Abstract
In all living organisms, the phosphorylation of proteins modulates various aspects of their functionalities. In eukaryotes, protein phosphorylation plays a key role in cell signaling, gene expression, and differentiation. Protein phosphorylation is also involved in the global control of DNA replication during the cell cycle, as well as in the mechanisms that cope with stress-induced replication blocks. Similar to eukaryotes, bacteria use Hanks-type kinases and phosphatases for signal transduction, and protein phosphorylation is involved in numerous cellular processes. However, it remains unclear whether protein phosphorylation in bacteria can also regulate the activity of proteins involved in DNA-mediated processes such as DNA replication or repair. Accumulating evidence supported by functional and biochemical studies suggests that phospho-regulatory mechanisms also take place during the bacterial cell cycle. Recent phosphoproteomics and interactomics studies identified numerous phosphoproteins involved in various aspect of DNA metabolism strongly supporting the existence of such level of regulation in bacteria. Similar to eukaryotes, bacterial scaffolding-like proteins emerged as platforms for kinase activation and signaling. This review reports the current knowledge on the phosphorylation of proteins involved in the maintenance of genome integrity and the regulation of cell cycle in bacteria that reveals surprising similarities to eukaryotes.
Keywords: protein phosphorylation, signaling, DNA replication, bacterial cell cycle, bacteria cell division
Introduction
In all living cells, many cellular processes are controlled through the reversible phosphorylation of proteins on serine, threonine and tyrosine (Ser/Thr/Tyr) which results from the opposing action of kinases and phosphatases. While the role of Ser/Thr and Tyr -protein kinases (STYKs) and phosphatases in cellular regulations is extensively documented in eukaryotes, the Ser/Thr/Tyr signaling network in bacteria has a more recent history (Hunter, 2000; Pereira et al., 2011; Dworkin, 2015; Kalantari et al., 2015; Manuse et al., 2015). Analysis of microbial genomes revealed that Hanks-type Ser/Thr kinases (eSTKs) and phosphatases (eSTPs) are also widespread in bacteria (Zhang et al., 2007; Zhu et al., 2014). However, prokaryotic Tyr phosphorylation is carried out by a different family of enzymes, the bacterial tyrosine kinases or BY-kinases, which do not share the structural homology with eukaryotic STYKs. Instead, BY-kinases possess structural features characteristic of P-loop ATPases. BY-kinases as well as eSTKs have been shown to regulate cell-cycle events, cell adaptation to environmental cues, and virulence in many species (Wright and Ulijasz, 2014; Zhu et al., 2014; Kalantari et al., 2015).
Phosphorylation provides a sensitive and dynamic way to regulate protein activity, stability, protein interaction and sub-cellular localization. Global regulators are often targeted by more than one kinase, altering the expression of many genes. The phosphorylation of the transition state transcription regulator AbrB of Bacillus subtilis by three Ser/Thr kinases, PrkC, PrkD, and YabT, leads to the deregulation of numerous target genes during the transition of vegetative to stationary growth (Kobir et al., 2014). In the human pathogen bacteria Staphylococcus aureus, the phosphorylation of the global regulator MgrA also modulates gene expression in a growth phase-dependent manner (Truong-Bolduc et al., 2008). By adding a local negative charge, phosphorylation can positively or negatively affect the DNA binding of transcriptional factors to their regulatory sequences, thus participating in eliciting the cellular response. A recent example of tyrosine phosphorylation-mediated positive regulation is illustrated by the regulator SalA, which undergoes a conformational change upon phosphorylation at tyrosine 327 that enhances its DNA binding affinity for scoC upstream regulatory sequences (Derouiche et al., 2015a). Conversely, phosphorylation of the virulence regulator SarA of S. aureus at serine 75 within the HTH-DNA binding region negatively modulates its ability to bind DNA leading to the negative regulation of gene expression (Didier et al., 2010).
It was also observed that modulation by phosphorylation of the DNA binding of transcriptional factors plays a role in the regulation of chromosomal replication. The two component regulators MtrA of Mycobacterium tuberculosis and CtrA of Caulobacter Crescentus are illustrative examples that phosphorylation acts to temporally restrain access of the replication initiator protein DnaA to the origin DNA sequence oriC (Didier et al., 2010; Pini et al., 2015). A similar strategy was proposed to account for the role of the master regulator Spo0A in negatively controlling DNA replication initiation by inhibiting the DnaA-dependent DNA duplex unwinding during sporulation in Bacillus subtilis (Castilla-Llorente et al., 2006; Xenopoulos and Piggot, 2011; Boonstra et al., 2013). Hence, taking advantage of the versatility of transcription factors combined with the limited time frame during which they contact their regulatory sequence, bacterial cells have evolved multifunctional regulatory proteins able to act in both gene expression and replication initiation control.
However, little is known about the role of phosphorylation in directly regulating the DNA-binding activity of proteins involved in other DNA-mediated processes. Large-scale phosphoproteome studies have been conducted in bacteria, covering gram-negative as well as gram positive species (Macek et al., 2007, 2008; Voisin et al., 2007; Soufi et al., 2008; Ravichandran et al., 2009; Prisic et al., 2010; Schmidl et al., 2010; Ge et al., 2011; Manteca et al., 2011; Misra et al., 2011; Bai and Ji, 2012; Elsholz et al., 2012; Esser et al., 2012; Hansen et al., 2013; Soares et al., 2013; Yang et al., 2013; Kennelly, 2014; Ortega et al., 2014; Ravikumar et al., 2014; Nakedi et al., 2015; Pan et al., 2015). These databases reveal an increasing number of bacterial proteins phosphorylated on Ser/Thr/Tyr, involved in a variety of cellular processes related to DNA metabolism (Pan et al., 2015). Recently, a protein-protein interaction (PPI) network centered on B. subtilis S/T/Y protein kinases and phosphatases emerged as a powerful tool in bacterial signal transduction research, highlighting the existence of many regulatory pathways controlled by phosphorylation during DNA replication, chromosomal segregation and cytokinesis (Shi et al., 2014b). As with transcription factors, we can anticipate that phosphorylation could modulate protein-binding properties to DNA as well as to other proteins. Therefore, a great challenge is now to understand the role of protein phosphorylation in the coordination and integration of the different DNA processes during the bacterial cell cycle. In this review we will focus on novel aspects of regulation in bacteria that resemble those taking place in eukaryotes. We will highlight the growing evidence for phosphorylation of proteins involved in many DNA-related processes and the existence of scaffold proteins that act as signaling integrators by facilitating interaction and co-localization of kinases and their targets.
Identification of phospho-proteins involved in DNA-related processes in bacteria
In living cells, the genome is continuously exposed to endogenous and exogenous damaging agents. If not repaired, the replication of damaged chromosomes can cause fork stalling or collapse and subsequent genome instability and possibly cell death (De Septenville et al., 2012; Merrikh et al., 2012; Cortez, 2015; Dungrawala et al., 2015). Sensing and repairing DNA damage is therefore necessary to ensure chromosome integrity. Upon detection of damage, eukaryotic cells relay information through signal transduction cascades to coordinate a biological response including cell cycle arrests and DNA repair pathways (Iyer and Rhind, 2013). These signaling cascades generally involve the successive activation of protein kinases, punctuated by the phosphorylation of protein effectors of DNA damage checkpoint and processing (Subramanian and Hochwagen, 2014). These checkpoints involve sensor proteins that identify DNA-related disorders, converting them into phosphorylation events that modulate the functions of specific target proteins. In eukaryotes, the evolutionary conserved ATM (ataxia-telangiectasia mutated) and ATR (ATM and Rad 3-related) Ser/Thr kinases are considered a paradigm for such transduction cascades (Cimprich and Cortez, 2008). These two master regulators act in response to double strand breaks (ATM), as well as in replication initiation control and repair of damaged forks (ATR) to coordinate cell cycle, repair, and replication (Shechter et al., 2004a,b; Cimprich and Cortez, 2008; Lee et al., 2015).
