Abstract
Objective
This study investigated whether pyrosequencing can be used to determine the methylation status of the MGMT promoter as a clinical biomarker using relatively old archival tissue samples of glioblastoma. We also examined other prognostic factors for survival of glioblastoma patients.
Methods
The available study set included formalin-fixed paraffin-embedded (FFPE) tissue from 104 patients at two institutes from 1997 to 2012, all of which were diagnosed histopathologically as glioblastoma. Clinicopathologic data were collected by review of medical records. For pyrosequencing analysis, the PyroMark Q96 CpG MGMT kit (Qiagen, Hilden, Germany) was used to detect the level of methylation at exon 1 positions 17–39 of the MGMT gene, which contains 5 CpGs.
Results
Methylation of the MGMT promoter was detected in 43 (41.3%) of 104 samples. The average percentage methylation was 14.0±16.8% overall and 39.0±14.7% for methylated cases. There was no significant pattern of linear increase or decrease according to the age of the FFPE block (p=0.687). In multivariate analysis, age, performance status, extent of surgery, method of adjuvant therapy, and methylation status estimated by pyrosequencing were independently associated with overall survival. Additionally, patients with a high level of methylation survived longer than those with low methylation (p=0.016).
Conclusion
In this study, the status and extent of methylation of the MGMT promoter analyzed by pyrosequencing were associated with overall survival in glioblastoma patients. Pyrosequencing is a quantitative method that overcomes the problems of MSP and a simple technique for accurate analysis of DNA sequences.
Keywords: MGMT promoter, Methylation, Glioblastoma, Prognosis, Pyrosequencing
INTRODUCTION
Glioblastoma (GBM) is the most frequent malignant brain tumor, accounting for approximately 12–15% of all intracranial neoplasms and 60–75% of astrocytic tumors. In most European and North American countries, the incidence of GBM is in the range of 3–4 new cases per 100000 populations per year11). In Korea, an analysis of 5208 patients who were diagnosed histologically with primary central nervous system (CNS) tumors in 2010 showed that GBM accounted for 5.2% of all CNS tumors and 40.6% of all glial tumors9,15). Although the overall incidence rate of primary brain tumors is very low among all human cancers, GBM is a highly invasive and aggressively growing tumor that responds poorly to radiation therapy and most forms of chemotherapy.
Temozolomide (TMZ) is an alkylating chemotherapeutic agent that causes DNA damage by transferring alkyl groups at several sites within DNA, including the O6 position of guanine. The MGMT encodes a DNA repair protein that removes alkyl groups from O6-guanine, and thus plays a fundamental role in maintaining genomic integrity20). Epigenetic silencing by promoter methylation results in decreased MGMT expression and correlates with improved survival in patients with GBM who are treated with alkylating agents4).
Although MGMT promoter methylation is used as a prognostic/predictive marker in patients with GBM, no consensus exists on the optimal analysis method. Methylation-specific polymerase chain reaction (PCR) (MSP) has been widely used, and enables cost-efficient analysis of MGMT promoter methylation. However, it is not a quantitative method and has the risk of false-positive or false-negative results, particularly when the DNA quality and/or quantity are low. In the EORTC 26981/22981 and NCIC CE.3 trials, Hegi et al.7) only collected data from 67% of the samples analyzed, representing 36% of cases from their archives. The success rate of MSP on formalin-fixed paraffin-embedded (FFPE) tumor samples was highly variable and the median success rate was 75.0% (range, 0% to 100%). They concluded that diagnostic MGMT testing requires sufficient and optimally preserved tumor tissue such as cryopreserved tumor specimens, thus avoiding fixation-related deterioration of tumor DNA quality. An alternative technique that overcomes the problems associated with MSP is pyrosequencing (PSQ), which is based on the sequencing-by-synthesis principle and involves a simple technique for accurate and quantitative analysis of DNA sequences. PCR followed by PSQ is a sensitive, highly reproducible, and cost-effective method for DNA methylation analyses13). It provides absolute quantitative information based on each interrogated CpG site, which is not possible with most other methods. Additionally, unlike other methods, the assay design allows inclusion of internal controls to address inaccuracies resulting from incomplete bisulfite conversion. A group of investigators tried to compare and optimize three quantitative MGMT promoter methylation techniques and concluded that the PSQ assay is the most accurate and is also robust when applied to archival samples, including those of GBM14).
The objective of this study was to determine whether PSQ could be used to determine quantitatively the methylation status of the MGMT promoter as a clinical biomarker in routine practice using 2- to 15-year-old archival tissue samples of GBMs. We also estimated the methylation status of the MGMT promoter using MSP, which is already known to be a prognostic marker for overall survival and a predictive marker for conventional treatment of GBM patients, and verified the results obtained with PSQ. In addition, we examined other prognostic factors for survival of GBM patients.
MATERIALS AND METHODS
Sample selection
This study was reviewed and approved by the Institutional Review Board of two individual institutes, and all patients or families provided written informed consent. The study set included FFPE brain tumor tissue of 129 patients from the Department of Pathology archives of one hospital (collected from 1997–2010) or from another hospital (collected from 2000–2012), of which all were diagnosed as GBM. All patients underwent surgical resection or biopsy sampling of their tumors. Adjuvant conventional radiotherapy and/or chemotherapy were performed after pathologic diagnosis in most of the cases. All hematoxylin and eosin-stained slides were reviewed using World Health Organization Classification (2007) by two pathologists without any information regarding clinical and pathologic parameters. Tumors were excluded if the tissue was almost entirely necrotized or the tumor proportion of the section was <80%14). Finally, 104 patients were selected for this study.
Clinical and radiological data
Epidemiologic characteristics [including sex, age at the time of surgery, and Karnofsky Performance Scale (KPS)], the type of postoperative treatment, therapeutic type after recurrence, duration of follow-up, and the time of death were retrospectively reviewed for each patient using medical records. The dose of irradiation and type of radiotherapy administered and the regimen and timing of chemotherapy were also investigated.
Radiologic evaluation of the extent of surgical resection and response to treatment was performed by two different neuroradiologists without any information on clinical and pathological parameters. Extent of surgical resection was estimated from magnetic resonance imaging (MRI) scans performed within 72 hours after surgery. Subtotal resection was defined as removal of more than 90% of the gadolinium-enhancing lesion in enhanced T1-weighted MRI, and gross total resection was defined as lack of detectable gadolinium-enhancing lesion. In terms of response, tumor measurement for determination of treatment response according to the Macdonald criteria was based on the product of orthogonal diameters on the image with the largest gadolinium-enhancing tumor area17). If multiple lesions were present, the sum of the products of individual measurable lesions was calculated. Radiologic studies were performed at regular 3-monthly intervals during follow-up and when there was clinical suspicion of disease progression.
