Summary
DNA that gains access to the cytoplasm generally serves as a danger signal for the hosts. An emerging paradigm for responding to cytosolic DNAs centres on the endoplasmic reticulum‐resident protein stimulator of interferon genes (STING, also known as MITA, ERIS or MPYS), the hub adaptor of the recently identified DNA sensors. Dynamic regulations of STING action are critical for shaping innate immune responses against microbial infections, as well as for preventing autoimmune diseases. STING is also indispensable for the detection of immunogenic tumours. A deeper understanding of STING modulations could be instrumental for developing novel immunotherapeutic strategies against infectious, autoimmune and cancerous diseases. In this review, we summarize the latest advances on the role of STING in the DNA‐triggered immune reactions, and underscore the critical issues that remain to be resolved in future studies.
Keywords: autoimmunity, cancer immunology, cytosolic DNA, innate immunity, STING, type I interferon
Introduction
Our bodies are constantly challenged by pathogenic microorganisms around us. Upon exposure to them, the immune systems are rapidly alerted to combat these dangerous invasions. Innate immunity triggers early and immediate host defences against microbes, and is critical for eliciting subsequent adaptive immune responses, including antibody production and cytotoxic T‐cell activation to further eradicate the infections.1 Host recognition of invading pathogens, a prerequisite for initiating immune responses, is realized through sensing the essential and conservative components of microbes, named pathogen‐associated molecular patterns, by an array of host germline‐encoded pattern recognition receptors.2, 3
As pathogen‐associated molecular patterns, nucleic acids species derived from microorganisms trigger an array of immune signalling pathways, ultimately inducing the nuclear factor‐κB (NF‐κB) ‐dependent and/or interferon regulatory factor 3 (IRF3) ‐dependent genes, including pro‐inflammatory cytokines and type I interferons (IFNs). Studies during the past decade have led to considerable advances in the mechanisms of microbial RNA‐triggered innate immune signalling.4, 5, 6, 7, 8 However, our understanding of the aberrant DNA‐provoked type I IFN induction is relatively limited. In contrast to Toll‐like receptor 9, which monitors unmethylated CpG‐DNA from topologically extracellular bacteria in immune cells, several proteins have recently been suggested to function as cytosolic DNA sensors that trigger IFN production during the primary host responses.9, 10, 11, 12, 13, 14, 15, 16 These DNA sensors include IFI16 (p204), the helicase DDX41, as well as cyclic GMP‐AMP synthase (cGAS, also known as MB21D1 or C6orf150). Recent breakthroughs characterized endoplasmic reticulum (ER) ‐resident protein stimulator of interferon genes (STING; also known as MITA, ERIS or MPYS) as the converging point of the identified DNA sensors, relaying the DNA‐triggered signalling transduction.17, 18, 19, 20 In addition, STING can also bind directly to bacterial secondary messenger cyclic dinucleotides (CDNs), including c‐di‐GMP and c‐di‐AMP.21 Consistently, STING has been demonstrated to be pivotal in a myriad of physiological and pathological processes. In this review, we summarize the current understanding on the functions of STING in immunity and diseases, highlighting important questions for future exploration.
The characteristics of STING and its role in immune responses
Using functional cDNA library screening, three groups independently identified a strong type I IFN stimulator, which they named, respectively, as STING, MITA or ERIS.17, 19, 20 In human, STING is a predicted 42 000 MW protein harbouring three functional domains: a cytoplasmic C‐terminal tail (CTT, 342–379 amino acids), a central globular domain (155–341 amino acids) and four putative N‐terminal transmembrane motifs (1–154 amino acids) that anchor STING to the ER.17, 20 Notably, an RIR motif at position 178–180 amino acids of STING has been identified to function as its ER retention or retrieval signalling, confirming its subcellular localization.20 Moreover, human STING (encoded 379 amino acids) and mouse STING exhibit approximate 68% sequence identity and 81% similarity at the amino acid level. Interestingly, its orthologues in other species also share high identity, especially at the C‐terminal region, suggesting evolutionary conservation of STING and its functional importance. In addition, STING is highly expressed in immune‐related cells and to a lesser extent in the non‐immune HEK293T, HeLa and Huh‐7 cells,19 implying its potential role in immunity.
