Analyses in Xenopus egg extracts show that the MRN complex, CtIP, BRCA1, and the interaction between CtIP and BRCA1 are required for the removal of Top2–DNA adducts, forsubsequent resection of Top2-adducted double-strand break ends, and for cellular resistance to etoposide during genomic DNA replication.
Abstract
Repair of DNA double-strand breaks (DSBs) with complex ends poses a special challenge, as additional processing is required before DNA ligation. For example, protein–DNA adducts must be removed to allow repair by either nonhomologous end joining or homology-directed repair. Here, we investigated the processing of topoisomerase II (Top2)–DNA adducts induced by treatment with the chemotherapeutic agent etoposide. Through biochemical analysis in Xenopus laevis egg extracts, we establish that the MRN (Mre11, Rad50, and Nbs1) complex, CtIP, and BRCA1 are required for both the removal of Top2–DNA adducts and the subsequent resection of Top2-adducted DSB ends. Moreover, the interaction between CtIP and BRCA1, although dispensable for resection of endonuclease-generated DSB ends, is required for resection of Top2-adducted DSBs, as well as for cellular resistance to etoposide during genomic DNA replication.
Introduction
DNA double-strand breaks (DSBs) are genotoxic lesions that arise by diverse mechanisms, including endogenous processes such as replication fork collapse and abortive DNA transactions by ligases, topoisomerases, or nucleases (Pommier et al., 2010; Symington and Gautier, 2011). Although DSBs are primarily repaired by nonhomologous end joining (NHEJ), they can also be resolved during S phase by homology-directed repair (HDR) using the sister chromatid as a DNA template (Symington and Gautier, 2011; Chapman et al., 2012; Andres et al., 2014; Aparicio et al., 2014). The decision to use NHEJ or HDR is governed in part by DNA resection, a nucleolytic process in which DSB ends are converted into 3′ single-strand DNA overhangs, an essential intermediate for the downstream steps of HDR and a potent inhibitor of NHEJ. DNA resection in eukaryotes is initiated by CtIP (Sae2 in yeast) and the MRN/X complex (Mre11, Rad50, and Nbs1/Xrs2 in yeast; Sartori et al., 2007; Huertas and Jackson, 2009; Qvist et al., 2011). Whereas MRN-CtIP mediates short-range 5′ to 3′ resection, exonuclease 1 (Exo1) can, after a lag, extensively resect DSBs independently of MRN-CtIP. The resection activity of CtIP is regulated in both a cell-cycle– and damage-dependent manner that is conserved among vertebrates (You et al., 2009; Peterson et al., 2011). Interestingly, CtIP also binds the BRCA1 tumor suppressor (Yu et al., 1998), an essential HDR protein. Whereas the effect of BRCA1 on CtIP-mediated DNA resection remains unclear (Reczek et al., 2013; Zhou et al., 2014), genetic data suggest a role for CtIP–BRCA1 interaction in cellular tolerance to camptothecin and, to a lesser extent, to etoposide (Nakamura et al., 2010).
Topoisomerases facilitate DNA transactions such as replication and transcription by relieving DNA topological stress. Type IB topoisomerases (Top1) remove supercoils by generating single-strand DNA breaks that allow DNA to rotate over its axis. Through a transesterification reaction, the catalytic tyrosine of the enzyme forms a transient phosphotyrosine covalent linkage, generating a nick in the DNA. After isomerization, the DNA phosphodiester backbone is restored when the 5′ OH of the broken DNA strand attacks the 3′ phosphotyrosine bond, liberating Top1 for subsequent cleavage and unwinding. Type IIA topoisomerases (Top2) remove topological constraints by generating staggered incisions, 4 bp apart, on both strands of DNA, which allow passage of a second DNA duplex through the DSB (Liu et al., 1983; Rowe et al., 1984; Wu et al., 2011). This reaction also entails formation of a transient protein–DNA adduct, in this case between a tyrosine residue at each active site of the Top2 dimer and the 5′ phosphates of DNA strands on both sides of the DSB. After isomerization, the resulting 3′-hydroxyl DNA ends direct the reversal of the phosphotyrosyl bonds, thereby enabling the release of the topoisomerase and religation of the DNA break (Pommier et al., 2010; Vos et al., 2011).
Given that DNA breaks are normal intermediates of topoisomerase activity, abortive topoisomerase reactions that stabilize the transient protein–DNA adduct represent a significant source of DNA damage (Vos et al., 2011). Moreover, the formation of such “trapped” protein–DNA adducts can be exacerbated by topoisomerase poisons such as etoposide (also known as VP-16-213), which increases the stability of Top2–DNA adducts (Pommier et al., 2010). These unprocessed Top2–DNA adducts block DNA replication and RNA transcription and generate lethal DSBs that can induce cell-death pathways. Because cancer cells rely more heavily on DNA repair than normal cells (Tewey et al., 1984; Treszezamsky et al., 2007; Nitiss, 2009), the cellular toxicity of etoposide has been exploited therapeutically for a variety of human malignancies, including small cell lung carcinoma, testicular cancer, and lymphomas.
Eukaryotic cells coordinate multiple pathways to eliminate trapped protein–DNA adducts. Whereas ubiquitin-dependent proteasome degradation can remove much of the adducted proteins, further enzymatic processing by a DNA repair pathway is required to release the residual adducted peptide. For example, formaldehyde-induced DNA–protein cross-links require nucleotide-excision repair or HDR for resolution (Ide et al., 2011). Similarly, Top2-adducted DNA ends can be converted into ligatable ends upon direct cleavage of the 5′-tyrosine phosphodiester bond by tyrosyl DNA phosphodiesterase 2 (TDP2; Mao et al., 2001; Zhang et al., 2006; Andres et al., 2014; Gao et al., 2014). In addition, biochemical studies in Escherichia coli and genetic analyses in yeast implicate the orthologous factors MRN/X and CtIP/Sae2 in nucleolytic removal of 5′-linked proteins from DNA including topoisomerase–DNA adducts (Neale et al., 2005; Hartsuiker et al., 2009; Cannavo and Cejka, 2014), as well as the Spo11–DNA adducts normally generated during meiotic recombination (de Massy et al., 1995; Keeney and Kleckner, 1995; Liu et al., 1995; Hartsuiker, 2011; Lee et al., 2012; Cannavo and Cejka, 2014; Makharashvili et al., 2014). Yeast Spo11, an enzyme related to DNA topoisomerases, is required to initiate meiotic recombination by generating stable 5′-end covalent protein–DNA intermediates (Keeney and Kleckner, 1995). Of note, rad50S and mre11S hypomorphic mutations, mre11 nuclease-deficient alleles, and null mutations in sae2 completely block 5′ DNA strand processing in meiosis, allowing the accumulation of Spo11–DNA complexes at DSBs. This suggests that Spo11 complexes are normally removed by nuclease activities of the MRX–Sae2 complex (Keeney et al., 1997; Keeney, 2001; Nitiss, 2009). In contrast, rad50S and mre11S hypomorphic mutations are compatible with mitotic cell growth and have a milder phenotype: delayed resection of endonuclease-generated DSB ends (Symington and Gautier, 2011). This implies that topoisomerase–DNA adducts are infrequent in mitotic cells and/or that alternative pathways remove 5′-end adducts in these cells. Finally, recent studies suggest that Top2 adducts could also be removed by the nucleolytic activities of the MRN complex (Mre11, Rad50, and Nbs1) and its associated protein CtIP (Cruz-García et al., 2014).
