Abstract
Mitochondria are renowned for their central bioenergetic role in eukaryotic cells, where they act as powerhouses to generate adenosine triphosphate from oxidation of nutrients. At the same time, these organelles are highly dynamic and undergo fusion, fission, transport, and degradation. Each of these dynamic processes is critical for maintaining a healthy mitochondrial population. Given the central metabolic function of mitochondria, it is not surprising that mitochondrial dynamics and bioenergetics reciprocally influence each other. We review the dynamic properties of mitochondria, with an emphasis on how these processes respond to cellular signaling events and how they affect metabolism.
Introduction
As the site for numerous biochemical processes—including oxidative phosphorylation (OXPHOS), the Krebs cycle, β-oxidation of fatty acids, calcium handling, and heme biosynthesis—the mitochondrion plays a central role in cellular metabolism. As a result, the dysfunction of mitochondria, particularly in their metabolic activities, has been associated with many disorders, including metabolic diseases, cancers, and neurodegenerative diseases, as well as the aging process (Carelli and Chan, 2014; Lightowlers et al., 2015).
To maintain their health, mitochondria engage in several dynamic behaviors. The main dynamic activities are fusion (the joining of two organelles into one), fission (the division of a single organelle into two), transport (directed movement within a cell), and mitophagy (targeted destruction via the autophagic pathway; Fig. 1). From yeast to mammals, these dynamic behaviors have been shown to be clearly important in both normal physiology and disease states (Labbé et al., 2014; Mishra and Chan, 2014). In an early example, deletion of Fzo1p, a yeast GTPase essential for mitochondrial fusion, resulted in mitochondrial fragmentation, complete loss of mitochondrial DNA (mtDNA), impairment of OXPHOS activity, and inability to grow on nonfermentable carbon sources (Hermann et al., 1998).
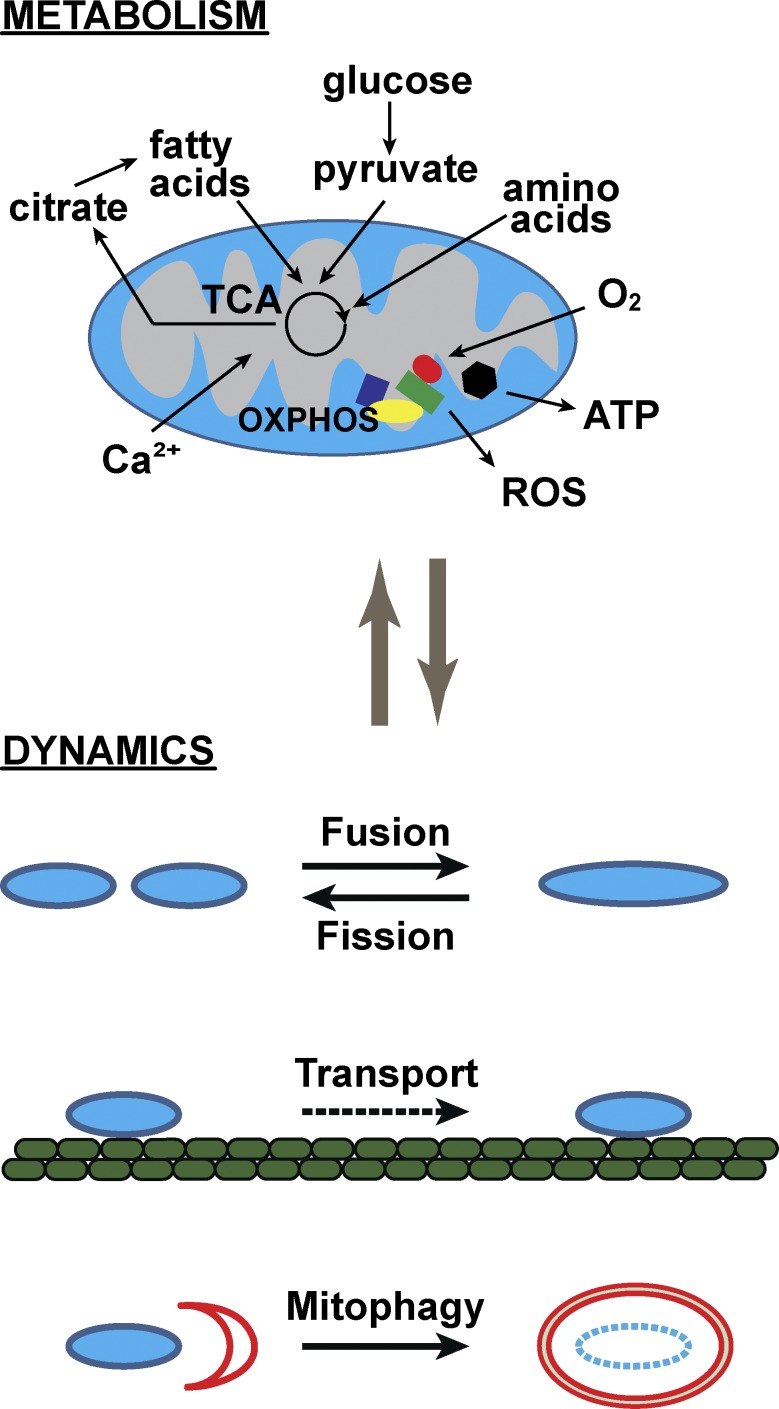
Figure 1.
Overview of mitochondrial metabolism and dynamics. The mitochondrion is central to metabolism, being involved in the catabolism of numerous substrates, generation of metabolic signals, and sensing of metabolic cues. The processes diagrammed are not meant to be exhaustive, but to illustrate the diversity of biochemical pathways that impinge on the organelle. Mitochondria participate in macroscopic behaviors (termed dynamics) including fusion, fission, transport, and mitophagy. Although these behaviors are molecularly distinct from the organelle’s bioenergetic reactions, recent studies suggest that metabolism and dynamics are highly linked and regulate one another. ROS, reactive oxygen species.
On the surface, these dynamic processes appear mechanistically distinct from the biochemical and metabolic processes occurring within the organelle. However, given the central role of mitochondria in bioenergetics, it is not surprising that in the last several years, multiple lines of evidence have emerged for a strong link between mitochondrial metabolism and dynamics. In this review, we discuss how metabolism regulates the key mitochondrial behaviors of fusion, fission, transport, and mitophagy.
