The C-terminal domain of NLG1 is sufficient to enhance spine and synapse number and to modulate synaptic plasticity, and it exerts these effects via its interaction with SPAR and the subsequent activation of LIMK1/cofilin-mediated actin reorganization.
Abstract
Neuroligin (NLG) 1 is important for synapse development and function, but the underlying mechanisms remain unclear. It is known that at least some aspects of NLG1 function are independent of the presynaptic neurexin, suggesting that the C-terminal domain (CTD) of NLG1 may be sufficient for synaptic regulation. In addition, NLG1 is subjected to activity-dependent proteolytic cleavage, generating a cytosolic CTD fragment, but the significance of this process remains unknown. In this study, we show that the CTD of NLG1 is sufficient to (a) enhance spine and synapse number, (b) modulate synaptic plasticity, and (c) exert these effects via its interaction with spine-associated Rap guanosine triphosphatase–activating protein and subsequent activation of LIM-domain protein kinase 1/cofilin–mediated actin reorganization. Our results provide a novel postsynaptic mechanism by which NLG1 regulates synapse development and function.
Introduction
Neuroligins (NLGs) are type I transmembrane proteins containing an extracellular cholinesterase domain, a transmembrane region, and a cytoplasmic C-terminal domain (CTD). Extensive studies have indicated that NLGs are important in synapse development and function (Baksh et al., 2005; Chubykin et al., 2007; Conroy et al., 2007; Dong et al., 2007; Futai et al., 2007). For example, expression of NLGs in nonneuronal cells cocultured with primary neurons induces synapse formation onto the nonneuronal cells, suggesting that NLGs may play a role in the initial establishment of the synapse (Scheiffele et al., 2000; Graf et al., 2004; Cline, 2005; Nam and Chen, 2005; Dean and Dresbach, 2006; Gottmann, 2008; Kwon et al., 2012). In addition, overexpression of NLGs in transfected neurons increases the number of spines and synapses (Chih et al., 2004; Boucard et al., 2005). Consistent with these in vitro studies, knockout (KO) mice lacking NLGs are severely impaired in synaptic transmission (Kattenstroth et al., 2004; Varoqueaux et al., 2006; Chubykin et al., 2007). Interestingly, the KO mice did not exhibit a reduction in synapse numbers, suggesting that NLGs are not essential for the initial formation of the synapses, but rather in their functional regulation during synaptic activity (Chubykin et al., 2007).
How NLGs exert their effects on spines and synapses remains unclear. Postsynaptic NLGs are thought to function by binding to and dimerizing presynaptic neurexins. Consistent with this idea, genetic deletions of neurexins have been shown to have a similar effect on synapse development and function (Tabuchi et al., 2007; Blundell et al., 2010). In addition, manipulations of presynaptic neurexin are sufficient to induce presynaptic specializations (Dean et al., 2003) and changes in postsynaptic glutamate receptors (Graf et al., 2004; Kattenstroth et al., 2004). Indeed, NLG1 mutants deficient in presynaptic neurexin binding are impaired in NLG-induced formation of neuronal synapses onto transfected nonneuronal cells (Varoqueaux et al., 2006). However, in transfected neurons, the effect of NLG1 on spine and synapse seems independent of either α- or β-neurexin. Therefore, overexpression of either wild-type (WT) or mutant NLG1 deficient in neurexin binding equally increases spine and synapse density and synaptic strength (Ko et al., 2009). These results suggest that the CTD of NLG1 may be sufficient in mediating some aspects of NLG1’s effects. In addition, recent studies have demonstrated that NLG1 undergoes activity-dependent proteolytic cleavage, releasing the CTD fragment to the cytosol (Peixoto et al., 2012; Suzuki et al., 2012), but the role of this process remains unknown.
In this study, we have investigated the role of the CTD of NLG1 in spine and synaptic regulation. We demonstrate that the CTD of NLG1 is sufficient to promote spine and synapse growth and that it does so through its interaction with spine-associated Rap GTPase-activating protein (SPAR) and subsequent activation of the LIM-domain protein kinase (LIMK)/cofilin pathway.
Results
NLG1 is a potent activator of cofilin phosphorylation
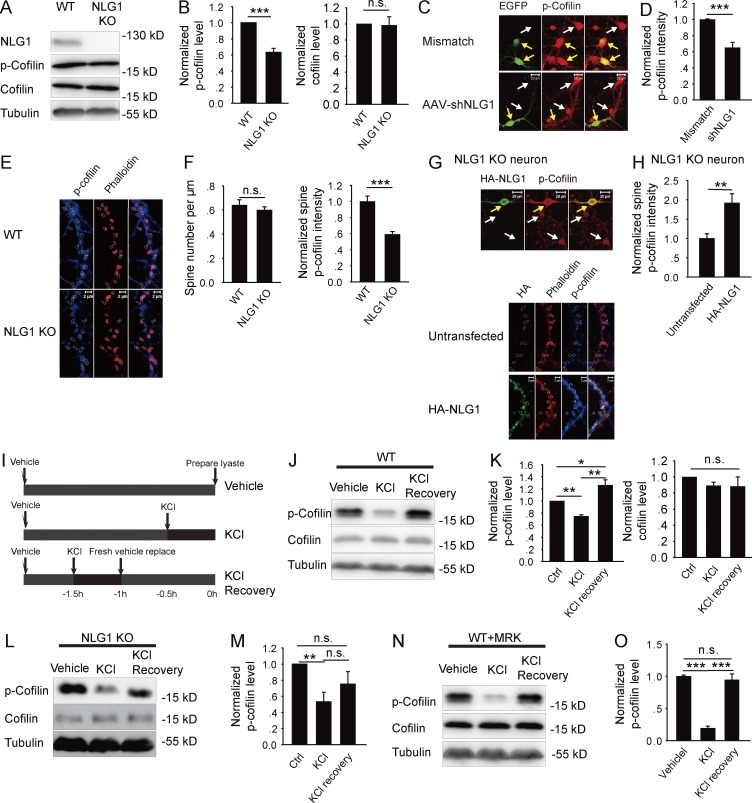
Although overexpression of NLG1 in cultured neurons increases spine and synapse growth (Ko et al., 2009; Kwon et al., 2012), how NLG1 exerts this effect remains unclear. Because both spine and synapse are regulated by the actin cytoskeleton mediated by the Rho GTPases (Govek et al., 2005) and LIMK1/cofilin (Bamburg, 1999; Bernard, 2007; Jia et al., 2009; Rust, 2015), we hypothesized that the effect of NLG1 may be mediated by LIMK1/cofilin. LIMK1 can directly phosphorylate cofilin at Ser3, rendering cofilin completely inactive, resulting in assembly and stabilization of F-actin (Bamburg, 1999; Meng et al., 2002; Bernard, 2007). Therefore, the level of p-cofilin is a reliable indicator of cofilin activity. First, we analyzed the effect of genetic deletion of NLG1 (KO) on p-cofilin. As shown in Fig. 1 (A and B), the level of p-cofilin (0.54 ± 0.05, n = 6, P < 0.001), but not total cofilin (0.98 ± 0.11, n = 6, P = 0.88), was significantly lower in NLG1 KO compared with the WT littermates. Because KO mice may suffer developmental compensations that could indirectly affect p-cofilin, we tested the effect of NLG1 knockdown by NLG1-shRNA (Xu et al., 2010). As shown in Fig. 1 (C and D), the level of p-cofilin was significantly lower in NLG1-shRNA compared with control shRNA-infected neurons (0.65 ± 0.07, n = 13, P < 0.001). The effectiveness of NLG1-shRNA was confirmed in both neurons and HEK293 cells (Fig. S1, A and B). To determine the effect of NLG1 on synaptic p-cofilin, we measured p-cofilin in the dendritic spine (Fig. 1, E and F) and showed that spine p-cofilin (0.59 ± 0.03, n = 10, P < 0.001), but not spine density (WT: 0.64 ± 0.04/µm, n = 11; NLG1 KO: 0.60 ± 0.03/µm, n = 10, P = 0.42), was significantly lower in NLG1 KO compared with WT neurons. Transfection of the KO neurons with HA-NLG1 (Fig. 1, G and H) rescued spine p-cofilin (1.97 ± 0.24, n = 5, P < 0.01 compared with untransfected NLG1 KO neurons). These results suggest that NLG1 is required for maintaining a basal level of p-cofilin. To determine whether NLG1 is involved in activity-dependent cofilin changes, we treated brain slices with KCl, and p-cofilin was analyzed immediately or 1 h after the treatment (Fig. 1 I). As shown in Fig. 1 (J and K), in WT slices p-cofilin was first decreased at the termination of the treatment (0.74 ± 0.03, n = 6, P < 0.001 compared with vehicle control group), but increased significantly after a 1-h recovery period, to a level higher than that of the control group (1.26 ± 0.09, n = 7, P < 0.05), indicating rapid and dynamic changes in cofilin activity induced by neuronal activity. The KCl treatment had no effect on total cofilin (KCl: 0.95 ± 0.06, n = 6, P = 0.43 compared with control group; KCl + recovery: 0.92 ± 0.1, n = 6, P = 0.44 compared with control group). In NLG1 KO slices (Fig. 1, L and M), however, although KCl induced an initial decrease in p-cofilin (0.53 ± 0.12, n = 6, P < 0.01 compared with control group), its subsequent increase after the recovery period was significantly impaired (0.76 ± 0.16, n = 6, P = 0.3 compared with control group). These results suggest that NLG1 is specifically required for activity-induced cofilin phosphorylation, but not cofilin dephosphorylation. Because KCl also induces proteolytic cleavage of NLG1 (Peixoto et al., 2012; Suzuki et al., 2012), which we confirmed (Fig. S2, A–D), we wondered whether the KCl-induced cofilin phosphorylation is related to this cleavage process. We treated cultured neurons with the γ-secretase inhibitor MRK (10 nM) followed by the KCl treatment. As shown in Fig. 1 (N and O), although MRK did not impair KCl-induced decrease in p-cofilin (0.19 ± 0.03, n = 5, P < 0.001 compared with control group), it blocked KCl-induced increase in p-cofilin (0.95 ± 0.09, n = 4, P = 0.57 compared with control group). Collectively, these results suggest that NLG1 is required for cofilin phosphorylation and that the proteolytically released CTD may play a key role in this process.
Figure 1.