BRCA1 tumor suppressor plays a role early in HDR, possibly by facilitating resection and antagonizing 53BP1 function (Moynahan et al., 1999; Bunting et al., 2012). However, analysis of BRCA1 inactivation on resection shows conflicting results, and how BRCA1 might regulate resection is unknown (Symington and Gautier, 2011).
Here, we show that MRN, CtIP, and BRCA1 are required for efficient removal of etoposide-induced Top2–DNA adducts from Xenopus laevis chromatin, as well as the subsequent resection of Top2-adducted DSB ends. We demonstrate enhanced recruitment of BRCA1 to etoposide-damaged DNA. Furthermore, unlike resection of endonuclease-generated DSBs, the processing of Top2 covalent complexes by CtIP is dependent on its interaction with BRCA1. These findings identify MRN–CtIP–BRCA1 as a major pathway for processing Top2–DNA adducts.
Results
Replication of etoposide-treated chromosomal DNA requires CtIP
CtIP promotes the resection of “clean” DSB ends generated by endonuclease cleavage (Sartori et al., 2007; You et al., 2009; Peterson et al., 2011). To assess its role in processing DSB ends that harbor a Top2–DNA cleavage complex, we first examined whether CtIP is required for chromosomal DNA replication in Xenopus cell-free extracts (Fig. 1 A). As shown in Fig. 1 (B and C, native genomic DNA), the kinetics of incorporation of a radiolabeled deoxynucleotide, α-[32P]dCTP, into chromosomal DNA was identical in control and CtIP-depleted extracts. Neither replication initiation nor replication elongation was reduced in the absence of CtIP, as indicated by the size and distribution of replication products after denaturation (Fig. 1 D). Thus, CtIP is dispensable for replication of undamaged DNA. We previously showed that Mre11 is also not required for DNA replication, although DSBs accumulate in Mre11-depleted extracts at a low frequency (Costanzo et al., 2001). To ascertain the requirements for CtIP and MRN in replication of DNA bearing Top2–DNA adducts, we examined DNA replication in the presence of a low concentration of the Top2 poison etoposide. Under these conditions, etoposide did not affect DNA replication in control extracts, but it strongly inhibited replication in CtIP-depleted extracts (Fig. 1, E and F, native genomic DNA). A similar result was observed in Mre11-depleted extracts, as well as in extracts depleted of both CtIP and Mre11 (Fig. S1, A and B). Significantly, DNA replication of etoposide-treated CtIP-depleted extracts was largely restored by addition of recombinant Xenopus CtIP protein purified from baculovirus-infected insect cells (see Fig. 6 B, bottom), thus establishing that the phenotype was specific to CtIP depletion. Interestingly, replication in CtIP-depleted extracts was partially rescued by the addition of caffeine at a concentration that completely inhibits the DNA damage response (10 mM; unpublished data). Because 10 mM caffeine inhibits both ATM and ATR, we then inhibited each kinase specifically. Although the ATR inhibitor VE-821 (Reaper et al., 2011) had limited effect, the ATM inhibitor KU55933 (Hickson et al., 2004) rescued DNA replication to nearly 50% of the level observed in the absence of etoposide (Fig. 1 F), indicating that Top2-adducted DSB ends trigger the ATM checkpoint. These inhibitors did not affect genomic replication in nondepleted or mock-depleted extracts (Fig. S1 C). Together, these results indicate that etoposide-induced inhibition of DNA replication is relieved through the action of CtIP and the MRN complex, and it is also mediated in part through checkpoint-dependent processes.
Figure 1.
Replication of etoposide-treated chromosomal DNA requires CtIP. (A) CtIP immunodepletion from LSS extracts. Control (ΔMock) and CtIP-depleted extracts (ΔCtIP) were probed with the indicated antibodies. (B) Mock and CtIP-depleted extracts were incubated with sperm nuclei. DNA replication was monitored by agarose gel electrophoresis after incorporation of α-[32P]dCTP into genomic DNA at the indicated time points. (C) Quantification of three independent experiments as shown in B. The mean is plotted, and error bars indicate the standard deviation. (D) Control (mock) and CtIP-depleted extracts were supplemented with sperm nuclei, and DNA replication of Xenopus sperm nuclei was monitored by alkaline gel electrophoresis after incorporation of α-[32P]dCTP into genomic DNA at the indicated time points. (E) DNA replication was monitored as in B in the presence of a low dose (2 µM) of etoposide in mock- or CtIP-depleted extracts, below quantification of three independent experiments (**, P = 0.003, two-tailed unpaired t test; n = 3). (F) Effect of ATM and ATR inhibitors in the sensitivity to low-dose etoposide (2 µM). Bar graph shows quantification of three independent experiments (*, P = 0.014, two-tailed unpaired t test; n = 3).
Figure 6.
CtIP–BRCA1 interactions are required for CtIP-dependent processing of etoposide-induced DSBs. (A) Control extract (no damage), extracts treated with etoposide (Etop.), or extracts treated with PflM1 were incubated with the CDK inhibitor roscovitine. Chromatin was isolated at 0 or 40 min and processed for Western blot with the indicated antibodies. Roscovitine inhibits genomic DNA replication (bottom). (B) Control (ΔMock) and CtIP-depleted extracts (ΔCtIP) with or without etoposide were supplemented with sperm nuclei, and replication was monitored by agarose gel electrophoresis after incorporation of α-[32P]dCTP into genomic DNA. CtIP-depleted extracts were supplemented with recombinant xCtIP WT or CtIP-S328A mutant (xCtIP S328A). Bar graph shows quantification of three independent experiments (*, P = 0.026, two-tailed unpaired t test). (C) Relative quantification of Top2–DNA adducts in CtIP-depleted extracts supplemented with buffer, xCtIP WT, or xCtIP S328A (*, P = 0.035, two-tailed unpaired t test; NS, not statistically significant [P = 0.08]; n = 3). (D) Control (ΔMock) and CtIP-depleted extracts were supplemented with sperm nuclei (5,000/µl) and treated with either PflM1 endonuclease or etoposide. CtIP-depleted extracts were supplemented with recombinant xCtIP WT or xCtIP S328A. Chromatin was isolated and processed for Western blotting with the indicated antibodies. NS, no sperm control; Ext., extract.
MRN and CtIP are involved in Top2 adduct removal during S phase
We then asked whether CtIP and MRN participate in the removal of etoposide-induced Top2 adducts during DNA replication. First, we confirmed that etoposide treatment increases Top2–DNA adduct concentrations. Chromatin-bound Top2 levels were monitored in replication-competent cytosol incubated with sperm nuclei as a template for chromatin assembly. As expected, addition of etoposide increased the amount of chromatin-associated Top2 (Fig. S2 A) in a dose-dependent manner. Note the accumulation of higher molecular weight Top2 intermediates corresponding to polyubiquitylated forms of Top2 in samples exposed to high etoposide concentrations (Fig. S2, A–C). Treatment with the proteasome inhibitor MG132 further enhanced the levels of polyubiquitylated Top2 (Fig. S2 D). DNA fragmented by exposure to PflM1 restriction enzyme or topotecan did not accumulate Top2 adducts (Fig. S2 C).