Metabolic control of mitochondrial fusion
Mitochondrial fusion is an evolutionarily conserved process that, in mammals, is mediated by three large GTPases of the dynamin superfamily (Chan, 2012; Labbé et al., 2014): Mitofusin 1 (Mfn1), Mfn2, and Optic Atrophy 1 (Opa1). Because mitochondria have double membranes, mitochondrial fusion is a two-step process requiring outer-membrane fusion followed by inner-membrane fusion. Mfn1 and Mfn2 are integral outer-membrane proteins that mediate outer-membrane fusion, whereas OPA1 has multiple isoforms associated with the inner membrane and mediates inner-membrane fusion. Mitochondrial fusion events occur frequently in numerous cell types cultured in vitro, although fusion rates are cell type dependent and often occur less frequently in tissues (Pham et al., 2012; Eisner et al., 2014). Because the balance between fusion and fission controls mitochondrial morphology, genetic deletion of the fusion genes results in severe fragmentation of the mitochondrial network and abolishes content exchange between mitochondria (Hermann et al., 1998; Chen et al., 2003, 2005). In humans, mutations in Mfn2 cause Charcot–Marie–Tooth disease type 2A, a peripheral neuropathy affecting long motor and sensory neurons (Züchner et al., 2004). Mutations in Opa1 cause dominant optic atrophy, a blindness caused by degeneration of retinal ganglion cells (Alexander et al., 2000; Delettre et al., 2000, 2002).
The fusion process is well known to be important for OXPHOS activity, particularly through the regulation of mtDNA levels. The sensitivity of cells to reduced mitochondrial fusion is context dependent. For example, mouse embryonic fibroblasts can tolerate a partial defect in mitochondrial fusion, such as loss of either Mfn1 or Mfn2, without much bioenergetic consequence. However, cerebellar Purkinje neurons cannot survive Mfn2 removal, because of loss of respiratory chain activity (Chen et al., 2007). Moreover, complete loss of mitochondrial fusion caused by removal of both mitofusins or Opa1 results in a dramatic decrease in mtDNA content, heterogeneous loss of mtDNA nucleoids and membrane potential, and reduced respiratory chain function in both cultured cells and mouse tissues (Chen et al., 2005, 2010). Other mechanisms also link these proteins with metabolism: Mfn2 maintains coenzyme Q levels (Mourier et al., 2015), and Opa1 maintains mitochondrial cristae structure and is critical for respiratory chain supercomplex assembly (Cogliati et al., 2013).
The energetic states of cells are often associated with specific mitochondrial morphologies. In yeast, nonfermentable culture conditions that force increased OXPHOS activity are accompanied by elongation of the mitochondrial network (Egner et al., 2002; Jakobs et al., 2003). An analogous study with human cells suggested that mitochondria elongate during growth in galactose media, which forces cells to rely more heavily on OXPHOS for ATP production (Rossignol et al., 2004). Elongated mitochondria have also been observed in other conditions associated with increased ATP production (Mitra et al., 2009; Tondera et al., 2009). These observations suggest that high OXPHOS activity correlates with mitochondrial elongation and is consistent with the proposal that elongated mitochondrial networks are more efficient at energy generation and capable of distributing energy through long distances (Amchenkova et al., 1988; Skulachev, 2001).
Another possibility, not mutually exclusive, is that increased OXPHOS activity stimulates mitochondrial fusion to cause elongation. The development of in vitro fusion assays using isolated organelles (Meeusen et al., 2004; Hoppins et al., 2011) allowed for more detailed investigations regarding the regulation of mitochondrial fusion. In isolated organelles, addition of respiratory chain substrates that promoted OXPHOS activity led to stimulation of mitochondrial inner-membrane fusion, whereas outer-membrane fusion was unaffected by the metabolic state (Mishra et al., 2014). OXPHOS activity stimulates the metalloprotease Yme1L to proteolytically process Opa1, leading to activation of its fusion activity.
Opa1 is expressed as a membrane-integrated long form, which can then be cleaved to a soluble short form by two distinct metalloproteases, the ATP-dependent protease Yme1L and the membrane potential–dependent protease Oma1. It has been well known that the presence of both long and short forms correlates with fusion-competent mitochondria (McQuibban et al., 2003; Song et al., 2007). In mitochondrial fusion intermediates that have undergone outer membrane fusion, proteolytic processing of Opa1 at the Yme1L or Oma1 cleavage site was sufficient to stimulate inner-membrane fusion (Mishra et al., 2014). The processing of Opa1 by two metalloproteases allows differential regulation of inner-membrane fusion. Proteolysis via Yme1L is responsible for OXPHOS-dependent stimulation of inner-membrane fusion (Fig. 2). In contrast, when membrane potential is dissipated, the long isoform of Opa1 is completely cleaved and inactivated (Fig. 2; Ishihara et al., 2006) owing to activation of Oma1 (Ehses et al., 2009; Head et al., 2009). A variety of cellular stresses can activate Oma1 to cleave Opa1 (Baker et al., 2014). This mechanism likely contributes to the mitochondrial fragmentation found in many forms of mitochondrial dysfunction (Duvezin-Caubet et al., 2006). Whereas large-scale depolarization of mitochondria clearly inactivates Opa1, transient depolarizations of mitochondria may partially activate Oma1 and be pro-fusogenic. Transient mitochondrial depolarizations have been reported to occur in cultured cells and are associated with fusion events (Santo-Domingo et al., 2013). It should be noted that cells lacking Yme1L and Oma1 do retain residual mitochondrial fusion activity (Anand et al., 2014) and more work will be needed to dissect how inner-membrane fusion is regulated in this situation.