NLG1 is required for cofilin phosphorylation. (A) Western blots of whole brain lysate of NLG1 KO and WT mice with indicated antibodies. (B) Summary graphs of A showing a significant reduction in p-cofilin but not total cofilin in NLG1 KO compared with WT mice. (C) Cultured hippocampal neurons infected with NLG1-shRNA or control mismatch shRNA and stained for p-cofilin. Yellow arrows indicate infected neurons (green) and white arrows indicate noninfected neurons. Note reduced p-cofilin intensity in NLG1-shRNA–infected neuron compared with noninfected neuron (bottom). No differences between mismatch shRNA-infected neurons and noninfected neurons (top). (D) Summary graph of C showing significantly reduced p-cofilin in NLG1-shRNA–treated neurons. (E) Cultured hippocampal neurons of NLG1 KO and WT costained for spine (phalloidin) and p-cofilin (circled). (F) Summary graphs of E showing significantly decreased p-cofilin in NLG1 KO compared with WT neurons, without changes in spine number. (G) Cultured hippocampal neurons of NLG1 KO transfected with HA-NLG1 and costained for spine (phalloidin, circled) and p-cofilin. Yellow arrows indicate infected neurons; white arrows indicate noninfected neurons. (H) Summary graph of G showing significantly increased p-cofilin in HA-NLG1–transfected NLG1 KO compared with untransfected neurons. (I) Schematic graph of KCl (30 mM) treatment and recovery. (J) Western blots of protein lysate of WT brain slices immediately or 1 h after KCl treatment. (K) Summary graph of J showing a transient reduction in p-cofilin immediately after KCl treatment and a sustained increase in p-cofilin after the 1-h recovery period in WT brain slices. Ctrl, control. (L) Western blots of protein lysate of NLG1 KO brain slices immediately or 1 h after KCl treatment. (M) Summary graph of L showing significantly reduced p-cofilin right after KCl treatment and no significant increase in p-cofilin after the 1-h recovery period in NLG1 KO brain slices. (N) Western blots of protein lysate of WT brain slices pretreated with MRK (10 nM) followed by the KCl treatment showing a transient decrease in p-cofilin right after the KCl treatment. (O) Summary graph of N showing a transient reduction in p-cofilin after KCl treatment, but no increase in p-cofilin above the baseline after the 1-h recovery period in the presence of MRK in WT slices. n.s., not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CTD of NLG1 is sufficient to induce cofilin phosphorylation
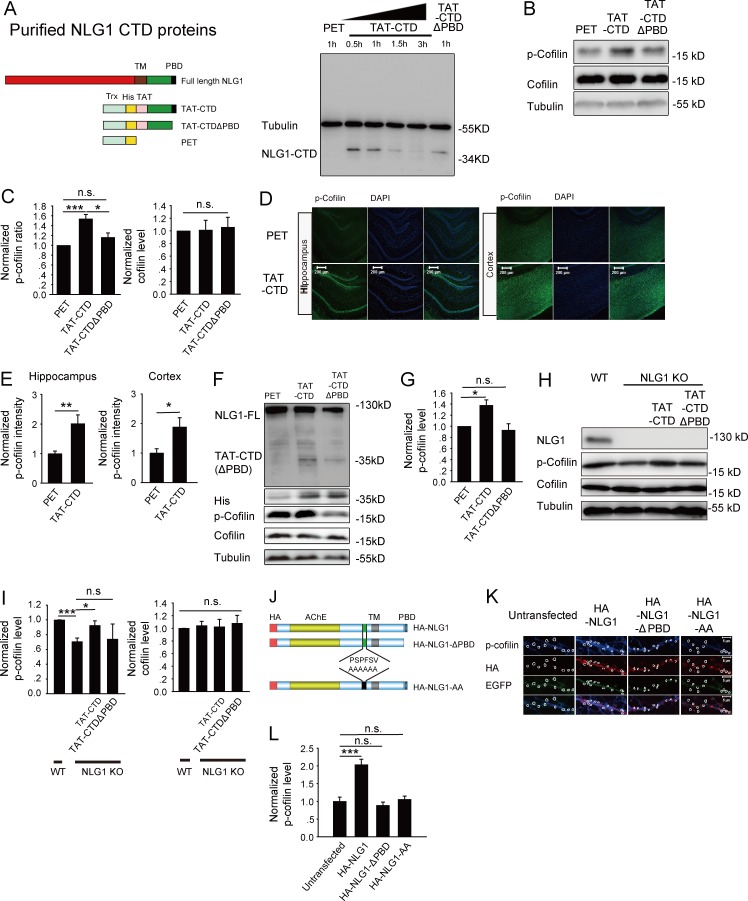
To directly test the role of the CTD of NLG1 in cofilin phosphorylation, we generated and purified three recombinant proteins using the human immunodeficiency virus TAT sequence (Becker-Hapak et al., 2001): TAT-tagged WT NLG1 CTD (TAT-CTD), TAT-tagged mutant CTD lacking the last four aa required for its binding to the PDZ domain (TAT-CTDΔPBD), and a control protein derived from the empty vector PET32a (PET; Fig. 2 A). To determine whether these TAT-tagged proteins could accumulate inside the cells, we treated cultured hippocampal neurons with these proteins (5 µM) for 1 h and analyzed the protein lysate prepared from these neurons using anti-His antibodies. As shown in Fig. 2 A, both TAT-CTD and TAT-CTDΔPBD with an expected molecular weight of 34 kD were detected, indicating that these proteins could enter the neurons. To confirm the cellular accumulation in brain tissues, we analyzed brain slices treated with these proteins and showed that an application of 1 or 5 µM of the recombinant proteins was sufficient to accumulate physiological levels of TAT-CTD (Fig. S3, A–D). Therefore, we used 5 µM of the proteins and 1-h treatment for the rest of the study. First, we examined their effect on cofilin phosphorylation in cultured neurons. As shown in Fig. 2 (B and C), p-cofilin was significantly higher in neurons treated with TAT-CTD compared with those treated with the PET control (1.53 ± 0.13, n = 6, P < 0.05). In contrast, TAT-CTDΔPBD had no significant effect (1.19 ± 0.12, n = 6, P = 0.2 compared with PET). Total cofilin was not affected by any of the recombinant proteins (TAT-CTD: 1.01 ± 0.13, n = 5, P = 0.93 compared with PET; TAT-CTDΔPBD: 1.06 ± 0.16, n = 5, P = 0.73 compared with PET). To rule out the possibility that TAT-CTD might have nonspecific effects on protein phosphorylation, we analyzed phosphorylated Akt but found no differences between TAT-CTD, TAT-NLG1ΔPBD, or PET-treated groups (Fig. S4, A and B). To test whether these proteins could have a similar effect in the brain, we intravenously injected the mice with either TAT-CTD or control PET, and the brain tissues of the injected mice were analyzed by immunohistochemical staining. As shown in Fig. 2 (D and E), p-cofilin was significantly higher in mice treated with TAT-CTD than those treated with control PET (2.01 ± 0.24, n = 5, P < 0.01 for the hippocampus; 1.88 ± 0.32, n = 5, P < 0.05 for the cortex). Western blot analysis of the injected mice (Fig. 2, F and G) also showed that the recombinant proteins were detected in the brain lysate and that p-cofilin was significantly higher in TAT-CTD (1.38 ± 0.1, n = 4, P < 0.05), but not TAT-NLG1ΔPBD (0.93 ± 0.12, n = 3, P = 0.58) compared with the PET-treated group. To test whether TAT-CTD can induce cofilin phosphorylation in the absence of NLG1, we treated the NLG1 KO slices with the recombinant protein, and the results showed that p-cofilin was restored to the WT level by TAT-CTD (0.98 ± 0.06, n = 5, P < 0.05), but not by TAT-NLG1ΔPBD (0.74 ± 0.2, n = 4, P = 0.81) compared with PET-treated NLG1 KO slices. To verify the effect of the recombinant proteins on p-cofilin using an independent approach, we transfected cultured neurons with various NLG1 constructs. As shown in Fig. 2 (J–L), HA-NLG1 (2.04 ± 0.15, n = 16, P < 0.001), but not HA-NLG1ΔPBD (0.89 ± 0.09, n = 10, P = 0.48) or HA-NLG1-AA (a proteolytic cleavage mutant known to decrease the production of NLG1 CTD; Peixoto et al., 2012; Suzuki et al., 2012; 1.06 ± 0.09, n = 6, P = 0.72), increased spine p-cofilin compared with untransfected neurons. Therefore, the CTD of NLG1 is sufficient to activate cofilin phosphorylation, and this process requires its terminal PDZ binding domain (PBD) motif and proteolytic cleavage.
Figure 2.
NLG1 CTD is sufficient to regulate cofilin activity. (A) Generation and purification of TAT-CTD, TAT-CTDΔPBD, and PET recombinant proteins. (left) Schematic graph of the full-length NLG1, TAT-NLG1CTD, TAT-CTDΔPBD, and PET. TM, transmembrane domain; PBD, PDZ binding domain; Trx, thioredoxin tag; His, 6× His tag; TAT, YGRKKRRQRRR. (right) Western blots of protein lysate of cultured neurons treated with TAT-CTD, TAT-CTDΔPBD, or PET for indicated time periods and probed with anti-His antibodies showing the accumulation of TAT-CTD and TAT-CTDΔPBD, but not PET inside the neurons. (B) Western blots of protein lysate of cultured neurons treated with various recombinant proteins. (C) Summary graph of B showing significantly increased p-cofilin, but not total cofilin, in neurons treated with TAT-CTD compared with those treated with TAT-CTDΔPBD or PET. (D) Hippocampal (left) and cortical (right) sections of the mice i.v. injected with TAT-CTD or PET and costained for p-cofilin and the nuclear marker DAPI. (E) Summary graph of D showing significantly increased p-cofilin in both hippocampus and cortex in mice treated with TAT-CTD. (F) Western blots of brain protein lysate from the mice injected with the recombinant proteins showing the presence of TAT-CTD and TAT-CTDΔPBD (as detected by anti-His and anti-NLG1 antibodies). (G) Summary graph of F showing significantly increased p-cofilin, but not total cofilin, in mice treated with TAT-CTD compared with those treated with TAT-CTDΔPBD or PET. (H) Western blots of protein lysate of WT and NLG1 KO brain slices treated with TAT-CTD or TAT-CTDΔPBD. (I) Summary graph of H showing rescued p-cofilin in NLG1 KO brain slices by TAT-CTD but not by TAT-CTDΔPBD treatment. (J) Schematic graph of full-length HA-tagged NLG1, HA-tagged NLG1ΔPBD, and HA-tagged NLG1-AA constructs. (K) Cultured hippocampal neurons transfected with HA-NLG, HA-NLG1ΔPBD, or HA-NLG1-AA and stained for spine p-cofilin (circled). (L) Summary graph of K showing significantly increased spine p-cofilin in HA-NLG1, but not in HA-NLG1ΔPBD– or HA-NLG1-AA–transfected neurons compared with untransfected neurons. n.s., not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CTD of NLG1 is sufficient to enhance spine and synapse growth
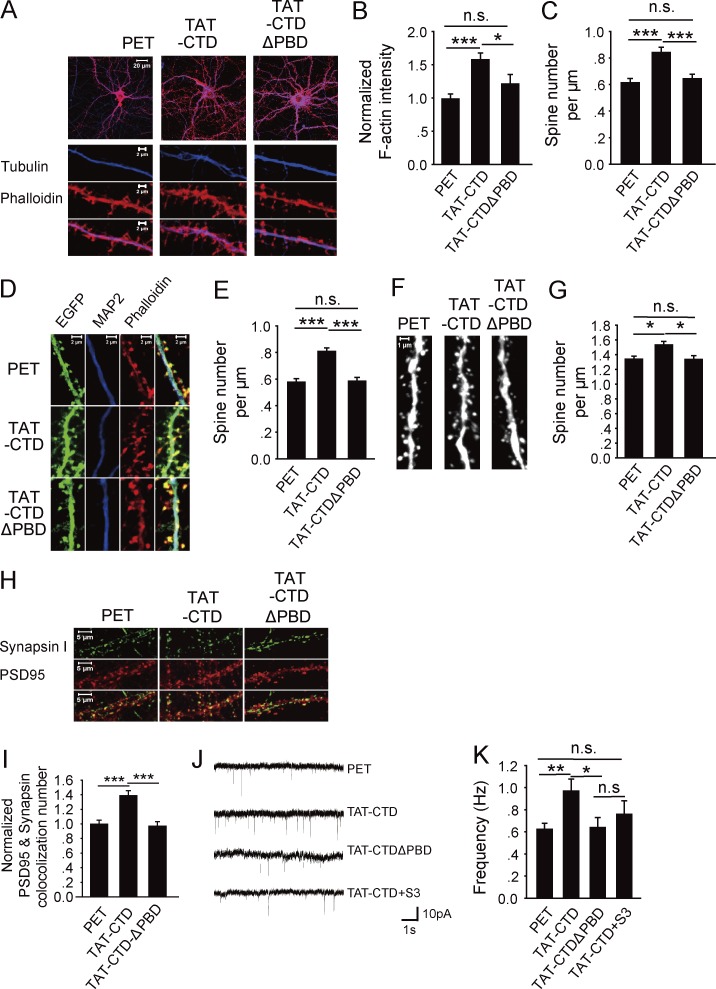
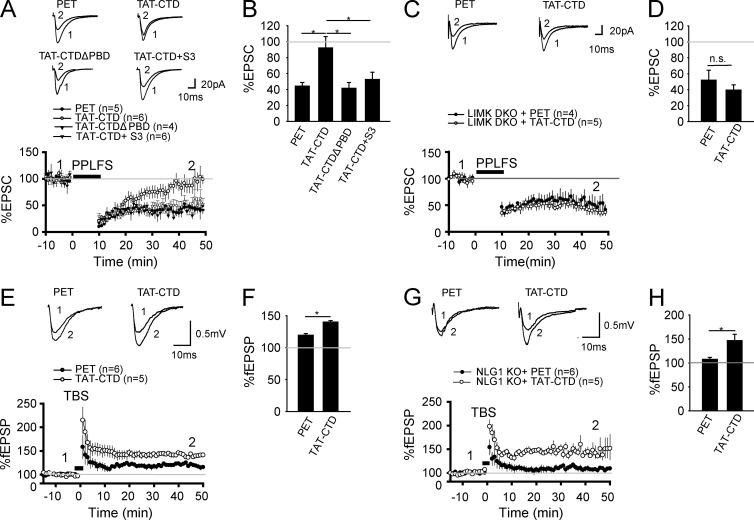
The result that the CTD of NLG1 is sufficient to induce cofilin phosphorylation suggests that it might also be sufficient to promote actin assembly and spine/synapse growth. To test this possibility, we first analyzed F-actin in cultured neurons treated with the recombinant proteins. As shown in Fig. 3 (A and B), TAT-CTD caused a significant increase in the amount of F-actin in both dendrites and spines compared with the control PET or TAT-CTDΔPBD (TAT-CTD: 1.6 ± 0.09, n = 19, P < 0.001 compared with PET; TAT-CTDΔPBD: 1.21 ± 0.14, n = 19, P = 0.13 compared with PET). The spine density measured based on the F-actin staining was also increased in TAT-CTD–treated neurons (Fig. 3 C; PET: 0.62 ± 0.03/µm, n = 21; TAT-CTD: 0.84 ± 0.04/µm, n = 31, P < 0.001 compared with PET; TAT-CTDΔPBD: 0.65 ± 0.03/µm, n = 22, P = 0.13 compared with PET). To verify the effect on spine density, we analyzed cultured hippocampal neurons infected with the adeno-associated virus (AAV)–EGFP virus. As shown in Fig. 3 (D and E), the spine density was increased in TAT-CTD but not TAT-CTDΔPBD–treated neurons compared with the control PET (PET: 0.58 ± 0.02/µm, n = 15; TAT-CTD: 0.81 ± 0.02/µm, n = 23, P < 0.001 compared with PET; TAT-CTDΔPBD: 0.59 ± 0.03/µm, n = 15, P = 0.83 compared with PET). To confirm whether TAT-CTD has the same effect on spines in vivo, we intravenously injected the Thy1-GFP transgenic mice with the recombinant proteins, and the CA1 neurons were analyzed for spine density. As shown in Fig. 3 (F and G), TAT-CTD but not TAT-CTDΔPBD significantly increased spine density compared with the PET group (PET: 1.34 ± 0.04/µm, n = 27; TAT-CTD: 1.53 ± 0.04/µm, n = 23, P < 0.05 compared with PET; TAT-CTDΔPBD: 1.34 ± 0.04/µm, n = 27, P = 0.96 compared with PET). Finally we examined the effect of CTD on synaptic properties. As shown in Fig. 3 (H–K), TAT-CTD treatment resulted in a significant increase in the number of synapses (i.e., colocalization puncta of synapsin I and PSD95; Fig. 3, H and I; TAT-CTD: 1.39 ± 0.07, n = 19, P < 0.001 compared with PET; TAT-CTDΔPBD: 0.97 ± 0.06, n = 18, P = 0.73 compared with PET) and the frequency of miniature excitatory postsynaptic currents (mEPSCs; Fig. 3, J and K; PET: 0.67 ± 0.08 Hz, n = 12; TAT-CTD: 0.97 ± 0.1 Hz, n = 9, P < 0.05 compared with PET; TAT-CTDΔPBD: 0.64 ± 0.09 Hz, n = 10, P = 0.87 compared with PET). Inclusion of the cofilin peptide S3, which inhibits cofilin phosphorylation (Aizawa et al., 2001), to the postsynaptic neurons via the recording pipette blocked the increased frequency induced by the TAT-CTD treatment (0.76 ± 0.12, n = 8, P = 0.27 compared with the PET group). These results indicate that the CTD of NLG1 is sufficient to promote F-actin assembly, spine/synapse growth, and synaptic strength, and this effect requires cofilin phosphorylation.