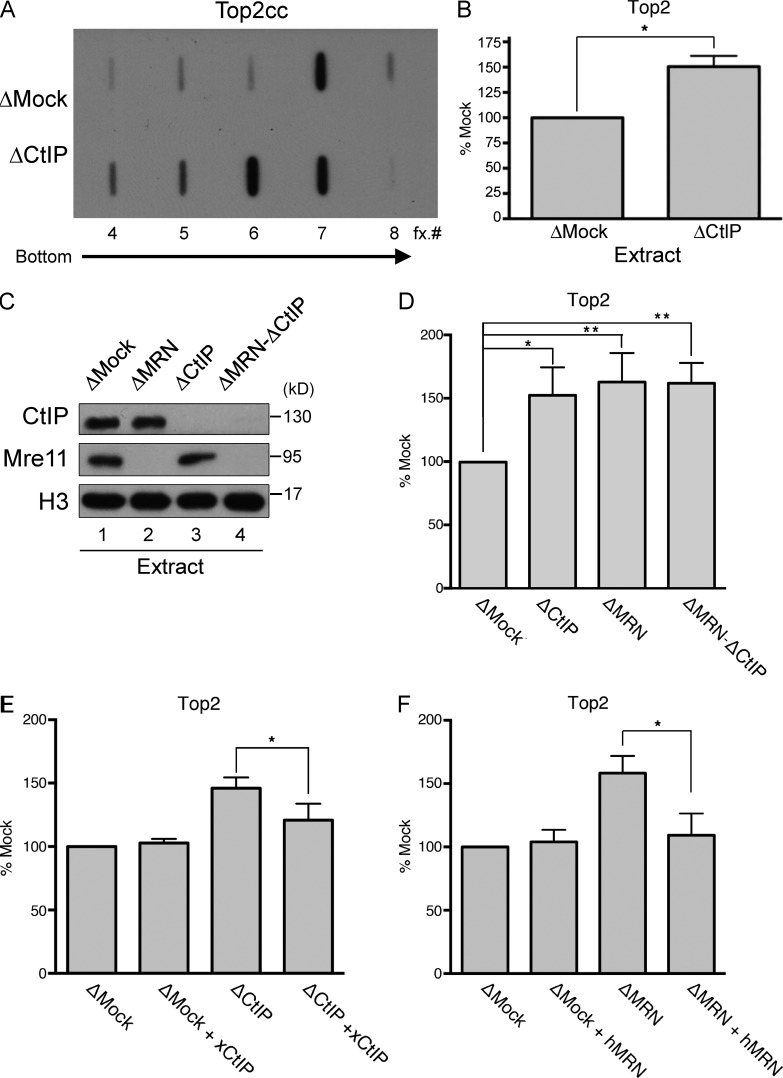
To quantify Top2–DNA adducts in Xenopus genomic DNA in the presence and absence of DNA repair factors, we adapted a protocol previously applied to fission yeast genomic DNA (Hartsuiker et al., 2009; Hartsuiker, 2011) and mammalian cells (Kiianitsa and Maizels, 2013; Figs. 2 A and S2, E–G). To this end, genomic DNA was isolated under denaturing conditions to remove all chromatin proteins except those covalently bound to DNA. By inhibiting DNA ligation and the release of topoisomerase, protein denaturation effectively captures all trapped Top2 molecules. Noncovalently bound proteins are separated from DNA (containing covalently bound proteins) on cesium chloride gradients. After centrifugation, fractions containing DNA–protein adducts are probed for Top2 by slot blot assay with anti-Top2 antibodies. As shown in Fig. 2 B, covalently bound Top2 concentrations increased by 50% in genomic DNA purified from CtIP-depleted extracts, suggesting that CtIP is required for processing Top2–DNA adducts. A previous study using a fluorescence-based method also implicated the MRN complex in removal of Top2 adducts from DNA (Lee et al., 2012). Similarly, we observed significant increases in levels of DNA-associated Top2 upon depletion of MRN or co-depletion of MRN and CtIP (Fig. 2, C and D; and Fig. S2 G), suggesting that MRN and CtIP operate in the same pathway to remove Top2 covalent complexes from DNA. Importantly, the accumulation of Top2 adducts in depleted extracts was reduced by addition of recombinant Xenopus CtIP (Figs. 2 E and S2 H) or recombinant human MRN complex (Figs. 2 F and S2 I), establishing the specificity of the antibody-mediated depletions and demonstrating a direct role for both factors in repair of Top2–DNA adducts.
Figure 2.
MRN and CtIP remove Top2 adducts during S phase. (A) Mock or CtIP-depleted extracts were supplemented with sperm nuclei (5,000/µl) and incubated in the presence of 100 µM etoposide for 40 min. Reactions were stopped and diluted in denaturing buffer, and genomic DNA was fractionated via CsCl gradients. The fractions were transferred and probed with anti-Xenopus Top2 antibodies. (B) Relative quantification of three independent experiments as shown in A. Error bars indicate standard deviations (*, P = 0.021, two-tailed unpaired t test). (C) CtIP, Mre11, or CtIP/Mre11 immunodepletion from LSS extracts. Control (ΔMock) and depleted extracts were blotted with the indicated antibodies. (D) Quantification of Top2–DNA adducts in Mre11-depleted, CtIP-depleted, or MRE11- and CtIP-depleted extracts as described in A. (*, P < 0.05; **, P < 0.01, two-tailed unpaired t test; n = 3). (E) Quantification of Top2–DNA adducts in CtIP-depleted extracts and CtIP-depleted extracts supplemented with recombinant CtIP protein (100 nM in extract; *, P = 0.048, two-tailed unpaired t test; n = 3). (F) Quantification of Top2–DNA adducts in Mre11-depleted extracts and Mre11-depleted extracts supplemented with recombinant MRN protein complex (500 nM in extract; *, P = 0.018, two-tailed unpaired t test; n = 3).
CtIP regulates DNA resection from topoisomerase adducts
To ascertain whether CtIP is required for nucleolytic processing and subsequent resection of Top2-adducted DSB ends, we compared the effects of CtIP depletion on resection of DNA breaks induced by endonuclease or etoposide in replication-competent S-phase extracts. Using replication protein A (RPA) binding to chromatin as a marker of single-strand DNA formation, we observed a modest reduction of DNA resection in CtIP-depleted extracts at early time points after treatment with PflMI endonuclease (Fig. 3, A [compare lanes 1–3 and 7–9] and B), consistent with previous results (Peterson et al., 2011). In contrast, a far more substantial reduction in RPA binding was obtained in CtIP-depleted extracts upon etoposide treatment (Fig. 3, A [lanes 4–6 and 10–12] and B; and Fig. S3 A), indicating a more stringent requirement for CtIP in removing Top2–DNA adducts and processing the damaged DNA. Similarly, MRN-depleted extracts (ΔMre11; Fig. S3 B) show a significant decrease in resection from Top2–DNA adducts. Importantly, the presence or absence of CtIP had no impact on RPA chromatin loading in extracts treated with ICRF-193, a Top2 inhibitor that does not produce Top2–DNA trapped intermediates (Fig. 3 C). Consistent with reduced RPA accumulation on chromatin, Chk1 kinase activation was impaired (Fig. S3 C). Thus, CtIP is required to remove Top2–DNA adducts induced by exposure to etoposide.
Figure 3.
CtIP regulates resection from Top2 adducts. (A) Control (ΔMock) and CtIP-depleted extracts were supplemented with sperm nuclei (5,000/µl) and incubated with 0.05 U/µl PflMI or 100 µM etoposide (Etop.). Chromatin was purified at the indicated times and processed for Western blot with Top2, CtIP, RPA70, Ku70, and H3 (loading control) antibodies. NS, no sperm control. (B) Plot of RPA binding as shown in A for control and CtIP-depleted extracts treated with PflMI or etoposide. Shown are mean RPA intensities. Error bars represent SD; n = 3. (C) Chromatin-bound RPA70 was monitored in control (ΔMock) and CtIP-depleted extracts treated with etoposide or ICRF-193. a.u., arbitrary units.