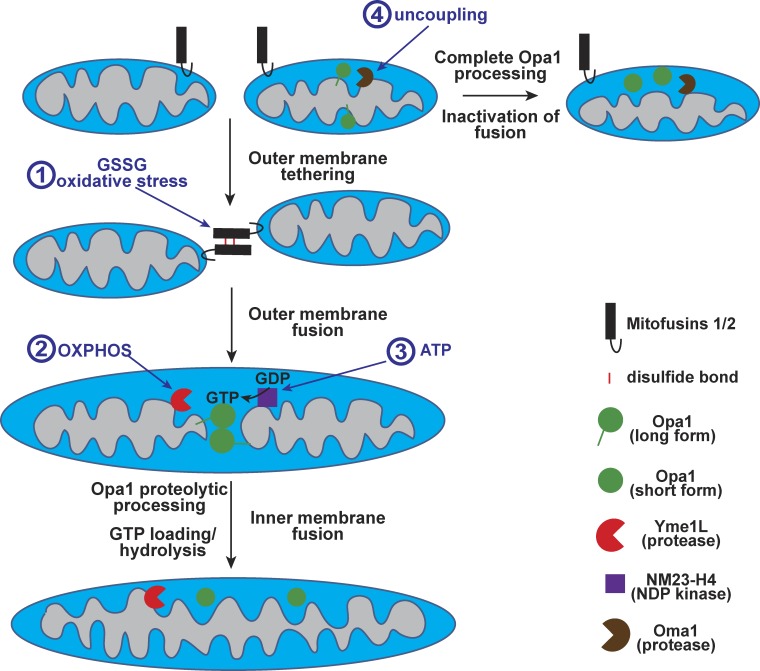
Figure 2.
Metabolic regulation of mitochondrial fusion. Mitochondrial fusion consists of outer membrane fusion, mediated by mitofusins, followed by inner membrane fusion, mediated by Opa1. Modes of regulation include the following: (1) Oxidative stress and high levels of oxidized glutathione (GSSG) promote trans complexes of mitofusins, facilitated by disulfide bonds (red bars), leading to organelle tethering and enhanced outer-membrane fusion. (2) Inner-membrane fusion is stimulated by OXPHOS activity, which enhances Yme1L-mediated proteolytic processing of Opa1 from the long form to the soluble short form. In isolated organelles, Opa1 proteolysis is necessary and sufficient to activate inner-membrane fusion. (3) Enhanced ATP levels are potentially linked to GTP-loading of Opa1 via the nucleotide diphosphate kinase NM23-H4. GTP loading and hydrolysis by Opa1 are required for inner-membrane fusion. (4) Metabolic stresses, including loss of membrane potential, activate the inner membrane protease Oma1 and result in complete proteolytic processing of Opa1. Short forms of Opa1, by themselves, are inactive for inner-membrane fusion.
Together, these studies suggest that the inner membrane proteases Yme1L and Oma1 serve as important sensors to link metabolic conditions to the inner-membrane fusion machinery. In particular, conditions that increase mitochondrial ATP function lead to enhanced fusion, whereas metabolic signals that grossly uncouple the mitochondria result in fusion inhibition. These regulatory modes appear to be in play in normal physiology and disease. In skeletal muscle, the more oxidative fiber types have enhanced mitochondrial fusion, presumably promoting health of the active mitochondrial population (Mishra et al., 2015). In mtDNA disease, defects in OXPHOS result in secondary defects in mitochondrial inner-membrane fusion (Mishra et al., 2014). This regulatory process may help to segregate dysfunctional mitochondria and prevent the spread of mtDNA mutations to wild-type mitochondria. In the case of skeletal muscle, the linkage of fusion to OXPHOS activity may serve to restrict mtDNA defects to a localized region of the muscle fiber, a phenomenon observed in older individuals and some patients with mitochondrial myopathy (Moraes et al., 1992; Elson et al., 2002; Bua et al., 2006). In addition, the inability of defective mitochondria to fuse with the remainder of the organelle population provides a means to segregate mutant organelles into small components ideal for autophagic destruction (Twig et al., 2008), as discussed later.
Other metabolic mechanisms to regulate mitochondrial fusion have also been proposed (Fig. 2). Oxidative stress can enhance fusion in both isolated organelles and cells. In this situation, elevated levels of oxidized glutathione promote disulfide-mediated dimerization of mitofusin molecules and organelle tethering, the first step in the fusion process (Shutt et al., 2012). Mfn1 is also regulated by phosphorylation by extracellular signal–regulated kinase, linking the MAPK pathway to mitochondrial fusion (Pyakurel et al., 2015). Other studies have suggested that fusion may be controlled via the local concentration of GTP. NM23-H4 is a mitochondrially localized nucleotide disphosphate kinase that can generate GTP (from GDP) in the presence of ATP. Interestingly, NM23-H4 can promote GTP loading onto Opa1, and knockdown of NM23-H4 leads to fusion defects reminiscent of Opa1 knockdown (Boissan et al., 2014). It remains possible that this mechanism allows cellular ATP levels to be linked to fusion via GTP-loading of Opa1. Thus, multiple steps of the fusion process may be independently targeted via distinct regulatory mechanisms, potentially providing an exquisite level of control over the fusion event (Fig. 2).
Metabolic control of mitochondrial fission
As a complement to fusion, fission of mitochondria is equally critical for cellular and organismal physiology (Chan, 2012; Labbé et al., 2014). Division of mitochondria is mediated by Dynamin-related Protein 1 (Drp1), a large GTPase that is recruited to the mitochondrial outer membrane via a collection of receptor proteins (Mff, Fis1, MiD49, and MiD50). Once on mitochondria, Drp1 assembles around the tubule and constricts it in a GTP-dependent manner to mediate scission. Besides influencing mitochondrial morphology, fission has been implicated in multiple functions, including the facilitation of mitochondrial transport, mitophagy, and apoptosis. In humans, two clinical studies have linked Drp1 mutation to microcephaly, neonatal lethality (Waterham et al., 2007), and refractory epilepsy (Vanstone et al., 2015), and another has linked Mff mutation to two cases of developmental delay with neuromuscular dysfunction (Shamseldin et al., 2012).
Perhaps the best-known regulatory mechanism for mitochondrial fission involves phosphorylation of Drp1. Multiple phosphorylation sites and kinases have been identified, and many of these events are linked to signaling pathways activated by metabolic events. In addition, phosphorylation can activate or inhibit Drp1, depending on the site involved. In this review, we focus on Drp1 phosphorylation, although it should be noted that Drp1 regulation is complex, and several other Drp1 posttranslational mechanisms have been identified, including S-nitrosylation, SUMOylation, and acetylation.