Figure 3.
NLG1 CTD is sufficient to regulate spine and synapse growth. (A) Cultured hippocampal neurons treated with the recombinant proteins and costained for F-actin (phalloidin) and tubulin. (B) Summary graph of A showing significantly increased F-actin in TAT-CTD compared with TAT-CTDΔPBD– or PET-treated neurons. (C) Summary graph of A showing significantly increased spine number in TAT-CTD compared with TAT-CTDΔPBD– or PET-treated neurons. (D) Cultured hippocampal neurons infected with the AAV-EGFP virus, treated with various recombinant proteins, and stained for F-actin and microtubule-associated protein 2 (MAP2). (E) Summary graph of D showing significantly increased spine number in TAT-CTD compared with TAT-CTDΔPBD– or PET-treated neurons. (F) CA1 dendritic spines of brain sections prepared from the Thy1-YFP transgenic mice i.v. injected with PET, TAT-CTD, or TAT-CTDΔPBD. (G) Summary graph of F showing significantly increased spine density in mice treated with TAT-CTD compared with those treated with PET or TAT-CTDΔPBD. (H) Cultured hippocampal neurons treated with the recombinant proteins and costained for synapsin I and PSD95. (I) Summary graph of H showing significantly increased synapsin I/PSD95 colocalization puncta in TAT-CTD compared with TAT-CTDΔPBD– or PET-treated neurons. (J) Sample traces of mEPSC whole-cell recordings of CA1 neurons of hippocampal slices treated with the recombinant proteins. (K) Summary graph of J showing significantly increased frequency of mEPSC in TAT-CTD compared with PET-, TAT-CTDΔPBD–, or TAT-CTD+S3 peptide–treated neurons. n.s., not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
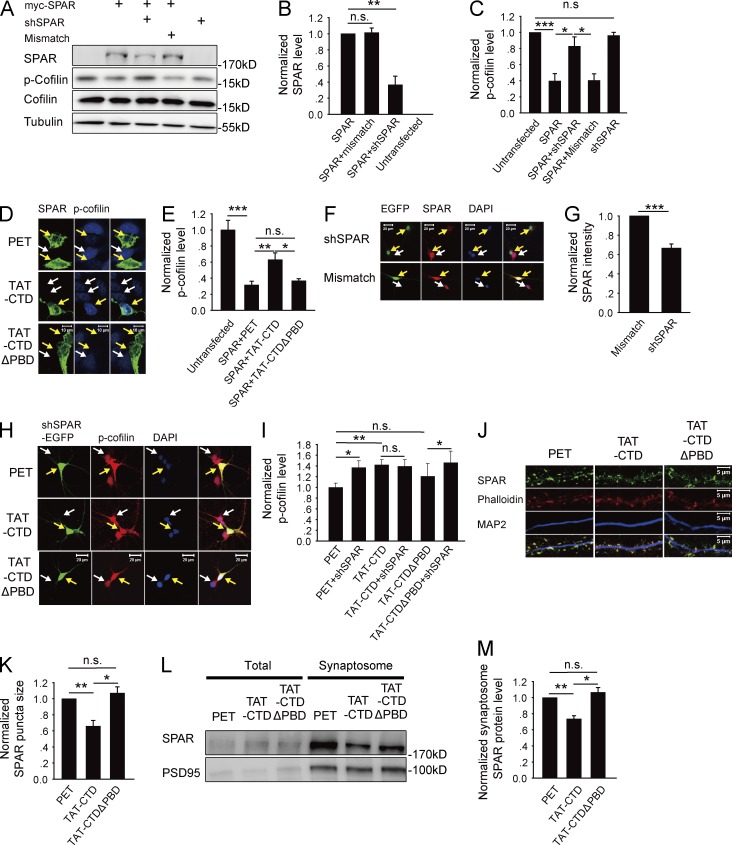
NLG1 regulates cofilin phosphorylation through its PBD interaction with SPAR
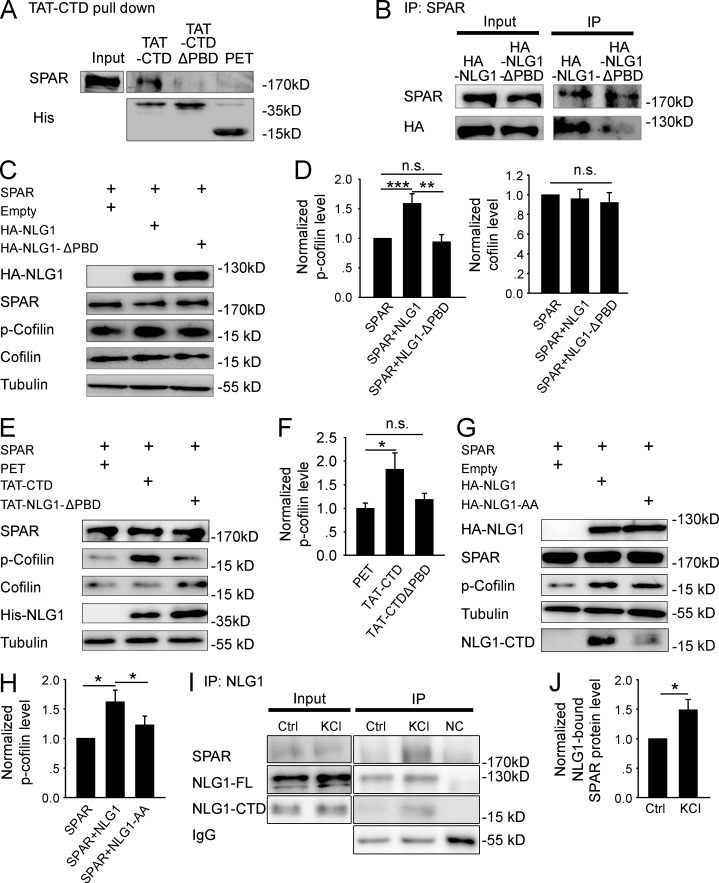
Having established that NLG1 is necessary and its CTD alone is sufficient to promote cofilin phosphorylation and synaptic strength, we next investigated how this process was achieved. We focused on SPAR because it was previously shown to be a potent regulator of the actin cytoskeleton (Irie et al., 1997; Pak et al., 2001; Hoe et al., 2009). Actin is the key structural component of the spine, essential for spine growth and synaptic function (Cingolani and Goda, 2008; Asrar and Jia, 2013). First, we showed that TAT-CTD, but not TAT-CTDΔPBD or PET, was able to pull down SPAR from the brain lysate (Fig. 4 A), confirming that the CTD of NLG1, via its PBD, is sufficient to interact with SPAR in an in vitro assay. To test whether NLG1 and SPAR interact in a heterologous cell system, we cotransfected HEK293 cells with myc-tagged SPAR and HA-NLG1 or HA-NLG1ΔPBD and immunoprecipitated the transfected cell lysate with anti-SPAR antibodies. As shown in Fig. 4 B, the anti-SPAR antibodies were able to coimmunoprecipitate with HA-NLG1 but not HA-NLG1ΔPBD. Finally, to test whether SPAR is required for cofilin phosphorylation induced by NLG1, we again cotransfected HA-NLG1 or HA-NLG1ΔPBD with or without SPAR into HEK293 cells, and 36 h later, p-cofilin was analyzed. As shown in Fig. S5 (A and B), neither HA-NLG1 nor HA-NLG1ΔPBD transfection alone altered p-cofilin. In contrast, as shown in Fig. 4 (C and D), cotransfection of myc-SPAR with HA-NLG1 (1.59 ± 0.17, n = 6, P < 0.01), but not with HA-NLG1ΔPBD (0.94 ± 0.12, n = 6, P = 0.64), significantly increased p-cofilin compared with myc-SPAR transfection alone. None of these transfections affected total cofilin (HA-NLG1: 0.96 ± 0.09, n = 6, P = 0.69 compared with SPAR alone; HA-NLG1ΔPBD: 0.92 ± 0.1, n = 6, P = 0.46 compared with SPAR alone). To further corroborate this result, we transfected HEK293 cells with myc-SPAR, and 36 h later the transfected cells were treated with TAT-CTD or TAT-CTDΔPBD. As shown in Fig. 4 (E and F), TAT-CTD (1.83 ± 0.34, n = 5, P < 0.05 compared with PET), but not TAT-CTDΔPBD (1.19 ± 0.13, n = 5, P = 0.18 compared with PET), significantly increased p-cofilin, consistent with the results obtained from myc-SPAR and HA-NLG1 cotransfection data. These results support the idea that NLG1 increases p-cofilin via its PBD interaction with SPAR. Finally, to determine whether the effect of NLG1 on p-cofilin is dependent on proteolytic cleavage of NLG1, we cotransfected HEK293 cells with myc-SPAR and HA-NLG1 or HA-NLG1-AA. As shown in Fig. 4 (G and H), the level of the full-length HA-NLG1 was lower and the level of the CTD fragment was higher in HA-NLG1–transfected cells compared with that of HA-NLG1-AA–transfected cells, confirming that the proteolytic generation of the CTD was blocked in the HA-NLG1-AA mutant. Importantly, p-cofilin was also significantly lower in HA-NLG1-AA than in HA-NLG1–transfected cells (HA-NLG1: 1.62 ± 0.2, n = 5, P < 0.01 compared with SPAR transfection alone; HA-NLG1-AA: 1.22 ± 0.15, n = 5, P = 0.17 compared with SPAR transfection alone). To test whether the endogenous NLG1 cleavage could promote NLG1CTD/SPAR interaction, we treated mouse brain slices with KCl and immunoprecipitated the slice lysate with anti–NLG1-CTD antibodies. As shown in Fig. 4 (I and J), more SPAR was immunoprecipitated with NLG1 (1.49 ± 0.17, n = 4, P < 0.05 compared with untreated control group). Collectively, these results suggest that the effect of NLG1 on p-cofilin requires the proteolytic release of the CTD of NLG1 and its subsequent binding to SPAR.
