Abstract
Debate exists about whether agricultural versus medical antibiotic use drives increasing antibiotic resistance (AR) across nature. Both sectors have been inconsistent at antibiotic stewardship, but it is unclear which sector has most influenced acquired AR on broad scales. Using qPCR and soils archived since 1923 at Askov Experimental Station in Denmark, we quantified four broad-spectrum β-lactam AR genes (ARG; blaTEM, blaSHV, blaOXA and blaCTX-M) and class-1 integron genes (int1) in soils from manured (M) versus inorganic fertilised (IF) fields. “Total” β-lactam ARG levels were significantly higher in M versus IF in soils post-1940 (paired-t test; p < 0.001). However, dominant individual ARGs varied over time; blaTEM and blaSHV between 1963 and 1974, blaOXA slightly later, and blaCTX-M since 1988. These dates roughly parallel first reporting of these genes in clinical isolates, suggesting ARGs in animal manure and humans are historically interconnected. Archive data further show when non-therapeutic antibiotic use was banned in Denmark, blaCTX-M levels declined in M soils, suggesting accumulated soil ARGs can be reduced by prudent antibiotic stewardship. Conversely, int1 levels have continued to increase in M soils since 1990, implying direct manure application to soils should be scrutinized as part of future stewardship programs.
Mass use of antibiotics for treating infectious disease over the 20th century has improved human health and agricultural productivity in the developed world, enhancing the quality of life of millions1. However, antibiotic use, especially in medicine and agriculture, also has fuelled increasing acquired antibiotic resistance (AR) in exposed organisms to a point where many antibiotics are compromised; multi-resistance pathogens are common; and new antibiotic development has become uneconomical2,3,4. Despite growing awareness of the AR crisis (e.g., ref. 5), debate continues over who and which activities are most responsible for increased AR in nature, largely because it is hard to link specific causative actions with explicit AR consequences6,7. AR is ancient with VanX proto-resistance and multi-resistance genes being detected in ~30,000-year old DNA from permafrost8. However, increased antibiotic use has clearly mobilised AR genes and accelerated bacterial AR evolution in strains not previously intrinsically resistant9,10, including human pathogens11. Antibiotic overuse and poor water quality in some parts of the world have further altered the environmental resistome (i.e., the pool of all AR genes; ARG)12,13, which increases the probability of AR acquisition in any exposed bacteria14.
Despite the above, debate continues about the relative role of each sectoral driver of acquired AR: medicine, agriculture or environmental pollution. This debate often follows parochial bias, but it also stems from substantial difficulties in identifying root causes of detected AR in most scenarios. AR often results from acquisition or mutation of a defence gene, which allows an organism to better protect itself. However, phenotypic detection of AR is only observed when a strain is exposed to an antibiotic. Therefore, organisms might acquire the potential for AR (e.g., an ARG or mutation) in one place, but phenotypic AR only becomes apparent when the “next” antibiotic treatment fails, possibly great distances away15. This reality has led many to falsely presume AR acquisition is primarily driven by factors at the point of detection (e.g., a hospital) when, in fact, original acquisition of ARGs or mutations might occur elsewhere, including the natural environment.
Within this context, long-term soil archives from the Netherlands were studied to assess relative ARG abundances in soils harvested since large-scale antibiotic manufacture increased in the 1940s16. This work showed relative soil ARG abundances, based on extracted DNA, have increased over the last 60 years. Although interesting, this work did not identify specific local causes for increasing ARG levels because detailed soil histories were not available. However, systematic soil archiving has been performed since 1923 at The Askov Long-Term Experiment (LTE) Station in Denmark, initiated in 1894 to study the role of animal manure versus inorganic fertilisers on soil fertility17. Detailed soil and field management records were kept for the Askov-LTE, which included fields that only received inorganic fertilizers (IF) over their entire history, whereas other fields received only animal manure (M). As such, the Askov-LTE soil archive provides a wholly unique platform for assessing long-term changes in in situ ARGs, especially relative impacts of animal versus non-animal factors on soil AR.
Here, DNA was harvested from archived Askov-LTE soils collected from 1923 to 2010, and relative abundances of four β-lactam ARGs (blaTEM, blaSHV, blaOXA and blaCTX-M) and class 1 integron genes (int1) were contrasted over time. These ARGs, which code for broad (blaTEM and blaSHV) and extended-spectrum (blaOXA and blaCTX-M) β-lactamases (ESBL), were chosen as “biomarkers” for the appearance of β-lactam resistance over time because they have distinct histories in hospitals between 1963 and 198918,19,20,21, and can confer resistance against an essential class of antibiotics for the treatment of infectious disease. With these data, one can contrast soil ARG levels associated with long-term fertiliser use and parallel observations in medicine, and also assess whether changes in Danish agricultural antibiotics practice has influenced relative ARG abundances in associated soils.
