11c+CD8α+Dy547+ cells were isolated by fluorescence-activated cell sorting. Total RNA was extracted and CD80 mRNA levels were determined by qPCR (b) and 5' RACE analysis was performed to confirm RNAi-specific mRNA cleavage (c). PCR products were resolved by gel electrophoresis. Lane 1 = siRNA CD80; Lane 2 = siRNA control (CN); Lane 3 = 100 bp ladder. Arrowhead shows ~190 bp product.
Interestingly, while the data in Figure 4 demonstrate the ability of our DEC-LNPs to effectively knockdown CD80 expression in DCs in vivo, we observed significant DC activation following injection of DEC-LNPs containing control siRNAs (Figure 4a, right panel). This observation was surprising considering these experiments were performed using siRNAs containing 2′F modified RNA, previously reported to have reduced immunostimulatory activity.24,25 To better assess this effect, we synthesized a series of luciferase-specific control siRNAs (LUC) containing 2′OH RNA (unmodified), 2′F pyrimidines or selected 2′OMe modifications (see Methods for modification strategy; based on refs. 25,26). LNPs were generated and their ability to stimulate immune responses using multiple assays was examined. First, B6 mice were injected with DEC-LNPs that contained either unmodified, 2′F or 2′OMe modified LUC-specific siRNAs in the absence of any adjuvant. One day later, splenic DCs were analyzed for the expression of costimulatory receptors (Supplementary Figure S2a). As expected, DCs isolated from mice that had received unmodified siRNAs demonstrated significant upregulation of CD40, CD80, and CD86. Consistent with our previous results (Figure 4a), animals treated with DEC-LNPs containing 2′F modified siRNAs also upregulated these costimulatory molecules. Importantly, and similar to work by other groups, we found that 2′OMe modified siRNAs induced minimal immune activation.21,26,27
To attempt to circumvent the need for a time and resource intensive in vivo assay, we wanted to determine whether an in vitro assay could be used to detect immunostimulatory siRNAs. However, when we cultured B6-derived BMDCs with DEC-LNPs (containing CD80-specific siRNAs) no difference between 2′OH modified siRNA, 2′F or 2′OMe modified siRNA was detected (Supplementary Figure S2b). We also used NT-LNPs to assess the effects of various siRNA formulations on human PBMCs and moDCs using standard preclinical assays (Supplementary Figure S2c). PBMCs or moDCs were cultured for 24 hours at which time cells were assessed for induction of apoptosis and culture supernatants were tested for the presence of proinflammatory cytokines (see Methods). Apoptosis was not observed under any conditions tested (data not shown). As expected, PBMCs cultured with the highest concentration of LNPs containing unmodified siRNAs elicited the secretion of several cytokines. 2′F and 2′OMe LUC siRNAs induced production of one cytokine (IL8), also at the highest concentration tested. No cytokine production was observed following culture of moDCs with LNPs encapsulating unmodified, 2′F or 2′OMe siRNAs. Taken together, these experiments suggest that the assays tested show marked differences in sensitivity. Surprisingly, BMDCs and moDCs were the least receptive for detecting immunostimulatory siRNAs. Conversely, DEC-LNP injection was useful for the identification of siRNAs with immune-activating potential. From these results, we used siRNAs containing selective 2′OMe substitutions for all subsequent studies.
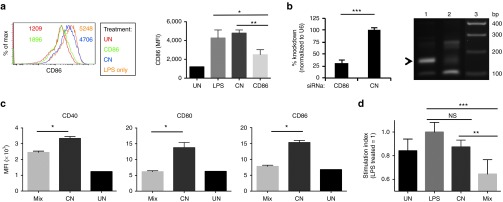
To target autoimmune diseases, reducing the expression of several costimulatory molecules will likely be more effective than knockdown of a single protein. We showed significant CD80 knockdown following DEC-LNP injection (Figure 4). Therefore, we next determined the ability of DEC-LNPs that encapsulated 2′OMe modified CD86 specific siRNAs, to reduce gene expression following i.v. administration (optimal siRNA identified as previously; Supplementary Figure S1). As the siRNAs were 2′OMe modified, mice were coinjected with LPS to stimulate expression of costimulatory molecules. Analysis of splenic DEC205+ DCs 1 day following DEC-LNP injection showed reduction of CD86 protein to near steady-state levels (Figure 5a). Total RNA was isolated from DEC205+ DCs that had taken up DEC-LNPs encapsulating either CD86 or control siRNAs. An approximately 70% reduction in CD86 mRNA levels was observed from DCs isolated from mice that received DEC-LNPs containing CD86 siRNA in comparison to DCs derived from control siRNA treated mice. To verify knockdown occurred via the RNAi pathway, 5′ RACE was used (Figure 5b). We next tested whether combining siRNAs in DEC-LNPs would induce gene silencing equivalent to that observed with single siRNAs. A combination of 2′OMe modified siRNAs targeting CD80, CD86 and CD40 were incorporated into DEC-LNPs and injected into B6 mice along with adjuvant (LPS) to activate the DCs. Reduction of each of the targeted costimulatory molecules was observed, with CD80 and CD86 knocked down to basal levels (Figure 5c). Gene silencing was similar to that achieved following treatment with DEC-LNPs containing single siRNAs (e.g., ~50% for CD86: compare CD86 DEC-LNP, Figure 5a with Mix DEC-LNP, Figure 5c right panel). To determine functional relevance of this knockdown, we used the robust MLR assay. B6 mice were injected with DEC-LNPs encapsulated with either a mix of CD80, CD86, and CD40 or control siRNAs. After 24 hours, splenic CD8α+ DCs that had taken up DEC-LNPs were isolated, irradiated, and cultured with splenic T cells derived from a BALB/c mouse. Maximal proliferation was observed when DCs were isolated from mice injected with LPS only, or LPS plus DEC-LNPs containing control siRNAs. In contrast, proliferation of T cells cultured with DCs derived from mice injected with LPS plus DEC-LNPs containing CD80, CD86, and CD40 siRNAs was significantly reduced (Figure 5d).