Studies on Drp1 phosphorylation have centered on two critical sites. Because studies designate these sites differently depending on the species studied, we consolidate the findings and refer to the sites as serine 616 (S616) and S637 based on the sequence of human Drp1, isoform 1. Protein kinase A (PKA) phosphorylation of Drp1 at S637 has been clearly shown to inhibit its activity in vitro, promoting overall elongation of the mitochondrial network in response to pharmacologic activation (e.g., forskolin), β-adrenergic stimulation, or forced exercise (Fig. 3; Chang and Blackstone, 2007; Cribbs and Strack, 2007). Phosphorylation at S637 is counteracted by the phosphatases calcineurin (Cribbs and Strack, 2007; Cereghetti et al., 2008) and PP2A/Bβ2 (Dickey and Strack, 2011). Negative regulation of Drp1 by phosphorylation at S637 also occurs during mTOR inhibition and nitrogen starvation, which increases cAMP levels and activates PKA (Gomes et al., 2011; Rambold et al., 2011). Even though autophagy is activated during starvation, inhibition of Drp1 results in enhanced mitochondrial tubulation that promotes mitochondrial ATP production and spares the organelles from degradation, because of their increased size.
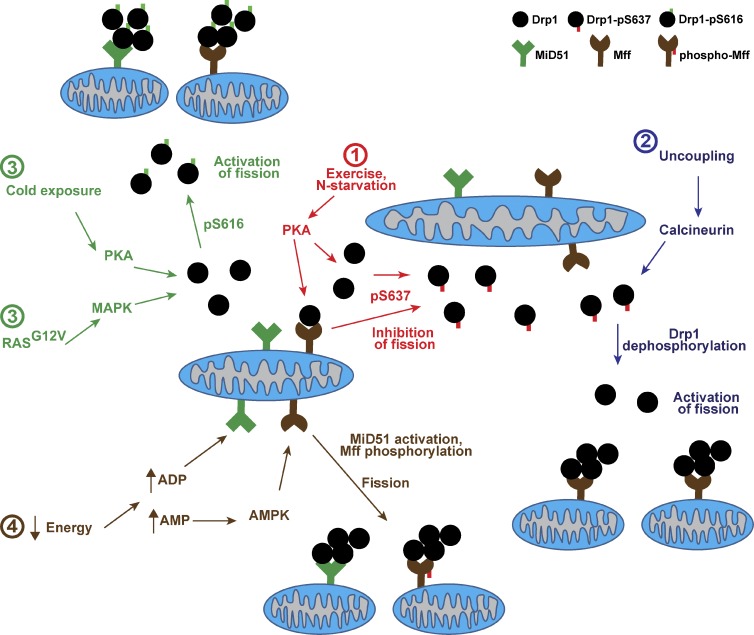
Figure 3.
Metabolic regulation of mitochondrial fission. Fission is mediated by the master regulator Drp1, which must be recruited from a cytosolic pool onto the mitochondrial surface. Receptor proteins on the outer membrane are required for Drp1 recruitment and activation of fission. For simplicity, only two receptor proteins, Mff and MiD51, are shown. Four modes of regulation are color-coded in the diagram: (1) Exercise and nitrogen starvation result in PKA activation, followed by phosphorylation of Drp1 at Ser637, which is inhibitory for fission because of sequestration of Drp1 in the cytosol. (2) Reversal of phosphoS637 can be achieved via calcineurin, which is activated by metabolic uncoupling of the organelle. These events lead to recruitment of Drp1 and rapid activation of fission. (3) Cold exposure and oncogenic RasG12V activate fission via Ser616 phosphorylation by PKA or MAPK, respectively. (4) Severe energy depletion can potentially activate fission via elevation of ADP and AMP levels. ADP binding to the MiD51 receptor is necessary for Drp1 recruitment and fission. AMP-sensing by AMPK results in phosphorylated Mff and activated fission.
In contrast, drug treatments that inhibit mitochondrial OXPHOS are generally associated with enhancement of fission. The most commonly used are the mitochondrial uncouplers (e.g., CCCP and FCCP), which result in a rapid and dramatic fragmentation of the organellar network in multiple cell types. As noted, uncouplers stimulate Oma1 to cleave Opa1, resulting in inactivation of mitochondrial fusion. On the fission side, dephosphorylation at Drp1 serine 637 via the Ca2+-dependent phosphatase calcineurin promotes Drp1 activation and recruitment to the mitochondrial surface (Fig. 3; Cribbs and Strack, 2007; Cereghetti et al., 2008). Calcineurin therefore relays metabolic stimuli associated with calcium changes into alterations in mitochondrial morphology. For example, dysfunction of the calcium-buffering activity of mitochondria would increase cytosolic calcium levels and potentially trigger calcineurin-mediated mitochondrial fission. In addition, mice with calcineurin knocked out in skeletal muscle have recently been shown to exhibit elongated mitochondria, increased respiratory chain activity, resistance to obesity under a high-fat diet, and diminished exercise performance (Pfluger et al., 2015).
In brown adipose tissue, thermogenesis has been shown to involve activation of Drp1 and mitochondrial fission. In this cell type, cold exposure results in the oxidation of fatty acids in uncoupled mitochondria to increase heat production, as opposed to ATP synthesis. Intriguingly, the norepinephrine-based signaling events initiated by cold exposure result in PKA activation and Drp1 phosphorylation at S616, which activates mitochondrial fission (Fig. 3; Wikstrom et al., 2014). Through unclear mechanisms, the fission event promotes enhanced uncoupling and sensitivity to fatty acids, thereby aiding heat generation. Thus, mechanisms to enhance fission may decrease OXPHOS efficiency and be useful in times of nutrient excess.
Drp1 phosphorylation at S616 has also been implicated in tumorigenesis by oncogenic Ras (Fig. 3; Kashatus et al., 2015; Serasinghe et al., 2015). Up-regulation of the MAPK pathway by Ras induces Erk1-mediated phosphorylation of Drp1 at S616, which results in enhanced mitochondrial fission. Remarkably, inhibition of Drp1 function attenuates the oncogenic activity of Ras in cell and xenograft models of tumorigenesis. In future studies, it will be interesting to determine whether mitochondrial dynamics play an important role in tumor cell metabolism.
Finally, regulation of fission may also occur at the level of the Drp1 receptor proteins. In cultured cells, Mff appears to be the primary receptor, because loss of Mff results in dramatic mitochondrial elongation (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Losón et al., 2013). Recent studies show that Mff is a phosphorylation substrate for AMP kinase (AMPK; Ducommun et al., 2015; Toyoma et al., 2016). This phosphorylation event activates Mff and mitochondrial fission, explaining how AMPK links energy deficiency to mitochondrial fragmentation (Fig. 3; Toyoma et al., 2016).