Materials and Methods
Field Experiment and Soil Archive
Soils assessed in this study were retrieved from the soil archive of the Askov-LTE initiated in 1894 at the Lermarken site17. The experimental site is located in South Jutland, Denmark and was first cultivated around 1800. It has mean annual temperature and precipitation of 7.7 °C and 862 mm, respectively, and is flat (gradient <2%) and well-sheltered by hedgerows and scattered woodlands. The soils are dominated by glacial moraine deposits, and Ap-horizons (0–18/20 cm) are characterized as coarse sandy loam.
Field experiments with different fertilization treatments have been performed on four sets of fields at the Lermarken site since 189417,22. Field treatments have included different levels (½x, 1x, 1½x, 2x) of animal manure, inorganic fertilisers only, and unamended plots (no manure or inorganic fertilisers). Absolute nutrient amendments were increased in 1923 and again in 1973 (to align with mainstream fertilization levels in Danish agriculture), but almost identical amounts of N, P, and K were applied to both M and IF fields throughout the experiment. The Askov-LTE experiment assessed classical four-course crop rotation, including winter cereals, row crops, spring cereals and grass-clover mixture. Typically, fertilization has been performed in late autumn to early spring; soils for archiving were sampled during the same period, but always before next fertilization. Soil samples have been collected from the Ap-horizon every four years since 1923, air dried, and archived at room temperature under dry conditions, creating one of the longest agricultural soil archives in Europe.
For this study, archived soil samples were only quantified from 1× IF and 1× M fields between 1923 and 2010 to ensure consistency in field amendments. Actual IF and M treatments were 70 kg total-N, 16–19 kg P and 58–70 kg K between 1923 and 1973, and 100 kg total-N ha−1, 19 kg P ha−1 and 87 kg K ha−1 since 1973. Further details are found in Christensen et al.17,22.
Chemical Soil Analyses
Total-C (TC) and -N (TN) in the soils were analysed by dry combustion and Kjeldahl digestion, respectively22. Extractable inorganic P (Ext P) was determined by extracting soil in 0.2 N sulphuric acid, whereas extractable K (Ext K) was extracted in 0.5 M ammonium-acetate17. Concentrations of heavy metals and trace elements were determined using ICP-MS following Aqua-Regia digestion.
DNA Extraction and Gene Quantification
DNA was extracted using the FastDNA SPIN (MP Biomedical, Cambridge, UK) columns for soils, following manufacturer’s protocols. Typically, 200–300 mg (as dry weight) of dried soil was aseptically transferred into centrifuge tubes containing phosphate buffer saline (PBS) and extraction beads (weighing tubes before and after soil addition). Samples were gently mixed in the PBS for 15–20 minutes for rehydration, and cells were then lysed using a FastPrep cell disruptor (6.0 setting, 30 seconds; MP Biomedicals). Cell lysis was repeated twice. Resultant DNA was temporarily stored at −20 °C until all soils were extracted, and then retained at −80 °C prior to qPCR analysis.
Four β-lactam ARGs (blaTEM, blaSHV, blaOXA and blaCTX-M) were chosen for quantification over time, which was based on previous experience assessing soils and sediments16,23,24,25 and their relative importance to therapeutic applications3. Primers for blaTEM26, blaSHV27,28, blaCTX-M29 and blaOXA30,31 target conservative regions among common β-lactamase genes (which encode for enzymes that inactivate penicillin and other β-lactam antibiotics). Int1 and 16S-rRNA bacterial gene abundances were quantified according to methods from Mazel et al.32 and Yu et al.33, respectively.
For qPCR analysis, two μl of DNA template and appropriate primers were combined with sso-Fast EvaGreen PCR reagent (Bio-Rad, Hercules, CA, USA) and molecular-grade water to create 20-μl reaction volumes. Analyses were performed using a Bio-Rad CFX96 system. Temperature cycles were 95 °C (30 sec) for enzyme activation, and then 40 cycles of 94 °C (5 sec), annealing temperatures (blaTEM: 50 °C, and 55 °C for blaSHV, blaCTX-M and blaOXA) for 10 seconds, and 72 °C for an additional 5 seconds. Samples were analysed in duplicate; any samples with a major discrepancy (high analytical variability) were re-analysed. Duplicate cycle-threshold values always ranged within ±0.3 units (log scale). A post-analytical temperature melt curve was used to verify reaction quality (50–95 °C, ΔT = 0.1 °C/second).