DNA extraction and bisulfite conversion
Genomic DNA was extracted from three 10-µm-thick slices of FFPE material using the QIAamp DNA FFPE extraction kit and the QIAcube automated DNA extraction machine (Qiagen, Hilden, Germany) and quantified by UV absorption (Nanodrop, Thermo Scientific, Wilmington, DE, USA), typically yielding a total of >1 µg of gDNA per specimen. gDNA (200 ng) was used in the bisulfite conversion reactions, in which unmethylated cytosine was converted to uracil with the EpiTect bisulfite kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Briefly, DNA was mixed with water, the DNA was protected by the buffer and bisulfite mix, and the conversion was run on an ABI 2727 Thermocycler (Applied Biosystems, Foster City, CA, USA) under the recommended cycle conditions. Converted DNA was purified and eluted in two steps in a total of 40 µL buffer EB and further diluted into 20-µL aliquots of 1000 cell-equivalents/µL (the cell calculations assumed 6 pg DNA per diploid cell). MSP was performed in a two-step approach using the methods of Palmisano et al.16).
PCR and PSQ
Primer sets with one biotin-labeled primer were used to amplify the above bisulfite-converted DNA samples. The PyroMark Q96 CpG MGMT kit5,10) (Ensembl ID : OTTHUMT00000051009) (Qiagen, Hilden, Germany) is available as a ready-to-use research kit; this kit was able to detect the level of methylation at positions 17–39 in exon 1 of the MGMT gene, which contains 5 CpGs. A cytosine not followed by a guanine, which was not methylated, served as an internal control and was programmed automatically by the PSQ96MA 2.1 software (Biotage, Uppsala, Sweden) to verify the efficiency of bisulfite conversion. Theoretically, the internal control must be zero, because all cytosines that are not followed by a guanine are converted to uracil during bisulfite conversion, and then uracil is converted to thymine after PCR. A value <5% was considered an acceptable value for the internal control according to the manufacturer's protocol. If the value of the internal control was >5%, all procedures were repeated again (from bisulfite conversion to PSQ).
PCR was performed using a converted gDNA equivalent of approximately 5000 cells and the PyroMark PCR kit (Qiagen, Hilden, Germany). Briefly, 12.5 µL master mix, 2.5 µL coral red, 5 pmol of each primer, 7 µL of water, and 2 µL sample were mixed for each reaction and run under the following thermal cycling conditions : 95℃ for 15 minutes, followed by 45 cycles of 30 seconds at 94℃, 30 seconds at the optimized primer-specific annealing temperature, and 30 seconds at 72℃, followed by a final extension for 10 minutes at 72℃. Amplification of the correct DNA product was confirmed by electrophoresis on a 2% low melting point agarose gel (Sigma-Aldrich, Steinheim, Germany) in tris-borate-EDTA buffer. A standard PSQ sample preparation protocol was applied5). Streptavidin beads (3 µL; GE Healthcare, Buckinghamshire, UK), 37 µL PyroMark binding buffer (Qiagen), 20 µL PCR product, and 20 µL water were mixed and incubated for 10 minutes on a shaking table at 1300 rpm. Amplicons were separated, denatured, washed, and added to 45 µL annealing buffer containing 0.33 µM of PSQ primer using the Biotage Q96 Vacuum Workstation. Primer annealing was performed by incubating the samples at 80℃ for 2 minutes and then cooling to room temperature prior to PSQ. PyroGold reagents were used for the PSQ reaction, and the signal was analyzed using the PSQ 96MA System (Biotage, Uppsala, Sweden). Target CpGs were evaluated by PSQ96MA 2.1 instrument software (Biotage, Uppsala, Sweden), which converts the pyrograms to numerical values for peak heights and calculates the proportion of methylation at each base as a C/T ratio. Methylation values ≤5% were considered potential background signals of questionable significance, as shown in other studies3,10,16). Unmethylated (300 ng; Qiagen, Hilden, Germany) and hypermethylated DNA (Millipore, Billerica, MA, USA) were used as standard controls, and were bisulfite converted as described above.
Data analysis and statistics
The main analyses converted individual C/T ratio data into mean values for the five CpGs analyzed in a gene segment using PSQ. The percentage of the mean value of the five CpGs was used to determine whether methylation affects the survival of GBM patients. Receiver operating characteristic (ROC) curve analysis was used to determine the cut-off value of mean percentage of methylation at the five CpGs for predicting the longer survival3). The area under the ROC curve (AUC) was used to determine the optimal threshold of the mean percentage of the methylation at the five CpGs. Sensitivity was calculated as the true positive rate (number of true positives divided by the sum of the number of true positives and number of false negatives); specificity as the true negative rate (number of true negatives divided by the sum of the number of true negatives and number of false positives); and accuracy as the sum of the number of true positives and true negatives, divided by the total number of positives and negatives. True positives mean that the methylation percentage above the cut-off value has an influence on the long survival, and true negatives mean that the methylation percentage below the cut-off value has an influence on the short survival.
