Abstract
Mesothelial cells (MCs) form a single layer of the mesothelium and cover the liver surface. A previous study demonstrated that, upon liver injury, MCs migrate inward from the liver surface and give rise to hepatic stellate cells (HSCs) in biliary fibrosis induced by bile duct ligation (BDL) or myofibroblasts in CCl4-induced fibrosis. The present study analyzed the role of transforming growth factor-β (TGF-β) signaling in mesothelial-mesenchymal transition (MMT) and the fate of MCs during liver fibrosis and its regression. Deletion of TGF-β type II receptor (Tgfbr2) gene in cultured MCs suppressed TGF-β-mediated myofibroblastic conversion. Conditional deletion of Tgfbr2 gene in MCs reduced the differentiation of MCs to HSCs and myofibroblasts in the BDL and CCl4 models, respectively, indicating that the direct TGF-β signaling in MCs is responsible to MMT. After BDL and CCl4 treatment, MC-derived HSCs and myofibroblasts were distributed near the liver surface and the thickness of collagen was increased in Glisson's capsule beneath the liver surface. Fluorescence-activated cell sorting analysis revealed that MC-derived HSCs and myofibroblasts store little vitamin A lipids and have fibrogenic phenotype in the fibrotic livers. MCs contributed to 1.4 and 2.0% of activated HSCs in the BDL and CCl4 models, respectively. During regression of CCl4-induced fibrosis, 20% of MC-derived myofibroblasts survived in the liver and deactivated to vitamin A-poor HSCs. Our data indicate that MCs participate in capsular fibrosis by supplying vitamin A-poor HSCs during a process of liver fibrosis and regression.
Keywords: capsular fibrosis, fibrosis regression, Glisson's capsule, myofibroblasts, vitamin A
chronic liver injury caused by alcohol abuse, drug intoxication, hepatitis virus infection, or obesity results in fibrosis and cirrhosis (10). Liver fibrosis is a scarring process associated with inflammation and deposition of extracellular matrixes such as collagen. Excessive deposition of collagen may compromise the regeneration potential of the liver and ultimately lead to advanced liver fibrosis and cirrhosis. Currently, there is no cure for liver cirrhosis except liver transplantation (27). Thus elucidation of mechanisms of liver fibrogenesis is essential to develop antifibrosis therapy.
Hepatic stellate cells (HSCs) reside in the space of Disse and store vitamin A (VitA) lipids in their cytoplasm (34). HSCs extend dendritic processes along the sinusoidal wall and express desmin (DES) and type I collagen (9, 34). Upon liver injury, various stimuli, such as reactive oxygen species generated from damaged hepatocytes and transforming growth factor-β (TGF-β) from Kupffer cells, induce the activation of HSCs that express α-smooth muscle actin (ACTA2). Activated HSCs acquire myofibroblastic phenotype and participate in fibrogenesis by synthesizing collagen and tissue inhibitor of metalloproteinase 1 (TIMP1). TGF-β binds to a heterodimer of two TGF-β type II (TGFBR2) and two type I receptors, activates phosphorylation of SMAD2/3, and regulates target gene expression (13). TGF-β induces the expression of type I collagen in HSCs and its signaling pathways may be potential therapeutic targets for suppression of HSC activation and liver fibrosis (32). However, TGF-β has divergent roles in activation of HSCs, suppression of inflammation, and growth of hepatocytes in the liver, and global inhibition of TGF-β signaling may not be beneficial for suppression of liver fibrosis.
Mesothelial cells (MCs) form an epithelial sheet on the liver surface and secrete a lubricating fluid to make a slippery surface to facilitate movement between liver lobes or internal organs in the peritoneal cavity (18). MCs originate from mesoderm and exhibit an intermediate phenotype between epithelial cells and mesenchymal cells (17). In mouse livers, MCs express numerous markers including glycoprotein M6a (GPM6A), keratin 8, mesothelin (MSLN), podoplanin, vimentin, and Wilms tumor 1 homolog (Wt1) (15, 21). A conditional cell lineage tracing study using Wt1CreERT2 mice revealed that Wt1+ MCs migrate inward from the liver surface and differentiate into HSCs and perivascular mesenchymal cells during liver development (3). A similar differentiation of MCs has been reported in developing heart, lung, and intestine (23, 30, 35). These data indicate that MCs act as mesenchymal progenitor cells in organogenesis.