Figure 5.
Knockdown of costimulatory molecules following injection of DEC-lipid nanoparticles (LNPs) containing 2'OMe-modified siRNAs is sufficient to inhibit a mixed lymphocyte response reaction. (a) B6 mice (3/group) were injected with siRNAs specific for CD86 (CD86) or a nontarget control (CN) followed by LPS, 6 hours later. One group was injected with LPS alone (LPS) or was left untreated (UN). One day later, splenocytes were stained for CD11c, CD8α, CD80, and CD86 and analyzed by flow cytometry. The left panel shows CD86 expression on CD11c+CD8α+ cells derived from mice that were untreated (red), CD86 siRNA (green), control siRNA (blue), or LPS only (orange). Numbers within histogram plot are mean fluorescence intensity values; each line represents one mouse. Data from all mice are quantified in the bar graphs. Significance was determined by one-way ANOVA. *P < 0.05; **P < 0.01; ns, not significant. (b) CD11c+CD8α+Dy547+ cells were isolated by FACS, total RNA extracted and CD86 mRNA levels determined by qPCR (left panel) and 5' RACE analysis was performed (right panel). PCR products were resolved by gel electrophoresis. Lane 1 = siRNA CD86; Lane 2 = siRNA control (CN); Lane 3 = 100 bp ladder. Arrowhead shows ~150 bp product. (c) B6 mice (3/group) were injected with DEC-LNPs containing a 1:1:1 mix of CD40, CD80, and CD86 siRNAs (Mix), or control siRNAs (CN), followed by LPS, 6 hrs later, or were left untreated (UN). One day later expression of CD80, CD86, and CD40 on CD11c+CD8α+ cells was determined by flow cytometry. (d) B6 mice were injected with DEC-LNPs and LPS (as in (c)). The following day CD11c+CD8α+ DCs were FACS sorted, irradiated and cocultured with carboxyfluorescein succinimidyl ester (CFSE)-labeled T cells purified from BALB/c spleens. Two days later T-cell activation was determined by CFSE dilution using flow cytometry. The graph shows the relative stimulation index, which compares the LPS only treated group (set to 1) to untreated (UN), DEC-LNPs containing control siRNA (CN) and DEC-LNPs containing a 1:1:1 mix of CD40, CD80, and CD86 siRNAs (Mix). (c and d) Statistical significance was calculated by student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; FACS, fluorescence-activated cell sorting; NS, not significant.
Discussion
Delivery remains the major obstacle for turning siRNAs into clinically relevant molecules. Currently, the most advanced LNP platforms use the ionizable cationic lipid DLinDMA, or its DLin-KC2-DMA and DLin-MC3-DMA derivatives.3,7,28 Following systemic administration, these LNPs accumulate in first pass organs, i.e., liver and spleen, making them attractive vehicles for hepatic gene knockdown. Furthermore, these LNPs have been shown to safely and effectively reduce expression of hepatocyte genes in clinical trials.5,29 However, there is only very limited data demonstrating targeted delivery of potentially clinically relevant LNPs to nonhepatic cells.14
We have shown that conjugation of a scFv, specific for the DC receptor DEC205, is sufficient to direct delivery of DLinDMA-formulated DEC-LNPs to splenic DCs. Similar to conventional NT-LNPs, DEC-LNPs incorporated siRNAs with high efficiency (>80%), and were of uniform diameter (~100 nmol/l). We found that coating LNPs with only ~50 scFv was sufficient for DC delivery (Figures 1, 2, and 3). We showed that DEC-LNP uptake correlated well with DEC205 receptor density: DCs > B cells, T cells and macrophages (DEC205 expression is 10–50-fold less than on DCs).15 Use of DEC205–/– mice further confirmed that uptake occurred via the DEC205 receptor (Figures 2a and 3d). We also showed that the intracellular pathway accessed by DEC-LNPs was consistent with the DEC205 pathway (Figure 2b).22 Importantly, we demonstrated that targeted delivery resulted in effective RNAi-mediated gene silencing of one, or several genes, to essentially basal expression levels (Figures 4 and 5a–c). This reduction in gene expression was sufficient to inhibit a robust MLR (Figure 5d).
A small number of studies have reported success in cell-specific siRNA delivery. Of relevance to our study, Zheng et al.30 coated LNPs formulated with the cationic lipid DDAB with full-length DEC205 antibody for siRNA delivery and demonstrated the ability to reduce expression of the costimulatory molecule CD40 in DEC205+ DCs. While some gene knockdown was achieved, this lipid is known for its adjuvant qualities, and is being developed for applications that require immune response stimulation.30,31,32 The LNPs also proved relatively inefficient at siRNA loading (≤10% compared with ≥85% for DLinDMA LNPs). The siRNAs used were also unmodified, which could serve as another possible source of immune stimulation. Therefore, the potential for this formulation appears to be limited.