All reactions were run with serially diluted plasmid-DNA standards derived from gene-positive bacteria. qPCR reaction efficiencies were determined by spiking sample with known amounts of DNA template, and results were compared with “neat” standards. Samples were typically diluted, either 1:100 or 1:1000, to reduce possible inhibitory effects from substances that co-elute when extracting soil DNA. Correlation coefficients for all standard curves were r > 0.98; and log gene-abundance values (except those below detection limits) were within the linear range of the calibration curves.
Data Processing and Analyses
ARG abundances were normalised to 16S-rRNA gene abundances to minimise variance caused by differential extraction and analytical efficiencies, and differences in background bacterial abundances. Two-sample tests were then employed to statistically compare different groupings of the normalised data (e.g., M vs IF fields), including the t-test for normally-distributed datasets or the Wilcoxon Ranked-Sum test for non-normally-distributed datasets. Bivariate correlation analyses across samples used the Spearman rank method because many of the variables were non-normally-distributed, even after log-transformation.
To better visualize gene-abundance changes over time, all relative gene abundances (i.e., normalized to 16S gene values) were further normalized relative to mean ARG levels detected in soils from 1923 and 1938, which represent a timeframe before mass-production of antibiotics commenced. To validate this assumption, ARG data were statistically compared between M and IF soils archived from 1923 and 1938, and no significant differences were observed between field treatments. Therefore, data from are 1923 and 1938 samples were combined to quantify pre-1940 baseline levels of ARGs (and heavy metals and nutrients). Unless otherwise noted, significance is defined as 95% confidence (i.e., p < 0.05). All statistics were conducted using SPSSTM version 22.
Results and Discussion
Antibiotics have been used in medicine and agriculture since the 1930s. Initially, antibiotics only were used for medical applications, but their use expanded to agriculture in the 1950s, including therapeutic and non-therapeutic applications2. As part of this trend, Denmark was among the leaders in employing antibiotics to increase agricultural productivity and industrial animal production, including non-therapeutic use (e.g., growth promotion)34. However, in the 1990s, antibiotics were banned for non-therapeutic use and were slowly phased out35,36, and Denmark has become a benchmark for prudent antibiotic stewardship. As such, the Danish experience provides an ideal template for studying the historic effects of antibiotic use, including pre-antibiotic conditions, large-scale antibiotic use in agriculture, and conditions after antibiotics were banned for non-therapeutic use since the 1990s.
Previous work16 showed that relative levels of ARGs in Dutch agricultural soils significantly increased between the 1940s and 2010. For example, selected broad-spectrum β-lactam ARG genes (blaTEM and blaSHV) increased by 15 times over this time, especially since the 1980s. Unfortunately, field data (e.g., type and amounts of fertilisers), irrigation sources, and other management factors varied across the Dutch study sites and soils, which made it difficult to determine specific field-level cause-and-effects relative to increased ARGs in the soils. Therefore, this earlier work was incomplete and more work was needed.
Here we used similar processing and ARG quantification methods as the Dutch study, but on soil archives with well-characterised field-management histories, particularly associated with M versus IF fertilization for over 100 years. Associated with M and IF fields, we quantified the selected bla ARGs, int1 and 16S rRNA gene abundances in archived soils since 1923. TC, TN, ExtP, ExtK, and selected heavy metal levels also were quantified over time to compare soil conditions with the ARG and int1 data. int1 levels were also quantified here because they are sometimes linked with acquired AR, especially due to animal waste releases10, and can imply an increased potential for horizontal gene transfer (HGT) in any ecosystem. As previously noted in the Methods, all post-1940 genetic and chemical data were normalised to pre-1940 values from 1923 and 1938 samples.
Mean gene-abundances in soils from before 1940 were as follows (log transformed values; see Tables S1 and S2): 16S rRNA = 9.50 ± 0.08 (±standard deviation), blaTEM = 5.09 ± 0.11, blaSHV = 4.27 ± 0.22, blaOXA = 4.33 ± 0.44, blaCTX-M = 4.61 ± 0.17, and int1 = 3.97 ± 0.24 gene copies g-1-soil dry-weight. From these data and data presented in Tables S1 and S2 (see Supporting Information; SI), relative abundances of blaTEM, blaSHV, blaOXA, blaCTX-M and int1 in post-1940 soils were determined and summarised in Fig. 1. As it can be seen, no major differences in relative abundances of the four ARGs were apparent in either M or IF soils prior to about 1960. However, blaTEM, blaSHV, and blaOXA levels all increased in M soils in 1976 samples, cresting in the mid-1980s, and then declining thereafter. No equivalent increase in ARG levels was seen in IF soils, implying detected increases were specifically associated with long-term manure treatment. Similarly, blaCTX-M levels did not vary over time in either M or IF soils prior to the 1980s, but then increased by almost 15× in M soils in 1988 samples and then progressively declined until 2010. Clearly, manure additions resulted in higher soil ARG levels of all four genes, but increases differed over time among genes. Interestingly, temporal levels of int1 genes in the soils differed somewhat from β-lactam ARGs, with a shallow increase occurring in parallel with increases in blaTEM, blaSHV, and blaOXA levels in the 1970 and 1980s, but then greatly increasing from the early 1990s through 2010.