In most bacteria, the replication of the circular chromosome requires the coordinated action of conserved proteins to perform the initiation, elongation and termination steps (Langston and O'Donnell, 2006; Langston et al., 2009). Initiation is mediated by the replication initiator protein DnaA, which binds to specific origin sequences. This forms a nucleoprotein-structure leading to the unwinding of the duplex DNA for subsequent loading of the replication machinery or replisome. A bacterial replisome is usually composed of a type III DNA polymerase, a DNA helicase, and a sliding clamp with an associated clamp loading protein complex. During elongation, the replication factory encounters numerous road blocks or damage that can cause fork arrests (Pomerantz and O'Donnell, 2010; Mettrick and Grainge, 2015). As in eukaryotes, mechanisms are required to repair and/or restart the ongoing replication fork to preserve genomic integrity (Merrikh et al., 2012). In all living organisms, chromosome replication must be coordinated with other cellular pathways (Sclafani and Holzen, 2007).
In bacteria, however, whether phosphorylation events play a role in sensing replication stresses remains to be ascertained. Recent studies highlight kinase activation by cross-phosphorylation in B. subtilis and M. tuberculosis, suggesting the existence of bacterial signaling cascades comparable to those found in eukaryotes (Baer et al., 2014; Shi et al., 2014a). Although the complement of proteins targeted by these potential regulatory cascades are not yet known, increasing numbers of large scale mass spectrometry-based proteomics studies find that proteins involved in various DNA-related processes are also phosphorylated in bacteria (Macek et al., 2007, 2008; Voisin et al., 2007; Soufi et al., 2008; Ravichandran et al., 2009; Prisic et al., 2010; Schmidl et al., 2010; Ge et al., 2011; Manteca et al., 2011; Misra et al., 2011; Bai and Ji, 2012; Elsholz et al., 2012; Esser et al., 2012; Hansen et al., 2013; Soares et al., 2013; Yang et al., 2013; Kennelly, 2014; Ortega et al., 2014; Ravikumar et al., 2014; Nakedi et al., 2015; Pan et al., 2015; Table 1). Another approach, based on yeast two-hybrid detection of protein binding partners of B. subtilis protein kinase and phosphatase, identifies numerous potential DNA-binding protein substrates, some of them already characterized as phosphoproteins in other studies, or further validated in vivo and/or in vitro (Shi et al., 2014b; Table 2, Figure 1). This study lays the foundation for investigating the role of phosphorylation in modulating DNA-related processes in this bacteria.
Table 1.
Phosphorylation of proteins involved in DNA-dependent machineries and processes in bacteria.
| Pathway | Target | KO | Function | Tyr-P | Ser/Thr-P |
|---|---|---|---|---|---|
| DNA repair and recombination | MUTL | K03572 | DNA Mismatch repair factor | MUTL_BACSU; B1XQB2_SYNP2 | B1XQB2_SYNP2 |
| MUTS | K03555 | DNA Mismatch repair protein | B1XM45_SYNP2 | ||
| MUTM | K10563 | FPG/DNA-N glycosylase | B1XQB2_SYNP2 | B1XQB2_SYNP2 | |
| UVRB | K03702 | UvrABC system protein B | UVRB_STRP2 | ||
| UVRD | K03657 | ATP-dep. DNA helicase (PcrA) | PCRA_BACSU; O26013_HELPY | O26013_HELPY; Q3K0V8_STRA1 | |
| RUVX | K07447 | Holliday junction resolvase | RUVX_ECO57 | ||
| RECA | K03553 | SOS repair/DNA processing | RECA_BACSU; RECA_ECO57 | RECA_BACSU; RECA_THET8 | |
| RECG | K03655 | ATP-dependent DNA helicase | RECG_MYCTO | ||
| MFD | K03723 | Transcription-repair factor | B1XL29_SYNP2 | Q04N58_STRP2; B1XL29_SYNP2 | |
| RECQ | K03654 | ATP-dependent DNA helicase | B1XQJ2_SYNP2 | ||
| REP | K03656 | ATP dependent DNA helicase | Q3K4P8_PSEPF | ||
| DINI | K12149 | DNA-damage-inducible | DINI_ECO57 | ||
| RECT | K07455 | DNA-Binding protein | RECT_ECOLI | ||
| EX1 | K01141 | Exodeoxyribonuclease I (sbsB) | Q8X8T9_ECO57; Q9HW85_PSEAE | Q9HW85_PSEAE | |
| EX3 | K01142 | Exodeoxyribonuclease III | EX3_ECOLI | ||
| EX7L | K03601 | Exodeoxyribonuclease VII (A) | EX7L_THETN | ||
| EX7S | K03602 | Exodeoxyribonuclease VII (B) | EX7S_STRA1 | ||
| RADA | K04485 | DNA repair protein (radA, sms) | Q8XB28_ECO57, B1XJC1_SYNP2 | B1XJC1_SYNP2 | |
| SSBB | Single-stand binding protein B | SSBB_BACSU | |||
| DNA replication | SSBA | K03111 | Single-stand binding protein A | SSBA_BACSU; | Q04JK9_STRP2; SSBA_BACSU |
| DPO1 | K02335 | DNA polymerase I | DPO1_BACSU; B1XL22_SYNP2 | B1XL22_SYNP2; DPO1_ECOLI | |
| LIGA | K01972 | DNA ligase A | DNLJ_LISMO; DNLJ_ECOLI | ||
| DNAA | K02313 | DNA replication initiator | DNAA_STRCO | ||
| DPO3 | K02337 | Replicative DNA polymerase PolC | HOLE_ECO57 | ||
| DNAB | K02314 | Replicative DNA helicase | B1XQ85_SYNP2 | B1XQ85_SYNP2; DNAC_BACSU | |
| DPO3A | K02337 | DNA polymerase III (θ su) | HOLE_ECO57 | ||
| TUS | K10748 | DNA replication terminus protein | TUS_ECO57 | ||
| Nucleoid structure and DNA condensation | GYRB | K02470 | DNA gyrase β subunit | GYRB_ECO57 | GYRB_BACSU |
| TOP1 | K03168 | DNA topoisomerase 1 | TOP1_MYCTO; Q8X7C5_ECO57 | TOP1_MYCPN | |
| PARC | K02621 | DNA topoisomerase 4 suA | PARC_PSEAE | ||
| NAP | K09747 | Nucleoid associated protein | B1XLF8_SYNP2; | ||
| DBH1 | K03530 | DNA-binding protein HupA | DBH1_BACSU; Q92A74_LISMO; DBH1_STRCO; B1XQQ7_SYNP2 | ||
| DPS | K04047 | DNA protection during starvation | DPS_ECOLI; DPS_ECO57 | DPS_ECOLI; DPS_CAMJE; DPS_LISMO; Q3K7V3_PSEPF | |
| DBHB | K03530 | DNA-binding protein HupB | DBHB_ECOLI; Q6N5M1_RHOPA | ||
| HNS | K03746 | DNA-binding protein H-NS | HNS_ECOLI; HNS_ECO57 | HNS_ECOLI | |
| STPA | K11685 | DNA-binding protein StpA | CNU_ECO57, STPA_ECOLI | ||
| SMC | K03529 | Chromosome partition protein | Q84EC7_SYNP2; SMC_MYCTO | ||
| Chrom. segregation | PARA | K03496 | ATPase, ParA-family | B1XQR5_SYNP2 | |
