Abstract
Poly (ADP-ribose) polymerase (PARP) inhibitors have emerged as promising therapeutics for many diseases, including cancer, in clinical trials1. One PARP inhibitor, olaparib (Lynparza™, AstraZeneca), was recently approved by the FDA to treat ovarian cancer with BRCA mutations. BRCA1 and BRCA2 play essential roles in repairing DNA double strand breaks, and a deficiency of BRCA proteins sensitizes cancer cells to PARP inhibition2,3. Here we show that receptor tyrosine kinase c-Met associates with and phosphorylates PARP1 at Tyr907. Phosphorylation of PARP1 Tyr907 increases PARP1 enzymatic activity and reduces binding to a PARP inhibitor, thereby rendering cancer cells resistant to PARP inhibition. Combining c-Met and PARP1 inhibitors synergized to suppress growth of breast cancer cells in vitro and xenograft tumor models. Similar synergistic effects were observed in a lung cancer xenograft tumor model. These results suggest that PARP1 pTyr907 abundance may predict tumor resistance to PARP inhibitors, and that treatment with a combination of c-Met and PARP inhibitors may benefit patients bearing tumors with high c-Met expression who do not respond to PARP inhibition alone.
Increased levels of reactive oxygen species (ROS) in cells can cause oxidative DNA damage that leads to genomic instability and tumor development4–7. ROS-induced DNA damage, such as single-strand breaks (SSBs), recruits poly (ADP-ribose) polymerase 1 (PARP1) to the lesion sites to orchestrate the DNA repair process through poly-ADP-ribosylation (PARylation) of itself and its target proteins, including histone proteins. PARylated histones destabilize the chromatin structure, allowing the DNA repair machinery to access the damaged DNA site8. Therefore, in theory, inhibiting PARP1 activity would prevent DNA repair and promote death of tumor cells. Tumor suppressors BRCA1 and BRCA2 play essential roles in repairing DNA damage. Notably, mutations in BRCA1 and BRCA2 genes have been associated with increased risk of ovarian and breast cancers9. Interestingly, tumor cells that lack functional BRCA1 or BRCA1 have demonstrated sensitivity to PARP1 inhibition in both pre-clinical and clinical studies2,3,10. PARP inhibitors were therefore initially investigated in clinical trials for both ovarian cancer and triple-negative breast cancer (TNBC), as this tumor type can harbor defective BRCA1 or BRCA211, and in other cancer types1. Recently, olaparib was approved by the FDA to treat BRCA mutant-carrying ovarian cancer12. TNBC is an aggressive subtype of breast cancer and closely related to basal-like breast cancer (BLBC)13 that initially responds to chemotherapy, but a majority of TNBCs eventually develop resistance to chemotherapy. There are no approved targeted therapies to treat TNBC14. While encouraging results were reported in one study of olaparib treatment of TNBC patients carrying tumors with BRCA mutations10, beneficial effects of olaparib treatment were not observed in another cohort15. These discrepant clinical observations raise the important question of how to increase the response rate of TNBC—and other cancer types—to PARP inhibitors. To address this question, we investigated the molecular mechanisms contributing to PARP inhibitor resistance in TNBC.
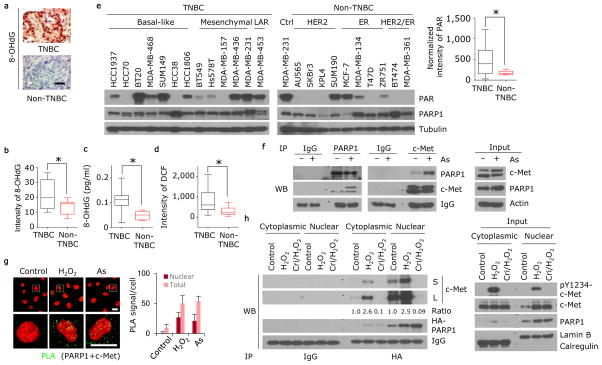
We first noticed that TNBC had higher oxidative damaged DNA than non-TNBC as indicated by immunohistochemical staining for the DNA damage marker 8-hydroxydeoxyguanosine (8-OHdG) on a human breast cancer tissue microarray (Fig. 1a and Supplementary Table 1) and in human breast cancer cell lines (Fig. 1b,c and Supplementary Fig. 1a) by immunofluorescence staining (1.9-fold difference TNBC vs non-TNBC, 95% confidence interval [CI] = 1.6–2.2) and ELISA assay (2.1-fold difference TNBC vs non-TNBC, 95% CI = 1.8–2.4). Oxidative DNA damage caused by ROS stimulates the activity of PARP116–20. In accordance with this, the abundance of ROS (Fig. 1d and Supplementary Fig. 1b,c, measured by the marker 2′,7′-dichlorofluorescein (DCF; intensity: 2.6- fold difference TNBC vs non-TNBC, 95% CI = 1.9–3.3; absorbance 1.33-fold difference, 95% CI = 1.3–1.4) and the level of PARP1 activity (Fig. 1e, right), measured by poly(ADP)-ribose (PAR; 2.7-fold difference TNBC vs non-TNBC, 95% CI = 2.3–3.2), were higher in most TNBC cell lines than in non-TNBC cell lines, suggesting a positive association between ROS and PARP1 activity in TNBC.
Figure 1. ROS induces the association of c-Met and PARP1.
(a) Human breast cancer tissue microarray was stained with 8-OHdG-specific antibody. Representative images of 216 non-TNBC and 90 TNBC cases are shown. Bar, 100 μm. (b) Human breast cancer cell lines shown in panel (e) were stained with 8-OHdG-specific antibody (see Supplementary Fig. 1a). Quantitation of 8-OHdG is shown. (c) Human breast cancer cell lines shown in panel (e) were subjected to ELISA assay to measure 8-OHdG abundance. (d) Human breast cancer cell lines shown in panel (e) were incubated with 10 μM of DCF-DA for 30 min. Quantitation of DCF is shown. (e) Western blot showing expression of PAR, PARP1, and tubulin in lysates of the indicated human breast cancer cell lines. Blots are representative of triplicate experiments. Right, band intensity of PAR normalized to tubulin. (f) MDA-MB-231 cells were treated with or without 20 μM sodium arsenite for 18 h. Left, endogenous PARP1 and c-Met association detected by immunoprecipitation (IP) and Western blot. Right, input control. (g) Detection of PARP1 and c-Met co-localization (green signals) in MDA-MB-231 cells treated with H2O2 or sodium arsenite (As), and in those cells not treated (control) by a Duolink assay. Bar, 20μm. Representative images and quantitation of PLA signals from 50 cells and three independent experiments are shown. (h) MDA-MB-231 cells with ectopic expression of HA-tagged PARP1 were treated with 20 μM H2O2 for 30 min with or without a one-hour pre-treatment with 2 μM c-Met inhibitor crizotinib. Left, IP and Western blot analysis of cytosolic and nuclear fractions with the indicated antibodies. The treatment-to-control ratio of c-Met/PARP1 interaction is shown below. Right, input control. TNBC, triple-negative breast cancer. LAR, luminal androgen receptor; As, sodium arsenite; L, long exposure; S, short exposure. Error bars represent s.d. *P < 0.05, t-test.