Intensive studies have established the essential role of STING in sensing nucleic acids, especially the cytosolic double‐stranded DNAs. Over‐expression of STING triggers the dimerization and nuclear‐translocation of IRF3, resulting in robust IFN induction.17, 19, 20 Compared with their wild‐type counterpart, STING‐deficient cells display profound defects in eliciting production of IFN‐β and pro‐inflammatory cytokines, in response to the transfection of IFN stimulatory DNA or challenges by DNA viruses (herpes simplex virus 1 or vaccinia virus) or by bacteria (Listeria monocytogenes or Mycobacterium tuberculosis) or the malaria parasite.18, 22, 23, 24, 25, 26, 27 Consistently, Tmem173 −/− mice were more susceptible to lethal infection after exposure to herpes simplex virus 1 than wild‐type mice, due to the deficiency of IFN production.18 In contrast, the loss of STING barely affects intracellular and extracellular poly(I:C)‐mediated type I IFN production, which are largely governed by melanoma differentiation‐associated protein 5 (MDA5) and Toll‐like receptor 3 (TLR3), respectively.18 In addition, STING deficiency exhibits no effect on either the cleavage of pro‐caspase 1 or interleukin‐1β maturation in response to cytosolic DNA, which is absent‐in‐melanoma‐2 (AIM‐2)‐dependent.18 Interestingly, both in vitro and in vivo evidence indicate that loss of STING also rendered hosts highly susceptible to vesicular stomatitis virus, a negative‐stranded RNA virus.17 Besides, knockout of STING results in defects in vesicular stomatitis virus‐induced or Sendai virus‐induced type I IFN production,19, 20 indicating that STING may be involved in facilitating viral RNA‐sensing retinoic acid‐inducible gene‐I (RIG‐I) signalling or may directly inhibit vesicular stomatitis virus and Sendai virus replication and propagation.
Remarkably, STING was also shown recently to be essential for IFN production stimulated by CDNs, such as c‐di‐GMP or c‐di‐AMP. As the ubiquitous second messengers, CDNs play important roles in regulating the diverse processes of bacteria, including biofilm formation, motility and virulence.28 Interestingly, c‐di‐GMP was first reported as an immune stimulatory molecule to enhance host immune responses.29 Further evidence substantiates that c‐di‐GMP is sensed in the cytosol of mammalian cells and triggers the induction of type I IFNs and the co‐regulated genes by activating IRF3, NF‐κB and mitogen‐activated protein kinases.30, 31 Additionally, Woodward et al. found that c‐di‐AMP secreted by intracellular L. monocytogenes is able to trigger the cytosolic host response, leading to type I IFN production.32, 33 These different lines of observation suggest that there is potentially a mammalian sensor for detecting and responding to CDNs in the cytoplasm. By conducting a forward genetic mutagenesis screen in C57BL/6 mice, Sauer et al. identified a novel mutant mouse strain, Goldenticket (Gt), which fails to produce type I IFNs upon L. monocytogens infection.34 Moreover, using genetic mapping, they found that Gt mice harbour a single nucleotide variant (T596A) in STING, which is responsible for this failure in IFN induction,34 suggesting that triggering of IFN production by CDNs is dependent on STING. The same group further observed that STING binds directly to c‐di‐GMP by using a UV cross‐linking assay, and the cytosolic region of STING is sufficient for this binding, demonstrating convincingly that STING is a direct sensor of the c‐di‐GMP in innate immunity.21 The crystal structures of the STING C‐terminal domain alone and its complex with c‐di‐GMP have been solved and analysed by several independent groups.35, 36, 37, 38, 39 It is shown that STING C‐terminal domain forms a V‐shaped dimer, which creates a c‐di‐GMP‐binding pocket at the dimer interface. Yin et al. proposed a plausible model of STING activation, in which c‐di‐GMP binding releases the C‐terminal tail of STING for recruiting and activating downstream signalling molecules.39 Similarly, antiviral small‐molecule drugs such as 10‐carboxymethyl‐9‐acridanone and 5,6‐dimethylxanthenone‐4‐acetic acid strongly induce type I IFN responses by directly activating STING.20, 40, 41, 42
Through this train of discoveries, the cyclic GMP‐AMP (cGAMP) synthase (cGAS, also known as MB21D1 or C6orf150) was identified as the sequence‐independent cytosolic DNA sensor. Unexpectedly, cGAS possesses the nucleotidyl‐transferase catalytic activity, which facilitates from ATP and GTP the synthesis of a non‐canonical CDN c[G(2′,5′)pA(3′,5′)p] (referred to as 2′3′cGAMP) upon binding cytosolic DNA.13, 43, 44, 45, 46, 47 Similar to bacterial CDNs, 2′3′cGAMP strongly induces the expression of IFNs via the STING–TANK‐binding kinase 1 (TBK1)–IRF3 signalling axis. Hence, the mechanisms by which STING recognizes the second messengers unify the host responses to the different forms of the intracellular nucleic acids.