| FTSK | K03466 | DNA translocase FtsK | FTSK_ECO57; FTSK_ECOLI | FTSK_MYCTO | |
| Restrictionmodification | B1XQZ6 | K01156 | Type III R/M-Helicase | B1XQZ6_SYNP2 | |
| ECO57IR | K00571 | Type IIS R/M-methyltransferase | O26046_HELPY | Q04K98_STRP2 | |
| HSDM | K03427 | Type I restriction enzyme M | O25953_HELPY | O33298_MYCTU | |
| DNA transcription | RPOB | K03043 | RNA polymerase β subunit | RPOB_BACSU; RPOB_ECO57 | RPOB_ECOLI; RPOB_MYCTORPOB_CLOAB; RPOB_STRP2; RPOB_BACSU |
| RPOC | K03046 | RNA polymerase β' subunit | RPOC_BACSU; RPOC_ECO57 | RPOC2_SYNP2; RPOC_CLOAB; RPOC_BACSU; C4WZY6_KLEPN RPOC_CLOAB; RPOC_STRP2 | |
| RPOBC | K13797 | RPOBC_HELPY; | RPOBC_HELPY | ||
| RPOA | K03040 | RNA polymerase α subunit | RPOA_ECO57; RPOA_ECOLI; C4X0R2_KLEPN | RPOA_STRP2; RPOA_CLOAB | |
| RPOE | K03088 | RNA polymerase σ factor H | RPOE_MYCPN; SIGH_MYCBO; SIGH_MYCTO | ||
| Other | K03722 | DnaQ exonuclease/DinG helicase | Q04L99_STRP2 | ||
| Putative helicase | B1XR86_SYNP2 | ||||
| K03578 | ATP dependent helicase HrpA | Q3KGE3_PSEPF | |||
| K04763 | Putative DNA integrase/recombinase | B1XQW1_SYNP2 |
Escherichia coli K12 and O157:H7 species (ECOLI and ECO57:H7) (Macek et al., 2008; Hansen et al., 2013; Soares et al., 2013), Helicobacter pylori (HELPY) (Ge et al., 2011), Campylocacter jejuni (CAMJE) (Voisin et al., 2007), Bacillus subtilis (BACSU) (Macek et al., 2007; Jers et al., 2010; Elsholz et al., 2012; Shi et al., 2014b), Clostridium acetobutilicum (CLOAB) (Bai and Ji, 2012), Listeria Monocytogenes (LISMO) (Misra et al., 2011), Pseudomonas fluorescens Pf0-1 (PSEPF) (Ravichandran et al., 2009), Streptococcus agalactiae serotype 1a (STRA1) (Burnside et al., 2011), Streptococcus pneumoniae serotype 2 (STREP2) (Sun et al., 2010), Streptococcus coelicolor (STREPCO) (Manteca et al., 2011), Thermus thermophilus HB8 (THET8) (Takahata et al., 2012), Thermoanaerobacter tengcongensis (THETN) (Lin et al., 2012), Mycobacterium tuberculosis (MYCTO and MYCTU) (Prisic et al., 2010), Mycobacterioum bovis (MYCBO) (Nakedi et al., 2015), Mycoplasma pneumoniae (MYCPN) (Schmidl et al., 2010), Synechococcus sp. (SYNP2) (Yang et al., 2013). http://dbpsp.biocuckoo.org/browse.php (Pan et al., 2015). Protein target names are given according to HAMAP standard (http://hamap.expasy.org) and UniProtKB (http://www.uniprot.org/uniprot). Phosphoproteins provided with links to the dbPSP database (http://dbpsp.biocuckoo.org/) are labeled in blue. Phosphoproteins identified by other approaches are indicated in black. KO identifiers are provided for each family of proteins with links to the Kegg database for functional information (Kanehisa et al., 2012).
Table 2.
DNA-binding proteins targeted for phosphorylation in B. subtilis.
| Acc | Name | Function | PK | PP | P-sites | Val | Orth | References |
|---|---|---|---|---|---|---|---|---|
| TYROSINE PHOSPHORYLATION | ||||||||
| P23477 | ADDB | ATP-dependent deoxyribonuclease (B) | PtkB | – | 1 | |||
| P49850 | MUTL | DNA mismatch repair factor | PtkB | PtpZ | a,d | Y | 1 | |
| O34996 | DPO1 | DNA polymerase I | PtkB | PtpZ | a,d | T | 1 | |
| O34580 | UVRD | ATP-dependent DNA helicase (PcrA) | PtkB | d | Y,S,T | 1 | ||
| P16971 | RECA | SOS repair factor/DNA processing | PtkA | b#,d | Y | 1,2,3 | ||
| P37870 | RPOB | RNA polymerase β subunit | PtkB | PtpZ | Y695 | a,c,d | Y,S,T | 1,4 |
| P37871 | RPOC | RNA polymerase β' subunit | Y338 | D | 4 | |||
| P37455 | SSBA | Single-Stand Binding protein A | PtkA | Y82 | b*#,c,d | S | 5,6 | |
| O31903 | YORK | Putative SPBc2 ss-DNA exonuclease | PtkA | Y3∕11∕168∕220∕368∕473 | d | 6,7 | ||
| SERINE/THREONINE PHOSPHORYLATION | ||||||||
| P17867 | CISA | Site-specific DNA recombinase | YabT | SpoIIE | a | 1 | ||
| P16971 | RECA | SOS repair factor/DNA processing | YabT | SpoIIE | S2 | a,b,c,d | Y | 1,2,3 |
| P45870 | RACA | Chromosome-anchoring protein | YabT | SpoIIE | a, b# | 1 | ||
| O08455 | SBCE | DSB repair | YabT | SpoIIE | a | 1 | ||
| P46344 | YQFF | Putative Phosphodiesterase | YabT | SpoIIE | a | 1 | ||
| P37469 | DNAC | Replicative DNA helicase DnaB-like | PrkD | b#,d | Y | 1 | ||
| P08821 | DBH1 | DNA-binding protein HU-1 | T4, T65, S74 | c,d | S,T | 6 | ||
| P05652 | GYRB | DNA gyrase subunit B | S400 | c,d | Y | 2 | ||
| P94590 | SSBB | Single-stand DNA binding protein B | PtkA | Y82,T52 | c,d | S | 4 | |
| P37871 | RPOB | RNA polymerase β subunit | S314 | c,d | S,T,Y | 4 | ||
| P37871 | RPOC | RNA polymerase β' subunit | S339 | c | 4 | |||
Validation (Val) is provided by (a) ability to interact with both a protein kinase (PK) and a cognate protein phosphatase (PP), (b) phosphorylation confirmed in vivo (*) and/or in vitro (#), (c) identified phosphosite(s), and (d) existence of phosphorylation at tyrosine (Y) serine (S) or threonine (T) in other bacterial orthologs (orth). 1 (Shi et al., 2014b); 2 (Soufi et al., 2008); 3 (Bidnenko et al., 2013); 4 (Elsholz et al., 2012); 5 (Mijakovic et al., 2006); 6 (Macek et al., 2007); 7 (Jers et al., 2010).
Figure 1.
PPI network centered on ser/thr (yellow nodes, bleu edges) and tyr (red nodes, black edges) kinases, phosphatases and modulators of B. subtilis reveals potential regulations by phosphorylation of various DNA-related pathways. Proteins are represented as nodes connected by edges. Potential substrates, defined as connected by both a kinase and cognate phosphatase, are represented by diamonds. HTH-containing proteins, connected by more than one kinase, are represented by squares; In vitro characterized phospho-proteins are labeled in red. Proteins found phosphorylated in other bacteria are indicated by an asterix. Other interactions between the proteins are illustrated by light gray edges (from Marchadier et al., 2011; Shi et al., 2014b). In vitro validated kinase-substrate phosphorylation is indicated by dashed lines.