ROS is also known to activate receptor tyrosine kinases (RTKs)21, which are druggable targets commonly overexpressed in TNBC22–24. To investigate the underlying molecular mechanisms regulating PARP1 response under ROS-induced oxidative stress and identify potential targets, we searched for RTKs that associate with PARP1 upon ROS stimulation. To this end, PARP1-knockdown MDA-MB-231 TNBC cells re-expressing HA-tagged PARP1 were treated with sodium arsenite to induce ROS production, and a human phospho-RTK antibody array analysis was performed on those whole cell lysates to determine the specific activated PARP1-interacting RTKs by an HA antibody. The top three candidates—defined according to the ratio of density of binding in sodium arsenite compared to control treated cells—were ERBB3, HGFR, and FLT3 (Supplementary Table 2). Analysis of the TCGA breast invasive carcinoma cohort (Supplementary Fig. 2a) indicated that only HGFR (encoding c-Met) expression was significantly higher (P = 1e-10) in TNBC than in non-TNBC (Supplementary Fig. 2b).
c-Met is a proto-oncogene, and c-Met expression correlates with poor survival of patients with TNBC23–25. We detected higher expression of c-Met in TNBC cell lines than in non-TNBC cell lines (Supplementary Fig. 2c). We next validated that c-Met and PARP1 co-immunoprecipitate in HEK293T cells (Supplementary Fig. 3a,b) and in MDA-MB-231 cells (Fig. 1f and Supplementary Fig. 3c). The interaction between c-Met and PARP1 was also detected in other human breast cancer cell lines, such as HCC1937 (endogenous, Supplementary Fig. 3d), and MDA-MB-436 and MCF-7 (with ectopic expression of c-Met, Supplementary Fig. 3e,f) in conditions of oxidative stress induced by H2O2 treatment. Because c-Met has been detected in the nucleus26,27 and PARP1 is a nuclear protein, we asked whether the c-Met-PARP1 interaction also occurs in the nucleus. Cellular fractionation analysis indicated that about 20–30% and 10–20% of total c-Met translocated into the nucleus upon H2O2 and sodium arsenite treatment, respectively (Supplementary Fig. 3g,h). Moreover, H2O2 and sodium arsenite treatment enhanced the interaction between c-Met and PARP1 in both the cytosol and nucleus as shown by a Duolink assay (Fig. 1g). As shown by treatment of MDA-MB-231 cells with the c-Met inhibitor crizotinib, the kinase activity of c-Met was required for the interaction between c-Met and PARP1, which was enhanced by H2O2 treatment (Fig. 1h). Nuclear trafficking of RTKs, including EGFR, from the cell surface has been proposed to utilize a vesicle membrane-associated pathway28–30, requiring the motor protein dynein and the SNARE protein syntaxin 631. Nuclear translocation of c-Met in response to H2O2 stimulation also required dynein and syntaxin 6 (Supplementary Fig. 3i,j), suggesting that c-Met might use a similar trafficking route. Together, these findings indicated that oxidative stress induces nuclear transport of c-Met and its interaction with PARP1.
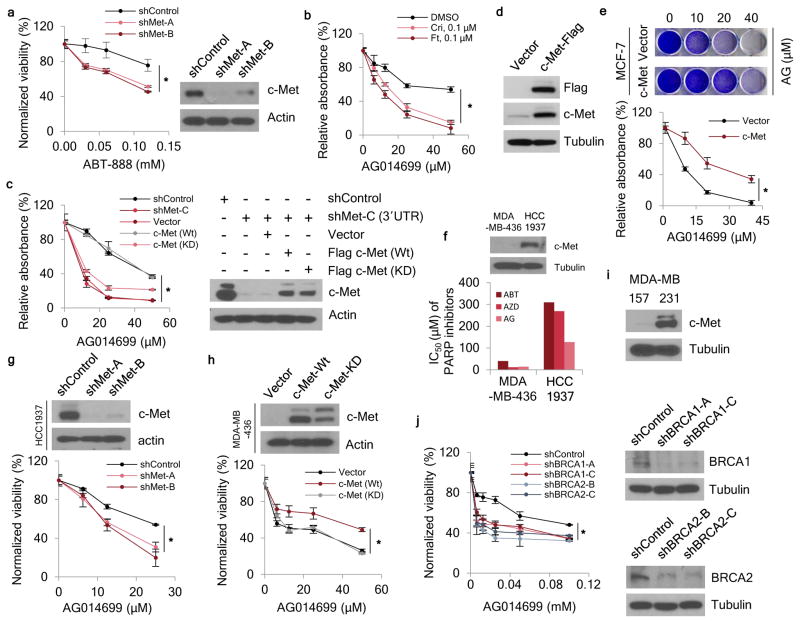
To determine whether c-Met influences tumor response to PARP inhibition, we examined TNBC cell line growth and colony formation in the presence of three different PARP inhibitors: the US FDA-approved olaparib (AZD2281), and veliparib (ABT-888) and rucaparib (AG014699), which are under evaluation in clinical trials32. shRNA-mediated knockdown c-Met expression rendered MDA-MB-231 cells more sensitive to all three PARP inhibitors, as indicated by decreased cell viability (Fig. 2a and Supplementary Fig. 4a–c). For example, c-Met knockdown cells showed 4.2- (shMet-A; 95% CI = 4.0–4.5) or 4.6-fold (shMet-B; 95% CI = 4.4–4.8) growth inhibition when treated with 60 μM ABT-888. Treatment with c-Met inhibitors crizotinib or foretinib also enhanced MDA-MB-231 sensitivity to the PARP inhibitors (Fig. 2b and Supplementary Fig. 4d,e); anchorage-independent cell growth also decreased when c-Met was knocked down (Supplementary Fig. 4f–h). Consistent with previous findings33, inhibition of c-Met either by shRNAs or small molecules reduced ROS abundance (Supplementary Fig. 4i,j), suggesting that a feed-forward mechanism regulating c-Met activation and ROS may be involved in the response to PARP1-mediated DNA damage and PARP inhibitor.
Figure 2. c-Met regulates resistance to PARP inhibitors.
(a) Left, c-Met-knockdown cells were treated with the indicated concentrations of ABT-888 for 72 h and subjected to a cell viability assay. Right, Western blot showing c-Met expression in c-Met-knockdown MDA-MB-231 cells. (b) MDA-MB-231 cells were treated with the indicated concentrations of AG014699 and crizotinib or foretinib for 8 days and subjected to clonogenic cell survival assay. Quantitation of clonogenic cells from three independent experiments is shown. (c) Left, c-Met-knockdown MDA-MB-231 cells were treated with the indicated concentrations of AG014699 for 8 days and subjected to clonogenic cell survival assay. Quantitation of clonogenic cell from three independent experiments is shown. Right, Western blot showing c-Met expression in c-Met- knockdown cells at the 3′-UTR (shMet-C) and re-expression of wild-type (Wt) and kinase dead (KD) c-Met in MDA-MB-231 cells. (d) Western blot analysis of c-Met expression in MCF-7 cells. (e) MCF-7-c-Met and vector control cells were treated with the indicated concentrations of AG014699 for 8 days and subjected to clonogenic cell survival assay. Quantitation of clonogenic cells from three independent experiments is shown. (f) Median inhibitory concentration (IC50) of PARP inhibitors in BRAC1-mutant TNBC cells (MDA-MB-436 and HCC1937). Top, Western blot showing c-Met expression. (g) c-Met-knockdown HCC1937 cells were treated with the indicated concentrations of AG014699 for 72 h and subjected to cell viability assay. Top, Western blot showing c-Met expression in c-Met-knockdown HCC1937 cells. (h) MDA-MB-436 cells with ectopic expression of c-Met were treated with the indicated concentrations of AG014699 for 72 h and subjected to cell viability assay. Top, expression of wild-type (Wt) and kinase dead (KD) c-Met in MDA-MB-436 cells. (i) Western blot showing expression of c-Met in MDA-MB-231 and MDA-MB-157 cells. (j) BRCA1- and BRCA2-knockdown MDA-MB-157 cells were treated with the indicated concentrations of AG014699 for 72 h and subjected to cell viability assay. Right, Western blot showing c-Met expression in c-Met-knockdown MDA-MB-157 cells. Error bars represent s.d. Cri, crizotinib; Ft, foretinib; ABT, ABT-888; AG, AG014699; AZD, AZD2281. *P < 0.05, rANOVA.