Figure 4.
NLG1 CTD regulates cofilin phosphorylation through interaction with SPAR. (A) Western blots of pull-down experiments of brain lysate showing that TAT-CTD, but not TAT-CTDΔPBD or PET, could pull down SPAR. (B) Western blots of anti-SPAR immunoprecipitates using protein lysate of HEK293 cells cotransfected with myc-SPAR plus HA-NLG1 or HA-NLG1ΔPBD showing that HA-NLG1, but not HA-NLG1-ΔPBD, was coimmunoprecipitated with SPAR. (C) Western blots of protein lysate of HEK293 cells cotransfected with myc-SPAR and HA-NLG1 or HA-NLG1ΔPBD. (D) Summary graphs of C showing significantly increased p-cofilin, but not total cofilin, in HEK293 cells cotransfected with myc-SPAR and HA-NLG1, compared with those transfected with myc-SPAR and HA-NLG1ΔPBD or with myc-SPAR alone. n.s., not significant. (E) Western blots of protein lysate of myc-SPAR–transfected HEK293 cells treated with the recombinant proteins. (F) Summary graph of E showing significantly increased p-cofilin in TAT-CTD, compared with TAT-CTDΔPBD– or PET-treated HEK293 cells pretransfected with myc-SPAR. n.s., not significant. (G) Western blots of protein lysate of HEK293 cells cotransfected with myc-SPAR plus HA-NLG1 or HA-NLG1-AA. (H) Summary graph of G showing significantly higher p-cofilin in myc-SPAR and HA-NLG1–cotransfected HEK293 cells compared with myc-SPAR and HA-NLG1-AA cotransfections. (I) Western blots of anti-NLG1 immunoprecipitates of hippocampal slices with or without KCl treatment. NC, negative IP control with rabbit IgG. (J) Summary graph of I showing significantly increased NLG1-bound SPAR after KCl treatment. Ctrl, control. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
NLG1/SPAR-induced cofilin phosphorylation requires Rap1 and LIMK1 activation
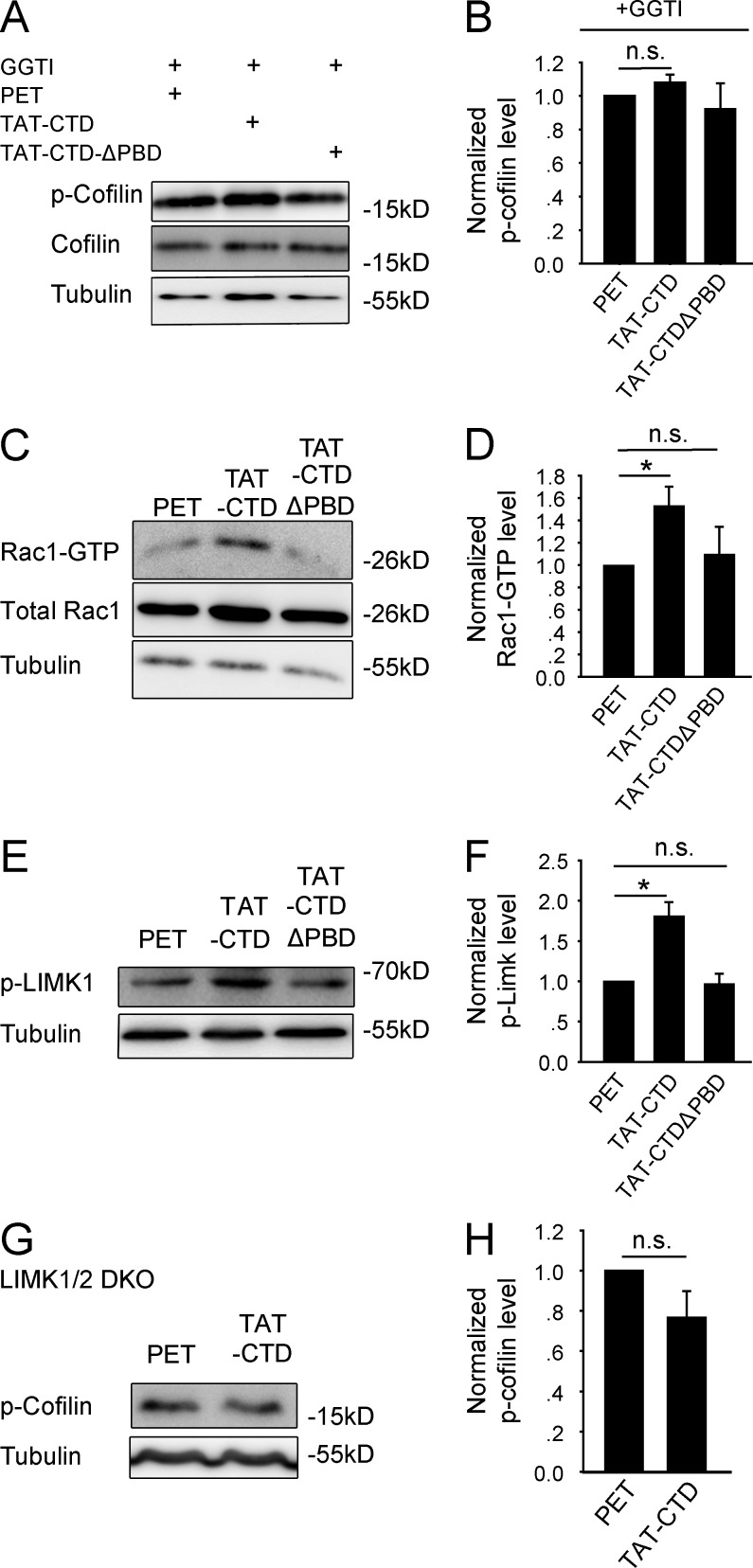
We next investigated how NLG1 CTD binding to SPAR regulates cofilin phosphorylation. Because SPAR is a RapGAP for the Rap GTPases (e.g., Rap1), which are known to be important for Rac1 activation and spine morphogenesis (Pak et al., 2001; Xie et al., 2005; Freeman et al., 2011), we reasoned that the effect of NLG1 might be mediated by activation of Rap1/Rac1 through inhibiting SPAR. To address this hypothesis, we first examined whether SPAR could act as a negative regulator of cofilin phosphorylation. As shown in Fig. 5 (A–C), transfection of HEK293 cells with myc-SPAR alone decreased p-cofilin compared with untransfected cells (SPAR: 0.40 ± 0.09, n = 4, P < 0.001 compared with untransfected cells) and cotransfections of HEK293 cells of myc-SPAR with SPAR-shRNA, but not with control shRNA, increased p-cofilin compared with myc-SPAR transfection alone (SPAR + SPAR-shRNA: 0.83 ± 0.12, n = 4, P < 0.01 compared with SPAR transfection alone; SPAR + mismatch shRNA: 0.40 ± 0.08, n = 3, P = 0.96 compared with SPAR transfection alone; SPAR-shRNA: 0.96 ± 0.04, n = 3, P = 0.38 compared with untransfected cells). The SPAR-shRNA was effective and specific because it decreased SPAR without affecting cofilin or tubulin (SPAR + mismatch shRNA: 1.01 ± 0.06, n = 3, P = 0.76 compared with SPAR transfection alone; SPAR + SPAR-shRNA: 0.37 ± 0.11, n = 3, P < 0.01 compared with SPAR transfection alone). Therefore, SPAR is a negative regulator of cofilin phosphorylation. To determine whether the effect of SPAR on cofilin phosphorylation is regulated by NLG1, we transfected HEK293 cells with myc-SPAR and subsequently treated these cells with the recombinant proteins. The results (Fig. 5, D and E) showed that TAT-CTD, but not TAT-CTDΔPBD, increased p-cofilin compared with the control PET group (SPAR + PET: 0.32 ± 0.05, n = 16, P < 0.001 compared with untransfected cells; SPAR + TAT-CTD: 0.63 ± 0.09, n = 20, P < 0.05 compared with the SPAR + PET group; SPAR + TAT-CTDΔPBD: 0.39 ± 0.03, n = 16, P = 0.33 compared with the SPAR + PET group; Fig. 4, C and D), supporting that NLG1 binding to SPAR negatively regulates SPAR, resulting in an increase in p-cofilin. To extend these studies to neurons, we transfected cultured neurons with the SPAR-shRNA and then treated these neurons with the recombinant proteins, and the results showed that SPAR knockdown (Fig. 5, F and G; 0.67 ± 0.04, n = 12, P < 0.001 compared with mismatch shRNA-transfected neurons) was sufficient to increase p-cofilin with or without TAT-CTD treatment (Fig. 5, H and I; PET + SPAR-shRNA: 1.37 ± 0.13, n = 10, P < 0.05 compared with PET-treated untransfected neurons; TAT-CTD–treated untransfected neurons: 1.42 ± 0.1, n = 12, P < 0.05 compared with PET-treated untransfected neurons; TAT-CTD + SPAR-shRNA: 1.39 ± 0.13, n = 12, P = 0.87 compared with TAT-CTD–treated untransfected neurons; TAT-CTDΔPBD–treated untransfected neurons: 1.21 ± 0.24, n = 9, P = 0.38 compared with PET-treated untransfected neurons; TAT-CTDΔPBD + SPAR: 1.46 ± 0.22, n = 9, P < 0.05 compared with TAT-CTDΔPBD–treated untransfected neurons), suggesting that SPAR is downstream of NLG1. To determine how SPAR might be inhibited by NLG1, we analyzed SPAR distribution at the synapse and found that TAT-CTD, but not TAT-CTDΔPBD, significantly decreased the amount of SPAR in both spines (Fig. 5, J and K; TAT-CTD: 0.69 ± 0.07, n = 13, P < 0.01 compared with PET; TAT-CTDΔPBD: 1.07 ± 0.08, n = 11, P = 0.43 compared with PET) and synaptosomes (Fig. 5, L and M; TAT-CTD: 0.74 ± 0.04, n = 4, P < 0.01 compared with PET; TAT-CTDΔPBD: 1.07 ± 0.06, n = 4, P = 0.22 compared with PET). Collectively, these results are consistent with the hypothesis that binding of NLG1-CTD to SPAR removes SPAR from the synapse, resulting in activation of the Rap GTPase and increased p-cofilin. To directly test if Rap1 activation is required for cofilin phosphorylation induced by TAT-CTD, we treated the cultured neurons first with the Rap1-specific inhibitor GGTI, then with the recombinant proteins, and found that TAT-CTD was no longer able to increase p-cofilin (Fig. 6, A and B; TAT-CTD: 1.08 ± 0.05, n = 4, P = 0.58 compared with PET; TAT-CTDΔPBD: 0.92 ± 0.15, n = 4, P = 0.71 compared with PET), suggesting that Rap1 activation is required for TAT-CTD–induced cofilin phosphorylation. To determine whether the downstream effectors of the Rap GTPases could be activated by NLG1, we performed a Rac1 activity assay (Fig. 6, C and D) and found that TAT-CTD, but not TAT-CTDΔPBD, increased the level of GTP-bound Rac1 (TAT-CTD: 1.53 ± 0.17, n = 3, P < 0.05 compared with PET; TAT-CTDΔPBD: 1.1 ± 0.25, n = 3, P = 0.63 compared with PET). Total Rac1 was not affected by these treatments. Because cofilin phosphorylation is predominantly mediated by LIMK1/2 in the brain (Meng et al., 2002, 2004), we tested whether TAT-CTD was able to activate LIMK1. As shown in Fig. 6 (E and F), TAT-CTD, but not TAT-CTDΔPBD, increased the amount of phosphorylated (active) form of LIMK1 (TAT-CTD: 1.81 ± 0.17, n = 6; P < 0.05 compared with PET; TAT-CTDΔPBD: 0.97 ± 0.12, n = 6; P = 0.91 compared with PET). Finally, to test whether LIMK1/2 is required for TAT-CTD–induced cofilin phosphorylation, we treated LIMK1/2 KO neurons with TAT-CTD and found that TAT-CTD was unable to induce cofilin phosphorylation (Fig. 6, G and H; 0.77 ± 0.13, n = 4, P = 0.22 compared with PET). These results together suggest that the CTD of NLG1 regulates cofilin phosphorylation through down-regulation of synaptic SPAR and subsequent activation of Rap/Rac1 and LIMK1.
