Summary
The Immune Epitope Database is uniquely positioned to assess the body of research related to immune epitopes, we have manually curated all such published data. Thus, we are able to make observations on the state of these fields of research, as well as aggregate the individual data points to present a clearer picture of the immune response to specific antigens in all studied hosts. Additionally, we are able to identify where conflicts in the literature exist and where publications fall short in terms of identifiable methods and in reproducibility. Here we present guidelines to improve the quality of immune epitope data, which will benefit journals and researchers alike.
Keywords: database, immune epitope, Immune Epitope Database, reproducibility
Abbreviation
- IEDB
Immune Epitope Database
Introduction
The Immune Epitope Database's (IEDB's) team of curators have read and analyzed >15,000 published articles, representing >98% of all publications describing immune epitopes.1 Once aggregated, this data can be used to perform meta‐analyses describing what is currently known in these fields2, 3, 4 To ensure accuracy and consistency, data is entered into the IEDB following strict curation guidelines.5 These guidelines require a minimal amount of data and unambiguous results. When curators cannot determine any of these details from the publication, they will refer to cited references and/or will contact the author(s). Through these efforts, it becomes clear that much needed data is not present in the original publication and many cited references lack the information as well. By aggregating all data related to similar antigens, it also becomes clear that different publications may refer to the same host, antigen, T‐cell receptor (TCR), major histocompatibility complex (MHC) or antibody in a variety of terms, creating confusion and errors. These issues are serious as they lead to the inability of one researcher to reproduce another's work, as well as make the unambiguous representation of the publication into the IEDB quite difficult.
Background
The IEDB data is derived from manuscripts published in 1474 different journals covering general immunology (e.g. Journal of Immunology), infectious diseases (e.g. Journal of Virology), and general biomedical research (e.g. Proceedings of the National Academy of Sciences of the USA), each with their own set of author guidelines. However, the reporting guidelines for each journal vary and the minimum information needed for the IEDB is not always required for inclusion in the publication.
In a recent study, Vasilevsky et al. found that regardless of the impact factor of the journal or the stringency of its reporting standards, the ability to uniquely identify research resources was hindered by lack of details reported in the publication.6 This study found that almost half of the 238 biomedical papers that were analysed failed to report sufficient information to uniquely identify all the resources reported in the methods section.6 Specific to the field of immunology, the results were particularly alarming with only 39% of antibodies and 38% of constructs found to be identifiable. As the IEDB tracks every publication it processes and cites where all data sources originate, it is uniquely positioned to perform a similar analysis.
Improved reporting standards for biomedical science have been proposed by a number of groups such as Force11 (www.force11.org), who developed formalized data citation principals (https://www.force11.org/datacitation) and Biosharing (biosharing.org), a curated web‐based searchable registry of linked information on content standards, databases and data policies, which began with the establishment of the Minimum Information for Biological and Biomedical Investigations (MIBBI).7 Nature Methods has recently adopted new author and reviewer guidelines aimed at improving the reproducibility and quality of manuscripts.8 In June 2014, the editors of approximately 40 journals came together at the National Institutes of Health to endorse new guidelines to improve reproducibility.9 In addition to improved author guidelines, recent initiatives have sought to improve how authors describe the reagents used in scientific studies. The Resource Identification Initiative10 is a project that aims to promote the use of unique identifiers for research resources to improve resource identification, discovery and reuse. These initiatives and efforts are very promising for the future of reproducibility, but each field of research should clearly specify its individual issues and needs to its journals and authors. The IEDB analysed the incidence of unavailable or incorrect critical information in relevant publications, as well as identifying conflicts across shared data or materials, and found similarly unacceptable results as to previously mentioned by the Vasilevsky et al. study. To remedy these issues, we suggest that journals publishing articles related to immune epitope data implement specific criteria to be required upon the submission of such articles.
Immune epitope data
As all data present in the IEDB represent the binding of an adaptive immune receptor, an antibody/B‐cell receptor (BCR) or T‐cell receptor and/or MHC, to an epitope, the specific identity of the epitope is crucial. To properly identify a peptidic epitope, its exact amino acid structure, as well as its position in the exact protein in question, must be known. This type of information is often reported incorrectly. For example, an author may refer to the hepatitis C virus NS3 epitope as ‘NS3 1037–1081’. This amount of information is too vague to identify the exact amino acid sequence. The residue positions of 1037–1081 can be interpreted as many different amino acids, depending upon which GenBank entry for hepatitis C virus (HCV) NS3 protein the author used. This is due to natural variability in the protein sequence as well as whether the N‐terminal methionine is included in the sequence entry. In other cases, the author may refer to an epitope by its name, such as ‘the well‐known 4P epitope’. Without a clear citation, this type of terminology provides no information regarding the epitope's sequence. In the IEDB curated data, approximately 85% of the time the epitope sequence was found within the publication and 15% of the time, the curator had to look into cited references and/or contact the author. These percentages reflect a fairly positive situation in regards to authors clearly stating the epitope sequence; however, because these manuscripts were primarily about epitope discovery – ideally all would provide a definitive amino acid sequence. If the epitope sequence could not be found, the manuscript will be deemed uncuratable. To date, 12% of papers found to be uncuratable were because of lack of epitope sequence.