Survival data were calculated from the date of diagnosis. Differences between subgroups were analyzed using the Student's t-test for normally distributed continuous values and the Mann-Whitney test for non-normally distributed continuous values. The χ2 test was used to analyze categorical variables. Variables that were found to be significantly associated with survival in GBM patients by univariate analyses were subjected to multivariate analyses. In addition, several variables that were of interest to the authors and were reported to be associated with survival of GBM patients in the literature also were included in multivariate analysis. We evaluated the impact of variables on survival by comparing the overall survival curves using the log-rank test. In multivariate analysis, Cox proportional hazard regression model was used to assess the independent effects of specific factors on overall survival and to define hazard ratios for significant covariates. Two-sided p values <0.05 were considered statistically significant. SPSS software version 12.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
RESULTS
Clinical characteristics
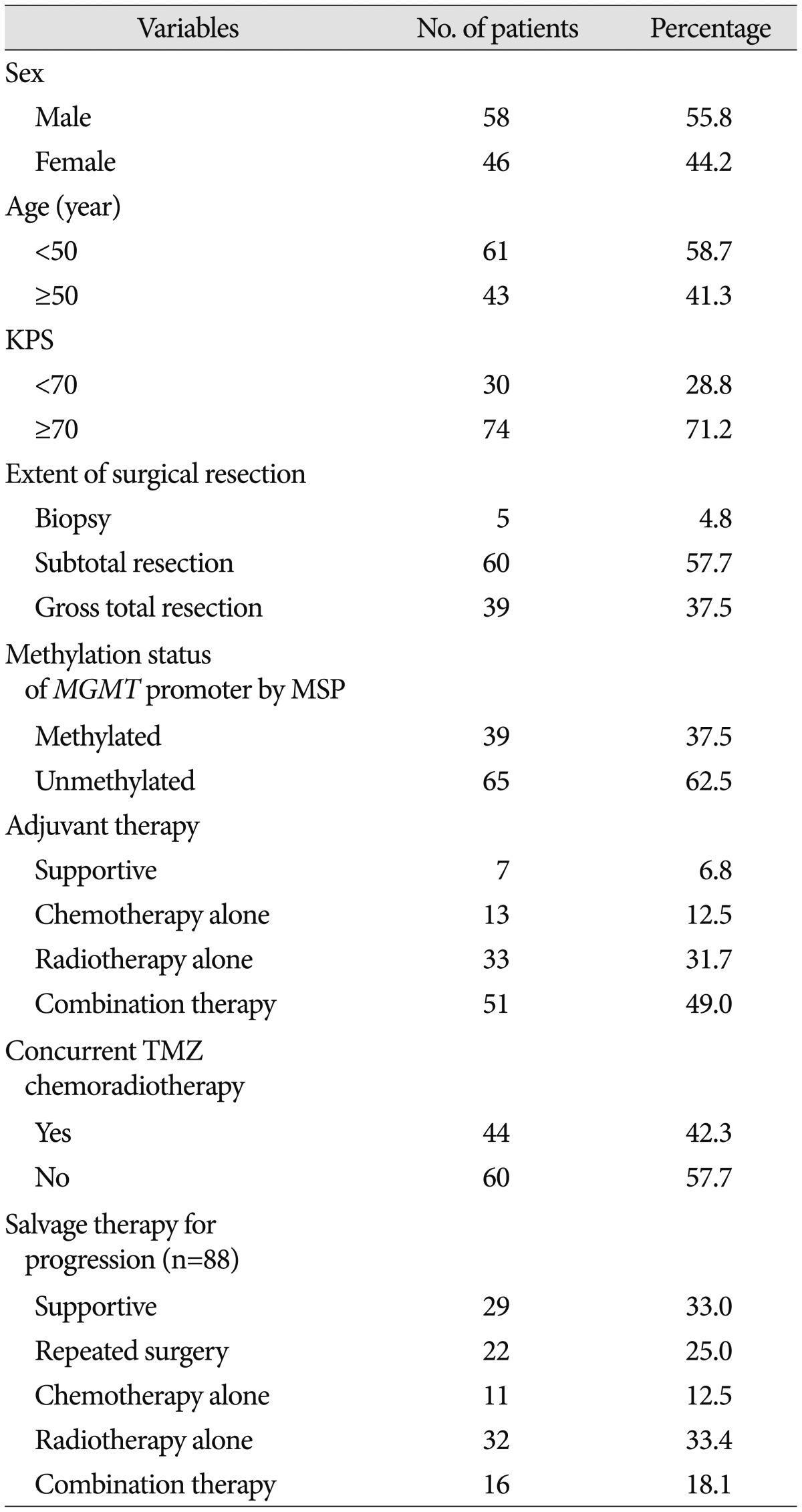
From a total of 129 GBM patients treated within the defined study period, 104 patients (58 male and 46 male) were eligible for analysis (Table 1). All patients were followed for at least 3 months, and the mean follow-up duration was 17.3 months (range, 3.2–41.5 months). During follow up, 79 patients (76.0%) died from GBM. The mean age at diagnosis was 51.4 years (range, 26.4–87.2 years). Seventy-four (71.2%) patients could perform activities of daily life independently (KPS scores ≥70). Ninety-four of the patients (90.4%) had clinical symptoms before diagnosis. The most frequent major complaints at presentation were headache (49 cases, 47.1%), seizure (16 cases, 15.4%), focal neurological deficits such as motor weakness and dysphasia (15 cases, 14.4%), and altered mentation (14 cases, 13.5%). Biopsy only was performed in 5 patients (4.8%), subtotal resection in 60 (57.7%), and gross total resection in 39 (37.5%).
Table 1. The characteristics of patients who were newly diagnosed with glioblastoma (n=104).
KPS : Karnofsky Performance Scale, MGMT : O6-methyl guanine methyltransferase, MSP : methylation specific polymerase chain reaction, TMZ : temozolomide
Most of the patients (97 patients, 93.3%) underwent active adjuvant treatment; 13 patients (12.5%) had chemotherapy alone, 33 patients (31.7%) had radiotherapy alone, and 51 patients (49.0%) had combination therapy with chemotherapy and radiotherapy. Forty-four patients (42.3%) were treated with concurrent temozolomide chemoradiotherapy as adjuvant treatment since 2005. During follow-up, 88 patients (84.6%) experienced disease progression. Mean time to disease progression was 11.4 months (range, 4.6–13.8 months). In terms of 44 patients who were treated with concurrent temozolomide chemoradiotherapy, mean time to progression was 12.2 months (range, 5.7–15.2 months). After progression, only 22 patients (25.0%) were able to undergo repeated surgical resection. For salvage therapy, 22 patients (25.0%) underwent repeated surgery, 11 patients (12.5%) were treated additionally with chemotherapy alone, 32 (33.4%) with radiotherapy alone, and 16 (18.1%) with both therapies.