More recently, Ramishetti et al.14 utilized a full-length antibody targeting CD4 to enhance T-cell uptake and induce siRNA-mediated gene silencing using a clinically relevant LNP formulation similar to the one we employed in our work. Interestingly, both this study and the work from Zheng et al. chose to utilize full-length antibodies for LNP targeting, which may limit their utility when considering the immunogenicity of whole antibodies and their rapid clearance from the circulation by Fc-mediated uptake by macrophages.14,33 In fact, in preliminary studies, we coated LNPs with full-length DEC205 antibody and failed to observe uptake by DEC205+ DCs.
Using our DEC-LNPs, we found that the concentration required for efficient gene knockdown (~0.6 mg/kg) comparable to that study which used N-acetylgalactosamine (GalNAc)-coated LNPs to target hepatocytes via the asialoglycoprotein receptor (ASGPR) in ApoE knockout mice.6 In contrast, when the ability of NT-LNPs to silence antigen presenting cells (APCs), including DCs, was investigated a dose of 5 mg/kg was required to achieve partial silencing: ~10-fold higher when compared with our DEC-LNPs.9 Furthermore, the authors used the DLinDMA derivative, DLin-KC2-DMA, which displays improved silencing ability in hepatocytes (Relative ED50 DLin-KC2-DMA: 0.1 mg/kg versus DLinDMA: 1 mg/kg).7 As these LNPs were formulated for effective gene silencing in hepatocytes, their relative inefficiency for knockdown in APCs is perhaps not surprising. Biodistribution studies show that NT-LNPs are found in the spleen, although at ~50-fold less than in the liver.3 While targeting ligands have been shown to have little effect on the overall biodistribution of nanoparticle formulations,34,35,36 they can enhance cell-specific uptake.34,35 Therefore, while high doses of NT-LNPs can confer partial gene silencing in APCs,9,10 at low LNP concentrations, we find that a targeted approach is required to achieve knockdown in a specific DC subset. Injection of low-dose NT-LNPs failed to result in detectable uptake/gene silencing.
As the RNAi pathway is resident in the cytoplasm, the intracellular pathway used following receptor ligation is an important consideration. We chose the well-characterized DEC205 receptor for targeting our LNPs. Following binding, the internalized receptor is routed to late endosomes and associated cargo gains access to the cytoplasm.22 The DLinDMA lipid fuses with anionic phospholipids present in the endosomal membrane resulting in release of encapsulated cargo into the cytoplasm.37 It remains unclear where in the endosomal pathway translocation into the cytoplasm takes place. However, it is likely that different routes of uptake will affect the efficiency of delivery and subsequent gene silencing. Thus, a logical extension to our approach is to determine whether we observe improved gene silencing using other DC specific targeting agents and/or more effective DLinDMA derivatives. We are currently investigating the efficacy of DEC-LNPs formulated with the DLin-MC3-DMA (ED50 0.03 mg/kg).28
Having demonstrated that we could efficiently target siRNAs to DCs, the lack of toxicity of these LNPs and their cargo had to be established. The DLinDMA platform has undergone extensive preclinical and clinical evaluation.5,29,38 We observed significant DC activation following injection of targeted LNPs containing unmodified and 2′F modified siRNAs, and incorporation of 2′OMe modifications at distinct residues was required to maintain costimulatory molecules analyzed at basal levels (Figure 4a and Supplementary Figure S2a). As injecting mice to determine immune stimulation is time and resource intensive, we assessed immune stimulation in murine and human tissue culture systems. Murine BMDCs were refractory to LNP stimulation suggesting they would not serve as a useful surrogate for gauging immunogenicity of LNPs. Human PBMCs and moDCs were also relatively insensitive to LNP stimulation. Taken together, these assays demonstrate a requirement for in vivo analysis to uncover the stimulatory capacity of the LNPs. Recently, a human whole blood cytokine assay has been described that showed similar activation profiles elicited by LNPs when compared with in vivo injection, potentially circumventing a requirement for cumbersome in vivo analyses.39
For the purposes of inhibiting a multifactorial immune response, such as that observed in graft rejection or autoimmunity, the simultaneous knockdown of several (co)stimulatory genes will likely be required. This approach has been used to demonstrate that combining antibodies targeting CD80 and CD86 can reduce disease severity in experimental autoimmune encephalomyelitis and in antigen-induced arthritis.40,41 Transfer of DCs treated ex vivo with CD40, CD80, and CD86 antisense oligonucleotides (AS-ODN) into NOD mice delayed type 1 diabetes (T1D) onset.42 A follow-up study demonstrated that multiple injections of microspheres containing CD40, CD80, and CD86 AS-ODNs were sufficient to prevent T1D, possibly due to reduced expression of the costimulatory molecules by splenic DCs.43 We achieve significant reduction in expression of CD40, CD80, and CD86 on splenic DCs using our DEC-LNPs following systemic delivery, and ongoing studies are addressing the durability of gene knockdown and the potential use of these DEC-LNPs for treating autoimmune diseases.
In summary, we have shown that a clinically relevant LNP can be modified for cell-specific delivery of siRNAs. In vivo administration did not produce overt inflammatory responses, and we achieved significant RNAi-mediated gene-specific knockdown at low siRNA doses. The knockdown achieved was sufficient to inhibit a robust MLR reaction. These attributes make this an attractive platform to warrant further investigation as a potential therapeutic agent for various autoimmune diseases. Furthermore, judicious selection of the ligand coating the LNP opens up the potential for targeting multiple cell types.