Figure 5.
SPAR inhibits cofilin phosphorylation. (A) Western blots of protein lysate of HEK293 cells cotransfected with myc-SPAR and/or SPAR-shRNA. (B) Summary graphs of A showing reduced SPAR protein by SPAR-shRNA compared with myc-SPAR transfection alone or cotransfection with control mismatch shRNA. (C) Summary graph of A showing significantly decreased p-cofilin in myc-SPAR–transfected compared with untransfected cells. SPAR knockdown rescued the effect of myc-SPAR (i.e., increased p-cofilin). (D) Immunostaining of cultured HEK293 cells transfected with myc-SPAR (green) and treated with various recombinant proteins. Arrows indicate transfected and untransfected cells. (E) Summary graph of D showing significantly decreased p-cofilin in myc-SPAR–transfected compared with untransfected cells and that TAT-CTD, but not TAT-CTDΔPBD, or PET treatment rescued this decrease. n.s., not significant. (F) Immunostaining of cultured hippocampal neurons transfected with SPAR-shRNA or mismatch control (green). Arrows indicate neurons. (G) Summary graph of F showing significantly reduced SPAR protein in SPAR-shRNA–transfected compared with mismatch-transfected neurons. (H) Immunostaining of cultured hippocampal neurons transfected with SPAR-shRNA (green) and treated with various recombinant proteins, showing that either SPAR knockdown or TAT-CTD treatment was sufficient to increase p-cofilin. Arrows indicate transfected (green) and adjacent untransfected neurons. Note that in PET- and TAT-CTDΔPBD–treated groups, the transfected neuron has higher p-cofilin than the untransfected neuron, and in the TAT-CTD–treated group, all neurons have increased p-cofilin. (I) Summary graph of H showing that knockdown of SPAR or TAT-CTD treatment was sufficient to enhance p-cofilin. (J) Immunostaining of cultured hippocampal neurons treated with the recombinant proteins and costained for SPAR, F-actin, and microtubule-associated protein 2 (MAP2). (K) Summary graph of J showing significantly reduced spine SPAR (punctum size), but not total SPAR in TAT-CTD compared with TAT-CTDΔPBD– or PET-treated neurons. (L) Western blots of total protein lysate and synaptosomal fraction of cultured neurons treated with various recombinant proteins. (M) Summary graph of L showing significantly reduced synaptosomal SPAR by TAT-CTD but not by TAT-CTDΔPBD or PET. n.s., not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 6.
NLG1 CTD induces cofilin phosphorylation via Rap1/LIMK1 pathway. (A) Western blots of protein lysate of cultured neurons treated with GGTI and various recombinant proteins. (B) Summary graph of A showing similar p-cofilin in TAT-CTD–, TAT-CTDΔPBD–, and PET-treated neurons. (C) Western blots of GTP-bound Rac1 of protein lysate of cultured neurons treated with various recombinant proteins. (D) Summary graph of C showing significantly elevated GTP-Rac1 in neurons treated with TAT-CTD compared with those treated with TAT-CTDΔPBD or PET. (E) Western blots of p-LIMK1 using protein lysate of cultured cortical neurons treated with various recombinant proteins. (F) Summary graph of E showing significantly increased p-LIMK1 in TAT-CTD compared with TAT-CTDΔPBD– or PET-treated neurons. *, P < 0.05. (G) Western blots of protein lysate of LIMK1/2 double-KO brain slices treated with TAT-CTD or PET recombinant proteins. (H) Summary graph of G showing similar p-cofilin in TAT-CTD– and PET-treated LIMK1/2 double-KO brain slices. n.s., not significant.
NLG1 CTD inhibits long-term depression (LTD) and facilitates long-term potentiation (LTP)
Finally, we aimed to investigate the functional significance of the CTD of NLG1 in the regulation of synaptic plasticity. We focused on hippocampal LTD and LTP because NLGs are shown to be involved in both forms of plasticity (Kim et al., 2008; Baudouin et al., 2012; Shipman and Nicoll, 2012; Jedlicka et al., 2015). Previous studies have also demonstrated that cofilin phosphorylation and dephosphorylation are associated with and required for LTP and LTD, respectively (Rust et al., 2010; Zhou et al., 2011; Wang et al., 2013; Bosch et al., 2014). Therefore, we hypothesized that the CTD of NLG1 would inhibit LTD and facilitate LTP by enhancing cofilin phosphorylation. To test this hypothesis, we performed whole-cell patch clamp recordings of CA1 neurons in hippocampal slices. First, we examined LTD induced by paired-pulse low-frequency stimulation (PP-LFS), known to trigger both NMDA receptor (NMDAR)- and mGluR-dependent LTD (Kemp and Bashir, 1999; Kemp et al., 2000). As shown in Fig. 7 (A and B), TAT-CTD, but not TAT-CTDΔPBD, blocked LTD (PET: 44.38 ± 4.48%, n = 5; TAT-CTD: 93.12 ± 14.24%, n = 6, P < 0.05 compared with PET; TAT-CTDΔPBD: 41.68 ± 7.16%, n = 4, P = 0.74 compared with PET). The inhibitory effect of TAT-CTD on LTD was blocked by the S3 peptide, which inhibits cofilin phosphorylation (52.56 ± 9.13%, n = 6, P = 0.47 compared with PET), suggesting that the effect of TAT-CTD is mediated by cofilin phosphorylation. To further test this genetically, we performed the same experiments in LIMK1/2 KO mice, which have impaired cofilin phosphorylation (Meng et al., 2002, 2004), and found that LTD in these mice was insensitive to the TAT-CTD treatment (Fig. 7, C and D; PET: 51.95 ± 12.56%, n = 4; TAT-CTD: 39.58 ± 6.62%, P = 0.38). Therefore, TAT-CTD inhibits LTD through cofilin phosphorylation, consistent with the aforementioned biochemical data. Then we tested the role of TAT-CTD in LTP induced by theta-burst stimulation (TBS). As shown in Fig. 7 (E and F), the magnitude of LTP was significantly enhanced in TAT-CTD–treated slices (PET: 119.65 ± 2.49%, n = 6; TAT-CTD: 140.24 ± 2.09%, n = 5, P < 0.05). Finally, we tested whether the impaired LTP in NLG1 KO mice (Kim et al., 2008; Blundell et al., 2010) could be rescued by TAT-CTD, given that TAT-CTD was able to restore p-cofilin to the WT level (Fig. 2, H and I). As shown in Fig. 7 (G and H), LTP was significantly higher in KO slices treated with TAT-CTD (PET: 107.48 ± 3.93%, n = 6; TAT-CTD: 146.36 ± 13.35%, n = 5, P < 0.05).
Figure 7.
NLG1 CTD inhibits LTD and facilitates LTP. (A) Whole-cell recordings of CA1 neurons of WT hippocampal slices pretreated with the recombinant proteins showing that TAT-CTD, but not PET or TAT-CTDΔPBD, blocked PP-LFS–induced LTD. TAT-CTD failed to block LTD when the S3 peptide was included in the recording electrode (TAT-CTD + S3). (B) Summary graph of A showing significant differences in PP-LFS–induced LTD between TAT-CTD and PET, TAT-CTDΔPBD, or TAT-CTD + S3–treated slices. (C) Whole-cell recordings of CA1 neurons of LIMK1/2 double-KO hippocampal slices pretreated with TAT-CTD or PET showing that TAT-CTD failed to block PP-LFS–induced LTD in these mice. (D) Summary graph of C showing similar LTD in TAT-CTD– and PET-treated slices of LIMK1/2 double-KO mice. n.s., not significant. (E) Field recordings in the CA1 region of WT hippocampal slices pretreated with TAT-CTD or PET showing that TAT-CTD enhanced TBS-induced LTP compared with PET. (F) Summary graph of E showing significantly higher LTP in TAT-CTD compared with PET-treated slices. (G) Field recordings of NLG1 KO hippocampal slices pretreated with TAT-CTD or PET showing that TAT-CTD enhanced TBS-induced LTP compared with PET. (H) Summary graph of G showing significantly higher LTP in TAT-CTD compared with PET-treated slices. *, P < 0.05.
Discussion
Although it is well established that NLG1 plays a critical role in spine and synapse development and function, the underlying mechanisms remain unclear. Here we show that the CTD is sufficient to enhance spine and synapse growth and that it does so via its PBD interaction with SPAR and activation of LIMK1/cofilin-mediated actin reorganization. Our results provide a novel postsynaptic mechanism by which NLG1 regulates synapse development and function.
First, we demonstrate that NLG1 is required for both basal and activity-dependent cofilin phosphorylation. Therefore, in both NLG1 KO and knockdown neurons, the level of p-cofilin, but not of total cofilin, is significantly reduced in both naive and KCl-treated conditions (Fig. 1). The reduced p-cofilin is not likely because of enhanced dephosphorylation, because the initial decrease in p-cofilin induced by the KCl treatment is not affected in NLG1 KO mice (Fig. 1). Therefore, we conclude that NLG1 is an important activator of cofilin phosphorylation. This conclusion is consistent with the results that the CTD of NLG1 is capable of activating LIMK1 (Fig. 6), the predominant kinase responsible for cofilin phosphorylation in the brain (Meng et al., 2002, 2004; Bernard, 2007; Bernstein and Bamburg, 2010; Rust, 2015), and that the CTD-induced increase in p-cofilin is abolished in LIMK1/2 KO mice (Fig. 5).
Second, we demonstrate that the CTD of NLG1 is sufficient to induce cofilin phosphorylation and spine/synapse growth (Figs. 2 and 3). We achieve this conclusion by analyzing the effect of TAT-tagged, cell-permeable recombinant proteins derived from the CTD of NLG1. We show that TAT-CTD treatment increases cofilin phosphorylation in both cultured neurons and brain tissues. The TAT-CTD–induced cofilin phosphorylation is not likely caused by nonspecific effects of the recombinant protein because (a) TAT-CTD, but not TAT-CTDΔPBD (which differs from TAT-CTD in only the last 4 aa), or control PET exerts this effect; (b) TAT-CTD does not induce pleiotropic changes in protein phosphorylation (e.g., p-Akt in Fig. S4); and (c) TAT-CTD–induced cofilin phosphorylation is abolished in LIMK1/2 KO mice (Fig. 6). Independent approaches by transfecting neurons with various NLG1 constructs also show that NLG1-induced cofilin phosphorylation requires the CTD and the proteolytic cleavage of NLG1 (Figs. 1 and 2).
Third, we show that the effect of NLG CTD on cofilin phosphorylation is mediated by its PBD interaction with SPAR (Fig. 4). We show that (a) purified TAT-CTD, but not TAT-CTDΔPBD recombinant protein, is able to pull down SPAR from the brain protein lysate, indicating that the CTD of NLG1 is sufficient to interact with SPAR, and this interaction requires the PBD of the CTD; (b) SPAR and the full-length NLG1, but not NLG1ΔPBD, coimmunoprecipitate in transfected HEK293 cells; (c) cotransfections of HEK293 cells with the full-length NLG1 and SPAR, but not with the full-length NLG1 alone, or with NLG1ΔPBD and SPAR, induce cofilin phosphorylation, suggesting that NLG1-induced cofilin phosphorylation requires the PBD of the NLG1 and SPAR; and (d) HEK293 cells expressing SPAR and treated with purified TAT-CTD, but not with TAT-CTDΔPBD recombinant protein, show an elevated level of p-cofilin. Therefore, NLG1 regulates cofilin phosphorylation via its PBD interaction with SPAR. It is important to note that in addition to SPAR, the CTD of NLG1 can interact with other proteins (Irie et al., 1997; Meyer et al., 2004), which may directly or indirectly affect cofilin phosphorylation and actin assembly (Woolfrey et al., 2009; Chen et al., 2014).