Exo1 cannot process Top2–DNA adducts
Resection in eukaryotes is initiated by MRN–CtIP and subsequently extended by the nucleolytic activities of Exo1 or Dna2 (Symington and Gautier, 2011). Using Xenopus egg extracts, we and others have shown that Exo1 can, after a lag, also initiate resection at endonuclease-generated DSBs in the absence of CtIP (Liao et al., 2011; Peterson et al., 2011). The ability of Exo1 to initiate resection in lieu of CtIP has also been observed for human proteins both in vitro and in vivo (Eid et al., 2010; Nimonkar et al., 2011; Tomimatsu et al., 2014). However, in contrast to resection of endonuclease-induced DSBs, the data presented in Fig. 3 suggest that Xenopus extracts lack a nuclease that can initiate resection of etoposide-induced DSBs in the absence of CtIP or MRN. Therefore, to assess directly the role of Exo1, we monitored the removal of Top2–DNA adducts in Exo1-depleted and CtIP-depleted extracts (Fig. 4 A). In contrast to CtIP depletion, depletion of Exo1 did not affect Top2 adduct removal (Fig. 4 B).
Figure 4.
Exo1 cannot process Top2–DNA adducts. (A) Mock, Exo1, or CtIP immunodepletions from LSS extracts. Control (ΔMock) and depleted extracts were blotted with the indicated antibodies. (B) Quantification of Top2–DNA adducts in CtIP-depleted or Exo1-depleted extracts treated with etoposide as described (*, P = 0.02, two-tailed unpaired t test; NS, not statistically significant). (C) Control (ΔMock) or CtIP-depleted replication-incompetent, membrane-free extracts (HSS) were supplemented with sperm nuclei (5,000/µl) and treated with etoposide. (D) Chromatin fractions from extracts in C were blotted with the indicated antibodies.
However, Exo1-CtIP co-depletion abolished DNA resection in etoposide-treated extracts to a greater degree than CtIP depletion alone (Fig. S4). This observation suggests that Exo1 can extend resection from DNA intermediates that have been processed by a CtIP-independent pathway. This might be the proteasomaldegradation and Tdp2-catalyzed release of Top2 (Cortes Ledesma et al., 2009). Furthermore, resection from etoposide-induced breaks was completely inhibited in CtIP-depleted membrane-free extracts (high-speed supernatants [HSSs]; Fig. 4, C and D, compare with Fig. 3 C), which are deficient in Exo1-mediated DNA end resection (Peterson et al., 2013).
BRCA1 is required to process Top2–DNA adducts
BRCA1 is thought to facilitate resection of endonuclease-generated breaks (Bunting et al., 2012) by a mechanism that is poorly understood. We sought to assess the role of BRCA1 in the processing of Top2 adducts. First, we examined chromosomal DNA replication in extracts BRCA1-depleted extracts using antibodies specific for Xenopus BRCA1 (Joukov et al., 2001; Long et al., 2014). Quantitative depletion of BRCA1 (Fig. 5 A) did not affect replication in untreated extracts (Fig. 5 B, compare lanes 1 and 3). In contrast, when we examined DNA replication in the presence of a low concentration of etoposide, we observed a profound effect of BRCA1 depletion. Etoposide did not affect DNA replication in control extracts (Fig. 5 B, lane 2) but strongly inhibited replication in BRCA1-depleted extracts (Fig. 5 B, lane 4).
Figure 5.
BRCA1 is required to process Top2–DNA adducts. (A) Mock and BRCA1 immunodepletions from LSS extracts; extracts were blotted with the indicated antibodies. (B) DNA replication in BRCA1-depleted extracts is sensitive to low-dose etoposide. Top: control (ΔMock) and BRCA1-depleted extracts were incubated with sperm nuclei. DNA replication was monitored by agarose gel electrophoresis after incorporation of α-[32P]dCTP into genomic DNA; bottom: quantification of three independent experiments. The mean is plotted, and error bars indicate the standard deviation (*, P < 0.026, unpaired two-tailed t test; n = 3). (C) Relative quantification of Top2–DNA adducts in BRCA1-depleted extracts (*, P = 0.02, two-tailed unpaired t test; n = 3). (D) Mock or BRCA1-depleted extracts were supplemented with sperm nuclei (5,000/µl), treated with PflMI or etoposide as indicated, and incubated for 40 min. Chromatin was isolated and immunoblotted with the indicated antibodies.
Next, we quantified Top2 DNA adducts in Xenopus genomic DNA in the presence and absence of BRCA1 (Fig. 5 C) using the protocol described for Fig. 2. We observed a reproducible and statistically significant increase in covalently bound Top2 in genomic DNA purified from BRCA1-depleted extracts, suggesting that BRCA1 is required for processing Top2–DNA adducts (Lee et al., 2012).
Finally, we determined whether BRCA1 was recruited to and subsequently required for nucleolytic processing and resection of Top2-adducted DSB ends. We first monitored BRCA1 association with chromatin in untreated (control) extracts or extracts treated with PflMI endonuclease or etoposide. BRCA1 associated with chromatin harboring clean DSBs, but the association was dramatically increased in presence of Top2–DNA adducts (Fig. 5 D, lanes 2, 4, and 6). Next, we compared the effects of BRCA1 depletion on resection of DNA breaks induced by endonuclease or etoposide in replication-competent S-phase extracts. Using RPA binding to chromatin as a marker of single-strand DNA formation, we observed a modest reduction of DNA resection in BRCA1-depleted extracts after treatment with PflMI endonuclease (Fig. 5 D, lanes 4 and 5). In contrast, etoposide induced a far more substantial reduction in RPA binding than was obtained in BRCA1-depleted extracts (Fig. 5 D, lanes 6 and 7), indicating a critical role for BRCA1 in removing Top2–DNA adducts and processing the damaged DNA.
CtIP–BRCA1 interaction is required for CtIP-dependent processing of etoposide-induced DSBs
To define the role of CtIP–BRCA1 interaction in DNA repair, we first assessed the impact of CDK phosphorylation. CDKs phosphorylate CtIP at two residues: S327 and T806 (S328 and T847 in human cells). Phosphorylation of S327 is required for the interaction between CtIP and the tandem BRCT domains of BRCA1 (Yu and Chen, 2004), whereas phosphorylation at residue T806 is required for CtIP-mediated resection (Huertas et al., 2008; Huertas and Jackson, 2009). Therefore, we compared the impact of the CDK inhibitor roscovitine on resection of DSBs generated by either endonuclease or etoposide treatment (Fig. 6 A). Roscovitine had little effect on resection of DSBs generated by PflMI, indicating that CDK phosphorylation of CtIP is dispensable for processing endonuclease-generated breaks, presumably because Exo1 can resect these DSBs in the absence of CtIP (Fig. 6 A, lanes 7 and 8). In contrast, roscovitine blocked resection from etoposide-induced DSBs, indicating that CDK activity, possibly through phosphorylation of CtIP, is essential for processing Top2-DNA ends (lanes 5 and 6).
To test directly whether the CtIP–BRCA1 interaction is important for processing etoposide-induced DSBs, we purified recombinant Xenopus CtIP polypeptides that do (xCtIP S328A) or do not (xCtIP WT) harbor S328A, a missense mutation that disrupts its interaction with BRCA1 in Xenopus extracts (Peterson et al., 2011). Because BRCA1 is required for genomic DNA replication in the presence of etoposide (Fig. 5 B), we assessed the impact of S328A substitution on DNA replication. As shown in Fig. 6 B, DNA replication in CtIP-depleted extracts exposed to etoposide was partly restored by addition of wild-type CtIP (xCtIP WT), but not xCtIP S328A (lanes 5 and 6).