Figure 1. Relative abundance of blaTEM, blaSHV, blaOXA, blaCTX-M and int1 gene levels in post-1940 archived soils from fields that have only received manure (M) or inorganic fertiliser (IF) applications since 1894.
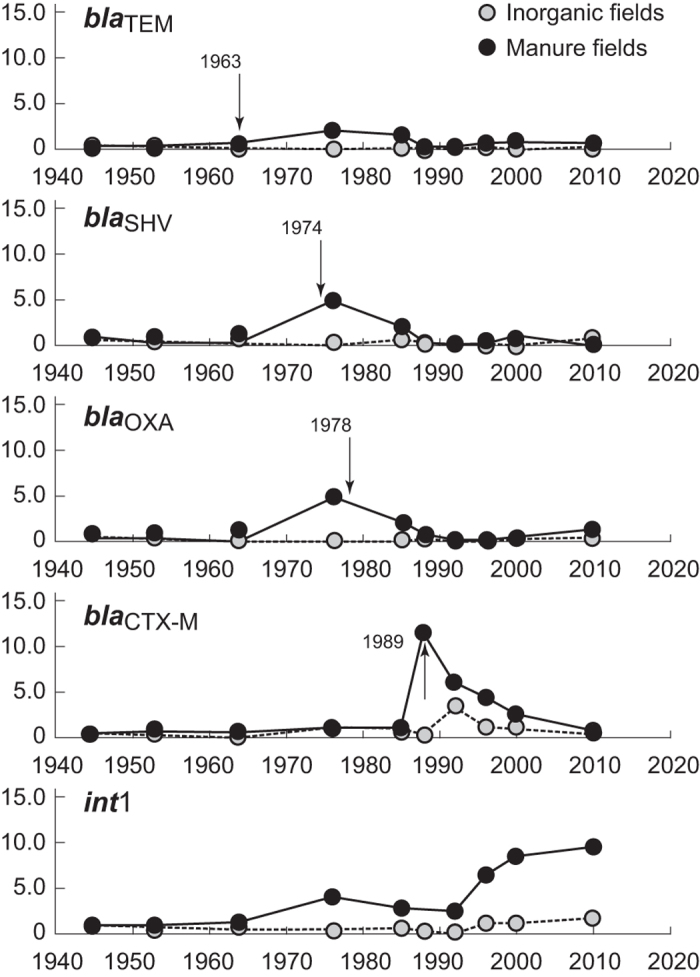
Data are reported as ratios of gene abundances (normalised to 16S-rRNA bacterial levels) in each sample relative to background levels determined from 1923 and 1938 samples. Dashed arrows correspond to dates when each gene was first reported in the literature associated with clinical isolates. Each point typically represents the mean of duplicate analyses for soil archive sample.
Four key questions arise from these data: do ARG levels significantly differ between M and IF soils; have ARGs significantly changed in “background” soils over time (i.e., IF; soil without manure addition); why do ARG patterns differ among genes in the M soils; and has reduced non-therapeutic antibiotic use in Danish agriculture changed soil ARG levels? To address the first question, normalised levels of blaTEM, blaSHV, blaOXA and blaCTX-M were combined for all IF and M samples post-1940 and statistically compared.
Mean “total bla levels” were found to be significantly higher in M than in IF soils (paired t-test; p = 0.001; see Fig. 2). Furthermore, individual bla gene and int1 levels were always greater in M versus IF soils, although differences were not always statistically significant (Wilcoxon Ranked Sum test; <0.05). Statistical comparisons were M > IF for blaTEM, p = 0.092; blaSHV, p = 0.051; blaOXA, p = 0.005; blaCTX-M, p = 0.037; and int1, p = 0.005. Despite these trends among ARGs, the greatest difference in detected gene abundances between M and IF soils was observed for int1 (Fig. 2), although this is not necessarily surprising given that int1 is not specific to β-lactam ARGs, and may be more associated with other factors, such as continued manure application onto the fields.
Figure 2. Mean relative abundances of the sum of all four bla genes, int1, blaTEM, blaSHV, blaOXA, and blaCTX-M in post-1940 archived soils that have received continue manure (M) or inorganic fertiliser (IF) application since 1894.