In primary culture, liver MCs form epithelial colonies (15). Upon treatment with TGF-β, MCs lose cell-cell contact, acquire fibroblastic morphology, downregulate the expression of MC markers such as GPM6A, and upregulate the expression of ACTA2 (15). We defined the change of MCs to myofibroblasts as mesothelial-mesenchymal transition (MMT) instead of epithelial-mesenchymal transition. Similar to liver development, MMT takes place during liver fibrosis in experimental mouse models (15). Conditional lineage tracing of liver MCs revealed that Wt1+ MCs migrate inward from the liver surface and give rise to ACTA2+ myofibroblasts in CCl4-induced liver fibrosis (15). In biliary fibrosis induced by bile duct ligation (BDL), MCs differentiate into ACTA2− DES+ HSCs. Treatment with a soluble form of TGFBR2 reduced differentiation of MCs to myofibroblasts or HSCs in these mouse models, indicating that TGF-β signaling is important for induction of MMT in liver fibrosis (15). However, it has remained unclear whether the direct TGF-β signaling in MCs is responsible to MMT in liver fibrosis. The present study examined the role of TGF-β signaling in MMT by conditional deletion of Tgfbr2 gene in MCs. We further traced the fate of MC-derived myofibroblasts during regression in liver fibrosis.
MATERIALS AND METHODS
Mouse models.
Wt1CreERT2 knockin and Collagen1a1 (Col1a1)-green fluorescent protein (GFP) transgenic mice were obtained from Dr. William Pu and Dr. David Brenner, respectively (33, 35). Rosa26mTmGflox (R26TGfl) and Tgfbr2fl mice were purchased from the Jackson Laboratory (Bar Harbor, ME) (14, 19). From these mice, we produced Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/+ (control Tgfbr2 fl/+ heterozygous) and Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/fl (Tgfbr2 fl/fl homozygous) mice. Tamoxifen (Sigma, St. Louis, MO) dissolved in ethanol was emulsified in sesame oil at 12.5 mg/ml and was injected intraperitoneally to the male mice at 100 μg/g body wt twice in a 3-day interval (15). Two weeks after tamoxifen injections, mice were injected subcutaneously with 1 ml/kg body wt of CCl4 (Sigma) mixed with mineral oil every 3 days 12 times (3 mice for each genotype) (15). To examine the recovery from liver fibrosis, mice were treated with CCl4 injections 12 times and then were kept without CCl4 treatment for 1 mo to examine liver fibrosis regression (4 mice for each condition). To induce biliary fibrosis, the mice were subjected to BDL and were killed 2 wk after the surgery (3 mice for each genotype). All animal experiments were performed in accordance with the NIH guidelines under the protocol approved by the IACUC at the University of Southern California.
Isolation of MCs and HSCs.
MCs were isolated from the liver surface as previously reported (15). After digestion of five mouse livers with 1 mg/ml pronase in DMEM/F-12 medium for 20 min, the cells were centrifuged at 1,700 g for 5 min and then suspended in DMEM containing 10% FBS. After washing, the cells were incubated with anti-GPM6A antibodies (MBL, Woburn, MA) at 1,500-fold dilution in DMEM for 15 min at 4°C. After washing, the cells were incubated with anti-rat IgG MicroBeads and MCs were purified by magnetic-activated cell sorting (MACS) using autoMACS (Miltenyi Biotech, Auburn, CA). MCs (2 × 104 cells) were plated on a collagen-coated 24-well plate in DMEM with low glucose containing 5% FBS. MCs were treated with an adenovirus vector carrying LacZ or Cre (Kerafast, Boston, MA, multiplicity of infection 50) from day 4 and treated with 10 ng/ml TGF-β1 (Sigma) from day 6 for 3 h to detect P-SMAD3 or 12 h to measure mRNA expression. Primary MCs were also treated with the adenovirus-Cre from day 2 and treated with TGF-β1 from day 4 to 8 to examine the myofibroblastic conversion.
HSCs were isolated by the nonparenchymal cell (NPC) core supported by NIH grant (R24AA012885) (15, 31). Mouse liver was perfused through the superior vena cava with EMEM for 10 min, followed by 0.5% pronase (Roche, Indianapolis, IN) for 20 min and 0.044% collagenase (Sigma) for 10 min. After agitation of the digested tissue with 10 μg/ml DNase for 15 min, the cell suspension was subjected to 50 g centrifugation for 30 s. The supernatant was centrifuged at 150 g for 5 min and the pellet was used as NPCs. To separate the HSCs, the NPCs were placed on the top of four OptiPrep gradients (1.034, 1.043, 1.058, 1.085) in Beckman ultracentrifuge tubes. The tubes were centrifuged in the SW-41Ti rotor at 20,000 rpm for 15 min. The HSCs were collected from the interfaces of medium/1.034/1.043 densities and cultured 1 × 106 cells in a 100-mm dish. Autofluorescence of VitA lipids in HSCs was captured under a fluorescent microscope (Axio Observer; Zeiss).