Along similar lines, recent crystal structures of the receptor protein MiD51 suggest a potential role for metabolic regulation (Losón et al., 2014; Richter et al., 2014). The cytosolic domain adopts an enzymatically dead nucleotidyltransferase fold, which contains a high-affinity binding site for the dinucleotides ADP and GDP. Indeed, dinucleotide binding appears to be required for receptor function, and the protein potentially serves as a sensor for ADP levels. Thus, MiD51 potentially links metabolic conditions to enhanced organelle fission (Fig. 3). Intriguingly, the paralogous receptor MiD49 does not contain this mode of regulation, as it does not bind nucleotides despite a similar fold and the presence of a binding pocket (Losón et al., 2015). It is unknown whether MiD49 binds an alternative class of ligands that regulates its activity.
Metabolic control of mitochondrial transport
Because of their ability to affect local ATP and calcium concentrations, the subcellular distribution of mitochondria can be extremely important. In a dramatic example, the mitochondria of the sperm cell are concentrated in the proximal region of the flagellum, where they appear ideally positioned to supply ATP for the force-generating motor proteins that drive sperm movement (Woolley, 1970). In a more dynamic example, the mitochondria of neurons can traffic to the axon terminals of neurons to fuel energy-consuming processes such as synaptic vesicle recycling. In many mammalian cells, mitochondrial transport is accomplished via the activity of motor proteins working along the microtubule network, although transport along other cytoskeletal elements can also occur (Saxton and Hollenbeck, 2012). Kinesin motor proteins, particularly the kinesin-1 family, mediate transport in the positive (or anterograde) direction, whereas dyneins mediate transport in the negative (or retrograde) direction (Pilling et al., 2006). The motor proteins are connected to the mitochondrial surface through receptor and adaptor proteins, whose functions were first revealed in Drosophila melanogaster neurons (Stowers et al., 2002; Guo et al., 2005). In mammals, the primary receptor proteins are Miro1 and Miro2, transmembrane GTPases localized in the mitochondrial outer membrane. The Miro proteins interact with kinesin via the Milton proteins, also known as Trak1 and Trak2. Together, the Miro–Milton–kinesin complex mediates anterograde transport of mitochondria along microtubules.
Regulation of mitochondrial transport is particularly important in neurons, and experiments in the neuronal system have been fruitful in identifying the relevant mechanisms. Mitochondrial transport behavior in axons is complex and involves a balance between movement and stalling. In axonal segments, a substantial fraction of the mitochondria is immobile at any given time. High calcium levels tend to cause pausing of mitochondria in axons, a phenomenon that may help to retain mitochondria at active sites along the axon. Because Miro contains regulatory EF-hands that bind calcium, it is ideally suited to sense local calcium levels near the mitochondrion. Multiple mechanistic models have been proposed, but they all center on conformational changes triggered by calcium binding to Miro (Macaskill et al., 2009; Wang and Schwarz, 2009; Chen and Sheng, 2013). In one model, high calcium causes the Miro–Milton–kinesin complex to be released from the microtubule (Fig. 4 A; Wang and Schwarz, 2009). In another model, high calcium causes the Miro–Milton complex to release kinesin, thereby freeing mitochondria from the microtubule (Macaskill et al., 2009). In the third, most recent model, mitochondrial pausing in axons is mediated by the protein syntaphilin, which serves to anchor axonal mitochondria to microtubules (Kang et al., 2008; Chen and Sheng, 2013). With high local calcium, kinesin is released by the Miro–Milton complex and binds syntaphilin, which substantially reduces its motor activity (Chen and Sheng, 2013).
Figure 4.
Metabolic regulation of mitochondrial transport and mitophagy. (A) In mammals, mitochondrial transport is primarily mediated by microtubule-dependent motors, such as kinesin for anterograde movement. Kinesin-1 attaches to mitochondria via its adaptor (Milton) and receptor (Miro). The Miro–Milton–kinesin complex allows for organelle movement under basal conditions. (1) At active synapses of neurons, increased Ca2+ levels result in pausing of mitochondria to supply local ATP to drive energy-intensive processes such as synaptic vesicle recycling. Depending on the model, Ca2+ loading of the EF-hands of Miro is followed by either release of the Miro–Milton–kinesin complex from the microtubule or anchoring of the mitochondrion via syntaphilin. (2) Elevated glucose levels also promote stalling, caused by O-GlcNAc transferase (OGT)-mediated glycosylation of Milton. Although glycosylated Milton is depicted in the syntaphilin model, this is for convenience; the precise method by which glycosylation of Milton mediates stalling is unclear. This regulatory pathway may allow mitochondria to be positioned at locations of nutrient abundance, increasing their efficiency of ATP generation. (B) Multiple mechanisms for regulation of mitophagy have been proposed: (1) Mitochondrial damage leading to loss of membrane potential (ΔΨm) causes Pink1 accumulation (not depicted), followed by Parkin recruitment and ubiquitination of multiple outer membrane proteins. These events activate the outer membrane for processing via the proteasome system (UPS), followed by targeting to autophagosome membranes. (2) Severe energy depletion leads to activation of AMPK, followed by phosphorylation and activation of the autophagy regulator ULK1. ULK1 is then able to activate generalized autophagy, including mitophagy. (3) Hypoxia is able to activate mitophagy via the dephosphorylation of FUNDC1 (on the outer membrane) by the PGAM5 phosphatase. Dephosphorylated FUNDC1 serves to recruit LC3 and autophagosomal membranes. (4) Through unknown mechanisms, enhanced OXPHOS activity in the mitochondrion recruits the autophagy regulator Rheb to the outer membrane receptor Nix. Mitochondrially-localized Rheb then promotes autophagy via recruitment of LC3 molecules.
Other mechanisms have been suggested to directly link mitochondrial transport to energy status. ATP depletion or hypoxia promotes anterograde mitochondrial movement into axons (Mironov, 2007; Li et al., 2009; Tao et al., 2014) via activation of AMPK or HIF-1α pathways. Alternatively, nutrient status has also been implicated in direct control of organelle transport. Milton interacts with and serves as a substrate for O-GlcNAc transferase, which glycosylates the adaptor at several residues. In the presence of high glucose, glycosylation of Milton results in immobilization of the mitochondria, although the precise mechanism is still unclear (Fig. 4 A; Pekkurnaz et al., 2014). This posttranslational modification may serve to enrich mitochondria at locations with high nutrients, promoting increased efficiency of ATP production.