An example of phosphorylated multifunctional protein playing a role in various DNA processes is the single-stranded DNA-binding protein SSB (Mijakovic et al., 2006; Vujaklija and Macek, 2012). In bacteria, SSB proteins assemble onto DNA as homodimers or tetramers, each unit composed of an N-terminal DNA binding domain and a C-terminal intrinsically disordered tail that mediates interaction with numerous proteins (Kozlov et al., 2015). The bacterial SSB proteins are mainly associated with the replication machinery at the replicating forks and serve as central hubs to coordinate DNA replication and repair (Lecointe et al., 2007; Costes et al., 2010; Antony et al., 2013; Bentchikou et al., 2015). As in bacteria, the eukaryotic single strand binding protein RPA has multiple roles in protecting ssDNA, sensing and promoting repair of DNA damage via PPIs (Oakley and Patrick, 2010; Maréchal and Zou, 2015). In particular, phosphorylation of RPA negatively regulates its binding to DNA as well as its interaction with protein partners (Binz et al., 2003; Oakley et al., 2003). B. subtilis possesses two single strand binding proteins SSBA and B, respectively phosphorylated at a tyrosine, identified as Tyr82 in SSBA (Mijakovic et al., 2006). Additionally, a phospho-site at Thr52 was also identified in SSBB (Table 1; Jers et al., 2010; Elsholz et al., 2012). Examination of the 3D- structure of the SSB/DNA complex shows that Tyr82 (as well as the Thr52 neighbor residue Trp54) are involved in stacking with DNA (Yadav et al., 2012). This strongly supports the observation that phosphorylation of SSBA at residue Tyr82 negatively modulates its binding onto DNA in vitro and regulates the cell response to DNA damage (Mijakovic et al., 2006). Therefore, the role of SSB protein phosphorylation in modulating the cellular DNA damage response might be evolutionarily conserved in eukaryotes and in prokaryotes.
Another strong case was made for the interaction of the DNA replicative helicase DnaC by the Ser/Thr kinase PrkD, validated by in vitro assays and high correlation of co-expressions (Shi et al., 2014b). Phosphorylation of numerous components of the DNA replication machinery, as well as of proteins involved in the different steps of the replication cycle, have been identified in one or more bacteria among Escherichia, Pseudomonas, Helicobacter, Camplylobacter, Listeria, Bacillus, Mycoplasma, Clostridium, Deinoccocus, Synechoccus, and Streptococcus genus (Tables 1, 2). Orthologs of the highly conserved DNA polymerase PolA, DNA helicase DnaC and single stranded binding protein SSB were found phosphorylated, potentially reflecting the existence of generic regulatory mechanisms to control their activity across species.
The conservation of phosphorylation within protein domain families is indicative of proteins with high regulatory potential (Maathuis, 2008). Remarkably, several phosphoproteomic analyses have reported phosphorylation of the two largest subunits β and β' of the RNA polymerase machinery at serine, threonine or tyrosine, in seven bacterial species (Table 1). In eukaryotes, the recruitment of factors to the elongating RNAPII is regulated by the differential and dynamic phosphorylation of serine, threonine and tyrosine residues within the C-terminal domain of the Rpb1 large subunit during the transcription cycle (Heidemann et al., 2013). Although the existence of phosphorylation of the main components of the RNA polymerase in both bacteria and eukaryotic cells does not imply similar regulatory mechanisms, it does suggest that evolutionary constraints on regulating gene expression led to targeting the transcription machinery for modulating its activity and its ability to interact with other factors.
Biological role of phospho-proteins in coordinating DNA- metabolism with transition state and cellular development in B. subtilis
A large panel of proteins involved in various DNA replication and repair pathways are identified with phospho-sites, suggesting that phosphorylation could play a role in coordinating the different cellular responses to DNA damage and integrity of the replicating chromosome (Table 1). Regulation by phosphorylation of DNA-related processes has been documented in B. subtilis. One process involves the DNA recombinase RecA, a multifunctional protein playing important roles in homologous recombination and repair of DNA double-strand breaks, as well as initiating the cellular SOS response to DNA damage in bacteria (Kuzminov, 2001; Chen et al., 2008; Butala et al., 2009). Recently, RecA was described as playing a role in monitoring and maintaining chromosome integrity at the onset of spore development (Bidnenko et al., 2013). This novel activity is regulated by phosphorylation by the ssDNA-dependent Ser/Thr kinase YabT, expressed during sporulation. Although the molecular mechanism underlying this activity is not yet understood, RecA phosphorylation at the N-terminal S2 plays a role in temporarily restraining sporulation furtherance upon encountering DNA damages. From the recent finding that RecA is also subjected to tyrosine phosphorylation by PtkA, we can anticipate a more complex picture for the regulation of its activities in B. subtilis (Shi et al., 2014b).
The DNA-binding developmental protein RacA illustrates a prospective role of phosphorylation in coordinating chromosome segregation with the asymmetric division taking place during sporulation in B. subtilis. During sporulation, RacA anchors the segregating chromosome at the cell pole of the forespore in a DivIVA-dependent manner (Ben-Yehuda et al., 2003; Wu and Errington, 2003; Ben-Yehuda et al., 2005; Perry and Edwards, 2006). In the B. subtilis PPI network, RacA is connected by both the Ser/Thr kinase YabT and cognate phosphatase SpoIIE, suggesting it may be a substrate of both (Shi et al., 2014b). This assumption, further confirmed in vitro, was also supported by the existence of a high positive correlation of expression of racA and spoIIE genes across numerous physiological conditions, indicating they are part of the same biological processes.
Nucleoid associated proteins (NAPs) are histone-like proteins involved in modifying the structural organization of chromosomes by changing its topology at a global or local level (Dillon and Dorman, 2010). As a consequence of their DNA-binding properties, NAP proteins are found to modulate different DNA-processes and act as global regulators of gene expression (Dorman, 2013). NAP proteins are diverse and present in all living cells (Dillon and Dorman, 2010; Dorman, 2013). The examination of phosphoproteomics data in the database of Phosphorylation Sites in Prokaryotes dbPSP reveals that many representatives of this class of proteins are phosphorylated, including HupA, HupB, H-NS (and H-NS like StpA), Dps and structural maintenance chromosome (SMC) proteins (Table 1). A particular feature of this class of proteins is the occurrence of phospho-sites across a broad spectrum of bacterial orthologs (Table 1), suggesting that they could be functionally regulated by phosphorylation. The recent discovery that phosphorylation of the histone-like protein HupB by STKPs kinases negatively modulates its ability to bind DNA in M. tuberculosis supports this assumption (Gupta et al., 2014). The phosphorylation state of the HupB protein correlates with the expression of pknE and pknF kinases. Their expression is high during the exponential growth phase and low when reaching the stationary phase. This differential phosphorylation strongly points to a potential mode of regulation of HupB which favors interaction with DNA during the stationary phase to promote compaction of the chromosome.
MinD, a signaling scaffold-like protein
Kinases are regulated by forming specific associations with arrays of different proteins, including kinase inhibitors and activators, as well as scaffold and anchoring proteins that target the kinase-complexes to specific subcellular sites. Scaffold proteins are versatile hubs that spatially and temporally tether signaling components in eukaryotic cells. Interactome-based research in yeast provides a definition of classical signaling scaffold proteins as (i) having an active role in signaling while being itself devoid of catalytic activity relevant for signaling, and (ii) being able to promote the interaction of multiple components of signaling cascades at a particular cellular location to regulate their activity (Zeke et al., 2009). A eukaryotic scaffold protein paradigm is the Ste5 protein of the mitogen-activated protein kinase (MAPK) pathway in yeast, which insulates three protein kinases MAPKKK Ste11, MAPKK Ste7, and MAPK Fus3 to promote a signaling cascade (Good et al., 2009).