Methylation status of MGMT promoter analyzed by pyrosequencing
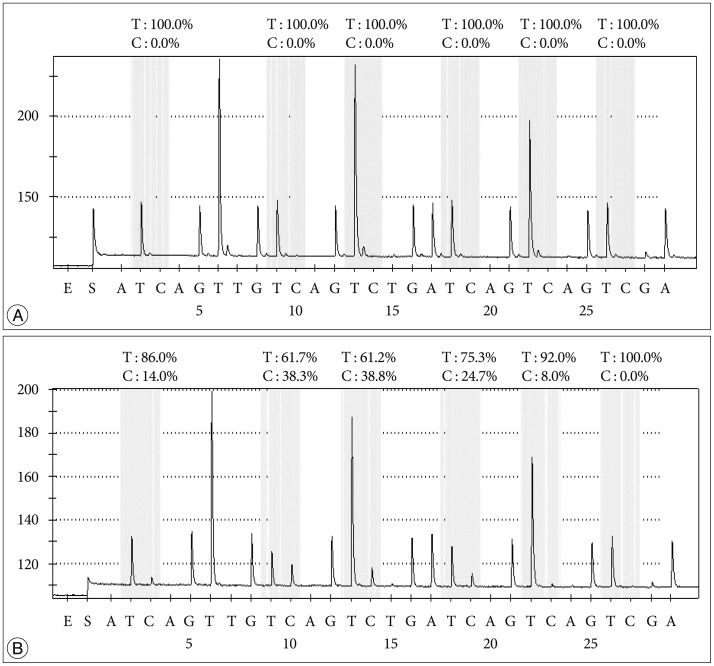
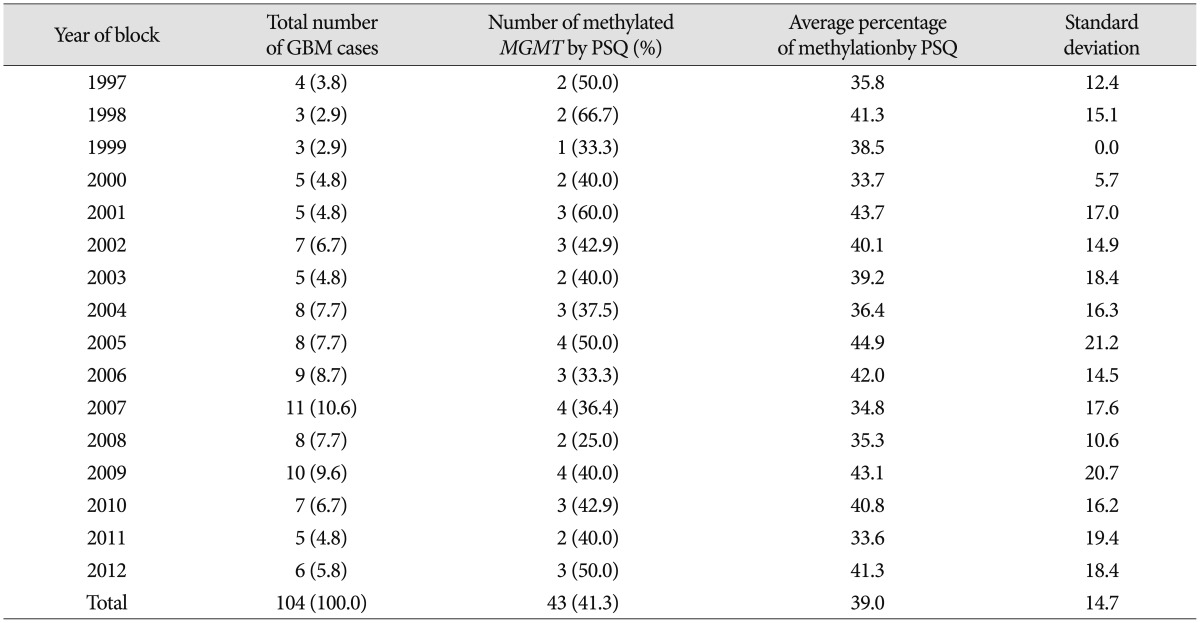
PSQ and MSP data were obtained for all 104 samples. For determining the methylation status of MGMT promoter methylation by PSQ, ROC curve analysis of using the mean percentage of methylation at the five CpGs to predict the longer survival showed that the AUC was 0.68. The optimal threshold of mean percentage of methylation for distinguishing between the patients with long survival and those with short survival was 9%, and this yielded a sensitivity of 53.8% [95% confidence interval (CI) 44.6–62.9%], a specificity of 66.7% (95% CI 52.9–80.5%), and an accuracy of 68.0%. Using PSQ analysis, MGMT promoter methylation was detected in 43 (41.3%) of the GBM samples. Cases of methylated and unmethylated promoters of GBMs were represented by a pyrogram (Fig. 1). The average percentage methylation for all samples of the methylated cases over the study period of 13 years was also investigated (Table 2). The average methylation percentage of all GBM samples was 14.0±16.8%; the percentage for methylated cases was 39.0±14.7% and that for unmethylated cases was 3.2±1.8% (p<0.001). The percentage methylation values for each year were not significantly different (p=0.687) and did not show an increasing or decreasing linear pattern according to the age of the FFPE block.
Fig. 1. Two representative glioblastoma pyrograms. Five consecutive positions are analyzed for CpGs; the last one is an internal control for bisulfite treatment. Each C peak in a yellow background represents the methylation percentage for each CpG. A : Unmethylated MGMT promoter of a glioblastoma sample. B : Methylated MGMT promoter of a glioblastoma sample.
Table 2. Average percentage of methylation in total and methylated cases from 1997 to 2011 which were analyzed by pyrosequencing.
GBM : glioblastoma multiforme, MGMT : O6-methyl guanine methyltransferase, PSQ : pyrosequencing
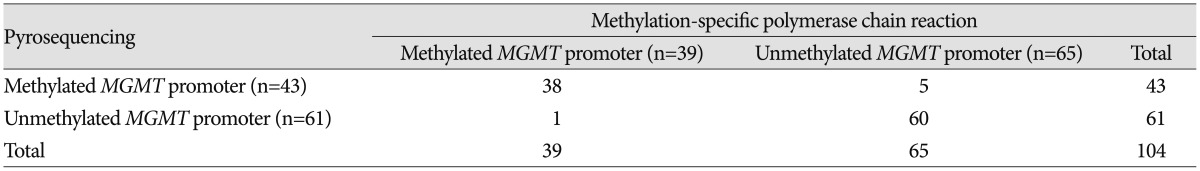
In comparison, MSP analysis showed that 39 samples (37.5%) had a methylated MGMT promoter and 65 samples (62.5%) had an unmethylated MGMT promoter. Six samples (5.8%) had an opposite methylation status of the MGMT promoter according to the two assays (Table 3); all 6 samples were obtained before 2003 and therefore were at least 10 years old and included one individual case for every year from 1997 to 2002.
Table 3. Comparison of the results of methylation status between using methylation-specific polymerase chain reaction and using pyrosequencing for the 104 glioblastoma samples.