Mechanisms of DNA‐driven assembly of STING signalosome and TBK1/IRF3 activation
It is known that CDNs and/or upstream DNA sensors directly and/or indirectly induce STING dimerization, causing its translocation from the ER, through Golgi apparatus, ultimately to the perinuclear microsome compartments, where TBK1 simultaneously congregates in a STING‐dependent manner.18, 48 Notably, the DNA‐driven assembly of STING–TBK1 complex is required for the activation of TBK1 and subsequent phosphorylation and nuclear translocation of the transcriptional factor IRF3 (Fig. 1). The precise mechanisms underlying the dynamic membrane traffic, such as the sequential translocation and assembly of the STING–TBK1 complex as well as the activation of TBK1 by STING, remain poorly understood. This represents a fast‐evolving frontier for further investigation. Wang et al. provide new mechanistic insights into the modulation of the STING–TBK1 axis during DNA virus infection.49, 50 They demonstrate that the ER‐resident autocrine motility factor receptor (AMFR) and insulin‐induced gene 1 (INSIG1) complex catalysed the K27‐linked poly‐ubiquitination of STING stimulated by aberrant DNAs. These poly‐ubiquitin chains are then required for proper engagement with TBK1 and efficient downstream signal transduction (Fig. 1). Notably, evidence indicates that the INSIG1–AMFR protein complex is specifically involved in cytoplasmic DNA or CDN‐triggered innate immune signalling, but not in RNA‐induced immune signalling. Wang's work provides a detailed example of the dynamic functions of poly‐ubiquitination in potentiating innate immunity against DNA microbes, highlighting a key role of ER membrane proteins AMFR and INSIG1 as well as K27‐linked poly‐ubiquitin chains in STING‐mediated signalling. Moreover, a previous study indicates that AMFR regulates the sterol‐dependent degradation of the enzyme HMG‐CoA reductase, so suppressing cholesterol biosynthesis in cell metabolism.51 The novel role of AMFR in STING‐dependent signalling potentially provides a new clue that AMFR serves as a link between immunity and metabolism. Another important question unanswered from this study is that of whether and how AMFR–INSIG1 complex modulates STING‐mediated NF‐κB activation. It will be interesting to investigate whether the formation of punctate structures is required for NF‐κB activation.
Figure 1.
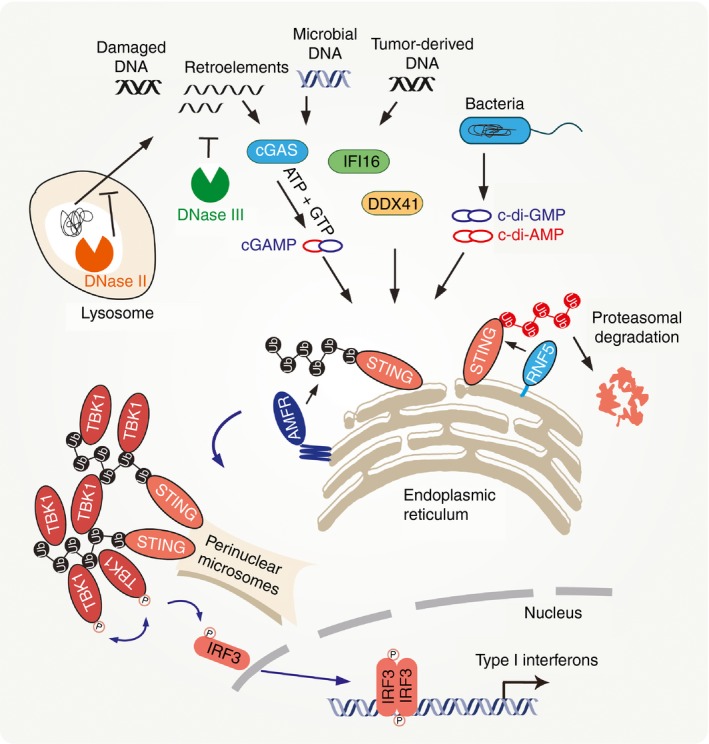
Stimulator of interferon genes (STING)‐mediated cytosolic DNA sensing pathway. Upon sensing cytosolic DNA or recognizing cyclic dinucleotides, STING is triggered to dimerize and translocate from the endoplasmic reticulum (ER), through the Golgi apparatus, to perinuclear microsomal compartments. Simultaneously, autocrine motility factor receptor (AMFR) catalyses K27‐linked poly‐ubiquitination of STING. TANK‐binding kinase 1 (TBK1) selectively binds to K27‐linked poly‐ubiquitin chains on STING, so congregating at perinuclear microsomes in a STING‐dependent manner. The DNA‐induced assembly of the STING–TBK1 complex is required for the activation of TBK1 and the subsequent phosphorylation and nuclear translocation of the transcriptional factor interferon regulatory factor 3 (IRF3), ultimately leading to expression of type I interferons.