To further investigate the function of c-Met during responses to PARP inhibitors, we re-expressed wild-type and kinase dead mutant c-Met in MDA-MB-231 cells subjected to c-Met shRNA-mediated knockdown (Fig. 2c, right); re-expression of wild-type but not kinase dead c-Met increased the cell survival (Fig. 2c, left and Supplementary Fig. 4k). Similarly, MCF-7 cells ectopically expressing c-Met had increased cell viability (Fig. 2d and Supplementary Fig. 5a–c), clonogenicity (Fig. 2e and Supplementary Fig. 5d), and anchorage-independent cell growth (Supplementary Fig. 5e,f) in the presence of PARP inhibitors. Of note, the doses used here for the in vitro assays were comparable to those used in previous studies14,34. Together, these results indicated that c-Met activity attenuates response to PARP inhibitors.
While BRCA mutations and loss of BRCA are thought to be the predictive markers for response to PARP inhibitors in ovarian and breast cancers2,3, a certain percentage of patients carrying BRCA mutations do not respond to PARP inhibition based on the reported objective response10,15. In agreement with these clinical findings, although both breast cancer cell lines, MDA-MB-436 and HCC1937, harbor BRCA mutations, MDA-MB-436 cells are sensitive and HCC1937 cells are resistant to PARP inhibition14. We speculated that the differences in PARP-inhibitor response observed in BRCA-mutated TNBC cells may be attributed to different expression of c-Met. Indeed, Western blot analysis indicated that HCC1937 cells, which expressed higher levels of c-Met than MDA-MB-436 cells (Fig. 2f, top), were also more resistant to PARP inhibition (Fig. 2f, bottom), and knocking down c-Met rendered HCC1937 cells more sensitive to PARP inhibition (Fig. 2g and Supplementary Fig. 6a,b). For example, when treated with 38 μM ABT-888, c-Met knockdown cells showed 2- (shMet-A; 95% CI = 1.5–2.5) or 1.9-fold (shMet-B; 95% CI = 1.3–2.5) growth inhibition. In contrast, increasing the ectopic expression of wild-type but not kinase dead mutant c-Met in MDA-MB-436 cells attenuated the effects of PARP inhibition on cell viability (Fig. 2h and Supplementary Fig. 6c,d). Knockdown or ectopic expression of c-Met had no effect on the abundance of BRCA1 and BRCA2 proteins (Supplementary Fig. 6e,f).
To further investigate the relationship between BRCA1, BRCA2 and c-Met, we knocked down BRCA1 and BRCA2 expression in a pair of wild-type-BRCA1 and -BRCA2 cell lines with high c-Met (MDA-MB-231) and low c-Met (MDA-MB-157) expression (Fig. 2i) and treated them with PARP inhibitors. Knocking down BRCA1 or BRCA2 sensitized only MDA-MB-157 cells expressing low levels of c-Met (Fig. 2j and Supplementary Fig. 6g–l). Collectively, these results suggest that enhanced expression of c-Met kinase renders cells resistant to PARP inhibitors in the context of BRCA inactivation, and provide a potential molecular explanation for discrepant clinical results.
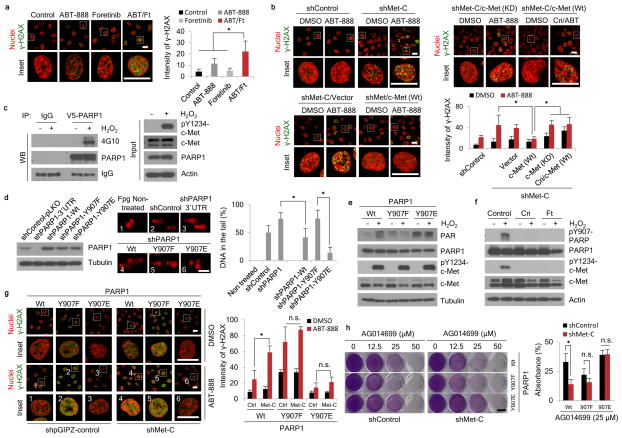
To address whether c-Met activates PARP1, we exposed MDA-MB-231 cells expressing control shRNA or c-Met shRNA to H2O2 and subjected them to a comet assay to evaluate the extent of DNA damage. c-Met-knockdown cells had higher tail intensity, which is indicative of increased oxidative DNA damage, than control cells (Supplementary Fig. 7a). Knockdown of c-Met also reduced their DNA repair activity, as measured by oxidative DNA damage (Supplementary Fig. 7b). Consistent with the shRNA results, inhibition of c-Met by foretinib increased the sensitivity of cells to PARP inhibitor-induced DNA damage, as indicated by enhanced γ-H2AX foci formation (Fig. 3a). DNA repair also required the kinase activity of c-Met as expression of wild-type but not kinase dead c-Met in MCF-7 cells reduced H2O2-induced DNA damage; this was restored by pre-treatment with a c-Met inhibitor (Supplementary Fig. 7c–e). Ectopic expression of c-Met in MCF-7 cells decreased ABT-888-induced γ-H2AX foci formation (Supplementary Fig. 7f). MDA-MB-231 cells expressing c-Met shRNA had higher γ-H2AX foci formation than those with vector control after ABT-888 treatment (Fig. 3b, top, left); re-expression of wild-type c-Met but not re-expression of vector control (Fig. 3b, bottom, left), kinase dead c-Met, or wild-type c-Met plus pre-treatment with c-Met inhibitor crizotinib restored this (Fig. 3b, top, right). These findings together suggest that c-Met kinase activity enhances the DNA repair function of PARP1.
Figure 3. c-Met mediates PARP1 functions through phosphorylation of PARP1 at Y907.
(a) MDA-MB-231 cells were treated with ABT-888 (50 μM), foretinib (1 μM), or the combination for 18 h. Representative images and quantitation of γ-H2AX (green) from three independent experiments are shown. Bar, 20 μm. (b) c-Met- or control-knockdown MDA-MB-231 cells as well as c-Met-knockdown MDA-MB-231 cells re-expressing wild-type (Wt) or kinase dead (KD) mutant were treated with the indicated drugs for 18 h. Representative images and quantitation of γ-H2AX (green) from three independent experiments are shown. Bar, 20 μm. (c) HEK293T cells were transfected with V5-PARP1 and Flag-c-Met expression plasmids, and the cells were treated with 10 μM H2O2 for 15 min. PARP1 was immunoprecipitated with V5 antibody, followed by Western blotting with 4G10 (anti phosphor-tyrosine antibody). (d) Left, Western blot showing expression of PARP1 and tubulin in PARP1-knockdown (shRNA targeting 3′-UTR) MDA-MB-231 cells and PARP1-knockdown MDA-MB-231 cells re-expressing PARP1 wild-type or Y907 mutant. Center, DNA damage as measured by comet assay with pre-incubation with formanidopyrimidine DNA glycosylase (Fpg) in PARP1-wild-type-, PARP1-Y907E-, PARP1-Y907F-expressing MDA-MB-231 stable cells treated with 20 μM H2O2 for 30 min. Right, quantitation of intensity of damaged DNA from three independent experiments. Bar, 100 μm. (e) Western blot showing poly-ADP ribosylation as indicated by PAR in MDA-MB-231 stable cells described in (d) treated with or without 20 μM H2O2 for 30 min. (f) MDA-MB-231 cells were treated with or without 20 μM H2O2 for 30 min or H2O2 plus 2 μM crizotinib or 1 μM foretinib pre-treatment 1h. Cell lysates were subjected to Western blot analysis using the indicated antibodies. (g) Re-expression of wild-type or mutant PARP1 in PARP1-knockdown MDA-MB-231 cells with or without c-Met knockdown were treated with 50 μM ABT-888 for 18 h. γ-H2AX (green) was detected by immunofluorescence confocal microscopy. Bar, 20 μm. Representative images and quantitation of three independent experiments are shown. (h) Stable cells described in (g) were treated with the indicated concentrations of AG014699 and subjected to clonogenic formation assay for 8 days. Representative images and quantitation of clongenic cells from three independent experiments are shown. Bar, 10 mm. Error bars represent s.d. *P < 0.05, t-test. n.s., not significant. ABT, ABT-888; Cri, crizotinib; Ft, foretinib.