Immunocytochemistry.
MCs cultured on a glass cover were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After bleaching of Tomato fluorescence with 3% H2O2 in methanol, the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min and then blocked with 5% serum for 30 min. After incubation with primary antibodies against GFP (Nacalai Tesque, 500-fold dilution), ACTA2-Cy3 (Sigma, 400-fold dilution), and P-SMAD3 (Abcam, ab52903, 100-fold dilution) for 1 h, the primary antibodies against GFP and P-SMAD3 were detected with secondary antibodies conjugated to AlexaFluor 488 and 568, respectively. The signals were captured with 90i microscope and DS-Qi1 digital camera (Nikon). The longitudinal diameters of GFP+ or GFP− MCs (120 MCs from Tgfbr2fl/+ or Tgfbr2fl/fl genotype) were measured by NIS-Element software (Nikon).
NPCs prepared in the 1.058 fraction were cultured on a glass cover for 24 h and were fixed with 4% paraformaldehyde. To detect lipids, cells were incubated with HCS LipidTOX Red neutral lipid stain (Life Technologies, 1,000-fold dilution) for 30 min. After washing with PBS, cells were further immunostained with GFP.
QPCR.
Total RNA was extracted with RNAqueous Micro (Life Technologies) and cDNA was synthesized by using a Maxima cDNA synthesis kit (Thermo Scientific, Waltham, MA) (2). Quantitative PCR (QPCR) was performed with SYBR FAST ROX (Kapa Biosystems, Wilmington, MA) in ViiA7 Real-Time PCR System (Life Technologies). The samples were run in triplicate. Each value was normalized against that of Gapdh. Primer sequences used are as follows: Ctgf, 5′-atc cca cca aag tga gaa cg and 5′-aca gct gga ctc agc ctc at; Cyr61, 5′- cac acc tct gca gac cag aa and 5′-atg ccc gct cca gta cta tg; Gfp, 5′-cct acg gcg tgc agt gct tca gc and 5′-cgg cga gct gca cgc tgc cgt cct c; Lrat, 5′-ctg gga gtc att tgc aag gt and 5′-cag att gca gga agg gtc at; Msln, 5′-ctt ggt cgc ctg cta tct tc and 5′-acg gac agg gct ttt atc ct; Neto1, 5′-tct tcc aaa tgc atc cat ga and 5′-aga gac cca atg agg cag aa; Reln, 5′-gaa acc gag aag caa agc tg and 5′-cag gtg atg cca ttg ttg ac; and Sectm1a, 5′-tta gag cct tcc aac gag ga and 5′-cac aga cct gga acc acc tt. The primer sequences for Acta2, Cd200, Col1a1, Gapdh, Gfap, Gpm6a, Tgfb1, Tgfbr2, and Timp1 were the same as described previously (2, 15).
FACS.
After isolation of MCs by MACS, the GPM6A+ MCs were further incubated with anti-rat IgG AlexaFluor 647 (Life Technologies). Then the cells were analyzed using fluorescence-activated cell sorting (FACS) Aria sorter (BD Bioscience, San Jose, CA) in USC-Flow Cytometry Core. After excluding cell debris, GFP+ AlexaFluor 647+ and GFP− AlexaFluor 647+ MCs were sorted and were subjected to QPCR. Cultured MCs treated with TGF-β1 were subjected to FACS for sorting GFP+ and GFP− MCs. Isolated HSCs were analyzed with a krypton laser and a 424-nm filter to detect VitA autofluorescence (16, 20, 31). Liver cells separated in the 1.034 or 1.058 density fractions were analyzed by FACS for the autofluorescence of VitA and GFP. FACS data were analyzed by FlowJo software.
Immunofluorescence staining and histology.
Liver tissues were fixed with 4% paraformaldehyde for 4 h, incubated with 30% sucrose in PBS overnight, and embedded in freezing medium. Cryosections were made at 7 μm in a cryostat (CM1900; Leica, Buffalo Grove, IL). For the immunostaining of R26TGfl mouse livers, Tomato fluorescence was bleached with 3% H2O2 in methanol for 10 min (15, 16). After blocking with 5% goat serum, the sections were incubated with primary antibodies against ACTA2-Cy3 (Sigma, C6198, 400-fold dilution), DES (Thermo Scientific, RB-9014, 300-fold dilution), GFP (Life Technologies, A11122, 800-fold dilution; Nacalai Tesque, 04404-84, 1,000-fold dilution), GPM6A (MBL, D055-3, 500-fold dilution), P-SMAD2/3 (Santa Cruz, sc-11769R, 50-fold dilution), and Ki-67 (Zymed, 18-0191Z, 100-fold dilution) as previously described (15). The primary antibodies were detected with secondary antibodies conjugated with AlexaFluor dyes (Life Technologies). The sections were counterstained with DAPI. Fluorescence signals were captured with 90i microscope (Nikon, Melville, NY). Deposition of collagen was detected by Sirius red staining (17).