Other studies have shown that K63‐linked poly‐ubiquitination signals also participate in the relay of the activation signal from STING to the downstream molecules TBK1/IRF3 in response to exogenous DNA. E3 ubiquitin ligases TRIM56 and TRIM32 have been proposed, respectively, to facilitate the K63‐linked poly‐ubiquitination of STING.52, 53 Notably, the indicated studies focused more on RNA‐sensing rather than DNA‐sensing pathways. In contrast, Wang et al. failed to detect any ubiquitination signal of STING in the presence of Trim56 or Trim32 by using the stringent two‐step immunoprecipitation that was not employed by the indicated studies.49 This suggests that Trim56 or Trim32 might catalyse the poly‐ubiquitination of other unknown factor(s) in the STING protein complex, and regulate the STING signalling indirectly.
Modulation of STING activity during host immune responses
Given the double‐edged functions of immune responses, the timing, strength and duration of STING‐dependent signalling must be rapidly and appropriately modulated, which ensures that the microbes are contained and eliminated, but the damage to the host is kept to a minimum.
Protein post‐translational modifications (phosphorylation and ubiquitination) have been reported to fine‐tune the spatial and temporary action of the immune signalling by way of multiple feedback mechanisms and STING‐dependent signalling is not an exception.54, 55, 56, 57 It has been found that upon cytosolic DNA stimulation, STING is phosphorylated at multiple serine residues (Ser358, Ser353 and Ser379), preceding the phosphorylation of IRF3 by TBK1.58 Silencing of TBK1 abrogates DNA‐induced STING phosphorylation as well as the interaction between STING and IRF3.58 This suggests that TBK1 mediates the phosphorylation of STING, which in turn enhances the association of STING with IRF3. However, Konno et al. reported that STING is phosphorylated at Ser366 by UNC‐51‐like kinase (ULK1, also known as ATG1) after STING trafficking out of ER and activation.59 This modification takes place after the CDN‐triggered ULK1 dissociation from its repressor AMPK.59 Notably, the phosphorylation of STING on Ser366 was found to specifically impair the STING‐dependent IRF3 activation, but not the NF‐κB activation. The mechanisms of the selectivity remain unknown.59 The discrepancy of the functional consequences of the STING phosphorylations needs further clarification.
The ubiquitin‐mediated protein degradation plays pivotal roles in the attenuation or termination of innate immune signalling pathways, effective for braking the detrimental effects of the excessive immune responses.60 The ER‐associated E3 ubiquitin ligase RING finger protein 5 (RNF5) was identified to catalyse the K48‐linked poly‐ubiquitination of STING and promote its proteasome‐dependent degradation, which results in down‐regulation of the STING‐dependent host antimicrobial response.61 Recently, it was reported that another ER‐resident E3 ligase RNF26 antagonizes the effect of RNF5 on STING, thereby protecting STING from K48‐linked poly‐ubiquitination and subsequent degradation.62 This is realized by RNF26‐mediated K11‐linked poly‐ubiquitination of STING at K150, which competes with the RNF5‐catalysed ubiquitination on the same lysine residue.62 In addition to stabilizing STING, RNF26 promotes the autophagic degradation of IRF3 in the later phase of virus infection, ultimately limiting the excessive type I IFN response.62 Therefore, RNF26 displays dual functions in the immune response by regulating the stability of both STING and IRF3 through different mechanisms and at different phases, so achieving a balanced activation of the STING signalling.
Several additional proteins (NLRC3 and ZDHHC1) have been identified to modulate the STING activity.63, 64 NLRC3 is a NOD‐like receptor (NLR) that directly associates with STING and inhibits the trafficking of STING to perinuclear puncta. NLRC3 could also bind TBK1 and interferes with the interaction between STING and TBK1. Therefore, NLRC3 functions as a brake to suppress cytosolic DNA‐driven immune responses. However, the precise mechanism by which NLRC3 regulates STING translocation awaits further exploration. In contrast, the ER‐resident protein ZDHHC1 promotes the STING‐dependent signalling by facilitating the oligomerization of STING and the recruitment of TBK1 and IRF3. Interestingly, ZDHHC1 belongs to the DHHC palmitoyl transferase family.65 It is intriguing to address whether the enzymatic activity of ZDHHC1 is required for its regulation of the STING‐dependent signalling.