Therefore, how does the interaction of NLG1 with SPAR enhance cofilin phosphorylation? Our experiments suggest that inhibition of SPAR and subsequent activation of Rap1/Rac1 and LIMK1/2 is essential (Figs. 5 and 6). This conclusion is supported by the following: (a) SPAR expression induces a decrease in p-cofilin, and this effect is partially prevented by TAT-CTD; (b) SPAR knockdown increases p-cofilin, which occludes the subsequent effect of TAT-CTD; (c) TAT-CTD decreases synaptic SPAR; (d) TAT-CTD–induced cofilin phosphorylation is blocked by the Rap1 inhibitor GGTI; (e) TAT-CTD induces activation of Rac1 and LIMK1; and (f) TAT-induced cofilin phosphorylation is absent in LIMK1/2 KO mice. These results suggest that the interaction between NLG1-CTD and SPAR removes SPAR from the synapse, resulting in activation of Rap1/Rac1/LIMK1 and increased cofilin phosphorylation. Our results are consistent with previous studies showing that Rap1 can cross talk with and activate the Rac1, an upstream activator of LIMK1/2 (Palsson et al., 2000; Zhu et al., 2002, 2005; Bernard, 2007; Knaus et al., 2007).
Finally, we demonstrate that the CTD of NLG1 is sufficient to regulate synaptic plasticity, and this process is mediated by LIMK1 and cofilin (Fig. 7). Previous studies have shown that NLG1 KO mice are impaired in hippocampal LTP (Kim et al., 2008; Kwon et al., 2012; Jedlicka et al., 2015), but how NLG1 regulates LTP remains unclear. Although NMDAR-mediated synaptic response is reduced in NLG1 KO mice, suggesting that the impaired NMDAR function may contribute to the LTP deficit (Jung et al., 2010), whether the downstream LTP expression processes are also altered remains unknown. We show here that TAT-CTD, but not TAT-CTDΔPBD, enhances hippocampal LTP. This effect is independent of the endogenous NLG1 because TAT-CTD also enhances LTP in NLG1 KO mice, again confirming that the CTD of NLG1 is sufficient to facilitate LTP. We also show that TAT-CTD is able to block LTD, and this blocking effect is abolished in LIMK1/2 doubleKO mice or WT slices acutely treated with the S3 peptide that inhibits cofilin phosphorylation, indicating that the effect on LTD is mediated by LIMK1/cofilin, consistent with our biochemical data. These results are consistent with previous results showing that manipulating cofilin affects both LTP and LTD (Meng et al., 2002; Gu et al., 2010; Rust et al., 2010; Huang et al., 2011; Zhou et al., 2011; Bosch et al., 2014).
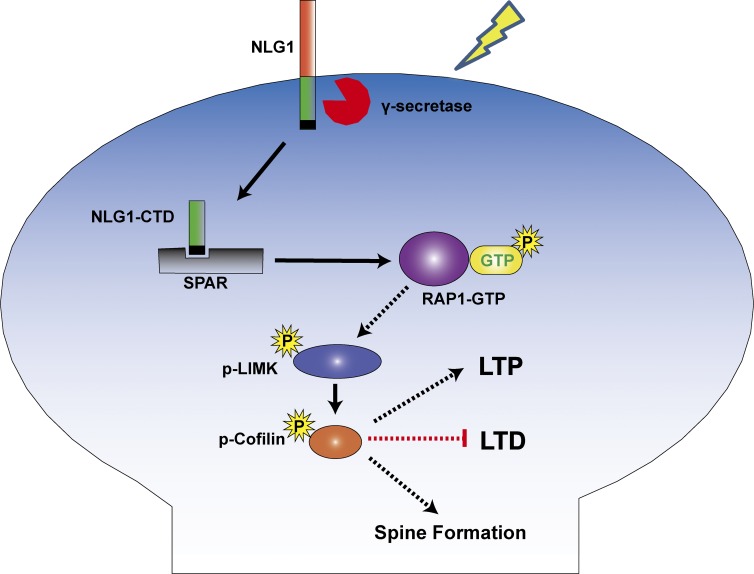
In summary, we have identified a novel process by which NLG1 regulates the actin cytoskeleton and synaptic function (Fig. 8). Because LIMK1/cofilin-dependent actin remodeling is central to spine morphology and plasticity (Jia et al., 2009; Bernstein and Bamburg, 2010; Rust, 2015), this process may also play a role in mediating other aspects of NLG1 function, including those requiring presynaptic neurexins. In addition, because LIMK1/cofilin is a common effector of the Rho and Rap family small GTPases, our results also highlight the cross talk between these GTPases and the cell-adhesion molecules. Given the importance of NLGs in psychiatric disorders, such as autism and schizophrenia, our results may also provide new therapeutic targets to treat NLG-related disorders by manipulating the LIMK1/cofilin pathway.
Figure 8.
Summary model. The CTD of NLG1, released from either proteolytically cleaved surface NLG1 or internalized NLG1, binds to and removes SPAR from the synapse, resulting in activation of Rap1/Rac1, which in turn stimulates LIMK1. The activated LIMK1 phosphorylates cofilin and promotes actin assembly, which facilitates spine/synapse formation/LTP and inhibits LTD.
Materials and methods
Mice, antibodies, and reagents
LIMK1 and LIMK2 KO mice were generated and genotyped as described previously (Meng et al., 2002, 2004). The LIMK1 KO mice were generated by replacing the coding exons of the LIM and PDZ domains with the neomycin cassette (Meng et al., 2002), whereas the LIMK2 KO mice were created by deleting exons 3–5 using the Cre/loxP system (Takahashi et al., 2002). The NLG1 KO mice, which were generated by inserting a neomycin resistance cassette to replace the first two coding exons of the NLG1 gene, were obtained from the Jackson Laboratory and genotyped using PCR techniques as previously described (Varoqueaux et al., 2006). The Thy1-YFP transgenic mice containing a YFP coding sequence linked to 5.6-kb regulatory elements from the 5′ portion of the mouse Thy1 gene were also obtained from the Jackson Laboratory and genotyped as described previously (Feng et al., 2000). All mice were maintained and used according to protocols approved by the Hospital for Sick Children (Toronto, Canada) and by Southeast University (Nanjing, China). The antibodies used in this study were as follows: anti-GFP (G1544, mouse, 1:1,000 dilution; Sigma-Aldrich), anti-tubulin (T9026, mouse, 1:5,000 dilution; Sigma-Aldrich), anti-cofilin (5175P, rabbit, 1:2,000 dilution; CST), anti–p-cofilin (sc-12912-R, rabbit, 1:2,000 dilution; Santa Cruz Biotechnology, Inc.), anti-LIMK1 (3842s, rabbit, 1:1,000 dilution; CST), anti–p-LIMK1 (BS4115, rabbit, 1:2,000 dilution; Bioworld), anti-His (66005–1-Ig, mouse, 1:2,000 dilution; Proteintech), anti-HA (3724s, rabbit, 1:2,000 dilution; CST), anti-myc (660021-Ig, mouse, 1:2,000 dilution; Proteintech), anti-NLG1 (129013, rabbit, 1:2,000 dilution; Synaptic System), anti-MAP2 (MAB3418, rabbit, 1:2,000 dilution; Millipore), anti-PSD95 (MABN68, mouse, 1:2,000 dilution; Millipore), and anti-synapsin I (BS4116, rabbit, 1:1,000 dilution; Bioworld). Secondary antibodies included: goat anti-rabbit (A00098, 1:2,000 dilution; Genscript), goat anti-mouse (A00160, 1:2,000 dilution; Genscript), Alexa Fluor 555 donkey anti–rabbit IgG (1:300; Invitrogen), Alexa Fluor 488 donkey anti–mouse IgG (1:300; Invitrogen), Alexa Fluor 488 goat anti–mouse IgG (1:300, Jackson ImmunoResearch Laboratories), Alexa Fluor 555 goat anti–mouse IgG (1:300; Invitrogen), Alexa Fluor 633 goat anti–mouse IgG (1:300; Invitrogen), Alexa Fluor 633 goat anti–mouse IgG (1:300; Invitrogen), and Alexa Fluor 633 goat anti–rabbit IgG (1:300; Invitrogen). Antibodies were validated for their intended purpose (immunoblotting, immunocytochemistry, immunohistochemistry, and coimmunoprecipitation) as outlined in the product sheet or in our laboratory. Rhodamine phalloidin was from Invitrogen (R415, 1:1,000 dilution) and DAPI was from Cayman Chemical (1:1,000). The specificity of the key antibodies used in this study (e.g., anti-NLG1, anti-LIMK1, anti-cofilin, anti–p-cofilin, and anti-SPAR) was tested either using KO mice or transfected HEK293 cells.
DNA plasmids
HA-NLG1, HA-NLG1ΔPBD, and HA-NLG1-aa were constructed by introducing HindIII and SalI fragments containing the mouse NLG1 coding sequence (HA tag sequence was inserted at the site 123 base after the start codon ATG) into the pCMV-tag2b vector. The original HA-NLG1 mouse cDNA was provided by A.M. Craig (University of British Columbia, Vancouver, BC, Canada). The myc-SPAR plasmid was described previously (Pak et al., 2001). The plasmids used for generating the recombinant fusion proteins (NLG1CTD, NLG1CTDΔPBD, and PET) were constructed by inserting TAT-tagged NLG1 CTD or NLG1ΔPBD CTD into the PET32a plasmid (LaVallie et al., 1993) using HindIII and EcoRI restriction sites. The NLG1-shRNA and control double-strand DNA sequences were made by annealing the following primer pairs: NLG1-shRNA: 5′-GATCTCCGGAAGGTACTGGAAATCTATTCAAGAGATAGATTTCCAGTACCTTCCTTTTTGGAAC-3′, 5′-TCGAGTTCCAAAAAGGAAGGTACTGGAAATCTATCTCTTGAATAGATTTCCAGTACCTTCCGGA-3′; control mismatch shRNA: 5′-GATCTCCGCAACGTTCTCGATATGTATTCAAGAGATACATATCGAGAACGTTGCTTTTTGGAAC-3′, 5′-TCGAGTTCCAAAAAGCAACGTTCTCGATATGTATCTCTTGAATACATATCGAGAACGTTGCGGA-3′. The DNAs were then cloned into the pSuper vector (gift from J. Xia; Hong Kong University of Science and Technology, Hong Kong, China) by BglII–XhoI to generate the pSuper-shNLG1 constructs. The recombinant AAVs and control virus AAV-EGFP were made by Bio-Miao-Biological Company. SPAR-shRNA and mismatch constructs were made by introducing specific sequences (SPAR-shRNA: 5′-ACCGCCATCTTACACGTTGGGAATTCAAGAGATTCCCAACGTGTAAGATGGCTTTTTTG-3′; mismatch: 5′-CACCGCACCCATTCACGCCTTGGAATCAAGAGTTCCAAGGCGTGAATGGGTGCTTTTTTG-3′) into pGenesil vector through HindIII and BamHI restriction sites.
Generation and purification of TAT-CTD, TAT-CTDΔPBD, and PET recombinant proteins
DNA constructs of TAT-CTD, TAT-CTDΔPBD, or PET32a empty vector were transformed into BL21, plated onto ampicillin-containing plates, and incubated overnight at 37°C. A single colony from each plasmid was inoculated in LB medium containing ampicillin and incubated at 37°C until the OD600 reached 0.4. Expression of the recombinant protein was induced by IPTG (0.5 mM) for 12 h. Cell pellets were collected by centrifugation and sonicated before purification. The recombinant proteins were then purified by using a Ni-NTAresin column (L00250; Genescript). In brief, Ni-NTA resin columns were equilibrated with IDA buffer before the recombinant protein extracts were added to the resin. IDA buffer, pH 7.9, contained the following: 20 mM Tris-HCl, 500 mM NaCl, and 10% glycerol. The columns were then washed with IDA buffer containing various concentrations of imidazole (20–250 mM), and the proteins were eluted using IDA buffer containing 500 mM imidazole. IDA buffer and imidazole were replaced with 0.01 M PBS through ultrafiltration.