We then analyzed the contribution of CtIP-BRCA1 interaction in Top2 adduct removal. Consistent with the defect in TOP2 removal in BRCA1-depleted extracts (Fig. 5 C), we observed that whereas xCtIP WT significantly restored TOP2 removal in CtIP-depleted extracts, xCtIP S328A did not reverse the defect in TOP2-removal (Fig. 6 C).
Finally, we compared the ability of CtIP polypeptides to restore resection of endonuclease- or etoposide-generated DSBs in CtIP-depleted extracts (Fig. 6 D). In accord with our previous observation (Peterson et al., 2011), resection of DSBs induced by PflMI nuclease was readily restored upon supplementation with either polypeptide (WT or S328A). In contrast, xCtIP WT, but not xCtIP S328A, supported resection of etoposide-induced DSBs (Fig. 6 D, compare lanes 6 and 12).
Together, these results establish that CtIP-BRCA1 interaction is specifically required for processing Top2–DNA adducts.
Discussion
Aberrant repair of DSBs is associated with multiple pathologies, including cancer, neurodegeneration, immunodeficiency, birth defects, and sterility (Huertas et al., 2008; Rulten and Caldecott, 2013). For example, mutations in the DSB-processing factors Nbs1, Mre11, and CtIP underlie the chromosome instability disorders Nijmegen breakage syndrome, ataxia telangiectasia–like disorder, and Seckel syndrome, respectively. This highlights the prominent role of the MRN–CtIP pathway in preserving genome stability in human cells. DNA–topoisomerase adducts are toxic lesions that result from normal cell physiology and chemotherapeutic treatments with topoisomerase inhibitors. These lesions block RNA transcription and DNA replication and must be removed to preserve genomic integrity and cell homeostasis. Genetic studies in yeast have identified proteins responsible for processing protein–DNA adducts, in particular DNA–topoisomerase adducts. Studies in mammalian cells have furthered our understanding of the cellular strategies used to deal with these adducts. Here, we have used cell-free extracts from Xenopus eggs to demonstrate the coordinated action of the MRN complex, CtIP, and BRCA1 (which is not found in yeast) in the processing of Top2–DNA adducts. Cell-free extracts uniquely allow assessment of the consequences of depleting these otherwise essential proteins in the presence of topoisomerase poisons on genomic DNA replication, DNA end processing, and adduct removal.
A previous study showed that human cells depleted of CtIP are hypersensitive to etoposide, a Top2 inhibitor that is widely used as a chemotherapeutic drug (Sartori et al., 2007). Nevertheless, the biochemical basis for this sensitivity was unclear. Here, we use Xenopus egg extracts to implicate CtIP, the MRN complex, and BRCA1 in removal of the 5′ DNA–protein adducts generated by etoposide. First, we show that CtIP, MRN, and BRCA1 are required for chromosomal replication in extracts treated with a low dose of etoposide. Interestingly, we also observed a similar replication sensitivity to low dose of topotecan, a derivative the Top1 inhibitor camptothecin (unpublished data). MRN and CtIP act in the same pathway of Top2 adduct tolerance, because co-depletion of MRN and CtIP did not further inhibit DNA replication in the presence of etoposide relative to depletion of either protein alone. We also show that CtIP is required to remove unprocessed Top2–DNA adducts and that resection of etoposide-induced DSBs is strongly inhibited in CtIP-depleted extracts. These biochemical observations in vertebrates are consistent with genetic studies of the MRN–CtIP orthologues in yeast (Neale et al., 2005; Hartsuiker et al., 2009; Cannavo and Cejka, 2014). In contrast to endonuclease-generated DSBs, which can be processed by Exo1 in the absence of CtIP, there is a strict requirement for CtIP and BRCA1 in processing protein–DNA adducts induced by etoposide. This lack of redundancy might be exploited clinically by enhancing the efficacy of etoposide chemotherapy with inhibitors of the MRN–CtIP pathway, such as mirin and its derivatives (Dupré et al., 2008; Shibata et al., 2014). Top2–DNA adducts are thought to be excised throughout the cell cycle. Indeed, we observe resection defects from Top2–DNA adduct sites in extracts that support DNA replication (Fig. 3) as well as in HSS, a cytosolic extract that cannot replicate DNA (Fig. 4, C and D). Therefore, processing of Top2–DNA adducts by CtIP is critical both during and outside of DNA replication.
Eukaryotic cells possess two redundant mechanisms to remove covalently trapped topoisomerases. On the one hand, topoisomerase adducts can be eliminated by proteasomal degradation of the topoisomerase along with excision of the residual adducted peptide by a tyrosyl-DNA phosphodiesterase such as TDP2. Additionally, direct removal can by achieved by the nucleolytic activities of MRN–CtIP. The latter pathway was initially described to remove adducts of Spo11, a Top2-related protein that generates the DSBs required for meiotic recombination (Neale et al., 2005). Nucleolytic processing by MRN–CtIP may represent a relatively rapid means of eliminating DSB adducts that might otherwise cause a catastrophic collapse of replication forks. Furthermore, this nucleolytic pathway generates a suitable substrate for resection initiation by either the Mre11 or Exo1 nucleases, thereby promoting subsequent DSB repair through HDR. Of note, repair of other classes of DNA–protein adducts, such as these generated after formaldehyde exposure, rely on HDR for repair (Ide et al., 2011; Duxin et al., 2014).
We have also elucidated a specific biochemical role for BRCA1 and for CtIP interaction with BRCA1 in processing protein–DNA adducts at DSB ends before resection (Figs. 5 and 6). Although CtIP and MRN are clearly implicated in resection of clean endonuclease-generated DSBs, it remained uncertain whether the resection activity of CtIP depends on its interaction with BRCA1. Indeed, our previous study in Xenopus extracts (Peterson et al., 2011) as well as work described in Fig. 6 D show that substituting CtIP 328A for CtIP WT does not affect resection of DSBs generated by endonucleases. BRCA1-deficient human cell lines and tumors are sensitive to etoposide, and this sensitivity is dependent on Top2 (Treszezamsky et al., 2007), suggesting that most etoposide-mediated DNA damage is repaired in S phase using homologous recombination. We establish that etoposide sensitivity in BRCA1-depleted extracts is associated with strong inhibition of DNA replication (Fig. 5 B) with reduction in Top2–DNA adduct removal (Fig. 5 C) and a marked defect in resection (Fig. 5 D). Strikingly, these defects are not observed with ligatable, endonuclease-generated DSBs (Fig. 5 D). The interaction with BRCA1 is dependent on CDK-mediated phosphorylation of human CtIP at amino acid S327 (Yu and Chen, 2004). Several studies have addressed the impact of replacing this residue (or its equivalent) with a nonphosphorylatable alanine in mouse cells, human cells, chicken DT40 cells, and Xenopus extracts (Yu and Chen, 2004; Hartsuiker et al., 2009; Yun and Hiom, 2009; Nakamura et al., 2010; Peterson et al., 2011; Reczek et al., 2013; Cruz-García et al., 2014; Polato et al., 2014). An early study in chicken DT40 cells found that a CtIP-S332A mutation enhanced cellular sensitivity to ionizing radiation (IR) and profoundly reduced HDR repair of endonuclease-generated DSBs (Yun and Hiom, 2009). In contrast, a subsequent study in DT40 cells observed no effects on HDR or IR sensitivity but instead reported that ablation of the CtIP–BRCA1 interaction increased sensitivity to DNA Top1 and Top2 inhibitors (Nakamura et al., 2010). Consistent with the latter, mouse cells expressing CtIP-S326A have no defects in IR sensitivity or HDR repair of endonuclease-generated DSB, but they display a modestly increased cellular sensitivity to camptothecin and etoposide (Reczek et al., 2013).