Recent report of a yeast two-hybrid based PPI network centered on tyrosine-kinase in B. subtilis provide a 2D map of potential BY-kinase interacting partners (Shi et al., 2014b). Numerous protein partners are contacted by cognate kinase/phosphatase pairs, supporting the hypothesis that they are substrates. However, there are exception to this rule. The bacterial cell division regulator MinD is contacted by both the tyrosine kinase PtkA and tyrosine phosphatase PtpZ in the PPI network, but not targeted for phosphorylation by PtkA (Shi et al., 2014b). Strikingly, PtkA has been found to phosphorylate the septum site selection protein DivIVA, which recruits MinD at the cell poles via MinJ. Subcellular localization studies reveal that PtkA localizes at the cell pole in a MinD-dependent fashion. Septum defect phenotypes are observed in absence of PtkA and aggravated in a zapA deficient background, strongly suggesting that PtkA could play a role during cell division. All these observations support the hypothesis that B. subtilis MinD, in addition to its characterized role in recruiting the FtsZ-negative regulator MinC to prevent the formation of a Z-ring at the cell poles (Gregory et al., 2008), also acts as an intracellular organizer of signaling components involved in the regulation of cytokinesis (Shi et al., 2014b).
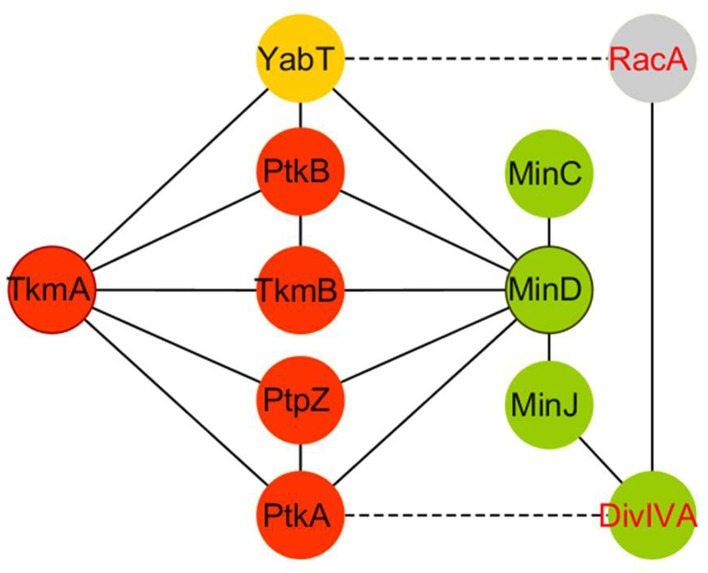
MinD belongs to the class of ParA-like AAA+ ATPases and bears structural homology to PtkA (Derouiche et al., 2015b). However, it lacks the tyrosine cluster determinant for autophosphorylation and is thus not a catalytically active kinase. Importantly, similar to TkmA, the transmembrane and cognate modulator of PtkA, necessary for the general activation of the kinase in the cell, MinD binds to and activates PtkA. In the PPI network, MinD is also found to interact with the second B. subtilis BY-kinase PktB, the Ser/Thr Hanks kinase YabT and a phosphatase (PtpZ), suggesting a potential ability to bring various components of tyrosine, but also serine/threonine phosphorylation pathways in close proximity (Figure 2). In many ways, MinD fulfills the definition of a signaling protein scaffold that allows several actors belonging to different pathways to co-localize at the cell poles. Remarkably, in the B. subtilis PPI phosphotyrosine network, MinD exhibits an interacting landscape similar to that of TkmA (Shi et al., 2014b; Figure 2). As a transmembrane protein, TkmA is classified as a receptor protein, activating PtkA upon sensing external inputs, while MinD perfectly matches with the definition of a classical scaffold, enforcing the proximity of kinase/phosphatase and substrate at a specific time and location during the bacteria cell cycle. However, the potential ability of TkmA to contact more than one kinase, a phosphatase and various PtkA substrates suggests that this receptor could also act as a docking platform to activate tyrosine kinase signaling events (Jers et al., 2010).
Figure 2.
PPI profile similarities between the protein modulators TkmA (anchor) and MinD (scaffold) in B. subtilis. Tyr and Ser/Thr phosphorylation pathways are filled in red and yellow, respectively. Cell division pathway is indicated in green.
MinD acting as a scaffold is therefore proposed to promote PtkA-mediated phosphorylation of DivIVA in B. subtilis (Shi et al., 2014b). Phosphorylation of DivIVA and other homologs by Ser/Thr kinases is described as playing a role in regulating apical growth and cell polarity in Actinobateria, as well as well as orchestrating peptidoglycan synthesis during cell elongation in streptococci (Fleurie et al., 2012; Hempel et al., 2012; Fleurie et al., 2014; Pompeo et al., 2015). A real question about the role of phosphorylation of DivIVA at tyrosine in regulating its dynamic from the cell poles to the future septum is now addressed in B. subtilis. In this bacterium, PtkA could modulate DivIVA at the cell pole through landing/activation by MinD. The potential presence of other signaling components, such as the phosphatase PtpZ, would also add to the fine-tuning of such regulations. The interaction of the sporulation YabT Ser/Thr kinase with MinD might also provide a clue toward understanding how DivVIA switches from a MinCD(J)-mediated control of FtsZ assembly during vegetative growth, to a RacA mediated chromosome anchorage to the pole of the fore spore compartment at the onset of sporulation (Ben-Yehuda et al., 2003; Wu and Errington, 2003; Ben-Yehuda et al., 2005; Perry and Edwards, 2006; Bramkamp et al., 2008). This novel function of MinD as a signaling scaffold is thus expected to participate in coordinating cytokinesis, the terminal step of the cell cycle, with other pathways during the different Bacillus life-styles.
MinD/ParA-like proteins are widely found distributed among bacteria. They are not only involved in different aspects of regulation of cell division and chromosome segregation, but also in polar localization of cellular machineries, such as those directing conjugative transfer or chemotaxis, as well as those organizing the polar positioning of Type IV pili (Kirkpatrick and Viollier, 2012; Lutkenhaus, 2012; Treuner-Lange and Søgaard-Andersen, 2014). ParA/MinD-like proteins are often described as protein-hubs responsible for the architectural and spatial organization of necessary components driving these processes (Lutkenhaus, 2012). In the asymmetrically dividing bacterium C. crescentus, the mechanisms underlying the segregation of the cell fate determinants to the two structurally distinct cell poles also requires MinD/ParA-like proteins to achieve polar localization of chromosomes and cell division (Thanbichler and Shapiro, 2006). The MinD-like ATPase MipZ forms a dynamic complex with the DNA partitioning protein ParB at the chromosomal origin of replication, and also negatively regulates the polymerization of FtsZ (Du and Lutkenhaus, 2012; Kiekebusch et al., 2012). Through ATP hydrolysis, MipZ acts as a molecular switch to couple chromosome segregation with the control of mid-cell positioning of the FtsZ ring in C. crescentus (Thanbichler and Shapiro, 2006). Although its role in signaling scaffold is not proven, recent studies highlight the importance of ParB phosphorylation in the localization and function of proteins involved in chromosome segregation in mycobacteria (Baronian et al., 2015). Lastly, the discovery that the ParA-like BY-kinase CspD, encoded by the cps capsular operon in S. pneumoniae, which plays a role in coordinating capsular polysaccharide with chromosome segregation through interaction with ParB, leads to the hypothesis that this kinase could act as molecular scaffold (Nourikyan et al., 2015). Although no proteins have so far been found to be phosphorylated by CpsD, ATP hydrolysis leading to CpsD autophosphorylation plays a major role in regulating the dynamic of localization of the ParB proteins bound to the chromosomal replication origin, and provides a molecular switch to couple the cell cycle with cell elongation (Nourikyan et al., 2015).