Peptidic epitopes are the portion of a protein that the adaptive immune receptor recognizes. Thus, the identity of the protein source of the epitope is critical. Authors often refer to proteins using common names, abbreviations or ambiguous names that can be interpreted as multiple possible proteins. With highly divergent strains of some viruses, like influenza, the exact strain is critical. These specific isolates and strains are very important to immunologists, as this sequence variety represents a key reason why some vaccines succeed while others fail. If the author fails to specify the strain used, the reader cannot assume to know the sequence of the protein nor the epitope that the paper describes. If a manuscript only states which residue(s) of the protein contact the antibody, such as K180 of influenza A virus hemagglutinin, it is unclear, when viewing the > 80 000 hemagglutinin protein entries for influenza A virus, where exactly that amino acid is found in the protein. Many of these protein entries do not have a lysine (K) at residue 180. The curator will not be able to tell if this is because the specific isolate differs at that residue or if it is due to a shift in amino acid numbering, and in fact the lysine appears at residue 182 instead. Thus, in order to accurately map antibody epitopes back to the three‐dimensional structure of the protein, detailed information within the original publication is crucial.
In the IEDB data, approximately 50% of the time, the author provides an unambiguous identifier for the protein source of the epitope. In 30% of the data, the curator must select a representative protein source for the epitope using the name that the author provides and 20% of the time, the certainty of the epitope source remains unclear. Thus, in half of the epitopes described, their exact protein source is ambiguously reported.
When proteins are described ambiguously without stable identifiers, in addition to the exact amino acid sequence, the organism source also becomes unclear. For example, when describing autoimmune epitopes, curators cannot assume the host species and the epitope source are the same. An epitope may be referred to in a manuscript as the ‘GAD65 epitope…’ without the specification of the source organism, such as rat, rabbit, mouse, or human GAD65 protein. The authors may have performed experiments in mice and also use human cells, making it unclear if two different proteins (mouse and human GAD65) were used, or if all experiments were performed using rat GAD65. Although many auto‐antigens are highly homologous between species, there are differences in the exact sequence between species and these differences may have a significant impact on the immune response. Without the author clearly stating the species and strain, and/or using Genpept or UniProt identifiers for the protein and NCBI taxonomy identifiers for the organism, such published data is not reliable.
Another critical feature for the immune response is the host where the response occurred. In the majority of cases, the species is known, but the reference to the strain is frequently omitted. For research models, the exact strain is very relevant. For example, non‐obese diabetic mice will spontaneously develop diabetes while BALB/c mice will not. Each inbred mouse strain expresses a highly specialized set of genes and may have a significantly different immune response. The variations in the immune responses between wild‐type mouse strains and transgenic mice are well documented in the Mouse Genome Informatics MGI website (http://www.informatics.jax.org/external/festing/mouse/STRAINS.shtml). Publications describing an immune response in a model organism must specify the specific strain of the organism in order for the data to be reproducible.
Forty per cent of the experiments in the IEDB reflect an antibody response to an epitope. Oftentimes multiple publications will utilize the same antibody, allowing the body of knowledge pertaining to that antibody to grow. Unfortunately, different authors may describe the same antibody as having a different immunogen or being raised in a different host, leaving one unable to know for certain exactly how the antibody was raised or if the authors are presenting data on the same antibody. Without this knowledge, the data become less valuable. A human antibody raised to a virus may be protective and have great value in vaccine development, while a mouse antibody raised to a small peptide might not. It is critical to know how the antibody was generated to interpret the results and to reproduce and reuse them. Discrepancies in antibody terminology represent an error of ambiguity. The same antibody may be referred to in many different ways, for instance as ‘mAb4.1'or ‘4‐1’ or ‘ab4.1’ or ‘4 1’, again causing the reader to be uncertain if these are the same or different antibody.