Given that c-Met and PARP1 physically associate in vivo (Fig. 1f,g and Supplementary Fig 3a–f), we speculated that c-Met could phosphorylate PARP1 under oxidative stress. Indeed, in HEK293T cells expressing Flag-tagged c-Met and V5-tagged PARP1, H2O2 induced PARP1 tyrosine phosphorylation (Fig. 3c). The software program NetworKIN (V2.0)35 predicted that Tyr907 (Y907), which is located on the H-Y-E motif in the catalytic domain of PARP136, is the c-Met phosphorylation site. An in vitro kinase assay showed that compared to wild-type PARP1, phosphorylation, as read out by γ-32P incorporation, was substantially reduced in the Y907F mutant but not in PARP1 bearing mutation of Y986, another Tyr residue in the H-Y-E domain (Supplementary Fig. 8a,b). These results suggest that Y907 is a bona fide c-Met phosphorylation site.
Since Y907 is located within the catalytic domain of PARP1, we next asked whether Y907 phosphorylation affects the function of PARP1. We stably expressed wild-type, Y907F (non-phosphorylatable), or Y907E (phosphomimetic) mutant PARP1 in PARP1-knockdown MDA-MB-231 cells (Fig. 3d, left) and measured H2O2-induced DNA damage by comet assay. PARP1 knockdown cells had more DNA damage than control cells (Fig. 3d, center and right). Re-expression of wild-type PARP1 but not the Y907F mutant reduced DNA damage, and cells expressing the Y907E mutant had the least amount of DNA damage (Fig. 3d, right). To determine whether phosphorylation of PARP1 at Y907 affects its activity, we compared the PARylation (PAR) levels in MDA-MB-231 expressing wild-type and mutant PARP1. Cells expressing wild-type PARP1 had increased PAR in response to H2O2 (Fig. 3e). Cells expressing the phosphomimetic Y907E mutant had higher levels of PAR than the non-phosphorylatable Y907F mutant; however, both mutants were no longer sensitive to the H2O2 treatment (Fig. 3e). To further investigate the functional importance of PARP1 phosphorylation at Y907, we generated an antibody to specifically detect phosphorylated Y907 (pY907) (Supplementary Fig. 8c–g). Treatment with either crizotinib or foretinib abolished H2O2-induced phosphorylation of PARP1 at Y907 (Fig. 3f). These results suggest that H2O2-induced wild-type PARP1 activity requires Y907 phosphorylation.
We then asked whether c-Met-mediated phosphorylation of Y907 of PARP1 affects PARP inhibitor response. MDA-MB-231 cells expressing wild-type or mutant PARP1 were treated with or without H2O2 and/or increasing concentrations of ABT-888 and then subjected to a PARP enzyme activity assay to measure the median inhibitory concentration (IC50) of ABT-888. The activity of the phosphomimetic Y907E mutant was similar to that of wild-type PARP1 treated with H2O2 (higher IC50) whereas the activity of the non-phosphorylatable Y907F mutant was similar to that of wild-type PARP1 without H2O2 (lower IC50) (Supplementary Fig. 8h). In addition, we measured the direct binding of wild-type and mutant (Y907F and Y907E) PARP1 to ABT-888 by an in vitro isothermal titration calorimetry (ITC) assay (Supplementary Fig. 8i). The results indicated a higher Kd value for the PARP1 Y907E mutant than either the wild-type or Y907F mutant, suggesting that phosphorylated PARP1 exhibited a lower binding affinity for ABT-888 than the non-phosphorylated form. Together, these results indicate that phosphorylation of PARP1 at Y907 attenuates the inhibitory effect of ABT-888.
Next, in MDA-MB-231 cells expressing PARP1 shRNA, we re-expressed wild-type, Y907F or Y907E mutant PARP1 (Supplementary Fig. 8j) and subjected them to control or c-Met shRNA knockdown and/or ABT-888 treatment. We then evaluated the extent of DNA damage by γ-H2AX foci formation (Fig. 3g). Knocking down c-Met sensitized cells to ABT-888-induced DNA damage in cells expressing wild type PARP1, but did not affect DNA repair in cells expressing Y907F or Y907E mutant PARP1. We observed similar results in clonogenic cell survival (Fig. 3h) and cell viability assays (Supplementary Fig. 8k,l), and using the inhibitor AG014699 instead of ABT-888.
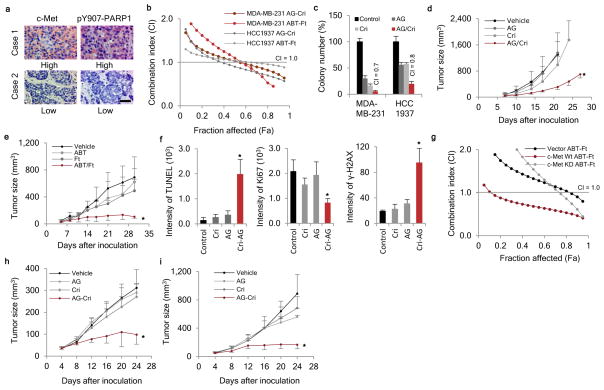
To evaluate the clinical relevance of our findings, we validated the specific antibody against pY907-PARP1 in formalin-fixed paraffin-embedded (FFPE) tumor tissues obtained from breast cancer patients (Supplementary Fig. 9a). Next, we used this antibody to measure pY907-PARP1 abundance in a human breast cancer tissue microarray by immunohistochemical (IHC) staining and observed a positive correlation between pY907-PARP1 and c-Met expression in both TNBC (Fig. 4a and Supplementary Table 3) and non-TNBC (Supplementary Fig. 9b). High ROS (8-OHdG) also correlated with high pY907-PARP1 abundance (Supplementary Fig. 9c). These results suggest that intracellular ROS may induce phosphorylation of PARP1 at Y907 in a c-Met-dependent manner.
Figure 4. Clinical relevance and potential therapeutic strategy targeting PARP1 and c-Met in TNBC.
(a) Representative images of immunohistochemical staining for pY907-PARP1 and c-Met in tissue microarrays of 77 cases of breast cancer (see Supplementary Table 3). (b) The synergistic effect of inhibiting c-Met and PARP in TNBC cell lines (MDA-MB-231 and HCC1937) was measured by cell viability assay following a 72-hour treatment. Fa, fraction affected. AG, AG014699; ABT, ABT-888; Cri, crizotinib; Ft, foretinib. (c) The synergistic effect of c-Met inhibitor crizotinib and PARP inhibitor AG014699 in MDA-MB-231 cells and HCC1937 cells was measured by soft agar assay following a 4-week treatment. (d) MDA-MB-231 cells were inoculated into the mammary fat pad of nude mice (10 mice per group) on day 0. When the tumor volume reached ~50 mm3, mice were orally administered crizotinib (5 mg/kg), AG014699 (5 mg/kg), or the combination five times per week for 21 days. Tumor volume was measured at the indicated time points. (e) MDA-MB-231 cells were inoculated into the mammary fat pad of nude mice (10 mice per group) on day 0. When the tumor volume reached ~50 mm3, mice were orally administered foretinib (5 mg/kg), ABT-888 (25 mg/kg), or the combination five times per week for 26 days. Tumor volume was measured at the indicated time points. (f) TUNEL, Ki67, and γ-H2AX staining of MDA-MB-231 xenograft tumor tissues after treatment. (g) The synergistic effect of c-Met and PARP inhibition on MCF-7/vector, MCF-7/c-Met wild-type, or MCF-7/c-Met KD cells was measured by cell viability assay following a 72-hour treatment. (h) MCF-7 cells with ectopic expression of c-Met were inoculated into the mammary fat pad of nude mice (10 mice per group) on day 0. When the tumor volume reached ~50 mm3, mice were orally administered crizotinib (5 mg/kg), AG014699 (5 mg/kg), or the combination five times per week for 21 days. Tumor volume was measured at the indicated time points. (i) H1993 cells were injected subcutaneously into the right flank of female nude mice (10 mice per group) on day 0. When the tumor volume reached ~50 mm3, mice were orally administered crizotinib (5 mg/kg), AG014699 (5 mg/kg), or the combination five times per week for 21 days. Tumor volume was measured at the indicated time points. Error bars represent s.d. *P < 0.05, t-test.