The importance of the STING signalling is echoed by some recent reports that viruses have developed multiple strategies to sabotage or inhibit this signalling. The hepatitis C virus NS4B displays a pronounced inhibitory effect on STING.66, 67 NS4B binds to STING and disrupts the association of STING with the downstream signalling effectors to block host antiviral responses. The viral oncogenes of the DNA tumour viruses, including E7 from human papillomavirus and E1A from adenovirus, have recently been identified as potent and specific inhibitors of the cGAS‐STING pathway by targeting STING via a Leu‐X‐Cys‐X‐Glu (LXCXE) protein motif on these oncogenes.68 Interestingly, Dengue virus NS2B3 protease was shown to cleave human STING, so inhibiting the host immune response.69, 70
Role of the STING‐dependent signalling in autoimmunity and cancers
In normal conditions, cellular DNA is restricted to the nucleus or mitochondria, whereby it is compartmentalized away from cytosolic DNA sensors. The DNases stationed in the cytosol, lysosome and ER are actively digesting the mis‐localized or unwanted DNA. It was recently established that inappropriate provocation of the immune system by aberrant self‐nuclear acids is responsible for the pathogenesis of certain autoimmune diseases, such as systemic lupus erythematosus, lupus‐like diseases and Aicardi–Goutieres syndrome.71
DNase II is a lysosome‐resident endonuclease that is ubiquitously expressed in various cells. DNase II knockout embryos exhibit abnormal over‐production of IFN and IFN‐inducible genes, and consequently develop severe anaemia and die in utero.72 Interestingly, the embryonic lethality in Dnase2a −/− embryos is rescued by further depleting IFN type I receptor (IFN‐IR),72 indicating that the production of type I IFN is responsible for the observed lethal anaemia. However, mice lacking both DNase II and IFN‐IR suffer from a chronic polyarthritis that resembles human rheumatoid arthritis as they age, which is ascribed to excessive tumour necrosis factor‐α production.73 The aberrant production of these cytokines is the result of accumulation of improperly digested DNA either from phagocytosed erythroid precursor cells and apoptotic cells or from damaged DNA in the nucleus.72, 74 Notably, loss of STING abolishes self‐DNA‐activated cytokine production in Dnase2a −/− embryos, which alleviates self‐DNA‐mediated polyarthritis. These observations reveal the critical role of STING in aberrant DNA‐induced inflammatory disorders.75
DNase III (also known as TREX1) is the most abundant 3′,5′‐exonuclease located in ER and degrades DNA originated from endogenous retro‐elements.76, 77 Loss‐of‐function mutations in TREX1 lead to a broad spectrum of inflammatory and autoimmune phenotypes, including Aicardi–Goutieres syndrome, familial chilblain lupus and systemic lupus erythematosus.78, 79, 80 Several studies demonstrated that these autoimmune disorders are associated with a cell‐intrinsic antiviral response to self‐DNA.77, 81, 82 Similarly, DNase III‐deficient mice showed dramatically reduced survival due to the development of inflammatory myocarditis, progressive cardiomyopathy and circulatory failure.83 Recent studies found that DNase III‐deficient mice lacking IRF3, the type I IFN receptor, or STING are all completely protected from autoimmune pathology and mortality,84 suggesting that the STING–IRF3 axis‐mediated type I IFN is responsible for the development of disease in DNase III‐deficient mice. These findings collectively highlight the key role of STING in the pathogenesis of autoimmune diseases, and yield a deeper understating of the STING‐related immune disorders.
Two independent groups recently reported that the STING‐dependent signalling is indispensable for CD8+ T‐cell priming against tumours by sensing tumour cell‐derived DNA.85, 86 Upon tumour implantation or radiation therapy, tumour‐derived DNA can be transferred into dendritic cell cytosol, where the immunogenic DNA activates STING‐dependent IFN‐β production. Afterwards, IFN‐β renders dendritic cells competent to present tumour antigens to promote the cross‐priming of CD8+ T cells and so control the tumours. This is further substantiated by the intra‐tumour injection of STING agonists, which generates a potent anti‐tumour immune T‐cell response and prominent therapeutic effects.87 Similarly, genome‐derived DNA species are found to be present in the cytosol of B‐cell lymphoma cells, which increases the level of type I IFN and contributes to IFN‐dependent immune rejection of tumour cells.88 However, the critical role of STING in this process remains unknown.
Concluding remarks and prospects
It is well established that STING plays a key role in anti‐microbial innate immunity, as well as in the pathogenesis of some autoimmune disorders, through mediating the aberrant cytosolic DNA‐triggered immune signalling. On the one hand, STING functions as a pattern recognition receptor by recognizing cyclic dinucleotides (e.g. c‐di‐GMP and c‐di‐AMP) directly and ultimately inducing the expression of IFNs and pro‐inflammatory cytokines; on the other hand, as a key scaffolding and adaptor protein, STING facilitates the signal transduction initiated from upstream cytosolic dsDNA receptors. In response to the corresponding stimuli, STING undergoes phosphorylation, ubiquitination and subcellular translocation, which ultimately relays the signal to and activates the downstream effectors TBK1 and IRF3, leading to the expression of type I IFNs.