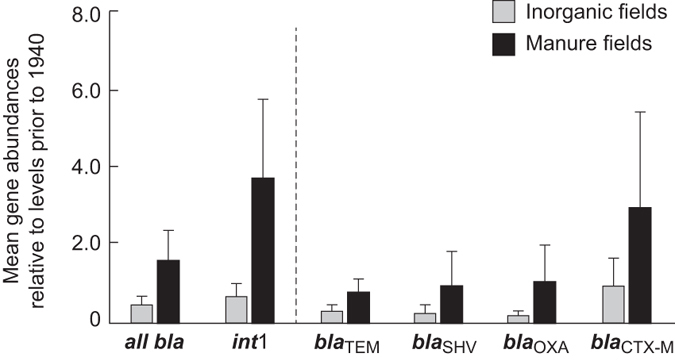
Data are mean ratios of each gene indicator (normalised to 16S-rRNA bacterial levels) in each sample relative to levels determined from 1923 and 1938 samples (n = 9 or 10). Error bars refer to 95% confidence levels in the means.
The second question is more general and relates to changes in ARG levels in M versus IF soils since 1923. This question is best answered using log-transformed mean abundance data behind Fig. 2 (i.e., Tables S1 and S2). If one statistically compares normalised ARG levels (to 16S-rRNA gene abundances) before and after 1940, no significant long-term change in ARGs was apparent in the IF soils (paired t-test; p = 0.912), whereas ARG levels were much higher after 1940 in the M soils, albeit on the fringe of significance (p = 0.063). These data confirm no long-term change in soil β-lactam ARG levels were seen over the last century unless manure was applied. Our data suggest manure use for 100 years has approximately doubled ARG abundances in Lermarken M soils, increasing the probably of broader ARG exposure in drainage water and fodder crops grown in the soils37. However, data also show int1 levels increased by 10× in the M soils since the 1920s, which implies manure application has increased the intrinsic potential of these soils for horizontal gene transfer (HGT). Although it is speculation, we suspect continued manure use (and resultant increased bacterial permissiveness38) may be explain why int1 levels have increased since the mid-1990s despite the Danish ban on non-therapeutic antibiotic use in agriculture. ARG levels would appear to have declined in the manure, but the manure still contains int1 that continues to be released in the soils.
These observations are consistent with previous observations of the impact of manure and wastewater applications on ARG and int1 levels in agricultural fields39, although the impact of 100 years of manure applications has not been reported. Jechalke et al.40 used chronosequence data from different Mexican fields irrigated with wastewater for up to 100 years, and found higher AR and int1 abundances in fields irrigated the longest, although their study did not assess broader spectrum β-lactam ARGs. Further, Udikovic-Kolic et al.41 showed additions of manure increased ARG levels in resident antibiotic-resistant soil bacteria, although their study was relatively short-term. In contrast, we show that prolonged manure additions increase basal levels of β-lactam ARGs and int1 relative to IF applications, although dominant ARGs vary over time (Fig. 1). This implies ARG levels detected at any moment are dominated by more immediate factors, such as prevalent ARGs in “contemporary” manure and other factors that might influence ARG presence or survival in soils42.
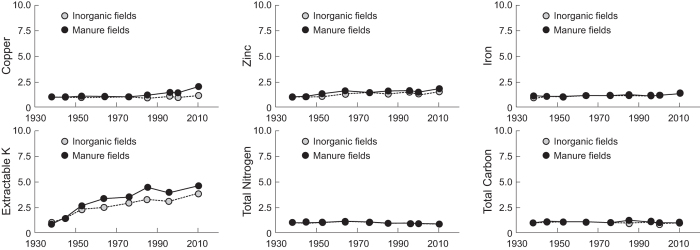
The third and fourth questions are related to factors that might explain the appearance and disappearance patterns of blaTEM, blaSHV, blaOXA and blaCTX-M in the soils. Clearly, co-selection by heavy metals or nutrients in the manure or soil, and/or sub-therapeutic antibiotic exposures are options25,42,43. However, if one examines metal and soil nutrient levels over time (see Figs 3 and S1, and Tables S3 and S4), different temporal patterns are apparent for metals and nutrients compared with β-lactam ARGs. Some bivariate correlations were seen between selected ARGs and soil parameters (see Table S5), but no overarching trends were observed, except isolated correlations in selected M versus IF soils. In fact, with the exception of increasing Cu levels in M soils after 1990, and Zn, P and K levels for longer periods, M and IF soils were historically very similar (see Figs 3 and S1). Temporary increases in Ni, Cr and Hg were seen for short windows in the M soils (see Figure S1), possibly related to co-contamination in the manure with metals from other sources, but these metals did not correlate with detected bla gene or int1 levels, except on an incidental basis. No antibiotics analyses were performed here because such results would be meaningless given long storage time of the archived soils44. Therefore, other explanations are needed for ARG patterns over time.
Figure 3. Relative mass of selected heavy metal and nutrients in archived soils compared with soils from before 1940 from manure (M) and inorganic fertiliser (IF) fields.