Next, we examined the effects of combining c-Met inhibitors (foretinib and crizotinib) and PARP inhibitors (ABT-888 and AG014699). Both the ABT-888-foretinib and AG014699-crizotinib combinations demonstrated synergistic cell growth inhibition in MDA-MB-231 and HCC1937 TNBC cells (Fig. 4b) but not in MCF10A mammary epithelial cells (Supplementary Fig. 10a). The combined treatment of AG014699 and crizotinib also synergized to suppress clonogenicity (Supplementary Fig. 10b,c) and anchorage-independent growth (Fig. 4c and Supplementary Fig. 10d). Similar inhibitory effects on clonogenic cell survival were observed for the ABT-888-foretinib combination (Supplementary Fig. 10e). Synergistic inhibition of c-Met and PARP1 was also observed in another breast cancer cell line, BT549 (Supplementary Fig. 10f). H2O2-induced phosphorylation of Y907-PARP1 was abolished by c-Met inhibition (Supplementary Fig. 10g). In addition to human breast cancer cell lines, we evaluated the effect of the combination treatment in two mouse mammary tumor cell lines derived from a TNBC transgenic mouse model expressing constitutively active human c-Met37. Combined treatment with c-Met and PARP inhibitors synergistically inhibited mouse tumor cell growth (Supplementary Fig. 10h,i). Also, pY907-PARP1 was stimulated by H2O2 and abolished by c-Met inhibition in these mouse cell lines (Supplementary Fig. 10j).
We also evaluated the effect of combining PARP and c-Met inhibitors in vivo in established TNBC xenograft models. In MDA-MB-231 xenograft tumor models, combination treatment (AG014699/crizotinib and ABT-888/foretinib) substantially reduced tumor growth compared to either inhibitor alone (Fig. 4d,e and Supplementary Fig. 11a,b). The AG014699-crizotinib combination also inhibited growth of mouse mammary tumor cells (A1034) in a syngeneic FVB mouse model and in an HCC1937 TNBC xenograft tumor model (Supplementary Fig. 11c,d). Increased apoptosis (TUNEL staining), reduced cell proliferation (Ki67 staining) and greater DNA damage (γ-H2AX staining) were observed in MDA-MB-231 xenograft tumor tissues harvested from mice within 24 hours after the last treatment (Fig. 4f, Supplementary Fig. 11e). In addition, the overall health of the animals was not adversely affected by the AG014699-crizotinib or ABT-888-foretinib combination compared to non- or single treatment (clinical chemistry analysis and body weight; Supplementary Fig. 11f–l).
Because PARP inhibitors have been used in clinical trials for multiples cancer types, including lung cancer, we also tested a non-TNBC cell line (MCF-7 with ectopic expression of c-Met) and two lung cancer cell lines (H1993 and A549) in vitro and in vivo. Synergistic inhibition of cell growth was observed in the MCF7/c-Met and H1993 cells (high c-Met expression) but not in MCF7/control, MCF7/c-Met KD, or A549 (low c-Met expression) cells (Fig. 4g and Supplementary Fig. 12a,b). c-Met inhibitor pre-treatment abolished H2O2-induced pY907-PARP1 in both MCF-7/c-Met and in H1993 cells (Supplementary Fig. 12c,d). Furthermore, the combined treatment of AG014699 and crizotinib demonstrated significant anti-tumor activity in MCF/c-Met breast cancer and H1993 lung cancer xenograft tumor models (Fig. 4h,i).
Taken together, our study revealed that c-Met-phosphorylated PARP1 at Y907 leads to PARP inhibitor resistance (Supplementary Fig. 12e) and identified c-Met as an important regulator of PAPR inhibitor response, suggesting that pY907-PARP1 may be a useful marker to stratify patients for PARP inhibitor treatment alone or in combination with a c-Met inhibitor. Of note, many studies have found aberrant c-Met activation and increased expression of c-Met in TNBC tumors38. Interestingly, we observed positive correlation between c-Met and pY907-PARP expression in TNBC patient samples (Fig. 4a). Based on our findings, about one-third (24/77 in Fig. 4a and Supplementary Table 3) of TNBC patients positive for pY907/c-Met positive would likely be resistant to PARP inhibitor alone and could benefit from the combined therapy of c-Met and PARP inhibition using pY907/c-Met as biomarkers.
It should be mentioned that the combined inhibition of EGFR and PARP induces synthetic lethality in TNBC39. However, the underlying mechanism of this combination is not yet clear, and given that EGFR was also identified from our phospho-RTK antibody array analysis, it is conceivable that EGFR may induce resistance to PARP inhibitors through a similar mechanism in a subpopulation of patients who do not respond to PARP inhibition. It will be important to further investigate the relationship between PARP1 and EGFR and between PARP1 and phosphatases that can dephosphorylate pY907 or other functionally important phosphorylation sites of PARP1. Moreover, investigating whether other protein kinases also regulate PARP inhibitor response may reveal a new perspective toward the development of combination therapy strategies to benefit a broader population of patients.
PARP inhibitors are being used in clinical trials for many cancer types in addition to TNBC1. c-Met is a proto-oncogene overexpressed in multiple cancer types40. Although we initiated our study using TNBC samples for historical (original synthetic lethality) and clinical (no effective target therapy for TNBC in the clinic) reasons, we also demonstrated that the combined treatment of c-Met and PARP inhibitors effectively reduced tumor growth in MCF-7/c-Met (Fig. 4h) and c-Met-expressing H1993 NSCLC xenograft tumor models (Fig. 4i). These results raise the interesting possibility that cancer patients with tumors that overexpress c-Met may benefit from this combination therapy regardless of the cancer type. Thus, it may be worthwhile to systematically test whether combined inhibition of both PARP and c-Met also exhibits synergistic therapeutic effects in other types of tumors.
Online Methods
Chemicals and antibodies
Hydrogen peroxide (#216763), cycloheximide (#C4859) and sodium arsenite solution (#35000) were obtained from Sigma-Aldrich (St. Louis, MO). Antibodies detecting tubulin (#T5168), flag (#F3165) and actin (#A2066) were also from Sigma-Aldrich (St. Louis, MO). Antibodies detecting γ-H2AX (#05–636) and phosphotyrosine (#05–321, 4G10) were from EMD Millipore (Billerica, MA). Antibodies detecting GST fusion protein (#sc-53909), HA-tag (#sc-805) and PARP1 (#sc-7150) for Western blot were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against PARP1 (#9532) for immunoprecipitation (IP) and for detecting cMet (#8198) and phosphorylated c-Met (#3077) were from Cell Signaling Technology (Danvers, MA). Antibody against 8-hydroxy-2′-deoxy guanosine (8-OHdG) was obtained from Genox Corporation (Baltimore, MD). All fluorescence-labeled secondary antibodies were obtained from Invitrogen (Carlsbad, CA). The mouse phospho-Y907-PARP1 antibody was generated against a phosphorylated synthetic peptide (ADMVSKSAN-Yp-CHTSQGD) at China Medical University, Center of Molecular Medicine. The horseradish peroxidase (HRP)-conjugated secondary antibodies for Western blotting were obtained from eBioscience (San Diego, CA); All primary antibodies were used according to the manufactory datasheet, or diluted to 1:1000 for Western blot or 1:100 for IP/Western. For secondary antibody, a 1:5000 dilution was used. c-Met kinase inhibitors crizotinib (#C-7900) and foretinib (#F-4185) were from LC Laboratories (Woburn, MA). PARP inhibitors ABT-888 (Veliparib, #CT-A888) and AG014699 (Rucaparib, #CT-AG01) were from ChemieTek (Indianapolis, IN); AZD2281 (Olaparib, #S1060) was from Selleck Chemicals (Houston, TX).