Metabolic control of mitophagy
The overall mitochondrial mass within a cell is likely regulated by a balance between biogenesis and degradation. When mitochondria are excessive, or become aged or defective, organelle clearance is thought to occur primarily through autophagy, a process termed mitophagy. The removal of mitochondria can be either random or selective. During bulk autophagy, mitochondrial degradation is included as part of a generalized autophagy program activated by the metabolic state of the cell. In other cases, mitophagy is a culling process that selectively degrades only defective mitochondria, thereby maintaining the overall health of the mitochondrial population.
Although there is widespread interest in mitophagy as a potential quality control process for mitochondria, it should be noted that in vivo evidence for the importance of mitophagy in mitochondrial homeostasis remains sparse. In particular, studies on the Pink1/Parkin system (described in the next paragraph) have usually relied on overexpressing Parkin and stressing the cells with an uncoupler. Although such approaches are extremely valuable in dissecting the biochemical pathway, further studies are required to determine the in vivo function of mitophagy. The recent development of a mouse reporter for tracking mitophagy in vivo (Sun et al., 2015) will be helpful in this regard.
The best-studied mitophagy pathway involves Pink1 and Parkin, genes responsible for some cases of familial Parkinson’s disease. Pink1, a mitochondrially localized kinase, is normally imported and degraded within the organelle. Because protein import is dependent on mitochondrial membrane potential, depolarization results in accumulation of Pink1 on the outer membrane (Matsuda et al., 2010; Narendra et al., 2010). The accumulated Pink1 phosphorylates numerous proteins, including ubiquitin, to recruit and activate Parkin, an E3 ligase (Okatsu et al., 2015). Activated Parkin results in widespread ubiquitination of mitochondrial outer-membrane proteins, whose degradation by the 26S proteasome (Chan et al., 2011; Sarraf et al., 2013) is required for targeting of the mitochondrion to autophagic membranes. Because Parkin is selectively enriched on dysfunctional mitochondria, healthy organelles are spared from autophagic degradation. Pink1 and Parkin have also been implicated in another mitochondrial quality control pathway, distinct from autophagy, in which vesicles bud off from mitochondria and are trafficked to the late endosome and lysosome (McLelland et al., 2014; Sugiura et al., 2014).
Mitophagy can be activated under certain cellular stresses. With energy stress, activation of AMPK results in phosphorylation of ULK1 and ULK2, mammalian protein kinases that are orthologues of the autophagy gene ATG1 (Egan et al., 2011). ULK1 and ULK2 promote autophagy, including the degradation of mitochondria. AMPK also inhibits the growth-promoting mTORC pathway, which normally inhibits ULK function. These interlinked mechanisms couple mitophagy to the nutrient status of the cell.
Hypoxic conditions are also able to trigger mitophagy via a distinct pathway. Activation of the mitochondrial phosphatase PGAM5 results in dephosphorylation of the mitochondrial autophagy receptor, FUNDC1. The dephosphorylation event promotes the interaction of FUNDC1 with ATG8 (also known as LC3), stimulating formation of the autophagic membrane (Liu et al., 2012; Chen et al., 2014). It is not clear how hypoxia activates PGAM5, and whether this mechanism is selective for individual mitochondria.
Finally, metabolic conditions that promote increased mitochondrial function are also associated with increased mitophagy (Melser et al., 2013). Under glucose-free (oxidative) conditions, mitochondrial OXPHOS is up-regulated, and bulk mitophagy is also enhanced. The small GTPase Rheb is proposed to be involved, as it partially localizes to the outer mitochondrial membrane under oxidative conditions and interacts with the mitochondrial autophagy receptor, Nix. The relocalization of Rheb promotes the recruitment of LC3 molecules, thereby enhancing mitophagy. The molecular signals that recruit Rheb are currently unclear. The promotion of mitophagy during oxidative conditions, when mitochondrial function is increased, appears to contrast with previous mitophagy models in which dysfunctional organelles are cleared. However, enhanced respiratory chain activity may promote damage to mitochondria (e.g., through increased production of reactive oxygen species), and it is possible that Rheb is specifically responding to these damaged mitochondria. Alternatively, this mechanism may be in place to increase bulk turnover of the population during conditions of increased functional demand. In either case, the increased mitophagic flux promotes overall energetic efficiency of the organelle population.
Future directions
It is becoming clear that the multiple functions and behaviors of the mitochondrion do not operate independently but instead influence each other and are subject to common regulatory pathways. For instance, depolarization of the organelle triggers fission, inhibits fusion, and promotes mitophagy, whereas hypoxia promotes transport, mitophagy, and fission. Predicting how the cell integrates multiple signals to regulate mitochondrial function is therefore complex and dependent on the specifics of the stimuli, as well as the cell type. It is clear, however, that the organelle is responsive to numerous types of metabolic stimuli, and this likely has resulting effects on the health and function of the organelle population. A future challenge will be to integrate the data from numerous studies, functions, and perturbations to further our understanding of the regulatory biology of mitochondria and its implication in normal physiology and disease states. Understanding how to regulate mitochondrial behavior may provide therapeutic approaches to modulate mitochondrial physiology in diseased states. We have only begun to investigate a few aspects of the dynamic behavior of mitochondria. Additional properties, such as its interaction with other organelles, including lipid droplets and the endoplasmic reticulum, are only beginning to be understood and likely will have clear implications on overall cellular function.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM110039) and the Muscular Dystrophy Association.
The authors declare no competing financial interests.