Materials and Methods
Mice. C57BL/6 (B6) and BALB/c mice were purchased from Taconic (Hudson, NY). B6.129P-Ly75tm1Mnz/J (DEC205+/-) (ref. 44) was purchased from Jackson Laboratories (Bar Harbor, ME). DEC205+/– mice were bred to homozygosity on the B6 background (DEC205–/–), and validated by PCR and flow cytometry. All mice used were 6–12 weeks old. Mice were housed in the barrier facility at the Institute for Animal Studies, Albert Einstein College of Medicine (Bronx, NY) according to Institutional Animal Care and Use Committee (IACUC) guidelines. This study was approved by the IACUC at the Albert Einstein College of Medicine.
scFv cloning and purification. The single-chain fragment variable (scFv) antibodies constructed for DEC205 (clone NLDC145) and isotype (clone GL117) were cloned from antibody constructs, as previously described.17 The variable regions of the heavy and light chain sequences were fused with a flexible linker (G4S)6 separating them and cloned into a pET28a (EMD Millipore, Billerica, MA) bacterial expression vector. Further modifications to the constructs included a Strep-tag-II (WSHPQFEK) at the N-terminus and a 10-residue histidine tag (H10) with a short flexible linker (G4S) near the C-terminus. A C-terminal unpaired cysteine residue was introduced for scFv conjugation to the lipid DSPE-PEG-MAL. The scFv constructs were transformed into BL21 (DE3) pLysS competent cells (Promega, Fitchburg, WI).
The scFv DEC205 or isotype were expressed in Escherichia coli strain BL21 (DE3) pLysS as insoluble inclusion bodies. Following bacterial growth in Luria broth (LB) medium and induction of protein expression with isopropyl 1-thio-D-galactopyranoside, cells were resuspended in Tris-HCl (pH 8.0) buffer. Cells were lysed, and insoluble protein pelleted by centrifugation. Solubilized inclusion bodies were passed over a Ni-NTA column and the bound protein was eluted with 8 M guanidine hydrochloride. The purified proteins were refolded by rapid dilution in refolding buffer,45,46 and purified by size-exclusion chromatography using Superdex 200 (GE Healthcare Life Sciences, Pittsburgh, PA). Proteins were buffer-exchanged in phosphate-buffered saline (PBS) and concentrated. scFvs were tested for endotoxin contamination using a kinetic chromogenic limulous amoebocyte lysate (LAL) assay (Kinetic-QCL LAL Assay, Lonza, Walkersville, MD).
scFv fluorophore conjugation. The free cysteine engineered onto the N-terminus of the scFv was reduced using Tris-(2-carboxyethyl) phosphine hydrochloride (Life Technologies, Carlsbad, CA). Following buffer exchange into PBS, the reduced scFv was reacted with a 10-fold molar excess of AlexaFluor488 (AF488)-C5-maleimide according to manufacturer instructions (Life Technologies, Carlsbad, CA). Unreacted AF488-C5-maleimide was removed by dialysis against PBS. Conjugates were confirmed via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining and Storm Molecular Phosphorimager scanning of the stained gel (GE Healthcare, Piscataway, NJ).
Preparation of LNPs. LNPs were formed using the following lipids at a 15:40:5:40 mol %: DSPC (Avanti Polar Lipids, Alabaster, AL), cholesterol (CHOL; Sigma-Aldrich, MO), 1,2-distearoyl-sn-glycero-3-phosphatidylethanolamine-N-(maleimide-(polyethylene glycol)-2000) (DSPE-PEG-MAL, Avanti Polar Lipids, Alabaster, AL), and 1,2-dilinoleyloxy-3-dimethylaminopropane (DLinDMA) synthesized by the Chemical Biology Core Facility at Albert Einstein College of Medicine as previously described.37 A fluorophore conjugated to a lipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (DOPE-rhodamine B; Avanti Polar Lipids, Alabaster, AL) was incorporated (0.1 %) to monitor LNP delivery. LNPs were formed using a modified version of the spontaneous vesicle formation by ethanol dilution method.47 The lipid mixture was extruded (Mini-Extruder and 1 ml gas-tight syringes, Avanti Polar Lipids, Alabaster, AL) sequentially with polycarbonate 200 and 80 nm pore size membranes. The extruded lipid mixture was mixed via rapid pipetting and vortexing with siRNA, followed by dialysis into PBS. For attachment of scFv, LNPs were mixed with reduced scFv at room temperature for 1 hour, and free maleimide groups were then quenched with β-mercaptoethanol (βME). Tangential flow filtration (MicroKros filter module) with a 500 kDa MWCO (Spectrum Laboratories, Rancho Dominguez, CA) was used for buffer exchange into PBS and concentration of LNPs. LNPs were assayed for size and polydispersity by dynamic light scattering (DynaPro Plate Reader, Wyatt Technology, Santa Barbara, CA). Cryo-electron microscopy confirmed LNP size, geometry and lamellarity (Analytical Imaging Facility; Albert Einstein College of Medicine, Bronx, NY). siRNA incorporation was determined by Quant-iT RiboGreen RNA assay (Life Technologies, Carlsbad, CA). To determine scFv conjugation efficiency, LNPs were resolved on SDS-PAGE gels. A 3 kDa difference in migration of the scFv was indicative of binding to DSPE-PEG-MAL (unconjugated scFv ~30 kDa, scFv-DSPE-PEG-MAL ~33 kDa). A cholesterol quantification kit (Abcam, Cambridge, MA) was use to determine the total lipid concentration. Based on lipid concentration, the surface area per lipid head group (~0.6 nm2), LNP diameter and their unilamellar structure, ~50–60 scFVs are conjugated to each LNP under the conditions outlined above.