Conclusions
Large-scale profiling of phosphoproteins and phospho-sites by mass spectrometry-based proteomics, differential phosphoproteomics across physiological conditions and kinase-related phosphoproteomes are helpful to identify kinase-specific substrates and understand the functional dynamics of phosphorylation networks. Additionally, interactomics identifies kinase-protein interactions revealing not only potential substrates, but also anchors and modulators. Hence, phosphoproteomics and interactomics are powerfull and complementary approaches to provide a blueprint of kinase and phosphatase-centered signaling networks. The ever growing data from large scale phosphoproteome studies in bacteria, coupled with identification of kinase, phosphatase, substrates, and modulators provides important insights into cellular regulations. It also provides a unique opportunity to understand how phosphorylation participates in the regulation of protein activities, their interactions with other proteins, and their localizations in the bacterial cell.
In many bacteria, phosphorylation of proteins involved in various DNA-related processes, together with biological evidence of their role during cell cycle, suggests the existence of checkpoint regulations akin to eukaryotes. In all living organisms, cell cycle events involve the temporal and spatial coordination of chromosome replication and segregation with cytokinesis, the entire process being also coordinated with cell growth. In eukaryotes, the existence of highly intricate signaling networks is ensured by quality control and checkpoint proteins that coordinate these sequential events and trigger cell cycle arrests when things go wrong. In bacteria, the different processes are more interwoven. Consequently, the accurate execution of each step relative to others in time and space is crucial and involves strict regulatory mechanisms. The finding, in B. subtilis, that cell division MinD may act as a signaling hub to reinforce the proximity of various components of the Tyr and Ser/Thr phosphorylation pathway, along with the septum site protein DivIA, again hints at the existence of scaffold-mediated signal transduction in bacteria. In addition, the recent discovery that bacterial kinase are themselves able to tune their activities through phosphorylation add to the complexity of the picture (Shi et al., 2014a). However, it also suggests that, just as in eukaryotes, such bacterial scaffolds could offer versatile strategies to dynamically coordinate cell processes via signaling.
Author contributions
TG, SP, LS, and AD analyzed the bibliographic data. TG and SP provided the tables. MN wrote the manuscript. IM revised it critically. All authors gave final approval of the version.
Funding
This work was supported by the grants from the Agence Nationale de la Recherche (2010-BLAN-1303-01) to MN and IM and the Vetenskapsrådet (2015-05319) and the Novo Nordisk Foundation to IM. TG is supported by the European Union, Marie Curie ITN AMBER, 317338.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Christophe Grangeasse for his valuable comments on the manuscript.
References
- Antony E., Weiland E., Yuan Q., Manhart C. M., Nguyen B., Kozlov A. G., et al. (2013). Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 425, 4802–4819. 10.1016/j.jmb.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C. E., Iavarone A. T., Alber T., Sassetti C. M. (2014). Biochemical and spatial coincidence in the provisional Ser/Thr protein kinase interaction network of Mycobacterium tuberculosis. J. Biol. Chem. 289, 20422–20433. 10.1074/jbc.M114.559054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Ji Z. (2012). Phosphoproteomic investigation of a solvent producing bacterium Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 95, 201–211. 10.1007/s00253-012-4156-3 [DOI] [PubMed] [Google Scholar]
- Baronian G., Ginda K., Berry L., Cohen-Gonsaud M., Zakrzewska-Czerwinska J., Jakimowicz D., et al. (2015). Phosphorylation of Mycobacterium tuberculosis ParB participates in regulating the ParABS chromosome segregation system. PLoS ONE 10:e0119907. 10.1371/journal.pone.0119907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentchikou E., Chagneau C., Long E., Matelot M., Allemand J. F., Michel B. (2015). Are the SSB-interacting proteins RecO, RecG, PriA and the DnaB-interacting protein rep bound to progressing replication forks in Escherichia coli? PLoS ONE 10:e0134892. 10.1371/journal.pone.0134892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S., Fujita M., Liu X. S., Gorbatyuk B., Skoko D., Yan J., et al. (2005). Defining a centromere-like element in Bacillus subtilis by Identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17, 773–782. 10.1016/j.molcel.2005.02.023 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S., Rudner D. Z., Losick R. (2003). RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299, 532–536. 10.1126/science.1079914 [DOI] [PubMed] [Google Scholar]
- Bidnenko V., Shi L., Kobir A., Ventroux M., Pigeonneau N., Henry C., et al. (2013). Bacillus subtilis serine/threonine protein kinase YabT is involved in spore development via phosphorylation of a bacterial recombinase. Mol. Microbiol. 88, 921–935. 10.1111/mmi.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz S. K., Lao Y., Lowry D. F., Wold M. S. (2003). The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 278, 35584–35591. 10.1074/jbc.M305388200 [DOI] [PubMed] [Google Scholar]
- Boonstra M., de Jong I. G., Scholefield G., Murray H., Kuipers O. P., Veening J. W. (2013). Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol. Microbiol. 87, 925–938. 10.1111/mmi.12141 [DOI] [PubMed] [Google Scholar]
- Bramkamp M., Emmins R., Weston L., Donovan C., Daniel R. A., Errington J. (2008). A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70, 1556–1569. 10.1111/j.1365-2958.2008.06501.x [DOI] [PubMed] [Google Scholar]
- Burnside K., Lembo A., Harrell M. I., Gurney M., Xue L., BinhTran N. T., et al. (2011). Serine/threonine phosphatase Stp1 mediates post-transcriptional regulation of hemolysin, autolysis, and virulence of group B Streptococcus. J. Biol. Chem. 286, 44197–44210. 10.1074/jbc.M111.313486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butala M., Zgur-Bertok D., Busby S. J. (2009). The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 66, 82–93. 10.1007/s00018-008-8378-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla-Llorente V., Muñoz-Espín D., Villar L., Salas M., Meijer W. J. (2006). Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J. 25, 3890–3899. 10.1038/sj.emboj.7601266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Yang H., Pavletich N. P. (2008). Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–484. 10.1038/nature06971 [DOI] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D. (2008). ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627. 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D. (2015). Preventing replication fork collapse to maintain genome integrity. DNA Repair 32, 149–157. 10.1016/j.dnarep.2015.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes A., Lecointe F., McGovern S., Quevillon-Cheruel S., Polard P. (2010). The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 6:e1001238. 10.1371/journal.pgen.1001238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche A., Shi L., Bidnenko V., Ventroux M., Pigonneau N., Franz-Wachtel M., et al. (2015a). Bacillus subtilis SalA is a phosphorylation-dependent transcription regulator that represses scoC and activates the production of the exoprotease AprE. Mol Microbiol. 97, 1195–1208. 10.1111/mmi.13098 [DOI] [PubMed] [Google Scholar]
- Derouiche A., Shi L., Kalantari A., Mijakovic I. (2015b). Evolution and tinkering: what do a protein kinase, a transcriptional regulator and chromosome segregation/cell division proteins have in common? Curr. Genet. 62, 67–70. 10.1007/s00294-015-0513-y [DOI] [PubMed] [Google Scholar]
- De Septenville A. L., Duigou S., Boubakri H., Michel B. (2012). Replication fork reversal after replication-transcription collision. PLoS Genet. 8:e1002622. 10.1371/journal.pgen.1002622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier J. P., Cozzone A. J., Duclos B. (2010). Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol. Lett. 306, 30–36. 10.1111/j.1574-6968.2010.01930.x [DOI] [PubMed] [Google Scholar]
- Dillon S. C., Dorman C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- Dorman C. J. (2013). Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 11, 349–355. 10.1038/nrmicro3007 [DOI] [PubMed] [Google Scholar]