T cells recognize epitopes in the context of MHC molecules. The specific molecule is of vital importance, as organisms not expressing that specific MHC molecule may not recognize the epitope. Authors often use abbreviated, outdated, or ambiguous names to describe these molecules. For example, an author may refer to a MHC molecule as ‘DR4’ throughout a publication, only for the curator to discover in a cited reference that the author meant ‘HLA‐DRB1*04:02’. The use of ‘DR4’ is ambiguous as it could have also meant ‘HLA‐DRB1*04:01’, a completely different MHC molecule. Journals should require authors to use complete formal nomenclature when describing any MHC molecule. The accepted formal nomenclature for human MHC molecules can be found at the International ImMunoGeneTics information system (www.ebi.ac.uk/ipd/imgt/hla/allele.html) and the nomenclature for other species can be found at the Immuno Polymorphism Database (www.ebi.ac.uk/ipd/mhc/).11 Otherwise, such information cannot be truly understood and used by other scientists.
Experiments defining immune epitopes often use cell lines, which are also often difficult to properly identify. Authors generally do not supply an unambiguous name or a stable identifier. Instead a shortened or common name for the cell line is used. Vasilevsky et al. found that cell lines were fully identifiable < 50% of the time. The specific antigen‐presenting cells processing and presenting epitopes to T cells must be fully identified in order to reproduce experimental results, as certain cell lines express specific MHC molecules, and deficiencies in processing machinery may critically alter the ability of the cell to process an antigen.
Discussion
Aggregating all published immune epitope data together allows the IEDB to draw new conclusions and present immunologists with a clearer view of the immune response. The Immunome Browser is a feature of the IEDB that plots data along the length of the epitopes’ protein sources, displaying how many subjects responded to each region, as well as how many did not respond, as shown in Fig. 1.
Figure 1.
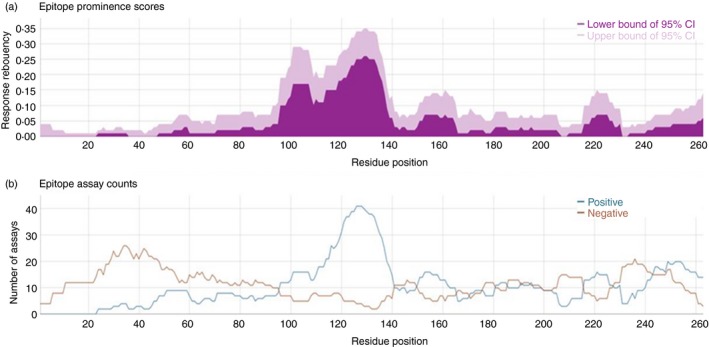
The human immune response to Timothy grass Phl p 1 protein. Four hundred assays were aggregated from 13 manuscripts. (a) Graphical representation of the human response frequency of each residue position of the Timothy grass Phl p 1 protein. (b) Positive and negative responses for each residue position of the Timothy grass Phl p 1 protein, depicted as the total number of assays performed for each residue.
The IEDB uses UniProt data to facilitate these mappings because UniProt provides complete reference proteomes for each species, supplying a stable identifier for each representative protein encoded by the species’ genome. This visual display allows immunologists to see the larger picture of the adaptive immune response. This summation of data makes new comparisons possible, such as differences in responses to the same protein in different species, differences in the antibody response compared to the T‐cell response, or determination if humans with a certain illness differ in their immune response to healthy individuals.
In addition to presenting the broad representation of the immune response, this aggregation of data also points out discrepancies between publications. Sometimes the differences in response are due to differences in experimental protocols, for example the route or dose of the immunization. Other times, the differences are due to errors on the part of the author or the interpretation of the curator. Subtle, but significant variables can be revealed to scientists wishing to build upon or reproduce other's work only if the responses are clearly known to be due to differences in the details of the experiment, rather than issues of clarity. For example, Fig. 2 demonstrates how important it is for authors to specify the strain of mouse used in their experiments. When authors state that ‘mice were immunized’ without providing which strain of mouse, interpretation of the immune response demonstrated in their work becomes impossible.
Figure 2.
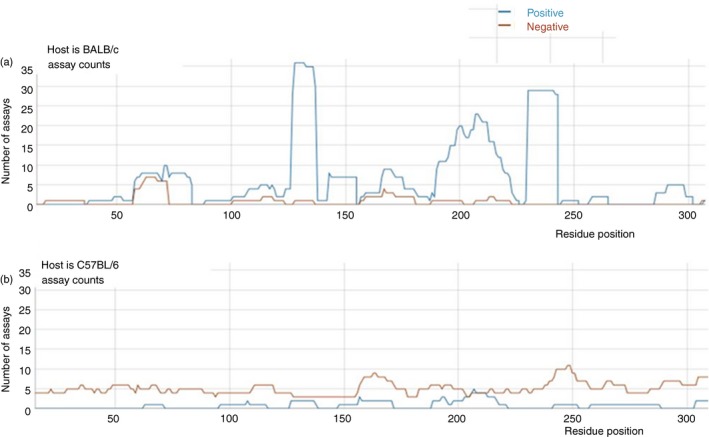
The immune response to the same region of influenza A virus haemagglutinin (HA) protein in two different strains of mice is significantly different. (a) Positive and negative immune response of the HA protein in BALB/c mouse strains. (b) Positive and negative immune response of the HA protein in C57BL/6 mouse strains.