Cell culture
All cells lines were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F12 or RPMI-1640 supplemented with 10% fetal bovine serum and antibiotics. A1034 and A1471 mouse cell lines were previously described37. Cell lines were validated by short tandem repeat (STR) DNA fingerprinting using the AmpFLSTR® Identifiler® PCR Amplification Kit (Life Technologies Grand Island, NY). The STR profiles were compared with ATCC fingerprints and the Cell Line Integrated Molecular Authentication database.
Plasmids and transfection
For stable knockdown of c-Met or PARP1 and c-Met or PARP1 overexpression studies, breast cancer cells were transfected with pGIPZ shRNA (control) vector (Thermo Fisher Scientific, Rockford, IL) or pLKOshRNA vector Sigma-Aldrich (St. Louis, MO) and pCDH-neo vector (System Biosciences, Mountain View, CA). shRNA sequences used in knockdown experiments are as follows (5′ to 3′):
MET (CCATCCAGAATGTCATTCT; GCATTAAAGCAGCGTATC; GCATTAAAGCAGCGTATC*; TGTGTTGTATGGTCAATAA; CCTTCAGAAGGTTGCTGAGTA);
PARP1 (TGGAAAGATGTTAAGCATTTA*);
BRCA1 (TTCATTTCTAATACCTGCC; TTAAGTCACATAATCGATC; TTCAGTACAATTAGGTGGG);
BRCA2 (TTGTTCAGCAGATTCCATG; TCTTTAAGACAGCTAAGAG; TATTAAATGACTCTTTGGC). *Targeting the 3′-UTR.
8-OHdG ELISA assay
Total DNA was purified from breast cancer cells by using DNeasy Blood & Tissue Kit (Qiagen,, Valencia, CA). 8-OHdG levels in breast cancer cells were measured by using 8-OHdG ELISA kit (Abcam, Boston, MA). Fluorescence intensity was measured by AxioVision software. The mean ± s.d. of 8-OHdG levels in each cell line was calculated.
ROS detection
Cells were seeded in the 12- or 96- well plates. After overnighter growth, cells were incubated with 10 μM 2′,7′-dichlorofluorescindiacetate (DCFDA) in PBS for 1 h. Cells were washed and the media replaced with PBS. 2′, 7′-dichlorofluorescein (DCF) was measured under a Zeiss microscope with spectra of 495EX nm/529EM nm. Fluorescence intensity was measured by AxioVision software. The mean ± s.d. of DCF intensityfrom five images in each cell line was calculated. Cells were also seeded in 96-well plate. After overnight incubation, cells were treated with 10 μM 2′,7′-dichlorofluorescindiacetate (DCFDA) in PBS. After an hour of incubation, medium was replaced with PBS. 2′, 7′-dichlorofluorescein (DCF) was measured under plate reader with spectra of 495EX nm/529EM nm. The mean ± s.d. of DCF levels in each cell line was calculated.
Receptor tyrosine kinase antibody array
A Human Phospho-RTK Array Kit (ARY001B) was purchased from R&D Systems (Minneapolis, MN). For PARP1-associated proteins study, we modified the manufacturer’s protocol. Briefly, MDA-MB-231 cells with endogenous PARP1 knockdown and re-expression of HA tagged wild-type PARP1 were treated with sodium arsenite (As) to induce ROS. Following the instructions of the protocol, cell lysates were incubated with the array membranes. The amounts of phospho-RTK were assessed with a horseradish peroxidase–conjugated HA antibody (#26183-HRP; Thermo Fisher Scientific, Rockford, IL) followed by chemiluminescence detection as described by the manufacturer. A GS-800 Calibrated Densitometer (Bio-Rad Laboratories, Hercules, CA) was used to quantify the density of the membranes.
Hierarchical clustering and display
Clustering of ERBB3, MET and FLT3 gene expression with TNBC signature genes (ERBB2, ESR1, and PGR) from The Cancer Genome Atlas database was analyzed using Cluster and TreeView41 program, as previously described24. Briefly, for any set of target receptor tyrosine kinases, an upper-diagonal similarity matrix was computed by using average-linkage clustering. This algorithm was determined by computing a dendrogram as described42. The heat map was represented graphically by coloring each cell on the basis of the measured fluorescence ratio. Log ratios of 0 (a ratio of 1.0 indicates that the genes are unchanged) were colored in black, positive log ratios were colored in red, and negative log ratios were colored in green.
Immunoprecipitation and immunoblotting
The immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses were performed as described previously31.
Confocal microscopy analysis of γ-H2AX foci
For fixed cells, confocal microscopy assay was performed as described previously31. Briefly, cells grown on chamber slides (Labtek, Scotts Valley, CA) were treated as described in the text. After washing with ice-cold PBS, cells were fixed, permeabilized, and incubated with γ-H2AX antibodies and fluorescence-labeled secondary antibodies. Immunostained cells were examined using Zeiss LSM 710 laser-scanning microscope (Carl Zeiss, Thornwood, NY) with a 63X/1.4 objective. The ZEN and AxioVison (Carl Zeiss) and ImageJ software programs (NIH, Bethesda, MD) were used for data analysis.
Fluorescence microscopy analysis of xenograft tumor tissues
Frozen MDA-MB-231 xenograft tumor slides were washed with ice-cold PBS. Cells were fixed, permeabilized, and incubated with Ki67 and γ-H2AX antibodies and fluorescent-labeled secondary antibodies. Immunostained cells were examined using Zeiss Axioplan 2 microscope (Carl Zeiss, Thornwood, NY) with a 20X objective. The AxioVison (Carl Zeiss) was used for data analysis.
TUNEL assay of xenograft tumor tissues
TUNEL assay of MDA-MB-231 xenograft tumor tissues was performed with DeadEnd™ Fluorometric TUNEL System Kit (Cat# G3250, Promega, Madison, WI) following the manufacturer’s protocol. Briefly, the frozen mouse tumor slides were washed with ice-cold PBS. Cells were fixed, permeabilized, and labeled with biotinylated nucleotide mix by terminal deoxynucleotidyl transferase (rTdT) reaction. Fragmented DNA was detected by streptavidin-conjugated fluorescence using Zeiss Axioplan 2 microscope. The AxioVison (Carl Zeiss) was used for data analysis.
Duolink assay
Cells were prepared for fluorescence microscopy analysis as described31. Primary antibodies were incubated with cells and a pair of oligonucleotide-labeled antibodies (PLA probes). Ligation and amplification were done according to the manufacturer’s protocol (Duolink Assay Kit, Sigma-Aldrich) before mounting the slide for measurement under confocal microscope. The mean ± s.d. of PLA signal intensity from 20 cells in each treatment group was calculated.
Cellular fractionation
Cytosolic and nuclear fractions were prepared as described previously31. The abundance of cytoplasmic to nuclear proteins is 5:1 after cellular fractionation. The same amount cytoplasmic or nuclear lysates was used for IP/Western, resulting in more proteins in the nuclear fraction.