MGMT : O6-methyl guanine methyltransferase
Survival of GBM patients
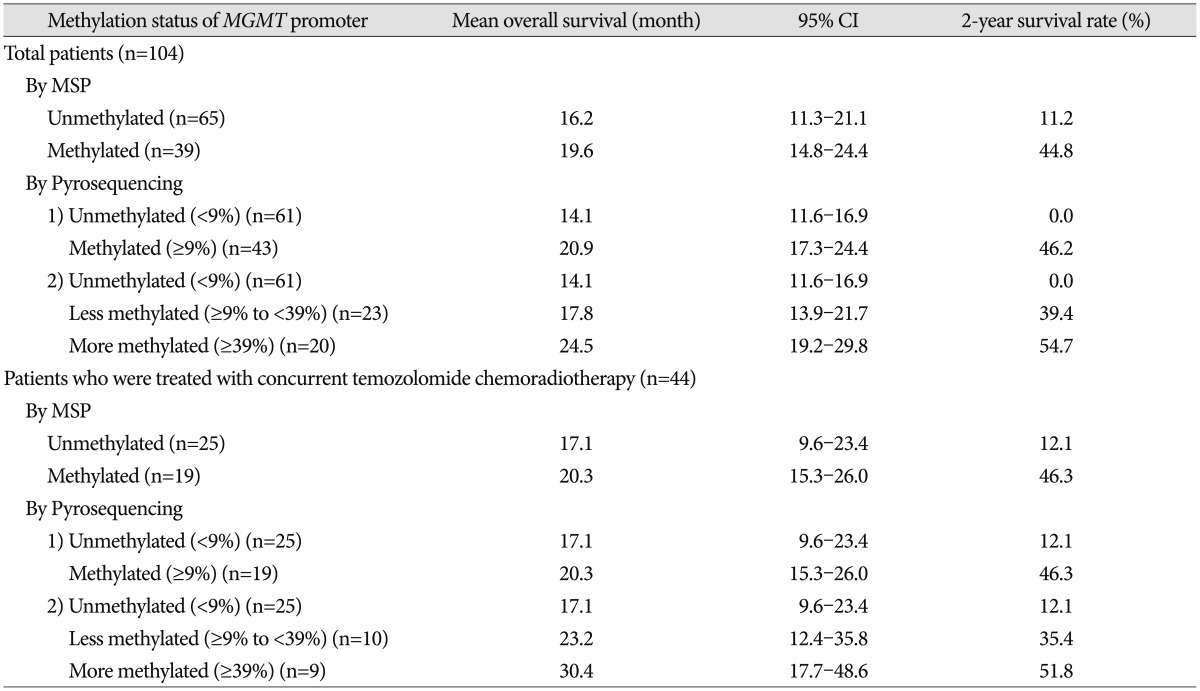
Mean overall survival (OS) of the total 104 GBM patients was 16.9 months (95% CI 12.4–21.5 months) and the 2-year survival rate was 25.2%. For analysis by MSP, the mean OS of GBM patients with an unmethylated MGMT promoter was 16.2 months (95% CI 11.3–21.1 months), and that of patients with a methylated MGMT promoter was 19.6 months (95% CI 14.8–24.2 months, p=0.004). In terms of 44 patients who were treated with concurrent temozolomide chemoradiotherapy, mean OS was 18.5 months (95% CI 12.8–24.5 months) and 2-year survival rate was 26.9%. For analysis by MSP, the mean OS of these patients with an unmethylated MGMT promoter was 17.1 months (95% CI 9.6–23.4 months), and that of patients with a methylated MGMT promoter was 20.3 months (95% CI 15.3–26.0 months, p=0.002) (Table 4).
Table 4. Overall survival according to the extent of MGMT promoter methylation analyzed by pyrosequencing.
CI : confidence interval, MGMT : O6-methyl guanine methyltransferase, MSP : methylation-specific polymerase chain reaction
According to analysis by PSQ, the mean OS of total 104 GBM patients with unmethylated MGMT promoter was 14.1 months (95% CI 11.6–16.9 months) and that of patients with a methylated MGMT promoter was 20.9 months (95% CI 17.3–24.4 months). There was a significant difference in OS between the two groups (p<0.001). More detailed categorization according to the percentage of methylation showed a significant difference in OS even among patients with a methylated MGMT promoter (p=0.012); the mean OS of GBM patients with a less methylated MGMT promoter (from 9–39%) was 17.8 months (95% CI 13.9–21.7 months), whereas that of patients with a more methylated MGMT promoter (above 39%) was 24.5 months (95% CI 19.2–29.8 months). Actually, in terms of 44 patients who were treated with concurrent temozolomide chemoradiotherapy, the methylation status analyzed by PSQ was completely identical to that by MSP in 44 patients who were treated with temozolomide concurrent chemoradiotherapy. Therefore, there was no difference of overall survival between two groups according to the analyzing technique; the patients with unmethylated MGMT promoter was 17.1 months (95% CI 9.6–23.4 months) and that of patients with a methylated MGMT promoter was 20.3 months (95% CI 15.3–26.0 months) (Table 4).