References
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. . 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215. 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- Amchenkova A.A., Bakeeva L.E., Chentsov Y.S., Skulachev V.P., and Zorov D.B.. 1988. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J. Cell Biol. 107:481–495. 10.1083/jcb.107.2.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E., and Langer T.. 2014. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204:919–929. 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.J., Lampe P.A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., and Langer T.. 2014. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33:578–593. 10.1002/embj.201386474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissan M., Montagnac G., Shen Q., Griparic L., Guitton J., Romao M., Sauvonnet N., Lagache T., Lascu I., Raposo G., et al. . 2014. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 344:1510–1515. 10.1126/science.1253768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bua E., Johnson J., Herbst A., Delong B., McKenzie D., Salamat S., and Aiken J.M.. 2006. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 79:469–480. 10.1086/507132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli V., and Chan D.C.. 2014. Mitochondrial DNA: Impacting central and peripheral nervous systems. Neuron. 84:1126–1142. 10.1016/j.neuron.2014.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti G.M., Stangherlin A., Martins de Brito O., Chang C.R., Blackstone C., Bernardi P., and Scorrano L.. 2008. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. USA. 105:15803–15808. 10.1073/pnas.0808249105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C. 2012. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46:265–287. 10.1146/annurev-genet-110410-132529 [DOI] [PubMed] [Google Scholar]
- Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., Hess S., and Chan D.C.. 2011. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20:1726–1737. 10.1093/hmg/ddr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.R., and Blackstone C.. 2007. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 282:21583–21587. 10.1074/jbc.C700083200 [DOI] [PubMed] [Google Scholar]
- Chen Y., and Sheng Z.H.. 2013. Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol. 202:351–364. 10.1083/jcb.201302040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Han Z., Feng D., Chen Y., Chen L., Wu H., Huang L., Zhou C., Cai X., Fu C., et al. . 2014. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 54:362–377. 10.1016/j.molcel.2014.02.034 [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chomyn A., and Chan D.C.. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280:26185–26192. 10.1074/jbc.M503062200 [DOI] [PubMed] [Google Scholar]
- Chen H., McCaffery J.M., and Chan D.C.. 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 130:548–562. 10.1016/j.cell.2007.06.026 [DOI] [PubMed] [Google Scholar]
- Chen H., Vermulst M., Wang Y.E., Chomyn A., Prolla T.A., McCaffery J.M., and Chan D.C.. 2010. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 141:280–289. 10.1016/j.cell.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L.C., et al. . 2013. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 155:160–171. 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs J.T., and Strack S.. 2007. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8:939–944. 10.1038/sj.embor.7401062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. . 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207–210. 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Pelloquin L., Belenguer P., and Hamel C.P.. 2002. OPA1 (Kjer type) dominant optic atrophy: A novel mitochondrial disease. Mol. Genet. Metab. 75:97–107. 10.1006/mgme.2001.3278 [DOI] [PubMed] [Google Scholar]
- Dickey A.S., and Strack S.. 2011. PKA/AKAP1 and PP2A/Bβ2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J. Neurosci. 31:15716–15726. 10.1523/JNEUROSCI.3159-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducommun S., Deak M., Sumpton D., Ford R.J., Núñez Galindo A., Kussmann M., Viollet B., Steinberg G.R., Foretz M., Dayon L., et al. . 2015. Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: Identification of mitochondrial fission factor as a new AMPK substrate. Cell. Signal. 27:978–988. 10.1016/j.cellsig.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N.G., et al. . 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281:37972–37979. 10.1074/jbc.M606059200 [DOI] [PubMed] [Google Scholar]
- Egan D.F., Shackelford D.B., Mihaylova M.M., Gelino S., Kohnz R.A., Mair W., Vasquez D.S., Joshi A., Gwinn D.M., Taylor R., et al. . 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 331:456–461. 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner A., Jakobs S., and Hell S.W.. 2002. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. USA. 99:3370–3375. 10.1073/pnas.052545099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.C., Westermann B., Rugarli E.I., and Langer T.. 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187:1023–1036. 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner V., Lenaers G., and Hajnóczky G.. 2014. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. J. Cell Biol. 205:179–195. 10.1083/jcb.201312066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson J.L., Samuels D.C., Johnson M.A., Turnbull D.M., and Chinnery P.F.. 2002. The length of cytochrome c oxidase-negative segments in muscle fibres in patients with mtDNA myopathy. Neuromuscul. Disord. 12:858–864. 10.1016/S0960-8966(02)00047-0 [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S., and van der Bliek A.M.. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412. 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.C., Di Benedetto G., and Scorrano L.. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Macleod G.T., Wellington A., Hu F., Panchumarthi S., Schoenfield M., Marin L., Charlton M.P., Atwood H.L., and Zinsmaier K.E.. 2005. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 47:379–393. 10.1016/j.neuron.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Head B., Griparic L., Amiri M., Gandre-Babbe S., and van der Bliek A.M.. 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187:959–966. 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., and Shaw J.M.. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373. 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Edlich F., Cleland M.M., Banerjee S., McCaffery J.M., Youle R.J., and Nunnari J.. 2011. The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol. Cell. 41:150–160. 10.1016/j.molcel.2010.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., and Mihara K.. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966–2977. 10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs S., Martini N., Schauss A.C., Egner A., Westermann B., and Hell S.W.. 2003. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell Sci. 116:2005–2014. 10.1242/jcs.00423 [DOI] [PubMed] [Google Scholar]
- Kang J.S., Tian J.H., Pan P.Y., Zald P., Li C., Deng C., and Sheng Z.H.. 2008. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 132:137–148. 10.1016/j.cell.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus J.A., Nascimento A., Myers L.J., Sher A., Byrne F.L., Hoehn K.L., Counter C.M., and Kashatus D.F.. 2015. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell. 57:537–551. 10.1016/j.molcel.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30:357–391. 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Li Y., Lim S., Hoffman D., Aspenstrom P., Federoff H.J., and Rempe D.A.. 2009. HUMMR, a hypoxia- and HIF-1alpha-inducible protein, alters mitochondrial distribution and transport. J. Cell Biol. 185:1065–1081. 10.1083/jcb.200811033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightowlers R.N., Taylor R.W., and Turnbull D.M.. 2015. Mutations causing mitochondrial disease: What is new and what challenges remain? Science. 349:1494–1499. 10.1126/science.aac7516 [DOI] [PubMed] [Google Scholar]
- Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., et al. . 2012. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14:177–185. 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- Losón O.C., Song Z., Chen H., and Chan D.C.. 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 24:659–667. 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O.C., Liu R., Rome M.E., Meng S., Kaiser J.T., Shan S.O., and Chan D.C.. 2014. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure. 22:367–377. 10.1016/j.str.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O.C., Meng S., Ngo H., Liu R., Kaiser J.T., and Chan D.C.. 2015. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci. 24:386–394. 10.1002/pro.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaskill A.F., Rinholm J.E., Twelvetrees A.E., Arancibia-Carcamo I.L., Muir J., Fransson A., Aspenstrom P., Attwell D., and Kittler J.T.. 2009. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 61:541–555. 10.1016/j.neuron.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. . 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221. 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland G.L., Soubannier V., Chen C.X., McBride H.M., and Fon E.A.. 2014. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban G.A., Saurya S., and Freeman M.. 2003. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 423:537–541. 10.1038/nature01633 [DOI] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J.M., and Nunnari J.. 2004. Mitochondrial fusion intermediates revealed in vitro. Science. 305:1747–1752. 10.1126/science.1100612 [DOI] [PubMed] [Google Scholar]
- Melser S., Chatelain E.H., Lavie J., Mahfouf W., Jose C., Obre E., Goorden S., Priault M., Elgersma Y., Rezvani H.R., et al. . 2013. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 17:719–730. 10.1016/j.cmet.2013.03.014 [DOI] [PubMed] [Google Scholar]
- Mironov S.L. 2007. ADP regulates movements of mitochondria in neurons. Biophys. J. 92:2944–2952. 10.1529/biophysj.106.092981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15:634–646. 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Carelli V., Manfredi G., and Chan D.C.. 2014. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19:630–641. 10.1016/j.cmet.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Varuzhanyan G., Pham A.H., and Chan D.C.. 2015. Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates organellar compartmentalization. Cell Metab. 22:1033–1044. 10.1016/j.cmet.2015.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K., Wunder C., Roysam B., Lin G., and Lippincott-Schwartz J.. 2009. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA. 106:11960–11965. 10.1073/pnas.0904875106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes C.T., Ricci E., Petruzzella V., Shanske S., DiMauro S., Schon E.A., and Bonilla E.. 1992. Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nat. Genet. 1:359–367. 10.1038/ng0892-359 [DOI] [PubMed] [Google Scholar]
- Mourier A., Motori E., Brandt T., Lagouge M., Atanassov I., Galinier A., Rappl G., Brodesser S., Hultenby K., Dieterich C., and Larsson N.G.. 2015. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 208:429–442. 10.1083/jcb.201411100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., and Youle R.J.. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Koyano F., Kimura M., Kosako H., Saeki Y., Tanaka K., and Matsuda N.. 2015. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J. Cell Biol. 209:111–128. 10.1083/jcb.201410050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., and Mihara K.. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz G., Trinidad J.C., Wang X., Kong D., and Schwarz T.L.. 2014. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 158:54–68. 10.1016/j.cell.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger P.T., Kabra D.G., Aichler M., Schriever S.C., Pfuhlmann K., García V.C., Lehti M., Weber J., Kutschke M., Rozman J., et al. . 2015. Calcineurin links mitochondrial elongation with energy metabolism. Cell Metab. 22:838–850. 10.1016/j.cmet.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Pham A.H., McCaffery J.M., and Chan D.C.. 2012. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 50:833–843. 10.1002/dvg.22050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling A.D., Horiuchi D., Lively C.M., and Saxton W.M.. 2006. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 17:2057–2068. 10.1091/mbc.E05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyakurel A., Savoia C., Hess D., and Scorrano L.. 2015. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol. Cell. 58:244–254. 10.1016/j.molcel.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter V., Palmer C.S., Osellame L.D., Singh A.P., Elgass K., Stroud D.A., Sesaki H., Kvansakul M., and Ryan M.T.. 2014. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J. Cell Biol. 204:477–486. 10.1083/jcb.201311014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., and Capaldi R.A.. 2004. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64:985–993. 10.1158/0008-5472.CAN-03-1101 [DOI] [PubMed] [Google Scholar]
- Santo-Domingo J., Giacomello M., Poburko D., Scorrano L., and Demaurex N.. 2013. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J. 32:1927–1940. 10.1038/emboj.2013.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., and Harper J.W.. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 496:372–376. 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.M., and Hollenbeck P.J.. 2012. The axonal transport of mitochondria. J. Cell Sci. 125:2095–2104. 10.1242/jcs.053850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasinghe M.N., Wieder S.Y., Renault T.T., Elkholi R., Asciolla J.J., Yao J.L., Jabado O., Hoehn K., Kageyama Y., Sesaki H., and Chipuk J.E.. 2015. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell. 57:521–536. 10.1016/j.molcel.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin H.E., Alshammari M., Al-Sheddi T., Salih M.A., Alkhalidi H., Kentab A., Repetto G.M., Hashem M., and Alkuraya F.S.. 2012. Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J. Med. Genet. 49:234–241. 10.1136/jmedgenet-2012-100836 [DOI] [PubMed] [Google Scholar]
- Shutt T., Geoffrion M., Milne R., and McBride H.M.. 2012. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 13:909–915. 10.1038/embor.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev V.P. 2001. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 26:23–29. 10.1016/S0968-0004(00)01735-7 [DOI] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., and Chan D.C.. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178:749–755. 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers R.S., Megeath L.J., Górska-Andrzejak J., Meinertzhagen I.A., and Schwarz T.L.. 2002. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 36:1063–1077. 10.1016/S0896-6273(02)01094-2 [DOI] [PubMed] [Google Scholar]
- Sugiura A., McLelland G.L., Fon E.A., and McBride H.M.. 2014. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Yun J., Liu J., Malide D., Liu C., Rovira I.I., Holmström K.M., Fergusson M.M., Yoo Y.H., Combs C.A., and Finkel T.. 2015. Measuring in vivo mitophagy. Mol. Cell. 60:685–696. 10.1016/j.molcel.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Matsuki N., and Koyama R.. 2014. AMP-activated protein kinase mediates activity-dependent axon branching by recruiting mitochondria to axon. Dev. Neurobiol. 74:557–573. 10.1002/dneu.22149 [DOI] [PubMed] [Google Scholar]
- Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., et al. . 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28:1589–1600. 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoma E.Q., Herzig S., Courchet J., Lewis T.L., Loson O.C., Helberg K., Young N.P., Chen H., Polleux F., Chan D.C., and Shaw R.J.. 2016. AMP-activated protein kinase mediates mitochondrial fission in response to energetic stress. Science. 351:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. . 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstone J.R., Smith A.M., McBride S., Naas T., Holcik M., Antoun G., Harper M.E., Michaud J., Sell E., Chakraborty P., et al. . 2015. DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur. J. Hum. Genet. 10.1038/ejhg.2015.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Schwarz T.L.. 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 136:163–174. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J., and Leonard J.V.. 2007. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356:1736–1741. 10.1056/NEJMoa064436 [DOI] [PubMed] [Google Scholar]
- Wikstrom J.D., Mahdaviani K., Liesa M., Sereda S.B., Si Y., Las G., Twig G., Petrovic N., Zingaretti C., Graham A., et al. . 2014. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO J. 33:418–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley D.M. 1970. The midpiece of the mouse spermatozoon: Its form and development as seen by surface replication. J. Cell Sci. 6:865–879. [DOI] [PubMed] [Google Scholar]
- Züchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J., et al. . 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36:449–451. 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]