siRNA synthesis. Modified and unmodified individual siRNA sequences were synthesized on an Expedite 8909 DNA synthesizer (Biolytic, Fremont, CA) and purified by standard protocols, as described.48 Dylight 547-phosphoramidite was attached to the 5′ end of the sense strand for synthesis of fluorescently labeled siRNA (Dy547-siRNA; Glen Research, Sterling, VA). Unmodified siRNA sequences used in this study are listed below. siRNAs were modified by 2′F substitutions at every pyrimidine or by 2′OMe substitutions at the bolded nucleotides:
CD40 Sense 5′-GAAGAUUAUCCCGGUCAUAUU-3′; CD40 Anti-Sense 5′-UAUGACCG; GGAUAAUCUUCUU-3′; CD80 Sense 5′-GAAUUACCUGGCAUCAAUAUU-3′; CD80 Anti-Sense 5′-UAUUGAUGCCAGGUAAUUCUU-3′; CD86 Sense 5′-CAACUGGACUCUACG ACUUUU-3′; CD86 Anti-Sense 5′-AAGUCGUAGAGUCCAGUUGUU-3′; Control Sense 5′-UAGCGACUAAACACAUCAAUU-3′; Control Anti-Sense 5′-UUGAUGUGUUUAGUC GCUAUU-3′. Luciferase Sense 5′-GAUUAUGUCCGGUUAUGUAUU-3′; Luciferase Anti-Sense 5′-UACAUAACCGGACAUAAUCUU-3′.
LNP binding, uptake, and intracellular localization. CHO cells, CHO cells that stably express DEC205 (CHO-DEC), a DEC205+ B lymphoma line (A20 cells) and BMDCs, derived from B6 and DEC205–/– mice, were used to assess LNP binding and uptake. Cells were grown in complete medium (Dulbecco's modified Eagle's medium, 5% FBS, 10 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 20 mmol/l L-glutamine). BMDCs were generated from bone marrow derived from the femur and tibia as previously described.49 Adherent cells (A20, CHO) were harvested using ethylenediaminetetraacetic acid to preserve receptor expression and were incubated at 4 °C, 30 minutes with 500 nmol/l scFv-LNPs (containing either 0.1% DOPE-rhodamine B or Cy3-labeled siRNAs). For uptake, additional 30 minutes incubation at 37 °C was performed. Following incubation(s) cells were washed with fluorescence-activated cell sorting buffer (PBS containing 0.5% BSA, 0.1% Na N3, 2 mmol/l EDTA) and analyzed by flow cytometry (LSRII, Becton Dickinson, Franklin Lakes, NJ; FlowJo, Ashland, OR).
To monitor intracellular localization, A20 cells were incubated with scFv-LNPs, containing siRNAs labeled with DyLight 547 (GE Dharmacon, Lafayette, CO), 1 hour, 37 °C. Following one wash with PBS, cells were pipetted onto poly-L-lysine coated coverslips and allowed to adhere. Adhered cells were fixed with 4% paraformaldehyde, washed with PBS and stained with anti-LAMP1 (clone 1D4B; eBioscience, San Diego, CA) and anti-EEA1 (clone C45B10; Cell Signaling Technology, Danvers, MA). Stained cells were mounted with Vectashield mounting media containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA) and samples were visualized on a Leica SP5 AOBS confocal microscope (Leica, Buffalo Grove, IL). Image processing (linear adjustments to brightness and contrast) was performed on image files using NIH ImageJ software (Bethesda, MD).
LNP uptake and gene knockdown in vivo. LNPs (0.6–1 mg/kg) were injected (retroorbital (r.o.)) into B6 mice and 25 ng ultrapure LPS (Invivogen, San Diego, CA) was injected r.o. ~4 hours later to induce expression of costimulatory molecules. One day later, spleens were harvested, collagenase treated to maximize DC yield,50 and single cell suspensions subjected to red cells lysis. Cells were stained with cell surface markers and analyzed by flow cytometry to determine LNP uptake and expression of costimulatory molecules. Antibodies were used for detection of the following cell surface markers: CD11c, CD8α, DEC205, CD40, CD80, CD86, CD19, B220, CD3ɛ, TCRβ, F4/80, CD11b, H2kd, DCIR2, CD49b, CD45.1. CD45.2, CD25 (eBioscience and BD Biosciences, San Diego, CA). Cell viability was monitored using a live/dead exclusion dye, i.e., live/dead BLUE (Life Technologies, Carlsbad, CA). Cells were acquired using an LSRII cytometer and data analyzed using FlowJo software.
To measure gene knockdown, CD11c+CD8α+ splenocytes that had taken up LNPs (i.e., siRNA-Dy547+) were sorted by flow cytometry (FACS Aria III, Becton Dickinson, Franklin Lakes, NJ) into RNAprotect (Qiagen, Valencia, CA) to stabilize RNA. Total RNA was isolated using the RNeasy RNA isolation kit (Qiagen, Valencia, CA), reverse transcribed using Superscript III (Life Technologies, Carlsbad, CA) and random hexamers (IDT, Coralville, IA), according to manufacturer's instructions. Quantitative PCR was performed using Platinum Taq polymerase (Life Technologies, Carlsbad, CA) and SYBR green to detect amplified products on an iQ5 iCycler (Biorad, Hercules, CA). Reactions were performed in triplicate with the following primers: U6 forward: 5′-CTCGCTTCGGCAGCACATATACTA-3′; U6 reverse: 5′-ACGAATTTGCGTGTCATCCTTGCG-3′; CD40 forward: 5′-CGGCTGTGCGCGCTATG-3′; CD40 reverse: 5′-GGCTGAGAATTCGCCTGAGTC-3′; CD80 forward: 5′-CTACTCTCTTATCATCCTGGGCCT-3′; CD80 reverse: 5′-CCCGGAAGCAAAGCAGGTAATCCT-3′; CD86 forward: 5′-GCCACCCACAGGATCAATTATCCT-3′; CD86 reverse: 5′-AAAGAGAGAGGCTGTTGGAGATAC-3′. Relative amounts of mRNA were normalized to U6 mRNA and primer specificity verified by melt curve analysis.