Electrophysiology
Standard procedures for electrophysiologic recordings for both brain slice and cultured neurons were followed (Meng et al., 2003; Zhou et al., 2011). In brief, the mouse brains from various genotypes were quickly removed, and sagittal 360-mm hippocampal slices were prepared in ice-cold artificial cerebrospinal fluid (ACSF) saturated with 95% O2/5% CO2. ACSF contained the following: 120 mM NaCl, 3.0 mM KCl, 1.2 mM MgSO4, 1.0 mM NaH2PO4, 26 mM NaHCO3, 2.0 mM CaCl2, and 11 mM d-glucose. The slices were recovered at RT for at least 2 h before a single slice was transferred to a submersion chamber perfused with 95% O2/5% CO2–saturated ACSF with (for whole-cell recordings) or without (for field recordings) 100 µM picrotoxin. Hippocampal CA1 neurons were visualized using an infrared differential interference contrast microscope (Olympus). Synaptic transmission was evoked by stimulation (at 0.05 Hz for field recordings or 0.1 Hz for whole-cell recordings) of Schaffer collaterals and recorded with glass pipettes (3–4 MΩ) filled with either ACSF (for field responses) or the intracellular solution (for whole-cell response) containing 130 mM CsMeSO4, 5 mM NaCl, 1 mM MgCl2, 0.05 mM EGTA, 10 mM Hepes, 3 mM Mg-ATP, 0.3 mM Na3GTP, and 5 mM QX-314, pH 7.25 (280–300 mOsm). PP-LFS consisting of 900 pairs of stimuli with 50-ms interstimulus intervals delivered at 1 Hz (PP-LFS–induced LTD) was used to induced LTD. Whole-cell series resistance was monitored throughout LTD experiments by applying a –3 mV step at the end of each response, and if it changed by >20%, the experiment was excluded from analysis. LTP was induced by two trains of TBS, each consisting of five bursts of four pulses at 100 Hz with an interburst interval of 200 ms and an intertrain interval of 10 s. LTD and LTP were calculated and statistically evaluated by comparing the mean values of the last 10 min of the recording and the mean values of the entire baseline. All data acquisition and analysis were done using pCLAMP 10.2 (Axon Instruments) and MiniAnalysis program (Synaptosoft). When mean data were plotted, data were normalized to the mean of the baseline responses unless indicated otherwise. In all electrophysiologic experiments, n represents the number of neurons or slices, and normally only one or two slices or neurons per animal were used.
Neuronal culture, immunostaining, and image analysis
Hippocampal (for staining experiments) and cortical (for biochemical experiments) neuronal cultures were prepared from postnatal day 1 pups as previously described (Meng et al., 2002). In brief, pups were euthanized, and hippocampal CA1 regions were dissected in ice-cold PBS. Tissues were trypsinized (0.25%) at 37°C for 15 min, dissociated by trituration, and plated onto glass coverslips coated with 50 µg/ml poly-d-lysine (60,000 cells/ml for immunostaining experiments and 180,000 cells/ml for Western experiments). The cultures were maintained by replacing half of the medium with fresh medium every 3–5 d. The maintenance medium contained Neurobasal A, 0.5 mM GlutaMax, and B27. At 17–18 d in culture, cultured neurons were treated with various recombinant proteins, inhibitors, and/or KCl as indicated in each experiment. Immediately, the treatment cells were either lysed for the preparation of the protein lysate or fixed with ice-cold 4% paraformaldehyde plus 4% sucrose for 30 min for immunostaining. The fixed cells were then permeabilized with 0.25% Triton X-100 for an additional 30 min. Cells were then blocked with 5% FBS in PBS for 1 h and incubated with primary antibodies overnight at 4°C followed by rhodamine-conjugated phalloidin and appropriate secondary antibodies for 1 h at RT. After washing with PBS, coverslips were mounted using DAKO mounting medium for image collections.
HEK cell culture, transfection, and treatments
HEK293 were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Invitrogen). Cells were grown to 80% confluence in six-well plates before being transiently transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. Cells were then maintained for either 24 or 48 h at 37°C before being harvested for biochemical analyses.
Slice treatment and biochemical assays
Hippocampal slices used for immunoprecipitation and Western immunoblotting experiments were prepared according to the same procedure as for electrophysiological recordings. Slices were recovered for 3–4 h at RT in ACSF saturated with 95% O2/5%CO2 and transferred to a treatment chamber for drug treatment. After the treatment, slices were rapidly placed into precooled cell lysis buffer. To prepare protein lysate, slice samples were lysed for 45 min in ice-cold cell lysis buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 20 mM NaF, and 1% protease inhibitor cocktail and phosphatase inhibitor (Roche). Debris was removed by centrifugation at 14,000 g (4°C) for 10 min. Synaptosomal protein lysate was prepared by using an extraction kit for synaptic proteins (87793; Pierce). Proteins were separated on 15% SDS-PAGE and electrotransfered to a polyvinylidene fluoride filter. Filters were then blocked with 5% dry milk TBST (20 mM Tris base, 9% NaCl, 1% Tween-20, pH 7.6) and incubated overnight at 4°C with appropriate primary antibodies in TBST. After washing and incubation with appropriate secondary antibodies, filters were developed using an enhanced chemiluminescence (Thermo Fisher Scientific) method of detection and analyzed using the AlphaEaseFC software(Alpha Innotech) per the manufacturer’s instruction. Protein loading was further controlled by normalizing each tested protein with tubulin immunoreactivity on the same blot.
Immunoprecipitation and pull-down experiments
300 µl of the protein lysate (200–300 µg, normally pooled from two to three hippocampal slices or over 106 transfected HEK293T cells) was incubated with appropriate primary antibodies or various recombinant proteins at 4°C with constant gentle rocking for 3–5 h followed by addition of 35 µl of Protein A/G agarose beads slurry (Santa Cruz Biotechnology, Inc.) or NTA-Ni beads (Beaver) and further incubation for 3 h at 4°C. The samples were microcentrifuged for 1 min at 4,000 g, the supernatant was carefully removed, and the beads were washed thoroughly with the lysis buffer five times and resuspended in loading buffer for Western blot analysis. n in the summary data represents the number of independent experiments.
Immunohistochemistry
Mice were anaesthetized and perfused with double-filtered saline and 4% PFA in PBS. Brains were collected and immersed in 4% PFA before being subjected to cryoprotection by 30% sucrose/PBS. After the brains had sunk, they were flash-frozen with dry ice before overnight freezing at −80°C. They were then sliced at 25 µm with a cryostat and stored at –20°C. Before staining, slices were washed three times for 10 min each with 0.1 M PBS, permeabilized and blocked in 0.1 M PBS with 10% BSA and 0.1% Triton X-100 for 2 h, and stained with anti–p-cofilin (1:1,000) at 4°C for 16 h. They were then washed and stained with Alexa 488 (1:1,000) and DAPI at 37°C for 2 h before washing and mounting.
Microscope setups and image analysis
For spine analysis, confocal images were obtained at RT on an LSM 700 (Carl Zeiss) at 2,048 × 2,048 pixels using a 63× (NA 1.4, oil immersion; Carl Zeiss) objective with the same settings and configurations for all samples within each experiment. Spines were defined as any dendritic protrusions 0.3–4 µm in length. For analysis of synaptic proteins, the fluorescence puncta (with an area >0.1 µm2) were automatically selected and counted and manually verified. For each treatment, ∼10–25 neurons from at least three independent cultures and a total of 100- to 150-µm linear dendrites per neuron were randomly selected, measured, and averaged. For neuronal cell body/dendrite and HEK293 staining, confocal images were obtained at RT on an LSM 700 at 2,048 × 2,048 pixels using a 40× (NA 0.95, dry; Carl Zeiss) objective with the same settings and configurations for all samples within each experiment. For each treatment, ∼10–25 cells from at least three independent cultures were randomly selected, measured, and averaged. For immunohistochemistry staining, confocal images were obtained at RT on an LSM 700 at 2,048 × 2,048 pixels using a 5× (NA 0.15, dry; Carl Zeiss) objective with the same settings and configurations for all samples within each experiment. All images were initially acquired through Zen 2010 software (Carl Zeiss). AimImageBrowser software (Carl Zeiss) was used to adjust the image brightness/contrast and extract a subregion. All measurements were performed using ImageJ software (National Institutes of Health).
Online supplemental material
Fig. S1 shows the knockdown of NLG1. Fig. S2 shows the endogenous NLG1-CTD and its regulation by MRK and KCl. Fig. S3 shows the effect of recombinant proteins at various concentrations. Fig. S4 shows the lack of effect of recombinant proteins on p-Akt. Fig. S5 shows that HA-NLG1 alone has no effect on p-cofilin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201509023/DC1.
Supplementary Material
Acknowledgments
This work was supported by grants from the China National Basic Research Program (973 Program 2012CB517903 to W. Xie and Z. Jia), Canadian Institutes of Health Research (MOP119421 to Z. Jia), National Natural Science Foundation of China and Canadian Institutes of Health Research Joint Health Research Initiative Program (81161120543 to W. Xie and CCI117959 to Z. Jia), National Natural Science Foundation of China (31430035 to W. Xie and 31200805 to Z. Zhou), Natural Science Foundation of Jiangsu Province (BK2012745 to Z. Zhou), and Natural Sciences and Engineering Research Council of Canada (RGPIN341498 to Z. Jia).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AAV
- adeno-associated virus
- ACSF
- artificial cerebrospinal fluid
- CTD
- C-terminal domain
- KO
- knockout
- LIMK
- LIM-domain protein kinase
- LTD
- long-term depression
- LTP
- long-term potentiation
- mEPSC
- miniature excitatory postsynaptic current
- NLG
- neuroligin
- NMDAR
- NMDA receptor
- PBD
- PDZ binding domain
- PET
- protein derived from the empty vector PET32a
- PP-LFS
- paired-pulse low-frequency stimulation
- SPAR
- spine-associated Rap GTPase-activating protein
- TBS
- theta-burst stimulation
- WT
- wild type
References
- Aizawa H., Wakatsuki S., Ishii A., Moriyama K., Sasaki Y., Ohashi K., Sekine-Aizawa Y., Sehara-Fujisawa A., Mizuno K., Goshima Y., and Yahara I.. 2001. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat. Neurosci. 4:367–373. 10.1038/86011 [DOI] [PubMed] [Google Scholar]
- Asrar S., and Jia Z.. 2013. Molecular mechanisms coordinating functional and morphological plasticity at the synapse: role of GluA2/N-cadherin interaction-mediated actin signaling in mGluR-dependent LTD. Cell. Signal. 25:397–402. 10.1016/j.cellsig.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Baksh M.M., Dean C., Pautot S., DeMaria S., Isacoff E., and Groves J.T.. 2005. Neuronal activation by GPI-linked neuroligin-1 displayed in synthetic lipid bilayer membranes. Langmuir. 21:10693–10698. 10.1021/la051243d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg J.R. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230. 10.1146/annurev.cellbio.15.1.185 [DOI] [PubMed] [Google Scholar]
- Baudouin S.J., Gaudias J., Gerharz S., Hatstatt L., Zhou K., Punnakkal P., Tanaka K.F., Spooren W., Hen R., De Zeeuw C.I., et al. . 2012. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 338:128–132. 10.1126/science.1224159 [DOI] [PubMed] [Google Scholar]
- Becker-Hapak M., McAllister S.S., and Dowdy S.F.. 2001. TAT-mediated protein transduction into mammalian cells. Methods. 24:247–256. 10.1006/meth.2001.1186 [DOI] [PubMed] [Google Scholar]
- Bernard O. 2007. Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 39:1071–1076. 10.1016/j.biocel.2006.11.011 [DOI] [PubMed] [Google Scholar]
- Bernstein B.W., and Bamburg J.R.. 2010. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20:187–195. 10.1016/j.tcb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J., Blaiss C.A., Etherton M.R., Espinosa F., Tabuchi K., Walz C., Bolliger M.F., Südhof T.C., and Powell C.M.. 2010. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 30:2115–2129. 10.1523/JNEUROSCI.4517-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Castro J., Saneyoshi T., Matsuno H., Sur M., and Hayashi Y.. 2014. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 82:444–459. 10.1016/j.neuron.2014.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard A.A., Chubykin A.A., Comoletti D., Taylor P., and Südhof T.C.. 2005. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron. 48:229–236. 10.1016/j.neuron.2005.08.026 [DOI] [PubMed] [Google Scholar]