Proposed guidelines
If authors would clearly identify all materials used in their experiments, a much broader and more meaningful picture of the immune response could be presented. By providing the exact amino acid sequence of all peptides studied, identifying each protein by a stable GenPept or Uniprot identifier, and supplying the species and strain of the protein, immune epitope data becomes much more reliable and reproducible. There exist many resources to assist authors in the clear identification of resources used in the course of experiments. The Resource Identification Portal (www.scicrunch.org/resources) is a searchable database that hosts stable identifiers for reagents, model organisms, and tools (software, databases, and services). The Antibody Registry (www.antibodyregsitry.org) assigns each antibody a persistent identifier, which will permanently link it back to any catalogue numbers associated with that reagent, its host and derivation. Table 1 presents the details required and which online resources should be used to fully describe the different aspects of immune epitope data. In order to best communicate one's scientific work and to facilitate the enhanced value through aggregation of related data sets, as described above, we suggest that authors and journals adopt these specific guidelines for the publishing of immune epitope data.
Table 1.
All published immune epitope data should include fully identifiable resources
| Resource type | Feature | Description | Type | Example | Resource |
|---|---|---|---|---|---|
| Peptide | Sequence | Linear string of amino acid abbreviations | String | SIINFEKL | www.ncbi.nlm.nih.gov/Class/MLACourse/Modules/MolBioReview/iupac_aa_abbreviations.html |
| Peptide | Start position | Position of first amino acid within the protein | Number | 257 | blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome |
| Peptide | Stop position | Position of last amino acid within the protein | Number | 264 | blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome |
| Protein | Name | Complete protein name | String | Ovalbumin | www.ncbi.nlm.nih.gov/protein, www.uniprot.org |
| Protein | Identifier | Genpept or UniProt identifier | String | 212505 | www.ncbi.nlm.nih.gov/protein, www.uniprot.org |
| Organism | Name | Taxonomic species name | String | Gallus gallus (chicken) | www.ncbi.nlm.nih.gov/taxonomy |
| Organism | Identifier | NCBI taxonomy identifier | Number | 9031 | www.ncbi.nlm.nih.gov/taxonomy |
| Antibody | Name | Complete antibody name | String | Mouse anti‐human GAD65/GAD67 | antibodyregistry.org |
| Antibody | Identifier | Antibody Registry identifier or catalogue number | String | AB_311783 | antibodyregistry.org |
| MHC molecule: human | Name | Formal accepted nomenclature | String | HLA‐A*02:01 | www.ebi.ac.uk/ipd/imgt/hla/allele.html |
| MHC molecule: non‐human | Name | Formal accepted nomenclature | String | BoLA‐DQA*0101 | www.ebi.ac.uk/ipd/mhc |
| Cell line | Name | Complete cell line name | String | JY3 [LASJY3] | Vendor's website |
| Cell line | Identifier | Catalogue number | String | ATCC: 77442 | Vendor's website |
Disclosures
We declare that we have no significant competing financial, professional or personal interests that might have influenced the performance or presentation of the work described in this manuscript.
References
- 1. Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR et al The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2014; 43(Database issue):D405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N et al Pre‐existing immunity against swine‐origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci USA 2009; 106:20365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim Y, Vaughan K, Greenbaum J, Peters B, Law M, Sette A. A meta‐analysis of the existing knowledge of immunoreactivity against hepatitis C virus (HCV). PLoS One 2012; 7:e38028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaughan K, Greenbaum J, Blythe M, Peters B, Sette A. Meta‐analysis of all immune epitopedata in the Flavivirus genus: inventory of current immune epitope data status in the context of virus immunity and immunopathology. Viral Immunol 2010; 23:259–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vita R, Peters B, Sette A. The curation guidelines of the immune epitope database and analysis resource. Cytometry A 2008; 73:1066–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vasilevsky NA, Brush MH, Paddock H, Ponting L, Tripathy SJ, Larocca GM et al On the reproducibility of science: unique identification of research resources in the biomedical literature. PeerJ 2013; 1:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M et al Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol 2008; 26:889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.URL http://www.nature.com/authors/policies/checklist.pdf [accessed on November 23, 2015].
- 9.URL https://chronicle.com/article/NIH-Presses-Journals-to-Focus/146951 [accessed on November 23, 2015].
- 10. Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S et al The resource identification initiative: a cultural shift in publishing. F1000Res 2015; 4:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 2015; 43:D423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]