Factors associated with overall survival
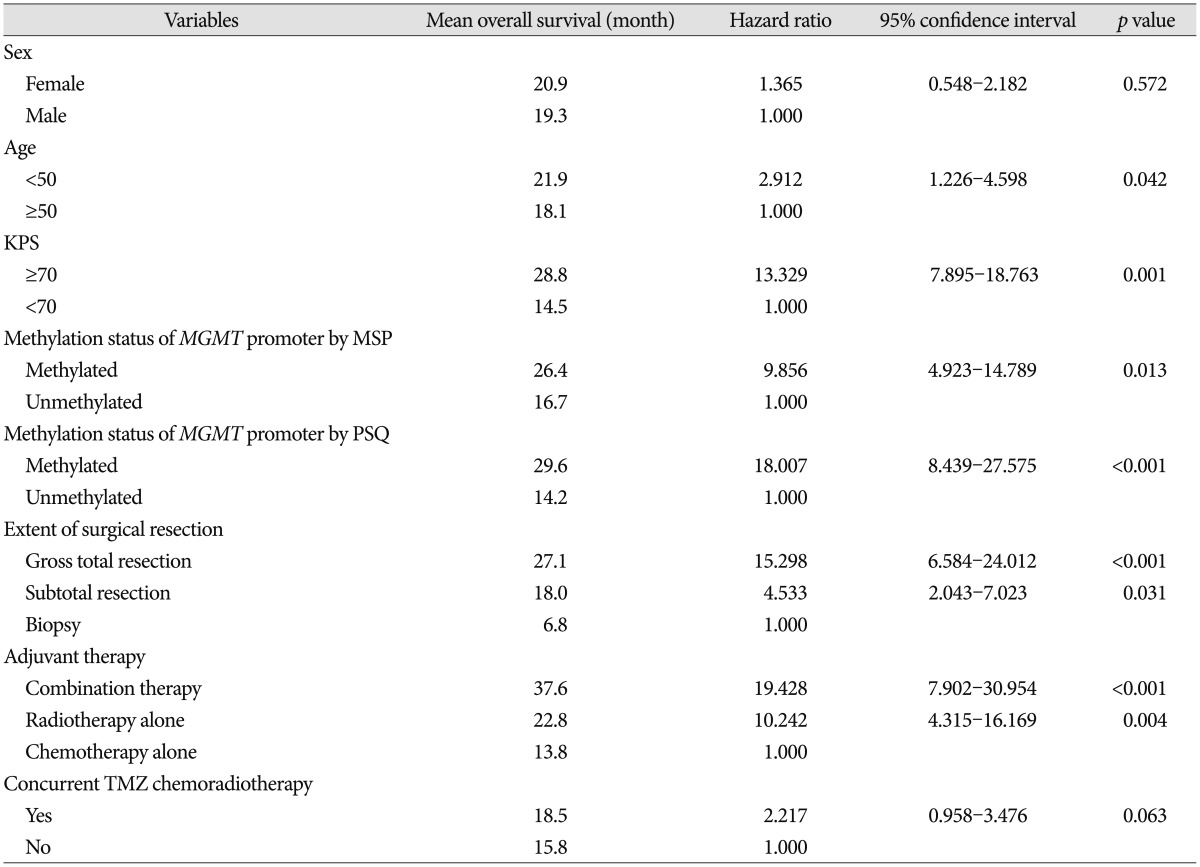
Univariate analysis for OS of all GBM patients was performed (Table 5). Age <50 [hazard ratio (HR) 2.912; p=0.042], KPS ≥70 (HR 13.329; p=0.001), extent of surgical resection (p<0.05), type of adjuvant therapy (p<0.05), methylation of MGMT promoter analyzed by MSP (HR 9.856; p=0.013), and methylation of MGMT promoter analyzed by PSQ (HR 18.007; p<0.001) were associated with OS. However, sex (p=0.572), concurrent temozolomide chemoradiotherapy (p=0.063) and method of salvage treatment (p>0.05) were not associated with OS.
Table 5. Univariate analysis for overall survival in patients who were newly diagnosed with glioblastoma according to the characteristics of patients.
KPS : Karnofsky Performance Scale, MGMT : O6-methyl guanine methyltransferase, MSP : methylation-specific polymerase chain reaction, PSQ : pyrosequencing, TMZ : temozolomide
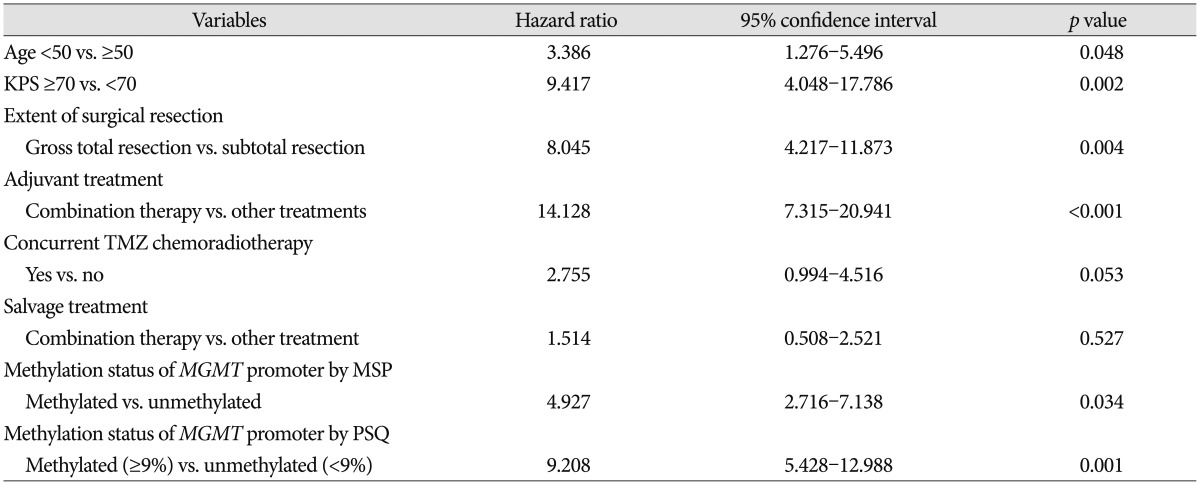
Multi-factor adjustment by the Cox-regression model was performed for the factors that were associated with OS in univariate analysis and additional factors of interest to the authors (such as method of salvage treatment) (Table 6). In multivariate analysis, we found that age (<50 years vs. ≥50 years; HR 3.386, p=0.048), KPS (≥70 vs. <70; HR 9.417, p=0.002), extent of surgical resection (gross total resection vs. subtotal resection, HR 8.045), method of adjuvant treatment (combination therapy vs. other treatment, HR 14.128, p<0.001), methylation status of MGMT promoter by MSP (methylated vs. unmethylated, HR 4.927, p=0.034), and methylation status of MGMT promoter by PSQ (methylated vs. unmethylated, HR 9.208, p=0.001) were independently associated with OS. Methods of salvage treatment and concurrent temozolomide chemoradiotherapy that were potentially associated with OS did not have any association in multivariate analysis (p>0.05). Kaplan-Meier survival curves according to each factor were estimated (Fig. 2).
Table 6. Multivariate analysis using Cox regression analysis for overall survival in patients who were newly diagnosed with glioblastoma according to the characteristics of patients.