- Du S., Lutkenhaus J. (2012). MipZ: one for the pole, two for the DNA. Mol. Cell 46, 239–240. 10.1016/j.molcel.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H., Rose K. L., Bhat K. P., Mohni K. N., Glick G. G., Couch F. B., et al. (2015). The Replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010. 10.1016/j.molcel.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J. (2015). Ser/Thr phosphorylation as a regulatory mechanism in bacteria. Curr. Opin. Microbiol. 24, 47–52. 10.1016/j.mib.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholz A. K., Turgay K., Michalik S., Hessling B., Gronau K., Oertel D., et al. (2012). Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 109, 7451–7456. 10.1073/pnas.1117483109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser D., Pham T. K., Reimann J., Albers S. V., Siebers B., Wright P. C. (2012). Change of carbon source causes dramatic effects in the phospho-proteome of the archaeon Sulfolobus solfataricus. J. Proteome Res. 11, 4823–4833. 10.1021/pr300190k [DOI] [PubMed] [Google Scholar]
- Fleurie A., Cluzel C., Guiral S., Freton C., Galisson F., Zanella-Cleon I., et al. (2012). Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol. Microbiol. 83, 746–758. 10.1111/j.1365-2958.2011.07962.x [DOI] [PubMed] [Google Scholar]
- Fleurie A., Manuse S., Zhao C., Campo N., Cluzel C., Lavergne J. P., et al. (2014). Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 10:e1004275. 10.1371/journal.pgen.1004275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R., Sun X., Xiao C., Yin X., Shan W., Chen Z., et al. (2011). Phosphoproteome analysis of the pathogenic bacterium Helicobacter pylori reveals over-representation of tyrosine phosphorylation and multiply phosphorylated proteins. Proteomics 11, 1449–1461. 10.1002/pmic.201000649 [DOI] [PubMed] [Google Scholar]
- Good M., Tang G., Singleton J., Reményi A., Lim W. A. (2009). The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136, 1085–1097. 10.1016/j.cell.2009.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory J. A., Becker E. C., Pogliano K. (2008). Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22, 3475–3488. 10.1101/gad.1732408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Sajid A., Sharma K., Ghosh S., Arora G., Singh R., et al. (2014). HupB, a nucleoid-associated protein of Mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J. Bacteriol. 196, 2646–2657. 10.1128/JB.01625-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. M., Chaerkady R., Sharma J., Díaz-Mejía J. J., Tyagi N., Renuse S., et al. (2013). The Escherichia coli phosphotyrosine proteome relates to core pathways and virulence. PLoS Pathog. 9:e1003403. 10.1371/journal.ppat.1003403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann M., Hintermair C., Voß K., Eick D. (2013). Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta 1829, 55–62. 10.1016/j.bbagrm.2012.08.013 [DOI] [PubMed] [Google Scholar]
- Hempel A. M., Cantlay S., Molle V., Wang S. B., Naldrett M. J., Parker J. L., et al. (2012). The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. U.S.A. 109, E2371–E2379. 10.1073/pnas.1207409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2000). Signaling–2000 and beyond. Cell 100, 113–127. 10.1016/S0092-8674(00)81688-8 [DOI] [PubMed] [Google Scholar]
- Iyer D. R., Rhind N. (2013). Checkpoint regulation of replication forks: global or local? Biochem. Soc. Trans. 41, 1701–1705. 10.1042/BST20130197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jers C., Pedersen M. M., Paspaliari D. K., Schütz W., Johnsson C., Soufi B., et al. (2010). Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol. Microbiol. 77, 287–299. 10.1111/j.1365-2958.2010.07227.x [DOI] [PubMed] [Google Scholar]
- Kalantari A., Derouiche A., Shi L., Mijakovic I. (2015). Serine/ threonine/tyrosine phosphorylation regulates DNA-binding of bacterial transcription regulators. Microbiology 161, 1720–1729. 10.1099/mic.0.000148 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly P. J. (2014). Protein Ser/Thr/Tyr phosphorylation in the Archaea. J. Biol. Chem. 289, 9480–9487. 10.1074/jbc.R113.529412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekebusch D., Michie K. A., Essen L. O., Löwe J., Thanbichler M. (2012). Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol. Cell 46, 245–259. 10.1016/j.molcel.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick C. L., Viollier P. H. (2012). Cell polarity: ParA-logs gather around the Hub. Curr. Biol. 22, R1055–R1057. 10.1016/j.cub.2012.10.053 [DOI] [PubMed] [Google Scholar]
- Kobir A., Poncet S., Bidnenko V., Delumeau O., Jers C., Zouhir S., et al. (2014). Phosphorylation of Bacillus subtilis gene regulator AbrB modulates its DNA-binding properties. Mol. Microbiol. 92, 1129–1141. 10.1111/mmi.12617 [DOI] [PubMed] [Google Scholar]
- Kozlov A. G., Weiland E., Mittal A., Waldman V., Antony E., Fazio N., et al. (2015). Intrinsically disordered C-terminal tails of E. coli single-stranded DNA binding protein regulate cooperative binding to single-stranded DNA. J. Mol. Biol. 427, 763–774. 10.1016/j.jmb.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. (2001). DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8461–8468. 10.1073/pnas.151260698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston L. D., Indiani C., O'Donnell M. (2009). Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8, 2686–2691. 10.4161/cc.8.17.9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston L. D., O'Donnell M. (2006). DNA replication: keep moving and don't mind the gap. Mol. Cell 23, 155–160. 10.1016/j.molcel.2006.05.034 [DOI] [PubMed] [Google Scholar]
- Lecointe F., Sérèna C., Velten M., Costes A., McGovern S., Meile J. C., et al. (2007). Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 26, 4239–4251. 10.1038/sj.emboj.7601848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Lan L., Peng G., Chang W. C., Hsu M. C., Wang Y. N., et al. (2015). Tyrosine 370 phosphorylation of ATM positively regulates DNA damage response. Cell Res. 25, 225–236. 10.1038/cr.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Wang J., Ying W., Qian X. (2012). Optimization of titanium dioxide enrichment of phosphopeptides and application in the Thermoanaerobacter tengcongensis phosphoproteome analysis. Se Pu 30, 763–769. 10.3724/SP.J.1123.2012.04027 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. (2012). The ParA/MinD family puts things in their place. Trends Microbiol. 20, 411–418. 10.1016/j.tim.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis F. J. (2008). Conservation of protein phosphorylation sites within gene families and across species. Plant Signal. Behav. 3, 1011–1013. 10.4161/psb.6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., et al. (2008). Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307. 10.1074/mcp.M700311-MCP200 [DOI] [PubMed] [Google Scholar]
- Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., et al. (2007). The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707. 10.1074/mcp.M600464-MCP200 [DOI] [PubMed] [Google Scholar]
- Manteca A., Ye J., Sánchez J., Jensen O. N. (2011). Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res. 10, 5481–5492. 10.1021/pr200762y [DOI] [PubMed] [Google Scholar]
- Manuse S., Fleurie A., Zucchini L., Lesterlin C., Grangeasse C. (2015). Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol. Rev. 40, 41–56. 10.1093/femsre/fuv041 [DOI] [PubMed] [Google Scholar]
- Marchadier E., Carballido-López R., Brinster S., Fabret C., Mervelet P., Bessieres P., et al. (2011). An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: exploration by an integrative approach. Proteomics 11, 2981–2991. 10.1002/pmic.201000791 [DOI] [PubMed] [Google Scholar]
- Maréchal A., Zou L. (2015). RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23. 10.1038/cr.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H., Zhang Y., Grossman A. D., Wang J. D. (2012). Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 10, 449–458. 10.1038/nrmicro2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettrick K. A., Grainge I. (2015). Stability of blocked replication forks in vivo. Nucleic Acids Res. 44, 657–668. 10.1093/nar/gkv1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I., Petranovic D., Macek B., Cepo T., Mann M., Davies J., et al. (2006). Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res. 34, 1588–1596. 10.1093/nar/gkj514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S. K., Milohanic E., Aké F., Mijakovic I., Deutscher J., Monnet V., et al. (2011). Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165. 10.1002/pmic.201100259 [DOI] [PubMed] [Google Scholar]