To detect RNAi-mediated cleavage products, 5′ RACE was used. Total RNA was isolated from CD11c+CD8α+ splenocytes that had taken up LNPs, i.e., siRNA-Dy547+ (as for gene knockdown by qPCR), ligated at the 5′ end to an adapter RNA oligonucleotide and first strand cDNA was synthesized using a gene-specific primer (GSP). PCR amplification using an adapter-specific primer and a nested GSP primer will amplify the cleavage product and identify the cut site which corresponds to position 10/11 of the siRNA guide strand if mediated via RNAi. The primers and oligonucleotide sequences used: Adapter RNA: 5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′; GSP forward: 5′-CGACTGGAGCACGAGGACACTGA-3′; CD80 GSP reverse: 5′-TGCGCCGAATCCTGCCCCAA-3′; CD80 reverse nested: 5′-ATCAGGAGGGTCTTCTGGGG-3′; CD86 GSP reverse: 5′-AGTAACTGAAGCTGTAATCTCCTTCCAATA-3′; CD86 reverse nested: 5′-CTGTGACATTATCTTGTGATATCTGCATGT-3′.
Analysis of immune stimulation. CD11c+ murine BMDCs (day 6; MACS purified, Miltenyi Biotec, San Diego, CA) were cultured with DEC-LNPs and monitored for activation (CD40, CD80, CD86) 1 day later by flow cytometry. In vivo stimulation was assessed by injection of DEC-LNPs r.o. into B6 mice, followed by analysis of activation markers (CD40, CD80, CD86) on splenic DCs after 24 hours by flow cytometry. Human PBMCs were obtained from three donors by standard ficoll separation.51 The PBMC fraction was used directly or following purification of CD14+ cells (Stemcell Technologies, Vancouver, British Columbia, Canada), monocytes were cultured for 5 days in the presence of granulocyte-macrophage colony-stimulating factor and IL4 to generate moDCs.52 NT-LNPs (5, 1, 0.2, 0.04, 0.008 μmol/l) were cultured with PBMCs or moDCs overnight and cell culture supernatants collected for cytokine analysis (IFNγ, IL1β, IL2, IL4, IL5, IL6, IL7, IL8, IL10, IL12, IL13, TNFα, granulocyte-macrophage colony-stimulating factor) using a Luminex multiplex format (Life Technologies Carlsbad, CA).
Mixed lymphocyte reaction. CD11c+CD8α+ splenic DCs were isolated by flow cytometry from B6 mice injected the previous day with DEC-LNPs containing either a 1:1:1 mix of CD40, CD80, and CD86 siRNAs or control siRNAs. The DCs were irradiated (2,000 rads) and used as stimulator cells. Splenic T cells from BALB/c mice were isolated by MACS using a Pan T cell isolation kit II (Miltenyi, San Diego, CA). MACS sorted BALB/c T cells were carboxyfluorescein succinimidyl ester labeled according to published protocols,53 and carboxyfluorescein succinimidyl ester-labeled T cells were cocultured with the irradiated B6 DCs. Three days later, dilution of carboxyfluorescein succinimidyl ester was monitored by flow cytometry to determine T-cell activation.
SUPPLEMENTARY MATERIAL Figure S1. Identification of siRNA sequences that confer maximal gene silencing. Figure S2. Immune stimulation by siRNAs is dependent on siRNA modification and experimental assay.
Acknowledgments
We thank Nahoko Dunlap (SRI International) for technical assistance with PBMC and moDC assays. We thank the Analytical Imaging Facility (AECOM) for generating cryo-EM images and the Flow Cytometry Facility (AECOM) for help with cell sorting. This work was supported in part by the NIH - R21 AI093539 and R01 AI099567 (D.P.); R21 CA157366, R21 CA157366 and Stand Up 2 Cancer (SU2C-AACR-IRG0809) (M.L.); R01 DK094327, R01 DK064315, and R03 AI119225 (T.P.D.); P60 DK020541, which supports the Diabetes Research Center of the Albert Einstein College of Medicine; and P30 CA013330, which supports the flow cytometry facility; T32 NS007098 (J.A.K.); and by the Juvenile Diabetes Research Foundation (Academic R & D Award 17-2010-789 to T.P.D. and D.P.). T.P.D. is the Diane Belfer, Cypres and Endelson Families Faculty Scholar in Diabetes Research.