The delineation of STING‐dependent signalling remains in its infancy. Further in‐depth studies and related structural analyses are needed to resolve the corresponding molecular mechanisms underlying the STING signal transduction. For example, the events that lead to STING translocation, and the mechanism whereby STING is activated remain to be elucidated. Additional critical signalling molecules in the STING signalling may be identified in the near future, and their functions are expected to expand and deepen our understanding of the cytosolic DNA‐induced signalling. In addition, some pathogen‐associated effectors have been reported to possess a unique and potent activity that interferes with the function of STING and is critical for pathogens to escape immune responses. The relevant studies will help to understand the microbial pathogenesis and host–microbe interactions. Insights from the investigations will surely provide new strategies and methods for the effective control and treatment of infectious and autoimmune diseases as well as tumours.
Disclosures
The authors have declared that no conflicts of interest exist.
Acknowledgements
The authors apologize to colleagues whose work is not cited here due to length limitations. We thank Dr Youdong Pan (Brigham and Women's Hospital, Harvard Medical School) and Sonal Jhaveri (Massachusetts Institute of Technology) for discussions and critical comments. This work was supported by grants from the National Natural Science Foundation of China (31030021, 31300735, 81161120542) and the Ministry of Science and Technology of China (2012CB910200, 2011CB910904).
References
- 1. Schenten D, Medzhitov R. The control of adaptive immune responses by the innate immune system. Adv Immunol 2011; 109:87–124. [DOI] [PubMed] [Google Scholar]
- 2. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805–20. [DOI] [PubMed] [Google Scholar]
- 3. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124:783–801. [DOI] [PubMed] [Google Scholar]
- 4. Loo YM, Gale M Jr. Immune signaling by RIG‐I‐like receptors. Immunity 2011; 34:680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakhaei P, Genin P, Civas A, Hiscott J. RIG‐I‐like receptors: sensing and responding to RNA virus infection. Semin Immunol 2009; 21:215–22. [DOI] [PubMed] [Google Scholar]
- 6. Weber M, Weber F. RIG‐I‐like receptors and negative‐strand RNA viruses: RLRly bird catches some worms. Cytokine Growth Factor Rev 2014; 25:621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oshiumi H, Miyashita M, Okamoto M, Morioka Y, Okabe M, Matsumoto M, et al DDX60 Is involved in RIG‐I‐dependent and independent antiviral responses, and its function is attenuated by virus‐induced EGFR activation. Cell Rep 2015; 11:1193–207. [DOI] [PubMed] [Google Scholar]
- 8. Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol 2011; 23:564–72. [DOI] [PubMed] [Google Scholar]
- 9. Paludan SR, Bowie AG. Immune sensing of DNA. Immunity 2013; 38:870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al A Toll‐like receptor recognizes bacterial DNA. Nature 2000; 408:740–5. [DOI] [PubMed] [Google Scholar]
- 11. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 2010; 11:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 2011; 12:959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee KG, Kim SS, Kui L, Voon DC, Mauduit M, Bist P, et al Bruton's tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Rep 2015; 10:1055–65. [DOI] [PubMed] [Google Scholar]
- 15. Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. Nucleic acid recognition orchestrates the anti‐viral response to retroviruses. Cell Host Microbe 2015; 17:478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ansari MA, Dutta S, Veettil MV, Dutta D, Iqbal J, Kumar B, et al Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN‐β responses. PLoS Pathog 2015; 11:e1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008; 455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA‐mediated, type I interferon‐dependent innate immunity. Nature 2009; 461:788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al The adaptor protein MITA links virus‐sensing receptors to IRF3 transcription factor activation. Immunity 2008; 29:538–50. [DOI] [PubMed] [Google Scholar]
- 20. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 2009; 106:8653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burdette DL, Monroe KM, Sotelo‐Troha K, Iwig JS, Eckert B, Hyodo M, et al STING is a direct innate immune sensor of cyclic di‐GMP. Nature 2011; 478:515–U111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, et al Mycobacterium tuberculosis differentially activates cGAS‐ and inflammasome‐dependent intracellular immune responses through ESX‐1. Cell Host Microbe 2015; 17:799–810. [DOI] [PubMed] [Google Scholar]
- 23. Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, et al The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to Induce type I interferons and activate autophagy. Cell Host Microbe 2015; 17:811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, et al Innate immune recognition of an AT‐rich stem‐loop DNA motif in the Plasmodium falciparum genome. Immunity 2011; 35:194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, et al DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti‐microbial innate immunity. Immunity 2015; 42:332–43. [DOI] [PubMed] [Google Scholar]