- Chen B., Brinkmann K., Chen Z., Pak C.W., Liao Y., Shi S., Henry L., Grishin N.V., Bogdan S., and Rosen M.K.. 2014. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 156:195–207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B., Afridi S.K., Clark L., and Scheiffele P.. 2004. Disorder-associated mutations lead to functional inactivation of neuroligins. Hum. Mol. Genet. 13:1471–1477. 10.1093/hmg/ddh158 [DOI] [PubMed] [Google Scholar]
- Chubykin A.A., Atasoy D., Etherton M.R., Brose N., Kavalali E.T., Gibson J.R., and Südhof T.C.. 2007. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 54:919–931. 10.1016/j.neuron.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani L.A., and Goda Y.. 2008. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 9:344–356. 10.1038/nrn2373 [DOI] [PubMed] [Google Scholar]
- Cline H. 2005. Synaptogenesis: a balancing act between excitation and inhibition. Curr. Biol. 15:R203–R205. 10.1016/j.cub.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Conroy W.G., Nai Q., Ross B., Naughton G., and Berg D.K.. 2007. Postsynaptic neuroligin enhances presynaptic inputs at neuronal nicotinic synapses. Dev. Biol. 307:79–91. 10.1016/j.ydbio.2007.04.017 [DOI] [PubMed] [Google Scholar]
- Dean C., and Dresbach T.. 2006. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 29:21–29. 10.1016/j.tins.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Dean C., Scholl F.G., Choih J., DeMaria S., Berger J., Isacoff E., and Scheiffele P.. 2003. Neurexin mediates the assembly of presynaptic terminals. Nat. Neurosci. 6:708–716. 10.1038/nn1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Qi J., and Chen G.. 2007. Molecular reconstitution of functional GABAergic synapses with expression of neuroligin-2 and GABAA receptors. Mol. Cell. Neurosci. 35:14–23. 10.1016/j.mcn.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Feng G., Mellor R.H., Bernstein M., Keller-Peck C., Nguyen Q.T., Wallace M., Nerbonne J.M., Lichtman J.W., and Sanes J.R.. 2000. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 28:41–51. 10.1016/S0896-6273(00)00084-2 [DOI] [PubMed] [Google Scholar]
- Freeman S.A., Lei V., Dang-Lawson M., Mizuno K., Roskelley C.D., and Gold M.R.. 2011. Cofilin-mediated F-actin severing is regulated by the Rap GTPase and controls the cytoskeletal dynamics that drive lymphocyte spreading and BCR microcluster formation. J. Immunol. 187:5887–5900. 10.4049/jimmunol.1102233 [DOI] [PubMed] [Google Scholar]
- Futai K., Kim M.J., Hashikawa T., Scheiffele P., Sheng M., and Hayashi Y.. 2007. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat. Neurosci. 10:186–195. 10.1038/nn1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottmann K. 2008. Transsynaptic modulation of the synaptic vesicle cycle by cell-adhesion molecules. J. Neurosci. Res. 86:223–232. 10.1002/jnr.21484 [DOI] [PubMed] [Google Scholar]
- Govek E.E., Newey S.E., and Van Aelst L.. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49. 10.1101/gad.1256405 [DOI] [PubMed] [Google Scholar]
- Graf E.R., Zhang X., Jin S.X., Linhoff M.W., and Craig A.M.. 2004. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 119:1013–1026. 10.1016/j.cell.2004.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Lee C.W., Fan Y., Komlos D., Tang X., Sun C., Yu K., Hartzell H.C., Chen G., Bamburg J.R., and Zheng J.Q.. 2010. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat. Neurosci. 13:1208–1215. 10.1038/nn.2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe H.S., Lee J.Y., and Pak D.T.. 2009. Combinatorial morphogenesis of dendritic spines and filopodia by SPAR and alpha-actinin2. Biochem. Biophys. Res. Commun. 384:55–60. 10.1016/j.bbrc.2009.04.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.H., Wang Y., Su Z.D., Geng J.G., Chen Y.Z., Yuan X.B., and He C.. 2011. Slit-2 repels the migration of olfactory ensheathing cells by triggering Ca2+-dependent cofilin activation and RhoA inhibition. J. Cell Sci. 124:186–197. 10.1242/jcs.071357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M., Hata Y., Takeuchi M., Ichtchenko K., Toyoda A., Hirao K., Takai Y., Rosahl T.W., and Südhof T.C.. 1997. Binding of neuroligins to PSD-95. Science. 277:1511–1515. 10.1126/science.277.5331.1511 [DOI] [PubMed] [Google Scholar]
- Jedlicka P., Vnencak M., Krueger D.D., Jungenitz T., Brose N., and Schwarzacher S.W.. 2015. Neuroligin-1 regulates excitatory synaptic transmission, LTP and EPSP-spike coupling in the dentate gyrus in vivo. Brain Struct. Funct. 220:47–58. 10.1007/s00429-013-0636-1 [DOI] [PubMed] [Google Scholar]
- Jia Z., Todorovski Z., Meng Y., Asrar S., and Wang L.Y.. 2009. LIMK-1 and actin regulation of spine and synaptic function. In The New Encyclopedia of Neurosciences. Squire L.R., editor. Academic Press, Oxford, England, UK: 467–472. [Google Scholar]
- Jung S.Y., Kim J., Kwon O.B., Jung J.H., An K., Jeong A.Y., Lee C.J., Choi Y.B., Bailey C.H., Kandel E.R., and Kim J.H.. 2010. Input-specific synaptic plasticity in the amygdala is regulated by neuroligin-1 via postsynaptic NMDA receptors. Proc. Natl. Acad. Sci. USA. 107:4710–4715. 10.1073/pnas.1001084107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenstroth G., Tantalaki E., Südhof T.C., Gottmann K., and Missler M.. 2004. Postsynaptic N-methyl-D-aspartate receptor function requires α-neurexins. Proc. Natl. Acad. Sci. USA. 101:2607–2612. 10.1073/pnas.0308626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp N., and Bashir Z.I.. 1999. Induction of LTD in the adult hippocampus by the synaptic activation of AMPA/kainate and metabotropic glutamate receptors. Neuropharmacology. 38:495–504. 10.1016/S0028-3908(98)00222-6 [DOI] [PubMed] [Google Scholar]
- Kemp N., McQueen J., Faulkes S., and Bashir Z.I.. 2000. Different forms of LTD in the CA1 region of the hippocampus: role of age and stimulus protocol. Eur. J. Neurosci. 12:360–366. 10.1046/j.1460-9568.2000.00903.x [DOI] [PubMed] [Google Scholar]
- Kim J., Jung S.Y., Lee Y.K., Park S., Choi J.S., Lee C.J., Kim H.S., Choi Y.B., Scheiffele P., Bailey C.H., et al. . 2008. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc. Natl. Acad. Sci. USA. 105:9087–9092. 10.1073/pnas.0803448105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus U.G., Bamberg A., and Bokoch G.M.. 2007. Rac and Rap GTPase activation assays. Methods Mol. Biol. 412:59–67. 10.1007/978-1-59745-467-4_5 [DOI] [PubMed] [Google Scholar]
- Ko J., Zhang C., Arac D., Boucard A.A., Brunger A.T., and Südhof T.C.. 2009. Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 28:3244–3255. 10.1038/emboj.2009.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.B., Kozorovitskiy Y., Oh W.J., Peixoto R.T., Akhtar N., Saulnier J.L., Gu C., and Sabatini B.L.. 2012. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat. Neurosci. 15:1667–1674. 10.1038/nn.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie E.R., DiBlasio E.A., Kovacic S., Grant K.L., Schendel P.F., and McCoy J.M.. 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N. Y.). 11:187–193. 10.1038/nbt0293-187 [DOI] [PubMed] [Google Scholar]
- Meng Y., Zhang Y., Tregoubov V., Janus C., Cruz L., Jackson M., Lu W.Y., MacDonald J.F., Wang J.Y., Falls D.L., and Jia Z.. 2002. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 35:121–133. 10.1016/S0896-6273(02)00758-4 [DOI] [PubMed] [Google Scholar]
- Meng Y., Zhang Y., and Jia Z.. 2003. Synaptic transmission and plasticity in the absence of AMPA glutamate receptor GluR2 and GluR3. Neuron. 39:163–176. 10.1016/S0896-6273(03)00368-4 [DOI] [PubMed] [Google Scholar]
- Meng Y., Takahashi H., Meng J., Zhang Y., Lu G., Asrar S., Nakamura T., and Jia Z.. 2004. Regulation of ADF/cofilin phosphorylation and synaptic function by LIM-kinase. Neuropharmacology. 47:746–754. 10.1016/j.neuropharm.2004.06.030 [DOI] [PubMed] [Google Scholar]
- Meyer G., Varoqueaux F., Neeb A., Oschlies M., and Brose N.. 2004. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 47:724–733. 10.1016/j.neuropharm.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Nam C.I., and Chen L.. 2005. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc. Natl. Acad. Sci. USA. 102:6137–6142. 10.1073/pnas.0502038102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak D.T., Yang S., Rudolph-Correia S., Kim E., and Sheng M.. 2001. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 31:289–303. 10.1016/S0896-6273(01)00355-5 [DOI] [PubMed] [Google Scholar]
- Palsson E.M., Popoff M., Thelestam M., and O’Neill L.A.. 2000. Divergent roles for Ras and Rap in the activation of p38 mitogen-activated protein kinase by interleukin-1. J. Biol. Chem. 275:7818–7825. 10.1074/jbc.275.11.7818 [DOI] [PubMed] [Google Scholar]
- Peixoto R.T., Kunz P.A., Kwon H., Mabb A.M., Sabatini B.L., Philpot B.D., and Ehlers M.D.. 2012. Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron. 76:396–409. 10.1016/j.neuron.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.B. 2015. ADF/cofilin: a crucial regulator of synapse physiology and behavior. Cell. Mol. Life Sci. 72:3521–3529. 10.1007/s00018-015-1941-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.B., Gurniak C.B., Renner M., Vara H., Morando L., Görlich A., Sassoè-Pognetto M., Banchaabouchi M.A., Giustetto M., Triller A., et al. . 2010. Learning, AMPA receptor mobility and synaptic plasticity depend on n-cofilin-mediated actin dynamics. EMBO J. 29:1889–1902. 10.1038/emboj.2010.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P., Fan J., Choih J., Fetter R., and Serafini T.. 2000. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 101:657–669. 10.1016/S0092-8674(00)80877-6 [DOI] [PubMed] [Google Scholar]
- Shipman S.L., and Nicoll R.A.. 2012. A subtype-specific function for the extracellular domain of neuroligin 1 in hippocampal LTP. Neuron. 76:309–316. 10.1016/j.neuron.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Hayashi Y., Nakahara S., Kumazaki H., Prox J., Horiuchi K., Zeng M., Tanimura S., Nishiyama Y., Osawa S., et al. . 2012. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron. 76:410–422. 10.1016/j.neuron.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Tabuchi K., Blundell J., Etherton M.R., Hammer R.E., Liu X., Powell C.M., and Südhof T.C.. 2007. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 318:71–76. 10.1126/science.1146221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Koshimizu U., Miyazaki J., and Nakamura T.. 2002. Impaired spermatogenic ability of testicular germ cells in mice deficient in the LIM-kinase 2 gene. Dev. Biol. 241:259–272. 10.1006/dbio.2001.0512 [DOI] [PubMed] [Google Scholar]
- Varoqueaux F., Aramuni G., Rawson R.L., Mohrmann R., Missler M., Gottmann K., Zhang W., Südhof T.C., and Brose N.. 2006. Neuroligins determine synapse maturation and function. Neuron. 51:741–754. 10.1016/j.neuron.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Wang Y., Dong Q., Xu X.F., Feng X., Xin J., Wang D.D., Yu H., Tian T., and Chen Z.Y.. 2013. Phosphorylation of cofilin regulates extinction of conditioned aversive memory via AMPAR trafficking. J. Neurosci. 33:6423–6433. 10.1523/JNEUROSCI.5107-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfrey K.M., Srivastava D.P., Photowala H., Yamashita M., Barbolina M.V., Cahill M.E., Xie Z., Jones K.A., Quilliam L.A., Prakriya M., and Penzes P.. 2009. Epac2 induces synapse remodeling and depression and its disease-associated forms alter spines. Nat. Neurosci. 12:1275–1284. 10.1038/nn.2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Huganir R.L., and Penzes P.. 2005. Activity-dependent dendritic spine structural plasticity is regulated by small GTPase Rap1 and its target AF-6. Neuron. 48:605–618. 10.1016/j.neuron.2005.09.027 [DOI] [PubMed] [Google Scholar]
- Xu J., Xiao N., and Xia J.. 2010. Thrombospondin 1 accelerates synaptogenesis in hippocampal neurons through neuroligin 1. Nat. Neurosci. 13:22–24. 10.1038/nn.2459 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Hu J., Passafaro M., Xie W., and Jia Z.. 2011. GluA2 (GluR2) regulates metabotropic glutamate receptor-dependent long-term depression through N-cadherin-dependent and cofilin-mediated actin reorganization. J. Neurosci. 31:819–833. 10.1523/JNEUROSCI.3869-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.J., Qin Y., Zhao M., Van Aelst L., and Malinow R.. 2002. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 110:443–455. 10.1016/S0092-8674(02)00897-8 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Pak D., Qin Y., McCormack S.G., Kim M.J., Baumgart J.P., Velamoor V., Auberson Y.P., Osten P., van Aelst L., et al. . 2005. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 46:905–916. 10.1016/j.neuron.2005.04.037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.