Quantification of immunostained images.
To quantify labeling efficiency of MCs in Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/+ or Tgfbr2fl/fl, the liver tissues were immunostained with GFP and GPM6A and images were captured with a ×20 objective (411 and 261 pictures from heterozygous and homozygous mice, n = 3 for each genotype). The length of the GFP+ mesothelium was divided by that of the GPM6A+ mesothelium.
To quantify MC-derived myofibroblasts or HSCs, we prepared cryosections from three mice in each group. After immunofluorescence with GFP and ACTA2 or DES, images including the liver surface were captured with a ×20 objective and GFP+ cells were counted as previously described (15). In the CCl4 model, 1,432 Tgfbr2 heterozygous and 481 homozygous GFP+ cells were counted. We calculated the ratio of ACTA2+ GFP+ myofibroblasts inside the liver per all GFP+ cells including myofibroblasts and MCs. In the BDL model, 530 Tgfbr2 heterozygous and 283 homozygous GFP+ cells were similarly counted.
Statistical analysis.
Statistical significance was assessed by using a Student's t-test between two samples or ANOVA followed by post hoc Tukey's honest significant difference test among multiple samples. A P value of less than 0.05 was considered statistically significant.
RESULTS
Induction of MMT by TGF-β1.
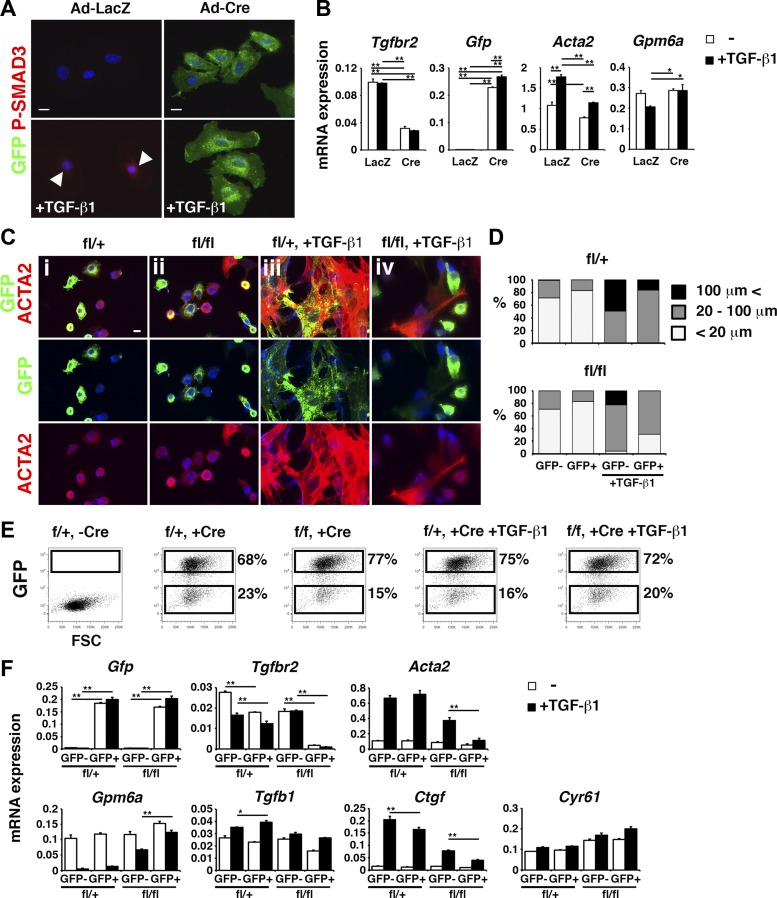
We previously reported that TGF-β induces the conversion of MCs to myofibroblasts (15). To further analyze the role of TGF-β in liver MCs, we isolated MCs from R26TGfl/fl;Tgfbr2fl/fl mouse livers by MACS using the anti-GPM6A antibody. After 4 days in culture, MCs were infected with an adenovirus vector carrying LacZ or Cre and treated with TGF-β1 from day 6 for 3 or 12 h. We reasoned that adenovirus-Cre deletes Tgfbr2 gene in MCs and switches the expression of Tomato to GFP. As expected, MCs began to express GFP upon infection with adenovirus-Cre (Fig. 1A). Three hours after TGF-β1 treatment, the control MCs infected with adenovirus-LacZ showed the nuclear staining of P-SMAD3 (Fig. 1A). In contrast, GFP+ MCs infected with adenovirus-Cre did not show the nuclear P-SMAD3 staining (Fig. 1A). QPCR confirmed the reduction of Tgfbr2 mRNA, during induction of Gfp mRNA by Cre (Fig. 1B). TGF-β1-induced expression of Acta2 mRNA was suppressed by the Cre expression (Fig. 1B). TGF-β1 weakly suppressed Gpm6a mRNA and its effect was blunted by the deletion of Tgfbr2 gene (Fig. 1B).