- Nakedi K. C., Nel A. J., Garnett S., Blackburn J. M., Soares N. C. (2015). Comparative Ser/Thr/Tyr phosphoproteomics between two mycobacterial species: the fast growing Mycobacterium smegmatis and the slow growing Mycobacterium bovis BCG. Front. Microbiol. 6:237. 10.3389/fmicb.2015.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourikyan J., Kjos M., Mercy C., Cluzel C., Morlot C., Noirot-Gros M. F., et al. (2015). Autophosphorylation of the bacterial tyrosine-kinase CpsD connects capsule synthesis with the cell cycle in Streptococcus pneumoniae. PLoS Genet. 11:e1005518. 10.1371/journal.pgen.1005518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley G. G., Patrick S. M. (2010). Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. 15, 883–900. 10.2741/3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley G. G., Patrick S. M., Yao J., Carty M. P., Turchi J. J., Dixon K. (2003). RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 42, 3255–3264. 10.1021/bi026377u [DOI] [PubMed] [Google Scholar]
- Ortega C., Liao R., Anderson L. N., Rustad T., Ollodart A. R., Wright A. T., et al. (2014). Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch. PLoS Biol. 12:e1001746. 10.1371/journal.pbio.1001746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Wang B., Zhang Y., Wang Y., Ullah S., Jian R., et al. (2015). dbPSP: a curated database for protein phosphorylation sites in prokaryotes. Database 2015:bav031. 10.1093/database/bav031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S. F., Goss L., Dworkin J. (2011). Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75, 192–212. 10.1128/MMBR.00042-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. E., Edwards D. H. (2006). The Bacillus subtilis DivIVA protein has a sporulation-specific proximity to Spo0J. J. Bacteriol. 188, 6039–6043. 10.1128/JB.01750-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pini F., De Nisco N. J., Ferri L., Penterman J., Fioravanti A., Brilli M., et al. (2015). Cell Cycle Control by the master regulator CtrA in Sinorhizobium meliloti. PLoS Genet. 11:e1005232. 10.1371/journal.pgen.1005232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz R. T., O'Donnell M. (2010). What happens when replication and transcription complexes collide? Cell Cycle 9, 2537–2543. 10.4161/cc.9.13.12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F., Foulquier E., Serrano B., Grangeasse C., Galinier A. (2015). Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis. Mol. Microbiol. 97, 139–150. 10.1111/mmi.13015 [DOI] [PubMed] [Google Scholar]
- Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C. M., et al. (2010). Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526. 10.1073/pnas.0913482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran A., Sugiyama N., Tomita M., Swarup S., Ishihama Y. (2009). Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9, 2764–2775. 10.1002/pmic.200800655 [DOI] [PubMed] [Google Scholar]
- Ravikumar V., Shi L., Krug K., Derouiche A., Jers C., Cousin C., et al. (2014). Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC. Mol. Cell. Proteomics 13, 1965–1978. 10.1074/mcp.M113.035949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl S. R., Gronau K., Pietack N., Hecker M., Becher D., Stülke J. (2010). The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol. Cell. Proteomics 9, 1228–1242. 10.1074/mcp.M900267-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R. A., Holzen T. M. (2007). Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41, 237–280. 10.1146/annurev.genet.41.110306.130308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. (2004a). ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6, 648–655. 10.1038/ncb1145 [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. (2004b). Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair 3, 901–908. 10.1016/j.dnarep.2004.03.020 [DOI] [PubMed] [Google Scholar]
- Shi L., Pigeonneau N., Ravikumar V., Dobrinic P., Macek B., Franjevic D., et al. (2014a). Cross-phosphorylation of bacterial serine/threonine and tyrosine protein kinases on key regulatory residues. Front. Microbiol. 5:495. 10.3389/fmicb.2014.00495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Pigeonneau N., Ventroux M., Derouiche A., Bidnenko V., Mijakovic I., et al. (2014b). Protein-tyrosine phosphorylation interaction network in Bacillus subtilis reveals new substrates, kinase activators and kinase cross-talk. Front. Microbiol. 5:538. 10.3389/fmicb.2014.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares N. C., Spät P., Krug K., Macek B. (2013). Global dynamics of the Escherichia coli proteome and phosphoproteome during growth in minimal medium. J. Proteome Res. 12, 2611–2621. 10.1021/pr3011843 [DOI] [PubMed] [Google Scholar]
- Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., et al. (2008). The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8, 3486–3493. 10.1002/pmic.200800069 [DOI] [PubMed] [Google Scholar]
- Subramanian V. V., Hochwagen A. (2014). The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 6:a016675. 10.1101/cshperspect.a016675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ge F., Xiao C. L., Yin X. F., Ge R., Zhang L. H., et al. (2010). Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J. Proteome Res. 9, 275–282. 10.1021/pr900612v [DOI] [PubMed] [Google Scholar]
- Takahata Y., Inoue M., Kim K., Iio Y., Miyamoto M., Masui R., et al. (2012). Close proximity of phosphorylation sites to ligand in the phosphoproteome of the extreme thermophile Thermus thermophilus HB8. Proteomics 12, 1414–1430. 10.1002/pmic.201100573 [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Shapiro L. (2006). MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126, 147–162. 10.1016/j.cell.2006.05.038 [DOI] [PubMed] [Google Scholar]
- Treuner-Lange A., Søgaard-Andersen L. (2014). Regulation of cell polarity in bacteria. J. Cell Biol. 206, 7–17. 10.1083/jcb.201403136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Ding Y., Hooper D. C. (2008). Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 190, 7375–7381. 10.1128/JB.01068-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin S., Watson D. C., Tessier L., Ding W., Foote S., Bhatia S., et al. (2007). The cytoplasmic phosphoproteome of the Gram-negative bacterium Campylobacter jejuni: evidence for modification by unidentified protein kinases. Proteomics 7, 4338–4348. 10.1002/pmic.200700483 [DOI] [PubMed] [Google Scholar]
- Vujaklija D., Macek B. (2012). Detecting posttranslational modifications of bacterial SSB proteins. Methods Mol. Biol. 922, 205–218. 10.1007/978-1-62703-032-8_16 [DOI] [PubMed] [Google Scholar]
- Wright D. P., Ulijasz A. T. (2014). Regulation of transcription by eukaryotic-like serine-threonine kinases and phosphatases in Gram-positive bacterial pathogens. Virulence 5, 863–885. 10.4161/21505594.2014.983404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. J., Errington J. (2003). RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49, 1463–1475. 10.1046/j.1365-2958.2003.03643.x [DOI] [PubMed] [Google Scholar]
- Xenopoulos P., Piggot P. J. (2011). Regulation of growth of the mother cell and chromosome replication during sporulation of Bacillus subtilis. J. Bacteriol. 193, 3117–3126. 10.1128/JB.00204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T., Carrasco B., Myers A. R., George N. P., Keck J. L., Alonso J. C. (2012). Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 40, 5546–5559. 10.1093/nar/gks173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. K., Qiao Z. X., Zhang W. Y., Xiong Q., Zhang J., Li T., et al. (2013). Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 12, 1909–1923. 10.1021/pr4000043 [DOI] [PubMed] [Google Scholar]
- Zeke A., Lukács M., Lim W. A., Remenyi A. (2009). Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 19, 364–374. 10.1016/j.tcb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao F., Guan X., Yang Y., Liang C., Qin S. (2007). Genome-wide survey of putative serine/threonine protein kinases in cyanobacteria. BMC Genomics 8:395. 10.1186/1471-2164-8-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhou J., Ni Y., Yu Z., Mao A., Hu Y., et al. (2014). Contribution of eukaryotic-type serine/threonine kinase to stress response and virulence of Streptococcus suis. PLoS ONE 9:e91971. 10.1371/journal.pone.0091971 [DOI] [PMC free article] [PubMed] [Google Scholar]