KPS : Karnofsky Performance Scale, MGMT : O6-methyl guanine methyltransferase, MSP : methylation-specific polymerase chain reaction, PSQ : pyrosequencing, TMZ : temozolomide
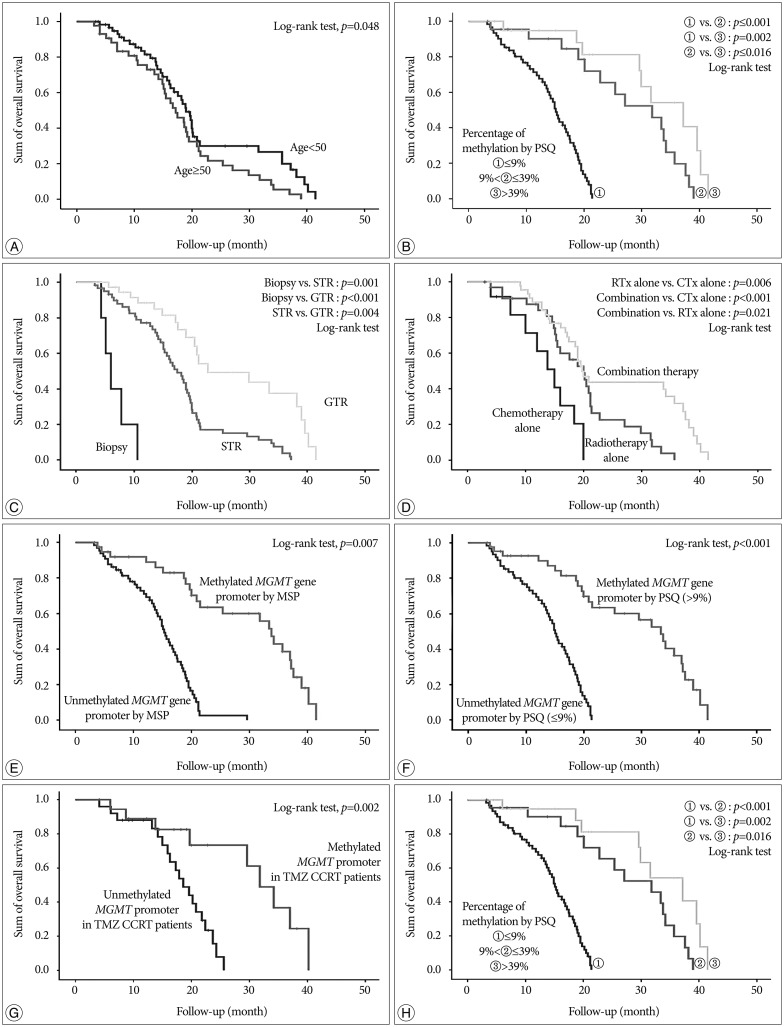
Fig. 2. Kaplan-Meier survival curves for patients with glioblastoma. A : Age <50 vs. age ≥50. B : KPS ≥70 vs. KPS <70. C : Extent of surgical resection. D : Method of adjuvant treatment. E : Methylation status of the MGMT promoter analyzed by methylation-specific PCR. F : Methylation status of the MGMT promoter analyzed by pyrosequencing. G : Methylation status of MGMT promoter in patients who were treated with concurrent temozolomide chemoradiotherapy (TMZ CCRT). H : Extent of methylation of the MGMT promoter analyzed quantitatively by pyrosequencing.
DISCUSSION
In this study, we performed PSQ analysis to determine MGMT promoter methylation status and estimate the extent of methylation using 1- to 13-year-old archival tissue samples of GBMs. We also investigated the prognostic significance of MGMT promoter methylation and other factors associated with OS in patients who were treated in the routine clinic.
Recently, Stupp et al.19) reported the results of 5-year analysis of the EORTC-NCIC trial for survival in GBM patients. Their randomized phase III study showed that the following factors had an association with outcome : concurrent chemoradiotherapy with TMZ, complete resection, age <50, good functional status (recursive partitioning analysis class III>IV>V), and methylated MGMT promoter. We obtained similar results in our study; age <50, good functional status (KPS ≥70), gross total resection, and methylated MGMT promoter analyzed by both MSP and PSQ were all associated with OS. Therefore, this study provided good verification of the published literature.
In terms of the frequency of MGMT promoter methylation in GBM, Hegi et al.7) reported that 45% of GBMs showed MGMT promoter methylation in their randomized trial, and that the median success rate of MSP analysis in FFPE samples was 75%. In contrast, the success rate of PSQ analysis in our study was 100% for 104 samples from 1- to 13-year-old archival specimens, and 41.3% of the GBMs were methylated. Recently, Lee et al.12) reported that 44.4% of 27 GBMs showed methylation. These results, and data from several other researchers, indicate that 40–50% of GBMs are potentially methylated, which might be of clinical benefit regarding treatment with alkylating agents.
Several methods for determining promoter methylation have been described, including the use of restriction enzymes18) or genomic bisulfite sequencing6). Among these methods, some are quantitative, some are semi-quantitative, and some are qualitative. The overwhelming majority of published data uses MSP following bisulfite treatment8). The MSP assay is highly sensitive and has been used widely in this context1,8). However, as there are no inbuilt measures of the adequacy of bisulfite treatment, the possibility of false positives due to inadequate conversion of non-methylated cytosine to uracil exists, particularly for DNA of poor quality or low quantity. Another potential source of false positives is mis-priming, which may be a greater problem when high numbers of PCR cycles or nested primers are used. An accurate quantitative approach is very important for DNA methylation analysis, and MSP has many weaknesses in this regard. One of the major concerns with MSP is the use of relative quantification by means of an external control using a different PCR for the control gene. The accuracy and reproducibility of this approach for the large diversity of genes has still not been adequately addressed. Development of DNA methylation risk prediction panels based on diverse genes will be required to develop and optimize an accurate and reproducible method.
In this study, we present results of PSQ analysis of 1- to 13-year-old archive specimens. As described above, PSQ is a simple technique based on the sequencing-by-synthesis principle and allows accurate and quantitative analysis of DNA sequences. The percentage of methylation was evenly distributed over the years of this study and no increasing or decreasing pattern of methylation with specimen age was observed, regardless of the age of the samples. This might represent a huge improvement in the success rate when compared with data reported using MSP by Hegi et al.7). Using the same method as in the current study, Dunn et al.2) analyzed 264 of 287 tumor samples from 121 cases by PSQ. Some failures occurred, most likely as a result of the small sample volume (<1 mm3) with low DNA yield or poor DNA integrity in the FFPE samples. Nonetheless, despite these failures the PSQ data were reproducible and showed a good correlation between duplicate PCR reactions from the same bisulfite modification and between two independent bisulfite modifications of the same DNA extract2). More recently, a quantitative real-time MSP assay in which the copy number of methylated MGMT alleles was calculated showed improvement over the gel-based MSP assay in terms of reproducibility and use with archival samples, but did not provide methylation data at individual CpG sites21). Therefore, PCR followed by the PSQ method is a stable technique that produces consistent data. These results support the need for a large nationwide retrospective trial to evaluate the MGMT promoter methylation status of old archived material from multiple institutions.