Fig. 1.
Deletion of Tgfbr2 gene in cultured mesothelial cells (MCs). GPM6A+ MCs were isolated from R26TGfl/fl;Tgfbr2fl/+ (fl/+) or R26TGfl/fl;Tgfbr2fl/fl (fl/fl) mouse livers. A and B: GPM6A+ MCs were isolated from R26TGfl/fl;Tgfbr2fl/fl mouse livers and were infected with an adenovirus vector (Ad) carrying LacZ or Cre (Ad-LacZ and Ad-Cre, respectively) and were treated with TGF-β1 for 3 (A) or 12 h (B). A: immunocytochemistry of MCs with green fluorescent protein (GFP; green) and P-SMAD3 (red). Arrowheads indicate P-SMAD3 in MCs treated with Ad-LacZ and TGF-β1. Nuclei were counterstained with DAPI. Bars: 10 μm. B: quantitative PCR (QPCR) of MCs infected with Ad-LacZ or Ad-Cre followed by treatment with TGF-β1. C–F: MCs were infected with the Ad-Cre on day 2 and were treated with TGF-β1 on day 4. C: immunocytochemistry of MCs with GFP and α-smooth muscle actin (ACTA2). D: quantification of the size of ACTA2+ MCs in C (n = 3). E: fluorescence-activated cell sorting (FACS) of GFP+ and GFP− MCs 2 days after TGF-β1 treatment. F: QPCR of GFP+ and GFP− MCs separated by FACS. *P < 0.05, **P < 0.01.
Next, MCs isolated from R26TGfl/fl;Tgfbr2fl/+ or R26TGfl/fl;Tgfbr2fl/fl mouse livers were infected with the adenovirus-Cre from day 2. After 8 days, ∼70% of MCs expressed GFP (Fig. 1, C, i and ii). When MCs were treated with TGF-β1 from day 4, Tgfbr2fl/+ MCs changed their morphology to large myofibroblastic cells and expressed ACTA2 on day 8 (Fig. 1, Ciii). Such myofibroblastic conversion of MCs was not prominent in Tgfbr2-deleted GFP+ MCs treated with TGF-β1 (Fig. 1, Civ). Quantification of ACTA2+ MCs with the expression of GFP showed suppression of myofibroblastic conversion of MCs by deletion of Tgfbr2 in GFP+ cells (Fig. 1, Civ and D). Moreover, Tgfbr2-wild-type GFP− MCs also showed less ACTA2 expression (Fig. 1, Civ and D). To examine the phenotypic changes of MCs by deletion of Tgfbr2 gene, we separated GFP+ and GFP− MCs on day 8 in culture by FACS (Fig. 1E). QPCR validated the increased expression of Gfp mRNA in GFP+ MCs in both genotypes (Fig. 1F). We confirmed the deletion of Tgfbr2 expression only in GFP+ MCs cells from the Tgfbr2fl/fl liver (Fig. 1F). TGF-β1 treatment increased Acta2 mRNA in Tgfbr2fl/+ MCs (Fig. 1F). Tgfbr2fl/fl GFP+ MCs did not increase expression of Acta2 mRNA by TGF-β1 (Fig. 1F). Similar to Acta2, Gpm6a mRNA was regulated inversely by TGF-β1 that suppressed its expression in MCs (Fig. 1F). Tgfbr2-null GFP+ MCs continued to express Gpm6a mRNA in the presence of TGF-β1 and Tgfbr2fl/fl GFP− MCs also moderately expressed its gene (Fig. 1F). Connective tissue growth factor (CTGF/CCN2) and cysteine-rich angiogenic inducer 61 (CYR61) are members of the CCN family of cysteine-rich matricellular proteins and mediators of fibrosis (1, 4, 6). TGF-β1 strongly induced Ctgf but not Cyr61 mRNAs in MCs, and the deletion of Tgfbr2 gene abolished TGF-β-induced expression of Ctgf mRNA (Fig. 1F). These data indicate that TGF-β induces the conversion of MCs to myofibroblasts in culture.
Role of TGF-β signaling in the conversion of MCs to myofibroblasts.