Specific metals or nutrients are noted on y-axes and reported as ratios (i.e., mass of metal per date/mean mass of metal in samples from 1923 and 1938 samples).
Although various unknown factors might explain our patterns, we hypothesised observed β-lactam ARG patterns over time might reflect influences of observed AR across broader society, such as antibiotic use across sectors. Unfortunately, until the 1990s when DANMAP was initiated34, little is publically known about absolute antibiotic use in agriculture. Broad and/or extended spectrum β-lactam antibiotics have been used in Denmark since the late 1950s, including agriculture, although anecdotal evidence hint early use of β-lactam antibiotics (e.g., penicillins, cephalosporins, oxyimino β-lactams, etc.)45 was primarily in medicine rather than agriculture. Therefore, to understand ARG patterns in our soils, one must better understand contemporary ARGs being detected in parallel clinical settings.
To test this hypothesis, the literature was surveyed to determine when each β-lactam ARG was first seen in clinical isolates, which is annotated on Fig. 1. As can be seen, the first clinical appearance of the four bla genes assessed here matches appearance data in our soil series. Literature data show the genes sequentially appeared in clinical isolates as follows: blaTEM (1963), blaSHV (1974), blaOXA (1978) and blaCTX-M (1989)18,19,20,21. Although these data are not specific to Denmark, they indicate rough appearance of these ARGs across the developed world because first isolation dates include European examples45,46. Therefore, we suggest detected ARGs in the archived soils reflect a broader expansion of these ARGs across society, implying clinical and agricultural antibiotic and resistance development are mutually influential.
Interestingly, the appearance of each gene over time in the soil samples (i.e., when it first became elevated relative to baseline) is quite consistent with the evolution of resistance development within a medical context. blaTEM and blaSHV code for structurally similar β-lactamases45 and were the dominant form of β-lactam resistance detected prior to 197947. However, as newer antibiotics were developed and employed, the prevalence of OXA-type β-lactamases increased (e.g., coded by blaOXA), and slow substitution of OXA β-lactams occurred in AR strains, possibly because they confer extended spectrum resistance and provide an ecological advantage over strains with blaTEM or blaSHV48. This does not mean blaTEM and blaSHV β-lactamases disappeared because different phenotypes continued to be identified47,49, but their association with resistant infections declined. However, in the late 1980s, CTX-M β-lactamases appeared which are structurally different from previous β-lactam enzymes, have broader spectrum, and have since come to dominate over most other β-lactam enzymes since the late 1990s. This expansion is possibly due to accelerated evolution, movement onto promiscuous plasmids, and-or dispersion of blaCTX-M genes across clinical and environmental settings46. For example, the emergence of CTX-M β-lactamases was simultaneously seen in Europe and South America21 and believed to have migrated on mobile genetic elements from environmental Kluyvera strains to organisms of clinical importance50,51.
Therefore, if manure-borne bla genes in archived soils and parallel clinical resistance are roughly synchronous, the question is which came first: environmental or clinical bla AR. Based on our data, one cannot say. However, if one considers the connectedness of environmental and human-associated AR, including cross-dissemination pathways via exposed water, food (vegetables, milk, meat, etc.) and even humans at the interface between agriculture and clinics, cross-dissemination is probable. In reality, our data suggest such a debate is now moot because these genes have become pervasive across nature. Most importantly, this connectivity suggests one cannot address broader problems of increasing AR by employing only medical, agricultural or environmental solutions because acquired AR (regardless of where it emerges) readily migrates across sectors. Therefore, having an antibiotic treatment fail in hospital may have nothing to do with the acquisition of AR in the hospital.
Despite these pessimistic observations, our results provide room for optimism. Although each bla gene increased in the archived M soils, they all subsequently declined to near baseline levels by 2010, presumably because causes of bla genes being present in the manure declined, possibly due to reduced non-therapeutic use in Danish agriculture, which other AR metrics have shown34,36. Therefore, our data imply reducing non-therapeutic antibiotic use can reduce some environmental AR legacies and an environment can recover given time. However, our data suggest recovery is not universal because int1 genes have increased over the same period. This suggests int1 accumulation may be more associated with manure use on soils rather than specific antibiotic use. Interestingly, we see an increase in Cu levels in the M soils (see Fig. 3), which inversely parallels the decrease in blaCTX-M gene levels, but also significantly correlates with increasing in int1 levels in the M soils (r = 0.673, p = 0.033; see Table S5). These data imply substitution of Cu and possibly Zn has occurred in Danish agriculture, which was recently admitted in new reporting data34. However, it also hints that substitution to Cu/Zn (in lieu of antibiotics) may be responsible for increasing int1 levels in the M fields, although this is not yet proven. Regardless, the value of such corroboration in our results is profound because it confirms that soil archives can tell important contemporary stories and also foretell cause-and-effects that would not be apparent otherwise.