- 26. Wu J, Cai B, Sun W, Huang R, Liu X, Lin M, et al Genome‐wide analysis of host–Plasmodium yoelii interactions reveals regulators of the type I interferon response. Cell Rep 2015; 12:661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, et al Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING‐mediated cytosolic DNA‐sensing pathway. PLoS Pathog 2014; 10:e1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 2007; 61:131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, et al Bacterial c‐di‐GMP is an immunostimulatory molecule. J Immunol 2007; 178:2171–81. [DOI] [PubMed] [Google Scholar]
- 30. McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, et al A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic‐di‐GMP. J Exp Med 2009; 206:1899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobayashi H, Kobayashi CI, Nakamura‐Ishizu A, Karigane D, Haeno H, Yamamoto KN, et al Bacterial c‐di‐GMP affects hematopoietic stem/progenitors and their niches through STING. Cell Rep 2015; 11:71–84. [DOI] [PubMed] [Google Scholar]
- 32. Woodward JJ, Iavarone AT, Portnoy DA. c‐di‐AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 2010; 328:1703–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Archer KA, Durack J, Portnoy DA. STING‐dependent type I IFN production inhibits cell‐mediated immunity to Listeria monocytogenes . PLoS Pathog 2014; 10:e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sauer JD, Sotelo‐Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, et al The N‐ethyl‐N‐nitrosourea‐induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 2011; 79:688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di‐GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 2012; 19:722–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, et al Crystal structures of STING protein reveal basis for recognition of cyclic di‐GMP. Nat Struct Mol Biol 2012; 19:725–7. [DOI] [PubMed] [Google Scholar]
- 37. Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, et al Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di‐GMP binding. Immunity 2012; 36:1073–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di‐GMP by STING. Nat Struct Mol Biol 2012; 19:728–30. [DOI] [PubMed] [Google Scholar]
- 39. Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, et al Cyclic di‐GMP sensing via the innate immune signaling protein STING. Mol Cell 2012; 46:735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species‐specific detection of the antiviral small‐molecule compound CMA by STING. EMBO J 2013; 32:1440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, et al Structure‐function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 2013; 154:748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao P, Zillinger T, Wang W, Ascano M, Dai P, Hartmann G, et al Binding‐pocket and lid‐region substitutions render human STING sensitive to the species‐specific drug DMXAA. Cell Rep 2014; 8:1668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS‐cGAMP signaling in antiviral defense and immune adjuvant effects. Science 2013; 341:1390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA‐activated cyclic GMP‐AMP synthase. Cell 2013; 153:1094–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, et al The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 2013; 3:1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al cGAS produces a 2′‐5′‐linked cyclic dinucleotide second messenger that activates STING. Nature 2013; 498:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al Cyclic GMP‐AMP containing mixed phosphodiester linkages is an endogenous high‐affinity ligand for STING. Mol Cell 2013; 51:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al Atg9a controls dsDNA‐driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 2009; 106:20842–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, et al The E3 ubiquitin ligase AMFR and INSIG1 Bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 2014; 41:919–33. [DOI] [PubMed] [Google Scholar]
- 50. Shu HB, Wang YY. Adding to the STING. Immunity 2014; 41:871–3. [DOI] [PubMed] [Google Scholar]
- 51. Goldstein JL, DeBose‐Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell 2006; 124:35–46. [DOI] [PubMed] [Google Scholar]
- 52. Tsuchida T, Zou JA, Saitoh T, Kumar H, Abe T, Matsuura Y, et al The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double‐stranded DNA. Immunity 2010; 33:765–76. [DOI] [PubMed] [Google Scholar]
- 53. Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63‐linked Ubiquitination. J Biol Chem 2012; 287:28646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu X, Wang Q, Pan YD, Wang C. Sensing and responding to cytosolic viruses invasions: an orchestra of kaleidoscopic ubiquitinations. Cytokine Growth Factor Rev 2015; 26:379–387. [DOI] [PubMed] [Google Scholar]
- 55. Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, et al Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol 2010; 30:2424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu X, Chen W, Wang Q, Li L, Wang C. Negative regulation of TLR inflammatory signaling by the SUMO‐deconjugating enzyme SENP6. PLoS Pathog 2013; 9:e1003480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi HX, Liu X, Wang Q, Tang PP, Liu XY, Shan YF, et al Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog 2011; 7:e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tanaka Y, Chen ZJJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 2012; 5:ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013; 155:688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu X, Wang Q, Chen W, Wang C. Dynamic regulation of innate immunity by ubiquitin and ubiquitin‐like proteins. Cytokine Growth Factor Rev 2013; 24:559–70. [DOI] [PubMed] [Google Scholar]