Cell viability assay
Cells (1,500) were seeded in a 96-well plate and treated with the indicated inhibitors for 72 h. Then cells were incubated in fresh media with 100 μM resazurin for 1 h. Cell viability was measured by fluorescent plate readers at spectra of 560EX nm/590EM nm. Survival curves were expressed as mean ± s.d. relative to DMSO-treated control from three independent experiments.
Clonogenic cell survival assay
Cells were plated into 12- or 24-well plates. After overnight incubation, cells were treated with inhibitors followed by 8 days of incubation. The colonies were fixed and stained with 0.5% crystal violet, washed, dried and imaged. Crystal violet was resolved from colonies by methanol and measured at 540 nm. Based on the absorbance at 540 nm, survival curves were expressed as a percentage ± s.d. relative to DMSO-treated control from three independent experiments.
Soft agar anchorage-independent cell growth assay
The base layer of cell growth matrix containing DMEM/F12 medium, 10% FBS, and 0.5% agar was paved in 6-well plates (1.5 ml/well). After solidification of the base layer, the top layer (1.5 ml/well) containing DMEM/F12 medium, 10% FBS, and 0.35% agarose, and cells were plated. Culture medium (1 ml) was added to each well and changed every 3 days. After 4-week culture, colonies were stained by 0.005% crystal violet. Colonies were counted by Image J software. Survival curves were expressed as mean ± s.d. relative to DMSO-treated control from three independent experiments.
Synergy quantification of drug combination
Cell growth was measured by cell viability, clonogenic cell survival, or soft agar anchorage-independent cell growth assay. Synergistic effects were determined by the Chou–Talalay method to calculate the combination index (CI)43.
Patient tissue samples and immunohistochemical staining
A human breast cancer tissue microarray was obtained from Pantomics (Richmond, CA). Human tumor tissue specimens were obtained from patients undergoing surgical resection of breast cancer as primary treatment at MD Anderson Cancer Center or Mackay Memorial Hospital (Taipei, Taiwan) between 1995 and 2009 under the guidelines approved by the Institutional Review Board at MD Anderson, and written informed consent was obtained from patients in all cases at the time of enrollment24. The tissue microarray (#BRC2281, #BRC1021; Pantomics, Richmond, CA) was incubated with primary antibody against 8-OHdG, c-Met or pY907-PARP1, detected with biotin-conjugated secondary antibody and avidin–peroxidase, and visualized by aminoethyl carbazole chromogen. Images were analyzed by ACIS (Dako North America, Carpinteria, CA). To validate the specificity of p-Y907-PARP1 antibody in IHC, we performed a peptide competition assay by staining human breast tumor samples with p-Y907-PARP1 antibody blocked with mock peptide, phospho-Y907-PARP1 peptide, non-phospho-Y907-PARP1 peptide, or another phospho-tyrosine peptide, p-Y986-PARP1. Patient tumor samples were deparaffinized and rehydrated. Antigen retrieval was carried out by heating in 0.01 M sodium-citrate buffer (pH 6.0) using a microwave oven. To block endogenous peroxidase activity, the sections were treated with 1% hydrogen peroxide in methanol for 30 min. After 1 h preincubation in 10% normal serum to prevent nonspecific staining, the samples were incubated with primary antibodies at 4 °C overnight. The sections were then treated with biotinylated secondary antibody, followed by incubations with avidinbiotin peroxidase complex solution for 1 h at room temperature. Color was developed with the 3-amino-9-ethylcarbazole solution. Counterstaining was carried out using Mayer’s hematoxylin.
Comet assay
A comet assay was performed as described previously44. Briefly, cells were treated with H2O2 for 10 min to induce DNA damage or with H2O2 and hydroxyurea/cytosine-β-arabinofuranoside (Hu/AraC) to induce DNA damage and allow DNA damage to accumulate to evaluate the extent of DNA damage repair. Trypsinized cells were washed with PBS and mixed with 1% low-melting point agarose (LMPA). LMPA-mixed cells were placed onto slides pre-coated with 1% LMPA and incubated on ice until agarose layer solidifies. A third 0.5% LMPA layer was then placed over the second layer. Prepared slides were washed three times in water for 5 min and incubated with formanidopyrimidine DNA glycosylase (Fpg) enzyme (2 U/slide, Enzymatics, Beverly, MA) at 37 °C for 1 h to digest oxidative damage DNA and induce comet tails which were imaged by fluorescence microscope and analyzed by using the Image J software. The mean ± s.d. of DNA intensity in the tail from 20 cells in each treatment group was calculated.
In vitro kinase assay
Recombinant glutathione S-transferase (GST)-Wt PARP1 (Ala374-Trp1014 of human PARP1) and mutants (GST-Y907F and GST-Y986E) were expressed by induction of isopropyl β-D-1-thiogalactopyranoside (IPTG) and purified with glutathione agarose beads. After cold-PBS washing 3 times, beads were suspended with 500 μl 1X kinase buffer, with 50 μl saved for Western blotting with GST. The beads were spun down and 100 μM ATP, 0.5 μg human recombinant active c-Met protein and 50 μCi [γ-32P]-ATP were added in 50 μl kinase buffer at 30 °C for 15–30 min. The kinase reaction was stopped by heating at 100 °C for 5 min in SDS loading dye. The samples were subjected to two identical SDS-PAGE assays. One was used for Coomassie blue staining of GST fusion PARP1 protein. The second gel was dried and used to detect phosphorylation of substrate by autoradiograph.
PARP enzyme activity assay
PARP1 enzyme activity was measured by using a commercial assay kit (Cat# 17–10149) from EMD Millipore according to the manufacturer’s protocol with the exception that cell lysates containing wild-type PARP1 or PARP Y907 mutant were used in place of the PARP1 protein included with the kit. Total lysate (500 ng) was added to each reaction. The dose course of PARP inhibitor ABT-888 was from 0.01 to 1,000 μM. PARP enzyme activity of wild-type and mutants was determined after incubation with the substrate was measured using a plate reader.
Isothermal titration calorimetry
ITC experiments were carried out using either a TA nano ITC instrument or an Auto ITC system (TA Instruments) at 25 °C. Titrations of 55–110 μM ABT-888 into the sample cell containing a 5–10 μM solution of either GST-WT PARP1, GST-Y907F PARP1, or GST-Y907E PARP1, dissolved in TIC buffer (19.8 mM HEPES, 148.5 mM NaCl, 0.495 mM TCEP and 1% (v/v) DMSO at pH 7.5). The heat of dilution control experiments was measured independently by titrating the same ABT-888 solution into the same buffer and subtracted from the observed heat measured for the titration of compound into proteins. Experimental data were fitted using the independent binding site model in NanoAnalyze v3.4.0 software (TA instruments).