By examining the effects of CtIP on processing etoposide-treated extract, we now demonstrate that the CtIP–BRCA1 interaction is required for MRN-CtIP–mediated removal of Top2–DNA adducts, a prerequisite for subsequent resection of the processed DSB ends. These biochemical observations explain the specific sensitivity of DT40 cells and mouse cells carrying CtIP-S332/326A alleles to topoisomerase inhibitors (Nakamura et al., 2010; Reczek et al., 2013), as well as the recent observation that CtIP–BRCA1 binding affects the speed of DNA resection, most prominently when DNA is blocked by a Top2–DNA adduct (Cruz-García et al., 2014). Interestingly, recent studies suggest that Top2 adduct removal can also be achieved by an intrinsic endonuclease activity of CtIP (Makharashvili et al., 2014; Wang et al., 2014). However, because this mechanism is independent of both MRN and the CtIP–BRCA1 interaction, it is clearly distinct from the MRN-CtIP-BRCA1–dependent processing of Top2–DNA adducts described here. In any event, the MRN–CtIP–BRCA1 pathway for resolving Top2–DNA adducts may be relevant to the tumor suppression activity of BRCA1, and its ablation could contribute to the cancer susceptibility of BRCA1-mutation carriers.
Whereas BRCA1 is required in vivo for HDR of endonuclease-generated DSBs (Moynahan et al., 1999), recent studies have uncovered additional requirements for BRCA1 in maintaining the function of DNA replication forks. For example, BRCA1 is proposed to alleviate replication fork stalling during replication-coupled repair of DNA interstrand cross-links (Bunting et al., 2012; Long et al., 2014), protect stalled replication forks from degradation (Schlacher et al., 2012), and dictate the outcome of homologous recombination at stalled replication forks (Willis et al., 2014). These findings raise the question of whether the CtIP–BRCA1 interaction also facilitates DNA transactions at blocked replication forks. A replication-specific role would be consistent with the observation that NHEJ repair of etoposide-induced damage in nonreplicating cells is independent of the CtIP–BRCA1 interaction (Quennet et al., 2011).
Finally, by demonstrating the requirement of CtIP in its interaction with BRCA1 in the repair of Top2 DNA adducts, our results explain why BRCA1-deficient tumor cells are sensitive to Top2 poisons such as etoposide.
Materials and methods
Extract preparation
Xenopus frogs were handled in accordance with guidelines provided by the Institutional Animal Care and Use Committee at Columbia University. Preparation of replication-competent cytosol (low-speed supernatant [LSS]), replication-competent membrane-free cytosol (HSS), and demembranated sperm nuclei (chromatin) were performed as described previously (Peterson et al., 2011, 2013). In brief, female Xenopus (Nasco) were injected subcutaneously with 50 U of pregnant mare serum gonadotropin (EMD Millipore) 3–7 d before extract preparation. Frogs were induced to lay eggs by subcutaneous injection of 800 U of human chorionic gonadotropin (Sigma-Aldrich). Eggs were collected overnight and rinsed in 0.25× MMR (20 mM Hepes-KOH, pH 7.8, 400 mM NaCl, 1 mM MgSO4, 2 mM CaCl2, and 0.1 mM EDTA). The jelly coat was dissolved in 10 mM cysteine in 0.25× MMR, and the eggs were washed with 0.25× MMR, activated with 1 µg/ml calcium ionophore A23187 (Sigma-Aldrich), and then washed with S buffer (50 mM Hepes-KOH, pH 7.8, 50 mM KCl, 2.5 mM MgCl2, 250 mM sucrose, and 2 mM β-mercaptoethanol). Eggs were crushed in polypropylene tubes at 16,500 g for 30 min at 4°C. The crude cytosol was collected with a needle, supplemented with 20 µg/ml cytochalasin B (Sigma-Aldrich), and homogenized by rotation for 5 min at 4°C. The extract was then subjected to a high-speed spin in an ultracentrifuge (Optima X-100; Beckman-Coulter) in a swing-bucket rotor (TLS-55) for 30 min at 200,000 g at 4°C. The cytosolic and lipid membrane fractions were collected (excluding mitochondria directly below the membrane layer) and supplemented with 30 mM creatine phosphate, 150 µg/ml phosphocreatine kinase, and 20 µg/ml cycloheximide. Similarly, HSS extract was prepared by spinning the crude extract prepared as described at 200,000 g for 2.5 h. The clear, membrane-free HSS extract (top layer) was recovered, carefully excluding the dense membrane layer below.
For chromatin isolation, 30-µl sperm-containing aliquots were diluted in 800 µl ice-cold chromatin isolation buffer (50 mM Hepes-KOH, pH 7.8, 100 mM KCl, and 2.5 mM MgCl2) supplemented with 0.125% Triton X-100 and overlaid onto 30% sucrose (wt/vol) chromatin isolation buffer. Samples were spun at 8,500 g for 30 min at 4°C in a swing-bucket rotor (HB-6; Sorval). Chromatin pellets were processed for Western blotting according to standard procedures (Peterson et al., 2011, 2013). Etoposide and ICRF-193 were purchased from Sigma-Aldrich. ATM inhibitor KU-55933 and ATR inhibitor VE-821 were purchased from Selleck Chemicals.
Antibodies, immunodepletions, and recombinant proteins
The following antibodies were used in these studies: mouse anti-CtIP (clone 11-1; Yu and Baer, 2000; Peterson et al., 2011), rabbit anti-Xenopus Mre11 (Di Virgilio and Gautier, 2005), mouse anti–H2AX-pS139 (05-636; Millipore), rabbit anti–histone H3 (9715; Cell Signaling Technology), mouse anti-Ku70 (SC-56129; Santa Cruz Biotechnology, Inc.), rabbit anti-Xenopus Exo1 (gift from J. Jirincy, University of Zurich, Zurich, Switzerland), rabbit anti-Xenopus topoisomerase IIα (gift from T. Hirano, Institute of Physical and Chemical Research Institute, Wako, Japan; Hirano and Mitchison, 1993), rabbit anti–CHK1-pS345 (#2341; Cell Signaling), and rabbit anti-Xenopus BRCA1 (gift from V. Joukov, Harvard Medical School, Boston, MA; Joukov et al., 2001).
Immunodepletions were performed by binding washed protein A–Sepharose CL-4B beads (GE Healthcare) with rabbit serum (Mre11, Exo1, and BRCA1) or 11–1 hybridoma (CtIP) supernatant overnight with constant rotation at 4°C in compact reaction columns (USB-Affimetrix). The antibody beads were then washed extensively in extract buffer, resuspended in extract, rotated at 4°C for 30 min, and collected for a second round of depletion. A 3:1 ratio of extract/beads (bed volume) was used for all immunodepletions. The amount of antibody used for each depletion (extract/antibody volume used to preload protein A beads) was as follows: mouse IgG (015-000-002; Jackson ImmunoResearch Laboratories) for mock depletions (1:0.044), anti-Xenopus Mre11 rabbit serum (1:0.5), rabbit serum anti-Exo1 (1:0.5), CtIP 11–1 monoclonal hybridoma supernatant (1:1.5), and anti-Xenopus BRCA1 (1:0.25).