- 61. Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, et al The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 2009; 30:397–407. [DOI] [PubMed] [Google Scholar]
- 62. Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al RNF26 temporally regulates virus‐triggered type I interferon induction by two distinct mechanisms. PLoS Pathog 2014; 10:e1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang L, Mo JY, Swanson KV, Wen HT, Petrucelli A, Gregory SM, et al NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 2014; 40:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou Q, Lin H, Wang SY, Wang S, Ran Y, Liu Y, et al The ER‐associated protein ZDHHC1 Is a positive regulator of DNA virus‐triggered, MITA/STING‐dependent innate immune signaling. Cell Host Microbe 2014; 16:450–61. [DOI] [PubMed] [Google Scholar]
- 65. Oku S, Takahashi N, Fukata Y, Fukata M. In silico screening for palmitoyl substrates reveals a role for DHHC1/3/10 (zDHHC1/3/11)‐mediated neurochondrin palmitoylation in its targeting to Rab5‐positive endosomes. J Biol Chem 2013; 288:19816–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano‐Kitazume A, et al Hepatitis C virus NS4B protein targets STING and abrogates RIG‐I‐mediated type I interferon‐dependent innate immunity. Hepatology 2013; 57:46–58. [DOI] [PubMed] [Google Scholar]
- 67. Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, et al Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol 2013; 59:52–8. [DOI] [PubMed] [Google Scholar]
- 68. Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS‐STING DNA sensing pathway. Science 2015; 350:568–571. [DOI] [PubMed] [Google Scholar]
- 69. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, et al DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 2012; 8:e1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, et al Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 2012; 8:e1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith S, Jefferies C. Role of DNA/RNA sensors and contribution to autoimmunity. Cytokine Growth Factor Rev 2014; 25:745–57. [DOI] [PubMed] [Google Scholar]
- 72. Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon‐beta produced in mouse embryos carrying undigested DNA. Nat Immunol 2005; 6:49–56. [DOI] [PubMed] [Google Scholar]
- 73. Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, et al Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 2006; 443:998–1002. [DOI] [PubMed] [Google Scholar]
- 74. Lan YY, Londono D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep 2014; 9:180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA‐dependent inflammatory disease. Proc Natl Acad Sci USA 2012; 109:19386–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′→5′ exonucleases. J Biol Chem 1999; 274:19655–60. [DOI] [PubMed] [Google Scholar]
- 77. Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell‐intrinsic initiation of autoimmunity. Cell 2008; 134:587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, et al Mutations in the gene encoding the 3′‐5′ DNA exonuclease TREX1 cause Aicardi–Goutieres syndrome at the AGS1 locus. Nat Genet 2006; 38:917–20. [DOI] [PubMed] [Google Scholar]
- 79. Lee‐Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, et al Mutations in the gene encoding the 3′‐5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet 2007; 39:1065–7. [DOI] [PubMed] [Google Scholar]
- 80. Hasan M, Fermaintt CS, Gao N, Sakai T, Miyazaki T, Jiang S, et al Cytosolic nuclease TREX1 regulates oligosaccharyltransferase activity independent of nuclease activity to suppress immune activation. Immunity 2015; 43:463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ablasser A, Hemmerling I, Schmid‐Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell‐autonomous immunity in a cGAS‐dependent manner. J Immunol 2014; 192:5993–7. [DOI] [PubMed] [Google Scholar]
- 82. Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, Zahn S, et al Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING‐dependent immune sensing. Immunity 2013; 39:482–95. [DOI] [PubMed] [Google Scholar]
- 83. Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, et al Gene‐targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 2004; 24:6719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gall A, Treuting P, Elkon KB, Loo YM, Gale M Jr, Barber GN, et al Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon‐dependent autoimmune disease. Immunity 2012; 36:120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, et al STING‐dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014; 41:830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al STING‐dependent cytosolic DNA sensing promotes radiation‐induced type I interferon‐dependent antitumor immunity in immunogenic tumors. Immunity 2014; 41:843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, et al Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep 2015; 11:1018–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shen YJ, Le Bert N, Chitre AA, Koo CX, Nga XH, Ho SS, et al Genome‐derived cytosolic DNA mediates type I interferon‐dependent rejection of B cell lymphoma cells. Cell Rep 2015; 11:460–73. [DOI] [PubMed] [Google Scholar]