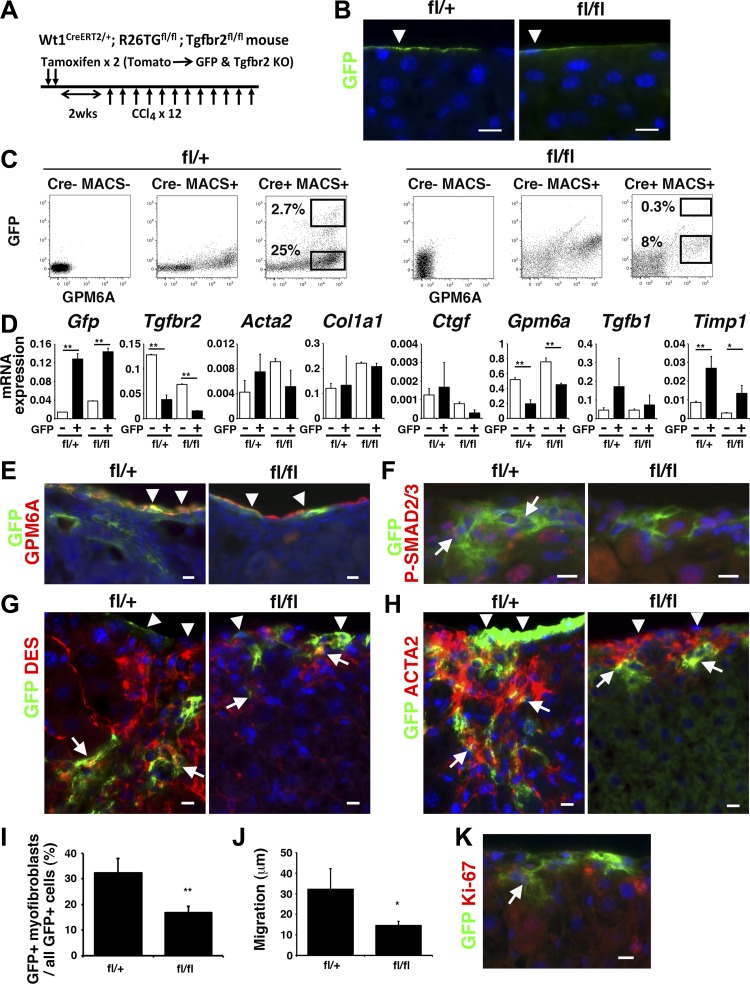
To test whether direct TGF-β signaling in MCs is required for MMT in liver fibrosis, we conditionally deleted Tgfbr2 gene in MCs using Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/+ (fl/+) or Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/fl (fl/fl) mice. We expected that tamoxifen-induced CreERT2 deletes Tgfbr2 gene and activates the expression of GFP only in Wt1+ MCs (Fig. 2A). After tamoxifen injection twice, GFP expression was observed only in MCs on the liver surface for both Tgfbr2 fl/+ and fl/fl mice (Fig. 2B). Immunofluorescence staining showed that 14.9 ± 1.9 and 10.7 ± 1.5% of MCs express GFP in the Tgfbr2fl/+ and Tgfbr2fl/fl livers, respectively (Fig. 2B). To confirm conditional deletion of Tgfbr2 gene in GFP+ MCs, we treated the mice with tamoxifen and isolated GPM6A+ MCs by MACS from the livers. Then the GPM6A+ MCs were subjected to FACS for sorting GFP+ GPM6A+ and GFP− GPM6A+ MCs (Fig. 2C). By QPCR, we confirmed the high expression of Gfp mRNA in GFP+ fractions in both Tgfbr2fl/+ and Tgfbr2fl/fl livers (Fig. 2D). The expression of Tgfbr2 mRNA was reduced to 77% in the GFP+ MCs compared with GFP− MCs in the Tgfbr2fl/fl liver (Fig. 2D), validating deletion of Tgfbr2 gene in the GFP+ MCs. Although the deletion of Tgfb2 gene changed the expression of Gpm6a and Timp1 mRNAs, the expression of fibrosis markers including Acta2, Col1a1, Ctgf, and Tgfb1 mRNAs was not changed significantly (Fig. 2D).
Fig. 2.