This study provides both optimistic and concerning results. A strong bridge between clinical and agricultural AR is apparent and to reduce globally increasing AR, antibiotic use and stewardship must improve across all sectors. If this is not done, AR from imprudent sectors will cross-contaminate the whole system. Optimistically, we show if one reduces the apparent pressure of antibiotic use in the environment, AR can be reduced (Fig. 1). A good starting point is the elimination of non-therapeutic antibiotic use in agriculture, which Denmark has done. However, recent increases in int1 and metal levels in the archived soils are concerning, and hint we may be solving one problem (reduced antibiotic use) by unintentionally creating another (increasing metals). Reducing non-therapeutic antibiotic use in agriculture is an important step, but one must consider the impact of alternate stewardship options within and across sectors or we will get nowhere relative to reducing AR in the future.
Additional Information
How to cite this article: Graham, D. W. et al. Appearance of β-lactam Resistance Genes in Agricultural Soils and Clinical Isolates over the 20th Century. Sci. Rep. 6, 21550; doi: 10.1038/srep21550 (2016).
Supplementary Material
Acknowledgments
Funding for the project were provided by ECOSERV, an E.U. Marie Curie Excellence Programme (#MEXT-CT-2006-023469), University of Strathclyde Research Development Fund (#2009-1551), and the UK Engineering and Physical Science Research Council (EPSRC EP/I002154/1). The Danish contribution was financially supported by the Ministry of Food, Agriculture and Fisheries.
Footnotes
Author Contributions Soil chemical analysis was performed by B.T.C. and qPCR was performed by C.W.K. and S.M. Data analysis was performed by D.W.G., C.W.K. and J.D. The manuscript was first drafted by D.W.G. and C.W.K. whereas revisions were performed by all authors. Final preparation for submission was done by D.W.G. and C.W.K.
References
- Carlet J. et al. Society’s failure to protect a precious resource: antibiotics. Lancet 378, 369–371 (2011). [DOI] [PubMed] [Google Scholar]
- Davies J. & Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation (WHO). Critically important antimicrobials for human medicine - 3rd revision. (2011). Available at: http://apps.who.int/iris/handle/10665/77376. (Accessed: 13th January 2016).
- Carlet J. et al. Ready for a world without antibiotics? Pensières antibiotic resistance call to action. Antibiotic Resist. Infect. Cont. 1, 11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Science and Technology Commission (STC). POST note 446: Antibiotic resistance in the environment. (2013) Available at: http://researchbriefings.parliament.uk/ResearchBriefings/Summary/POST-PN-446. (Accessed: 13th January 2016).
- Ashbolt N. J. et al. Human Health Risk Assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Persp. 121, 993–1001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley R. L. et al. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57, 704–710 (2013). [DOI] [PubMed] [Google Scholar]
- D’Costa V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011). [DOI] [PubMed] [Google Scholar]
- Forsberg K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M. R. Integrons: Past, Present, and Future. Microbiol. Mol. Biol. Rev. 78, 257–277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Kampfer P. & Nordmann P. Chromosome-encoded ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nature Rev. Microbiol. 5, 175–186 (2007). [DOI] [PubMed] [Google Scholar]
- Wright G. D. Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594 (2010). [DOI] [PubMed] [Google Scholar]
- Dantas G. & Sommer M. O. A. Context matters - the complex interplay between resistome genotypes and resistance phenotypes. Curr. Opin. Microbiol. 15, 577–582 (2012). [DOI] [PubMed] [Google Scholar]
- Kennedy K. & Collignon P. Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1501–1506 (2010). [DOI] [PubMed] [Google Scholar]
- Knapp C. W., Dolfing J., Ehlert P. A. I. & Graham D. W. Evidence of Increasing Antibiotic Resistance Gene Abundances in Archived Soils since 1940. Environ. Sci. Technol. 44, 580–587 (2010). [DOI] [PubMed] [Google Scholar]
- Christensen B. T., Petersen J. & Trentemøller U. The Askov Long-Term Experiments on Animal Manure and Mineral Fertilizers: The Lermarken site 1894–2004. 104 (Danish Institute of Agricultural Sciences, 2006).