In terms of the relationship between the extent of methylation estimated by PSQ and clinical outcome in GBM, this study showed that GBM patients with a more methylated MGMT promoter had longer overall survival than those with a less methylated MGMT promoter. Similarly, Dunn et al.2) reported an interesting clustering analysis based on the extent of methylation. Progression-free survival was significantly different between low (mean percentage of methylation 9–29%) and high (≥29%) methylation groups of GBM, and between high methylated and unmethylated cases. Log-rank tests showed significant differences in OS between unmethylated and methylated groups and between cases with a low versus high percentage of methylation. This result suggested that qualitative and quantitative data should be simultaneously obtained from the MGMT promoter methylation analysis to predict progression-free survival, overall survival, or chemoresponsiveness in clinical practice. Although we did not obtain exactly the same results as those of Dunn, we found a similar tendency for a higher percentage of methylation to be associated with longer overall survival. The discrepancy in results probably originates from different sample size and different follow-up duration. Moreover, our archives included a relatively small number of samples with a low percentage of methylation.
Another issue for MGMT promoter methylation analysis is the relevant CpG sites for methylation status. The pattern of MGMT promoter methylation is very heterogeneous from tumor to tumor, and the specific CpG sites that need to be methylated to silence transcription and provide a favorable outcome have not been established. Several studies have investigated the methylation status at individual CpGs within MGMT CGIs and compared them with gene expression. Watts et al.22) studied the methylation status of 108 CpGs across MGMT CGIs using bisulfite sequencing and identified three distinct regions within the CGI where high methylation levels were found, but only in the MGMT non-expressing cell line. Recently, Karayan-Tapon et al.10) assessed MGMT promoter methylation status by five different methods including PSQ, and the PSQ primer sets used included 5 CpGs in the DMR2 region that were also used in this study. They concluded that PSQ using FFPE samples predicted overall survival well in patients who were initially treated with radiotherapy plus TMZ.
The limitations of this study can be summarized as follows : First, the cut-off value for methylation used in this study should be verified in more detail and in a larger number of samples. Second, we studied limited sites of CGI methylation using the Pyro Mark Q96 CpG MGMT kit (which evaluates only 5 CGIs). Therefore, further evaluation of other sites of CGI methylation is essential to increase the accuracy of the status and extent of MGMT promoter methylation, and to commercialize this kit. Third, the relatively small number of GBM samples made it impossible for us to find any critical methylation status value for predicting the outcome of these tumors. Fourth, this study is neither a randomized study nor a prospective study. Finally, progression-free survival could not be estimated because there was no specific protocol for follow-up and management.
CONCLUSION
This study showed that both the status and extent of methylation of the MGMT promoter analyzed by PSQ were found to be associated with OS in patients with GBM. To date, MGMT promoter methylation analysis is one of the most recommended molecular assays in clinical neurooncology. A range of new methodologies are available for MGMT testing that potentially allow higher levels of sensitivity, specificity, robustness, and reproducibility. In this regard, among many methods for determining the characteristics of gene promoter methylation, PSQ is a quantitative approach that overcomes the problems associated with MSP, and is a simple technique for accurate analysis of DNA sequences. As a result, PSQ might be a promising technique for MGMT evaluation in daily practice. Furthermore, the development of new biomarkers for malignant brain tumors using molecular methods such as PSQ might be helpful for improvement of therapeutic outcome in patients with malignant brain tumors.
Acknowledgements
This study was financially supported by a Samsung Biomedical Research Institute grant (SMR-112061). The authors would like to thank Yun Gyu Song, M.D. (Department of Radiology, Samsung Changwon Hospital) and Sun Sup Choi, M.D. (Department of Radiology, Dong-A University Medical Center) for their review of the neuroradiological images; Young Jin Song, M.D., (Department of Neurosurgery, Dong-A University Medical Center) for preparing clinical data; Mi Hyun Jin, B.S. (Department of Biostatistics, Samsung Changwon Hospital) for assistance with statistical analysis; Mee Seon Kim, M.D. (Department of Pathology, Samsung Changwon Hospital) for performing the immunohistochemical staining for this research.
References
- 1.Derks S, Lentjes MH, Hellebrekers DM, de Bruïne AP, Herman JG, van Engeland M. Methylation-specific PCR unraveled. Cell Oncol. 2004;26:291–299. doi: 10.1155/2004/370301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eng J. Receiver operating characteristic analysis : a primer. Acad Radiol. 2005;12:909–916. doi: 10.1016/j.acra.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 5.Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, et al. DNA methylation in glioblastoma : impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 8.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR : a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung KW, Ha J, Lee SH, Won YJ, Yoo H. An updated nationwide epidemiology of primary brain tumors in republic of Korea. Brain Tumor Res Treat. 2013;1:16–23. doi: 10.14791/btrt.2013.1.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 11.Lantos PL, Louis DN, Rosenblum Mk, Kleihues P. Tumors of the Nervous System. London: Oxford University Press; 2002. [Google Scholar]
- 12.Lee SH, Hwang TS, Koh YC, Kim WY, Han HS, Kim WS, et al. A consideration of MGMT gene promotor methylation analysis for glioblastoma using methylation-specific polymerase chain reaction and pyrosequencing. Korean J Pathol. 2011;45:21–29. [Google Scholar]
- 13.Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007;373:15–24. doi: 10.1385/1-59745-377-3:15. [DOI] [PubMed] [Google Scholar]
- 14.Mikeska T, Bock C, El-Maarri O, Hübner A, Ehrentraut D, Schramm J, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JW, Hwang JH. The long-term survival in patients with anaplastic astrocytoma or glioblastoma. J Korean Neurosurg Soc. 2003;34:514–520. [Google Scholar]
- 16.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 17.Provenzale JM, Ison C, Delong D. Bidimensional measurements in brain tumors : assessment of interobserver variability. AJR Am J Roentgenol. 2009;193:W515–W522. doi: 10.2214/AJR.09.2615. [DOI] [PubMed] [Google Scholar]
- 18.Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M, et al. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990;10:4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study : 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 20.Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br Med Bull. 2008;85:17–33. doi: 10.1093/bmb/ldm036. [DOI] [PubMed] [Google Scholar]
- 21.Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]