Cloning of Xenopus CtIP into pFastBac1 vector and its mutagenesis was described previously (Peterson et al., 2011, 2013). Protein expression in Sf9 insect cells was performed using the Bac-to-Bac System (Invitrogen) following the manufacturer’s instructions. For purification of recombinant xCtIP-WT and xCtIP-S328A, cells were collected 96 h after infection Sf9 and homogenized in buffer A (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5% Triton X-100, and 10% glycerol) supplemented with phosphatase inhibitors (250 mM NaF, 50 mM sodium vanadate, 50 mM β-glycerophosphate, and 50 mM tetrasodium pyrophosphate). Lysate was cleared by centrifugation and incubated with equilibrated anti-FLAG M2 monoclonal antibody–conjugated beads (Sigma-Aldrich) for 3 h at 4°C. Beads were washed extensively in buffer A. Proteins ware eluted in 25 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 20% glycerol plus 300 µg/ml 3× FLAG peptide (Sigma-Aldrich). Small aliquots were flash-frozen in liquid nitrogen and stored at −80°C. T. Paull (University of Texas, Austin, TX) provided recombinant human Mre11-Rad50 complex and Nbs1 proteins. MRN complex was reconstituted by mixing equimolar amounts at the time of use.
DNA replication assays
Reactions containing 10 µl LSS extracts and sperm nuclei (2,000 nuclei/µl) were supplemented with α-[32P]dCTP and incubated at 21°C. Extract was then diluted in 0.1% SDS, 50 mM Tris-HCl, pH 8.0, and 5 mM EDTA. Genomic DNA was isolated by proteinase K digestion at 37°C for 2 h before phenol/chloroform extraction and ethanol precipitation. The DNA pellet was then resuspended in 20 µl TE buffer and run on a 0.8% agarose gel, fixed in 30% trichloroacetic acid, dried by pressing between Whatman 3MM Chromatography Paper and paper towels overnight, and exposed for autoradiography (Srinivasan and Gautier, 2011). Signal quantification was performed using the “Analyze Gel” tool in ImageJ v.1.48. Statistical analyses were performed using GraphPad Prism v5.0c (GraphPad Software).
Top2 covalent complex detection assay
For detection of covalently bound Top2–DNA complexes, we adapted a method previously described (Hartsuiker et al., 2009; Hartsuiker, 2011) with several modifications. In brief, 10-µl extract reactions containing 5,000 sperm/µl were diluted in 300 µl DNAzol reagent (Life Technologies) and incubated at 65°C for 1 h. After cooling to room temperature and clarification at 14,000 g for 15 min, supernatants containing equal amounts of genomic DNA were loaded onto 2.4-ml CsCl gradients (1.50–1.82 g/ml) and centrifuged at 36,000 g for 18 h (Optima X-100, rotor TLS-55; Beckman-Coulter). Ten 250-µl fractions were collected from the bottom of the tube with the aid of a peristaltic pump discarding top and bottom fractions. DNA was sheared in a water bath sonicator for 15 min and slot-blotted onto activated PDVF membrane (Thermo Fisher Scientific). Membranes were washed extensively in PBS–Tween 20 and immunoblotted against topoisomerase IIα. Signal quantification was performed using the Analyze Gel tool in ImageJ v.1.48 for fractions 4–8; means were calculated and plotted using GraphPad Prism v5.0c.
Online supplemental material
Fig. S1 shows the sensitivity of CtIP- and MRN-depleted extracts to low doses of etoposide. Fig. S2 describes the assay to detect etoposide-induced Top2–DNA adducts in egg extracts and purified recombinant CtIP proteins used in this study. Fig. S3 shows impaired DNA-end resection and checkpoint activation after MRN and CtIP depletion. Fig. S4 shows that Exo1 and CtIP initiate resection at etoposide-induced DNA breaks. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504005/DC1.
Supplementary Material
Acknowledgments
We thank members of the Gautier laboratory for helpful discussions, Tanya Paull for the recombinant human MRN complex, Tatsuya Hirano for Xenopus anti–topoisomerase IIα antiserum, Dr. Josef Jirincy for Xenopus anti-Exo1 antiserum, and Dr. Vladimir Joukov for Xenopus anti-BRCA1 antibodies.
This work was supported in part by National Cancer Institute grants CA092245, CA167826, and CA174653 to J. Gautier.
The authors declare no competing financial interests.
Author contributions: T. Aparicio carried out all experiments; T. Aparicio, R. Baer, M. Gottesman, and J. Gautier conceived experiments; and T. Aparicio, M. Gottesman, and J. Gautier wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- DSB
- double-strand break
- HDR
- homology-directed repair
- HSS
- high-speed supernatant
- IR
- ionizing radiation
- LSS
- low-speed supernatant
- NHEJ
- nonhomologous end joining
- RPA
- replication protein A
- Top2
- topoisomerase II
- WT
- wild type
References
- Andres S.N., Schellenberg M.J., Wallace B.D., Tumbale P., and Williams R.S.. 2014. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio T., Baer R., and Gautier J.. 2014. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst.). 19:169–175. 10.1016/j.dnarep.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Kozak M.L., Kim J.M., Wong N., López-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A., et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 46:125–135. 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., and Cejka P.. 2014. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 514:122–125. 10.1038/nature13771 [DOI] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R., and Boulton S.J.. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 47:497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Cortes Ledesma F., El Khamisy S.F., Zuma M.C., Osborn K., and Caldecott K.W.. 2009. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 461:674–678. 10.1038/nature08444 [DOI] [PubMed] [Google Scholar]
- Costanzo V., Robertson K., Bibikova M., Kim E., Grieco D., Gottesman M., Carroll D., and Gautier J.. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 8:137–147. 10.1016/S1097-2765(01)00294-5 [DOI] [PubMed] [Google Scholar]
- Cruz-García A., López-Saavedra A., and Huertas P.. 2014. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Reports. 9:451–459. 10.1016/j.celrep.2014.08.076 [DOI] [PubMed] [Google Scholar]
- de Massy B., Rocco V., and Nicolas A.. 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 14:4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M., and Gautier J.. 2005. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J. Cell Biol. 171:765–771. 10.1083/jcb.200506029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré A., Boyer-Chatenet L., Sattler R.M., Modi A.P., Lee J.H., Nicolette M.L., Kopelovich L., Jasin M., Baer R., Paull T.T., and Gautier J.. 2008. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4:119–125. 10.1038/nchembio.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin J.P., Dewar J.M., Yardimci H., and Walter J.C.. 2014. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 159:346–357. 10.1016/j.cell.2014.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid W., Steger M., El-Shemerly M., Ferretti L.P., Peña-Diaz J., König C., Valtorta E., Sartori A.A., and Ferrari S.. 2010. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 11:962–968. 10.1038/embor.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Schellenberg M.J., Huang S.Y., Abdelmalak M., Marchand C., Nitiss K.C., Nitiss J.L., Williams R.S., and Pommier Y.. 2014. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2·DNA and Top2·RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2). J. Biol. Chem. 289:17960–17969. 10.1074/jbc.M114.565374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E. 2011. Detection of covalent DNA-bound Spo11 and topoisomerase complexes. Methods Mol. Biol. 745:65–77. 10.1007/978-1-61779-129-1_5 [DOI] [PubMed] [Google Scholar]
- Hartsuiker E., Neale M.J., and Carr A.M.. 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 33:117–123. 10.1016/j.molcel.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., and Smith G.C.. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159. 10.1158/0008-5472.CAN-04-2727 [DOI] [PubMed] [Google Scholar]
- Hirano T., and Mitchison T.J.. 1993. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol. 120:601–612. 10.1083/jcb.120.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., and Jackson S.P.. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284:9558–9565. 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Cortés-Ledesma F., Sartori A.A., Aguilera A., and Jackson S.P.. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 455:689–692. 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H., Shoulkamy M.I., Nakano T., Miyamoto-Matsubara M., and Salem A.M.. 2011. Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 711:113–122. 10.1016/j.mrfmmm.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Joukov V., Chen J., Fox E.A., Green J.B., and Livingston D.M.. 2001. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. USA. 98:12078–12083. 10.1073/pnas.211427098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1–53. 10.1016/S0070-2153(01)52008-6 [DOI] [PubMed] [Google Scholar]