Mouse xenograft models and toxicity study
All animal procedures were conducted under the approval of the Institutional Animal Care and Use Committee (IACUC) at MD Anderson Cancer Center (protocol number 10-14-07231). MDA-MB-231 (0.5 × 106), HCC1937 (2 × 106) or MCF-7 (5 × 106) cells were injected into the mammary fat pads of female nude (Swiss Nu/Nu) mice of 6–8 weeks of age (Department of Experimental Radiation Oncology Breeding Core, The University of Texas MD Anderson Cancer Center). A1034 (0.5 × 106) cells were injected into the mammary fat pads of female FVB/NJ mice (The Jackson Laboratory, Stock No. 001800) of 6–8 weeks of age. H1993 (0.5 × 106) cells were injected subcutaneously into the right flank of female nude (Swiss Nu/Nu) mice (Department of Experimental Radiation Oncology Breeding Core, The University of Texas MD Anderson Cancer Center) of 6–8 weeks of age. When the tumor volume reached ~50 mm3, crizotinib (5 mg/kg) and foretinib (5 mg/kg), AG014699 (5 mg/kg) and ABT-888 (25 mg/kg), dissolved in aqueous 50 mM sodium acetate, pH 4, were administered to mice five times per week as single agents or in combination for the number of days specified in the figure legend. Tumor was measured at the indicated time points, and tumor volume was calculated by the formula: π/6 × length × width2. For MDA-MB-231 and A1034 xenograft mouse models, mice were imaged before and after treatment using the IVIS Imaging System to assess tumor growth. Mice were injected with 100 μl of D-luciferin (Xenogen; 15 mg/ml in PBS). After 10 min, mice were anesthetized with a mixture of oxygen and isoflurane (Inhalation Anesthesia System; Matrix Medical, Orchard Park, NY) and imaged using the IVIS Imaging System. Imaging parameters were maintained across experiments for comparative analyses.
Tumor samples were collected after final treatment and analyzed by immunofluorescence staining. For toxicity assessment, mice were weighed before and after treatment (on day 21 for AG014699 and crizotinib, and on day 16 for ABT-888 and foretinib. Blood samples were collected from the orbital sinus using a microhematocrit tube after each treatment and subjected to biochemical analysis for liver marker enzymes alanine transaminase (ALT) and aspartate transaminase (AST) and kidney marker by-products creatinine and blood urea nitrogen to evaluate treatment toxicity by COSBA INTERGRA 400 plus (Roche Diagnostics, Rotkreuz, Switzerland) at The Department of Veterinary Medicine & Surgery. All in vivo experiments were conducted with 10 mice for each treatment and control group. No statistical methods were used to predetermine sample size.
Statistical analysis
Unless otherwise noted, each sample was assayed in triplicate. For in vitro analyses, each experiment was repeated at least three times. All error bars represent standard deviation (s.d.). Student’s t test was used to compare two groups of independent samples. Repeated measure ANOVA analysis was used to evaluate the statistical significance of dose curve response. Correlations were analyzed using the Pearson chi-square test. A P value < 0.05 was considered statistically significant. No statistical methods were used to determine sample size.
Supplementary Material
Acknowledgments
This study was funded in part by the following: U.S. National Institutes of Health grants [CA109311 (M.-C. Hung), CA099031 (M.-C. Hung), and Cancer Center Support Grant CA16672 (R.A. DePinho)]; Susan G. Komen Foundation [SAC100016 (M.-C. Hung)]; Breast Cancer Research Foundation; Patel Memorial Breast Cancer Endowment Fund; The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund; Ministry of Science and Technology, International Research-intensive Centers of Excellence in Taiwan [I-RiCE; MOST 104-2911-I-002-302 (M.-C. Hung)]; Ministry of Health and Welfare, China Medical University Hospital Cancer Research Center of Excellence [MOHW104-TDU-B-212-124-002 (M.-C. Hung)]; Center for Biological Pathways at MD Anderson; and Ting Tsung and Wei Fong Chao Research Fund. We also thank S. Patterson of the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for her editorial assistance.
Footnotes
Author contributions
Y.D. designed and performed the experiments, analyzed data, and wrote the manuscript; H.Y., Y.W., Y.-H.H., W.-C.L., W.-H.Y., P.L., G.R.L. W.-C.C., C.-H.-C., and M.-K.C. performed experiments and analyzed data; H.-L.W. generated the antibody; M.-C. Hsu, C.-T.C., K.N., and Y.S. performed experiments; Y.W., W-.C.H., C.-L.L., and Y.-C.C. provided patient tissue samples; H.Y. and J.L.H. provided scientific input and wrote the manuscript; M.P. provided transgenic mouse cell lines. G.N.H. provided scientific and clinical input. P.J. provided scientific input. M.-C. Hung supervised the entire project, designed the experiments, analyzed data, and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Feng FY, de Bono JS, Rubin MA, Knudsen KE. Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function. Mol Cell. 2015;58:925–934. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 3.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 4.Irani K, et al. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 5.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 6.Radisky DC, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 9.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 10.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 11.Lips EH, et al. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer. 2013;108:2172–2177. doi: 10.1038/bjc.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 13.Badve S, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelmon KA, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 16.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swindall AF, Stanley JA, Yang ES. PARP-1: Friend or Foe of DNA Damage and Repair in Tumorigenesis? Cancers (Basel) 2013;5:943–958. doi: 10.3390/cancers5030943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robaszkiewicz A, et al. Hydrogen peroxide-induced poly(ADP-ribosyl)ation regulates osteogenic differentiation-associated cell death. Free Radic Biol Med. 2012;53:1552–1564. doi: 10.1016/j.freeradbiomed.2012.08.567. [DOI] [PubMed] [Google Scholar]
- 21.Chiarugi P, Cirri P. Redox regulation of protein tyrosine phosphatases during receptor tyrosine kinase signal transduction. Trends Biochem Sci. 2003;28:509–514. doi: 10.1016/S0968-0004(03)00174-9. [DOI] [PubMed] [Google Scholar]
- 22.Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nature reviews Cancer. 2012;12:387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speers C, et al. Identification of novel kinase targets for the treatment of estrogen receptor-negative breast cancer. Clin Cancer Res. 2009;15:6327–6340. doi: 10.1158/1078-0432.CCR-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu YH, et al. Definition of PKC-alpha, CDK6, and MET as therapeutic targets in triple-negative breast cancer. Cancer Res. 2014;74:4822–4835. doi: 10.1158/0008-5472.CAN-14-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zagouri F, et al. High MET expression is an adverse prognostic factor in patients with triple-negative breast cancer. Br J Cancer. 2013;108:1100–1105. doi: 10.1038/bjc.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30:937–945. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- 27.Gomes DA, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008;283:4344–4351. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YN, et al. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J Biol Chem. 2012;287:16869–16879. doi: 10.1074/jbc.M111.314799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang YN, et al. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem Biophys Res Commun. 2010;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Y, et al. Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene. 2014;33:756–770. doi: 10.1038/onc.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonnenblick A, de Azambuja E, Azim HA, Jr, Piccart M. An update on PARP inhibitors--moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- 33.Jagadeeswaran R, Jagadeeswaran S, Bindokas VP, Salgia R. Activation of HGF/c-Met pathway contributes to the reactive oxygen species generation and motility of small cell lung cancer cells. American journal of physiology. Lung cellular and molecular physiology. 2007;292:L1488–1494. doi: 10.1152/ajplung.00147.2006. [DOI] [PubMed] [Google Scholar]
- 34.Anders CK, et al. Pharmacokinetics and efficacy of PEGylated liposomal doxorubicin in an intracranial model of breast cancer. PLoS One. 2013;8:e61359. doi: 10.1371/journal.pone.0061359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc Natl Acad Sci U S A. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight JF, et al. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc Natl Acad Sci U S A. 2013;110:E1301–1310. doi: 10.1073/pnas.1210353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho-Yen CM, Jones JL, Kermorgant S. The clinical and functional significance of c-Met in breast cancer: a review. Breast Cancer Res. 2015;17:52. doi: 10.1186/s13058-015-0547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowsheen S, Cooper T, Stanley JA, Yang ES. Synthetic lethal interactions between EGFR and PARP inhibition in human triple negative breast cancer cells. PloS one. 2012;7:e46614. doi: 10.1371/journal.pone.0046614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6:637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokal RR, Michener CD, Kansas Uo. A Statistical Method for Evaluating Systematic Relationships. University of Kansas; 1958. [Google Scholar]
- 43.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 44.Lee HJ, et al. Tyrosine 370 phosphorylation of ATM positively regulates DNA damage response. Cell Res. 2015;25:225–236. doi: 10.1038/cr.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.