Supplementary Material
References
- Davidson, BL and McCray, PB Jr (2011). Current prospects for RNA interference-based therapies. Nat Rev Genet 12: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer, A and Ginhoux, F (2014). Organization of the mouse and human DC network. Curr Opin Immunol 26: 90–99. [DOI] [PubMed] [Google Scholar]
- Zimmermann, TS, Lee, AC, Akinc, A, Bramlage, B, Bumcrot, D, Fedoruk, MN et al. (2006). RNAi-mediated gene silencing in non-human primates. Nature 441: 111–114. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky, M, Grefhorst, A, Anderson, NN, Racie, TS, Bramlage, B, Akinc, A et al. (2008). Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci USA 105: 11915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, K, Frank-Kamenetsky, M, Shulga-Morskaya, S, Liebow, A, Bettencourt, BR, Sutherland, JE et al. (2014). Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc, A, Querbes, W, De, S, Qin, J, Frank-Kamenetsky, M, Jayaprakash, KN et al. (2010). Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther 18: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple, SC, Akinc, A, Chen, J, Sandhu, AP, Mui, BL, Cho, CK et al. (2010). Rational design of cationic lipids for siRNA delivery. Nat Biotechnol 28: 172–176. [DOI] [PubMed] [Google Scholar]
- Barros, SA and Gollob, JA (2012). Safety profile of RNAi nanomedicines. Adv Drug Deliv Rev 64: 1730–1737. [DOI] [PubMed] [Google Scholar]
- Basha, G, Novobrantseva, TI, Rosin, N, Tam, YY, Hafez, IM, Wong, MK et al. (2011). Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol Ther 19: 2186–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novobrantseva, TI, Borodovsky, A, Wong, J, Klebanov, B, Zafari, M, Yucius, K et al. (2012). Systemic RNAi-mediated Gene Silencing in Nonhuman Primate and Rodent Myeloid Cells. Mol Ther Nucleic Acids 1: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X, Kuipers, F, Havekes, LM, Havinga, R, Dontje, B, Poelstra, K et al. (2005). The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem Biophys Res Commun 328: 57–62. [DOI] [PubMed] [Google Scholar]
- Wilner, SE, Wengerter, B, Maier, K, de Lourdes Borba Magalhães, M, Del Amo, DS, Pai, S et al. (2012). An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids 1: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, C, Guo, B, Wu, H, Shao, N, Li, D, Liu, J et al. (2015). Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat Med 21: 288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramishetti, S, Kedmi, R, Goldsmith, M, Leonard, F, Sprague, AG, Godin, B et al. (2015). Systemic Gene Silencing in Primary T Lymphocytes Using Targeted Lipid Nanoparticles. ACS Nano 9: 6706–6716. [DOI] [PubMed] [Google Scholar]
- Inaba, K, Swiggard, WJ, Inaba, M, Meltzer, J, Mirza, A, Sasagawa, T et al. (1995). Tissue distribution of the DEC-205 protein that is detected by the monoclonal antibody NLDC-145. I. Expression on dendritic cells and other subsets of mouse leukocytes. Cell Immunol 163: 148–156. [DOI] [PubMed] [Google Scholar]
- Dudziak, D, Kamphorst, AO, Heidkamp, GF, Buchholz, VR, Trumpfheller, C, Yamazaki, S et al. (2007). Differential antigen processing by dendritic cell subsets in vivo. Science 315: 107–111. [DOI] [PubMed] [Google Scholar]
- Hawiger, D, Inaba, K, Dorsett, Y, Guo, M, Mahnke, K, Rivera, M et al. (2001). Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J Exp Med 194: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz, L, Bonnyay, D, Mahnke, K, Rivera, M, Nussenzweig, MC and Steinman, RM (2002). Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J Exp Med 196: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz, LC, Bonnyay, DP, Charalambous, A, Darguste, DI, Fujii, S, Soares, H et al. (2004). In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med 199: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, M, Judge, A and MacLachlan, I (2009). siRNA and innate immunity. Oligonucleotides 19: 89–102. [DOI] [PubMed] [Google Scholar]
- Morrissey, DV, Lockridge, JA, Shaw, L, Blanchard, K, Jensen, K, Breen, W et al. (2005). Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol 23: 1002–1007. [DOI] [PubMed] [Google Scholar]
- Mahnke, K, Guo, M, Lee, S, Sepulveda, H, Swain, SL, Nussenzweig, M et al. (2000). The dendritic cell receptor for endocytosis, DEC-205, can recycle and enhance antigen presentation via major histocompatibility complex class II-positive lysosomal compartments. J Cell Biol 151: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vremec, D, Zorbas, M, Scollay, R, Saunders, DJ, Ardavin, CF, Wu, L et al. (1992). The surface phenotype of dendritic cells purified from mouse thymus and spleen: investigation of the CD8 expression by a subpopulation of dendritic cells. J Exp Med 176: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan, M, Akinc, A, Pandey, RK, Qin, J, Hadwiger, P, John, M et al. (2011). Unique gene-silencing and structural properties of 2'-fluoro-modified siRNAs. Angew Chem Int Ed Engl 50: 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekaite, L, Furset, G, Hovig, E and Sioud, M (2007). Gene expression analysis in blood cells in response to unmodified and 2'-modified siRNAs reveals TLR-dependent and independent effects. J Mol Biol 365: 90–108. [DOI] [PubMed] [Google Scholar]
- Judge, AD, Bola, G, Lee, AC and MacLachlan, I (2006). Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 13: 494–505. [DOI] [PubMed] [Google Scholar]
- Morrissey, DV, Blanchard, K, Shaw, L, Jensen, K, Lockridge, JA, Dickinson, B et al. (2005). Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41: 1349–1356. [DOI] [PubMed] [Google Scholar]