- Keeney S., and Kleckner N.. 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA. 92:11274–11278. 10.1073/pnas.92.24.11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., and Kleckner N.. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 88:375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kiianitsa K., and Maizels N.. 2013. A rapid and sensitive assay for DNA-protein covalent complexes in living cells. Nucleic Acids Res. 41:e104 10.1093/nar/gkt171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.C., Padget K., Curtis H., Cowell I.G., Moiani D., Sondka Z., Morris N.J., Jackson G.H., Cockell S.J., Tainer J.A., and Austin C.A.. 2012. MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol. Open. 1:863–873. 10.1242/bio.20121834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Toczylowski T., and Yan H.. 2011. Mechanistic analysis of Xenopus EXO1‘s function in 5’-strand resection at DNA double-strand breaks. Nucleic Acids Res. 39:5967–5977. 10.1093/nar/gkr216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu T.C., and Lichten M.. 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: Evidence for a covalently linked DNA-protein intermediate. EMBO J. 14:4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F., Rowe T.C., Yang L., Tewey K.M., and Chen G.L.. 1983. Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 258:15365–15370. [PubMed] [Google Scholar]
- Long D.T., Joukov V., Budzowska M., and Walter J.C.. 2014. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol. Cell. 56:174–185. 10.1016/j.molcel.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N., Tubbs A.T., Yang S.H., Wang H., Barton O., Zhou Y., Deshpande R.A., Lee J.H., Lobrich M., Sleckman B.P., et al. 2014. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell. 54:1022–1033. 10.1016/j.molcel.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Desai S.D., Ting C.Y., Hwang J., and Liu L.F.. 2001. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 276:40652–40658. 10.1074/jbc.M104009200 [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., and Jasin M.. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell. 4:511–518. 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. 2010. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6:e1000828 10.1371/journal.pgen.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Pan J., and Keeney S.. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 436:1053–1057. 10.1038/nature03872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., and Kowalczykowski S.C.. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–362. 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J.L. 2009. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 9:338–350. 10.1038/nrc2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.E., Li Y., Chait B.T., Gottesman M.E., Baer R., and Gautier J.. 2011. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J. Cell Biol. 194:705–720. 10.1083/jcb.201103103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.E., Li Y., Wu-Baer F., Chait B.T., Baer R., Yan H., Gottesman M.E., and Gautier J.. 2013. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell. 49:657–667. 10.1016/j.molcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polato F., Callen E., Wong N., Faryabi R., Bunting S., Chen H.T., Kozak M., Kruhlak M.J., Reczek C.R., Lee W.H., et al. 2014. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 211:1027–1036. 10.1084/jem.20131939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Leo E., Zhang H., and Marchand C.. 2010. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17:421–433. 10.1016/j.chembiol.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quennet V., Beucher A., Barton O., Takeda S., and Löbrich M.. 2011. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 39:2144–2152. 10.1093/nar/gkq1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvist P., Huertas P., Jimeno S., Nyegaard M., Hassan M.J., Jackson S.P., and Børglum A.D.. 2011. CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet. 7:e1002310 10.1371/journal.pgen.1002310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper P.M., Griffiths M.R., Long J.M., Charrier J.D., Maccormick S., Charlton P.A., Golec J.M., and Pollard J.R.. 2011. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 7:428–430. 10.1038/nchembio.573 [DOI] [PubMed] [Google Scholar]
- Reczek C.R., Szabolcs M., Stark J.M., Ludwig T., and Baer R.. 2013. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 201:693–707. 10.1083/jcb.201302145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T.C., Tewey K.M., and Liu L.F.. 1984. Identification of the breakage-reunion subunit of T4 DNA topoisomerase. J. Biol. Chem. 259:9177–9181. [PubMed] [Google Scholar]
- Rulten S.L., and Caldecott K.W.. 2013. DNA strand break repair and neurodegeneration. DNA Repair (Amst.). 12:558–567. 10.1016/j.dnarep.2013.04.008 [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., and Jackson S.P.. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F., et al. 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 53:7–18. (published erratum appears in Mol. Cell. 2014. 53:361.) 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S.V., and Gautier J.. 2011. Study of cell cycle checkpoints using Xenopus cell-free extracts. Methods Mol. Biol. 782:119–158. 10.1007/978-1-61779-273-1_10 [DOI] [PubMed] [Google Scholar]
- Symington L.S., and Gautier J.. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271. 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- Tewey K.M., Rowe T.C., Yang L., Halligan B.D., and Liu L.F.. 1984. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 226:466–468. 10.1126/science.6093249 [DOI] [PubMed] [Google Scholar]
- Tomimatsu N., Mukherjee B., Catherine Hardebeck M., Ilcheva M., Vanessa Camacho C., Louise Harris J., Porteus M., Llorente B., Khanna K.K., and Burma S.. 2014. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat. Commun. 5:3561 10.1038/ncomms4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treszezamsky A.D., Kachnic L.A., Feng Z., Zhang J., Tokadjian C., and Powell S.N.. 2007. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 67:7078–7081. 10.1158/0008-5472.CAN-07-0601 [DOI] [PubMed] [Google Scholar]
- Vos S.M., Tretter E.M., Schmidt B.H., and Berger J.M.. 2011. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12:827–841. 10.1038/nrm3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y., He J., Do J., Cho M.J., Li H., Negrete A., et al. 2014. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 54:1012–1021. 10.1016/j.molcel.2014.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis N.A., Chandramouly G., Huang B., Kwok A., Follonier C., Deng C., and Scully R.. 2014. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 510:556–559. 10.1038/nature13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.C., Li T.K., Farh L., Lin L.Y., Lin T.S., Yu Y.J., Yen T.J., Chiang C.W., and Chan N.L.. 2011. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 333:459–462. 10.1126/science.1204117 [DOI] [PubMed] [Google Scholar]
- You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.-W., Basilio A., Tonnu N., Verma I.M., Berns M.W., and Hunter T.. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 36:954–969. 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., and Baer R.. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275:18541–18549. 10.1074/jbc.M909494199 [DOI] [PubMed] [Google Scholar]
- Yu X., and Chen J.. 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24:9478–9486. 10.1128/MCB.24.21.9478-9486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., Aronheim A., and Baer R.. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388–25392. 10.1074/jbc.273.39.25388 [DOI] [PubMed] [Google Scholar]
- Yun M.H., and Hiom K.. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463. 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Lyu Y.L., Lin C.P., Zhou N., Azarova A.M., Wood L.M., and Liu L.F.. 2006. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 281:35997–36003. 10.1074/jbc.M604149200 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Caron P., Legube G., and Paull T.T.. 2014. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 42:e19 10.1093/nar/gkt1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.