Conditional deletion of Tgfbr2 gene in MCs in CCl4-induced liver fibrosis. A: the tamoxifen-induced CreERT2 in MCs activates expression of GFP, while deleting Tgfbr2 gene in Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/fl mice. After deletion of Tgfbr2 gene, liver fibrosis was induced by CCl4 injections 12 times. B: immunofluorescence staining of normal Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/+ (fl/+) or Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/fl (fl/fl) mouse livers. Tamoxifen treatment caused specific expression of GFP in MCs. Bars: 10 μm. C: FACS of MCs isolated from normal Tgfbr2fl/+ (fl/+) or Tgfbr2fl/fl (fl/fl) mouse livers. After isolation of MCs using the anti-GPM6A antibody by magnetic-activated cell sorting, the cells were separated into GFP+ GPM6A+ and GFP− GPM6A+ MCs by FACS. MCs isolated from the Wt1+/+;R26TGfl/fl;Tgfbr2fl/+ and Tgfbr2fl/fl mouse were used as negative controls. D: QPCR of the GFP+ GPM6A+ and GFP− GPM6A+ MCs separated in C. GFP+ GPM6A+ MCs decrease Tgfbr2 mRNA expression, while increasing Gfp mRNA in both genotypes. E–H: immunofluorescence staining of the liver from the fl/+ and fl/fl mice treated with CCl4 by using antibodies against GFP (green) and GPM6A (E), P-SMAD2/3 (F), desmin (DES; G), or ACTA2 (H, red). Arrowheads indicate MCs. Arrows indicate GFP+ cells migrating inside the liver. Nuclei were counterstained with DAPI. I: quantification of GFP+ myofibroblasts in all GFP+ cells in the CCl4-treated fl/+ and fl/fl mouse livers. The Tgfbr2 knockout decreases the percentage of GFP+ myofibroblasts. **P < 0.01 (n = 3 for each group). J: migration of ACTA2+ GFP+ myofibroblasts in all GFP+ cells in the CCl4-treated fl/+ and fl/fl mouse livers. *P < 0.05. K: immunofluorescence staining of CCl4-treated fl/+ livers using antibodies against GFP (green) and Ki-67 (red). An arrow indicates Ki-67+ GFP+ MC-derived myofibroblasts.
After labeling MCs as GFP+ cells by tamoxifen treatment, liver fibrosis was induced by CCl4 injections 12 times to the mice (Fig. 2A). In the CCl4 model, GPM6A+ MCs remained on the liver surface in the both genotypes (Fig. 2E). Tgfbr2fl/+ mice showed expression of GFP not only in GPM6A+ MCs, but also beneath the mesothelium (Fig. 2E), and these GFP+ cells showed the nuclear localization of P-SMAD2/3 (Fig. 2F). In contrast, Tgfbr2fl/fl livers did not show the nuclear staining of P-SMAD2/3 in GFP+ cells (Fig. 2F). GFP+ cells inside the Tgfbr2fl/+ liver expressed DES and 32.4% were positive for ACTA2 (Fig. 2, G–I), demonstrating the differentiation of MCs to myofibroblasts. In contrast, the Tgfbr2fl/fl mouse showed a few GFP+ cells beneath the mesothelium and 16.8% of GFP+ cells coexpressed ACTA2 (Fig. 2, G–I). In the Tgfbr2fl/+ mouse, ACTA2+ GFP+ myofibroblasts migrated average 32.1 μm (n = 3, maximum 162 μm) from the liver surface (Fig. 2J). In contrast, the migration was reduced to 14.5 μm (n = 3, maximum 53.0 μm) in the Tgfbr2fl/fl mouse. Immunofluorescence staining revealed that 4.9 ± 1.3% of GFP+ HSCs are positive for Ki-67 in fibrosis (Fig. 2K). These results indicate that direct TGF-β signaling in MCs is required for their differentiation to myofibroblasts and migration of MC-derived myofibroblasts in liver fibrosis.
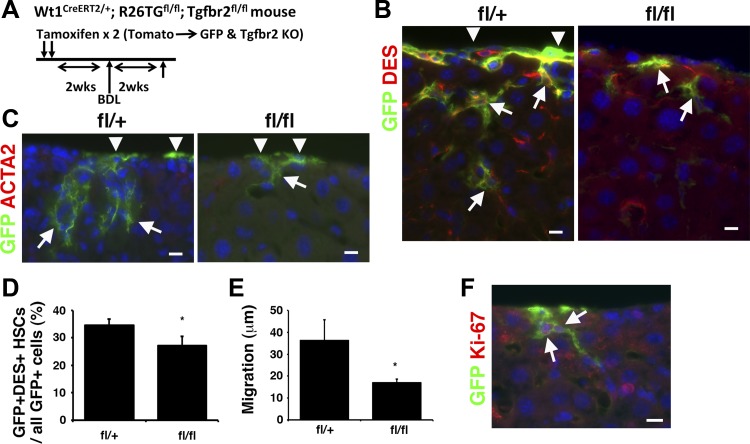
TGF-β induces the differentiation of MCs to HSCs in biliary fibrosis.