- Jayaraman, M, Ansell, SM, Mui, BL, Tam, YK, Chen, J, Du, X et al. (2012). Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl 51: 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, T, Adams, D, Silva, A, Lozeron, P, Hawkins, PN, Mant, T et al. (2013). Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med 369: 819–829. [DOI] [PubMed] [Google Scholar]
- Zheng, X, Vladau, C, Zhang, X, Suzuki, M, Ichim, TE, Zhang, ZX et al. (2009). A novel in vivo siRNA delivery system specifically targeting dendritic cells and silencing CD40 genes for immunomodulation. Blood 113: 2646–2654. [DOI] [PubMed] [Google Scholar]
- van Dissel, JT, Joosten, SA, Hoff, ST, Soonawala, D, Prins, C, Hokey, DA et al. (2014). A novel liposomal adjuvant system, CAF01, promotes long-lived Mycobacterium tuberculosis-specific T-cell responses in human. Vaccine 32: 7098–7107. [DOI] [PubMed] [Google Scholar]
- Korsholm, KS, Agger, EM, Foged, C, Christensen, D, Dietrich, J, Andersen, CS et al. (2007). The adjuvant mechanism of cationic dimethyldioctadecylammonium liposomes. Immunology 121: 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothdiener, M, Beuttler, J, Messerschmidt, SK and Kontermann, RE (2010). Antibody targeting of nanoparticles to tumor-specific receptors: immunoliposomes. Methods Mol Biol 624: 295–308. [DOI] [PubMed] [Google Scholar]
- Park, JW, Hong, K, Kirpotin, DB, Colbern, G, Shalaby, R, Baselga, J et al. (2002). Anti-HER2 immunoliposomes: enhanced efficacy attributable to targeted delivery. Clin Cancer Res 8: 1172–1181. [PubMed] [Google Scholar]
- Choi, CH, Alabi, CA, Webster, P and Davis, ME (2010). Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proc Natl Acad Sci USA 107: 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, DW, Su, H, Hildebrandt, IJ, Weber, WA and Davis, ME (2007). Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci USA 104: 15549–15554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes, J, Palmer, L, Bremner, K and MacLachlan, I (2005). Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release 107: 276–287. [DOI] [PubMed] [Google Scholar]
- Barros, SA and Gollob, JA (2012). Safety profile of RNAi nanomedicines. Adv Drug Deliv Rev 64: 1730–1737. [DOI] [PubMed] [Google Scholar]
- Kumar, V, Qin, J, Jiang, Y, Duncan, RG, Brigham, B, Fishman, S et al. (2014). Shielding of Lipid Nanoparticles for siRNA Delivery: Impact on Physicochemical Properties, Cytokine Induction, and Efficacy. Mol Ther Nucleic Acids 3: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvin, AM, Dal Canto, MC, Rhee, L, Salomon, B, Sharpe, A, Bluestone, JA et al. (2000). A critical role for B7/CD28 costimulation in experimental autoimmune encephalomyelitis: a comparative study using costimulatory molecule-deficient mice and monoclonal antibody blockade. J Immunol 164: 136–143. [DOI] [PubMed] [Google Scholar]
- Odobasic, D, Leech, MT, Xue, JR and Holdsworth, SR (2008). Distinct in vivo roles of CD80 and CD86 in the effector T-cell responses inducing antigen-induced arthritis. Immunology 124: 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machen, J, Harnaha, J, Lakomy, R, Styche, A, Trucco, M and Giannoukakis, N (2004). Antisense oligonucleotides down-regulating costimulation confer diabetes-preventive properties to nonobese diabetic mouse dendritic cells. J Immunol 173: 4331–4341. [DOI] [PubMed] [Google Scholar]
- Phillips, B, Nylander, K, Harnaha, J, Machen, J, Lakomy, R, Styche, A et al. (2008). A microsphere-based vaccine prevents and reverses new-onset autoimmune diabetes. Diabetes 57: 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M, Gong, S, Maric, S, Misulovin, Z, Pack, M, Mahnke, K et al. (2000). A monoclonal antibody to the DEC-205 endocytosis receptor on human dendritic cells. Hum Immunol 61: 729–738. [DOI] [PubMed] [Google Scholar]
- Zhang, X, Schwartz, JC, Almo, SC and Nathenson, SG (2002). Expression, refolding, purification, molecular characterization, crystallization, and preliminary X-ray analysis of the receptor binding domain of human B7-2. Protein Expr Purif 25: 105–113. [DOI] [PubMed] [Google Scholar]
- Garboczi, DN, Ghosh, P, Utz, U, Fan, QR, Biddison, WE and Wiley, DC (1996). Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384: 134–141. [DOI] [PubMed] [Google Scholar]
- Jeffs, LB, Palmer, LR, Ambegia, EG, Giesbrecht, C, Ewanick, S and MacLachlan, I (2005). A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res 22: 362–372. [DOI] [PubMed] [Google Scholar]
- Wincott, F, DiRenzo, A, Shaffer, C, Grimm, S, Tracz, D, Workman, C et al. (1995). Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res 23: 2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, K, Inaba, M, Romani, N, Aya, H, Deguchi, M, Ikehara, S et al. (1992). Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med 176: 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, K, Swiggard, WJ, Steinman, RM, Romani, N and Schuler, G (2001). Isolation of dendritic cells. Curr Protoc Immunol Chapter 3: Unit 3.7. [DOI] [PubMed] [Google Scholar]
- Fuss, IJ, Kanof, ME, Smith, PD and Zola, H (2009). Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol Chapter 7: Unit7.1. [DOI] [PubMed] [Google Scholar]
- Nair, S, Archer, GE and Tedder, TF (2012). Isolation and generation of human dendritic cells. Curr Protoc Immunol Chapter 7: Unit7.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish, CR, and Warren, HS (2002). Use of the intracellular fluorescent dye CFSE to monitor lymphocyte migration and proliferation. Curr Protoc Immunol. John Wiley & Sons, Inc 49: 4.9:4.9.1–4.9.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.