- Datta N. & Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208, 239–241 (1965). [DOI] [PubMed] [Google Scholar]
- Matthew M., Hedges R. & Smith J. Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 138, 657–662 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M. Properties of the β-lactamase specified by the Pseudomonas plasmid R151. FEMS Microbiol. Lett. 4, 241–244 (1978). [Google Scholar]
- Bauernfeind A., Grimm H. & Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infect. 18, 294–298 (1990). [DOI] [PubMed] [Google Scholar]
- Christensen B. T. & Johnston A. Soil organic matter and soil quality - lessons learned from long-term experiments at Askov and Rothamsted. Dev. Soil Sci. 25, 399–430 (1997). [Google Scholar]
- Graham D. W. et al. Antibiotic Resistance Gene Abundances Associated with Waste Discharges to the Almendares River near Havana, Cuba. Environ. Sci. Technol. 45, 418–424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp C. W. et al. Seasonal variations in antibiotic resistance gene transport in the Almendares River, Havana, Cuba. Front. Microbiol. 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp C. W. et al. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. Plos One 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gheldre Y., Avesani V., Berhin C., Delmee M. & Glupczynski Y. Evaluation of Oxoid combination discs for detection of extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 52, 591–597 (2003). [DOI] [PubMed] [Google Scholar]
- Haeggman S., Lofdahl S., Paauw A., Verhoef J. & Brisse S. Diversity and evolution of the class a chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 2400–2408 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson N. D. et al. Molecular characterization of a multiply resistant Klebsiella pneumoniae encoding ESBLs and a plasmid-mediated AmpC. J. Antimicrob. Chemother. 44, 377–380 (1999). [DOI] [PubMed] [Google Scholar]
- Birkett C. I. et al. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum beta-lactamases. J. Med. Microbiol. 56, 52–55 (2007). [DOI] [PubMed] [Google Scholar]
- Brinas L. et al. Beta-lactamase characterization in Escherichia coli isolates with diminished susceptibility or resistance to extended-spectrum cephalosporins recovered from sick animals in Spain. Microb. Drug Resist.-Mech. Epidemiol. Dis. 9, 201–209 (2003). [DOI] [PubMed] [Google Scholar]
- Moland E. S. et al. Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 44, 3318–3324 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D., Dickinson D., Webb V. A. & Davies J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44, 1568–1574 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Lee C., Kim J. & Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 (2005). [DOI] [PubMed] [Google Scholar]
- DANMAP (Danish Integrated Antimicrobial Resistance Monitoring Programme). Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. (2014). Available at: http://www.danmap.org/downloads/reports.aspx. (Accessed: 13th January 2016).
- Aarestrup F. et al. Antibiotic use in food-animal production in Denmark. APUA Newsletter 18, 1–3 (2000). [Google Scholar]
- World Health Organisation (WHO). Impact of antimicrobial growth promoter termination in Denmark, 57 (2011). Available at: http://apps.who.int/iris/handle/10665/68357. (Accessed: 13th January 2016).
- Marti R. et al. Impact of Manure Fertilization on the Abundance of Antibiotic-Resistant Bacteria and Frequency of Detection of Antibiotic Resistance Genes in Soil and on Vegetables at Harvest. Appl. Environ. Microbiol. 79, 5701–5709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musovic S., Klümper U., Dechesne A., Magid J. & Smets B. F. Long-term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 6, 125–130 (2014). [DOI] [PubMed] [Google Scholar]
- Heuer H., Schmitt H. & Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243 (2011). [DOI] [PubMed] [Google Scholar]
- Jechalke S. et al. Effects of 100 years wastewater irrigation on resistance genes, class 1 integrons and IncP-1 plasmids in Mexican soil. Front. Microbiol. 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udikovic-Kolic N., Wichmann F., Broderick N. A. & Handelsman J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Nat. Acad. Sci. USA 111, 15202–15207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. & LaPara T. M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1, 191–203 (2007). [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., Wright M. S., Stepanauskas R. & McArthur J. V. Co-selection of antibiotic and metal resistance. Trend. Microbiol. 14, 176–182 (2006). [DOI] [PubMed] [Google Scholar]
- Diaz-Cruz M. S., de Alda M. J. L. & Barcelo D. Environmental behavior and analysis of veterinary and human drugs in soils, sediments and sludge. Trac-Trend. Anal. Chem. 22, 340–351 (2003). [Google Scholar]
- Medeiros A. A. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin. Infect. Dis. 24, S19–S45 (1997). [DOI] [PubMed] [Google Scholar]
- Canton R., Jose M. G. A. & Galan J. C. CTX-M enzymes: origin and diffusion. Front. Microbiol. 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford P. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A. & Munoz-Price L. S. Mechanisms of disease: The new beta-lactamases. New England J. Med. 352, 380–391 (2005). [DOI] [PubMed] [Google Scholar]
- Paterson D. L. et al. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob. Agents Chemother. 47, 3554–3560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decousser J. W., Poirel L. & Nordmann P. Characterization of a chromosomally encoded extended-spectrum class A beta-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45, 3595–3598 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeniuk C. et al. beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46, 3045–3049 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.