To test whether TGF-β signaling drives MMT in biliary fibrosis, we similarly analyzed effects of Tgfbr2 deletion in MCs (Fig. 3A). Two weeks after BDL, GFP+ MCs migrated inward and differentiated into DES+ HSCs in the Tgfbr2fl/+ mice (Fig. 3B). In contrast, Tgfbr2fl/fl mice showed less DES+ GFP+ cells inside the liver (Fig. 3B). Differing from the CCl4-induced liver fibrosis, MC-derived GFP+ cells did not express ACTA2 in the BDL model (Fig. 3C). In these GFP+ cells inside the Tgfbr2fl/+ liver, 34.5% were positive for DES (Fig. 3, B and D). In contrast, the Tgfbr2fl/fl mouse showed a few GFP+ cells beneath the mesothelium and 27.1% of GFP+ cells coexpressed DES (Fig. 3, B and D). Tgfbr2fl/fl mice showed a decreased migration of MC-derived HSCs inside the liver compared with that in Tgfbr2fl/+ mice (Fig. 3E). DES+ GFP+ HSCs migrated average 36.2 μm (n = 3, maximum 107.2 μm) from the liver surface in the Tgfbr2fl/+ mouse (Fig. 3, B and E). In contrast, the migration was reduced to 17.1 μm (n = 3, maximum 47.5 μm) by Tgfbr2 deletion. In biliary fibrosis, 5.8 ± 2.6% of GFP+ MC-derived HSCs were positive for Ki-67 (Fig. 3F). Collectively, these data suggest that the direct TGF-β signaling in MCs is required for differentiation and migration of MC-derived HSCs in biliary fibrosis.
Fig. 3.
Conditional deletion of Tgfbr2 gene in MCs in biliary fibrosis induced by BDL. A: the tamoxifen-induced CreERT2 in MCs activates expression of GFP, while deleting Tgfbr2 gene in Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/fl mice. After deletion of Tgfbr2 gene, biliary fibrosis was induced by bile duct ligation (BDL). B and C: immunofluorescence staining of the liver from the fl/+ and fl/fl mice 2 wk after BDL. The liver tissues were immunostained with antibodies against GFP (green) and DES or ACTA2 (red). Arrowheads and arrows indicate GFP+ MCs and HSCs, respectively. Nuclei were counterstained with DAPI. Bars: 10 μm. D: quantification of GFP+ HSCs in all GFP+ cells in the fl/+ and fl/fl mouse livers after BDL. The Tgfbr2 knockout decreases the percentage of GFP+ myofibroblasts. *P < 0.05 (n = 3 for each group). E: migration of GFP+ DES+ HSCs in all GFP+ cells in the fl/+ and fl/fl mouse livers. F: immunofluorescence staining of fl/+ mouse livers after BDL using antibodies against GFP (green) and Ki-67 (red). Arrows indicate Ki-67+ GFP+ MC-derived HSCs.
Phenotype of MC-derived HSCs in biliary fibrosis.
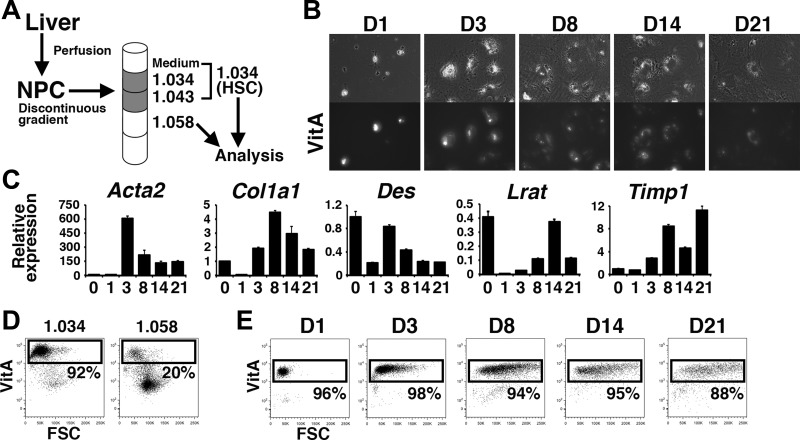
We further analyzed phenotype of MC-derived HSCs in the BDL model. One of the unique features of HSCs is storage of VitA lipids as retinyl ester in their cytoplasm (28). Using FACS (20), we detected autofluorescence of VitA in HSCs. By discontinuous gradient ultracentrifugation, VitA-laden HSCs were enriched at interfaces of medium/1.034/1.043 densities (abbreviated as 1.034 fraction thereafter) (Fig. 4A). VitA-poor HSCs and Kupffer cells were enriched in the 1.058 density (25). In culture, primary HSCs from the 1.034 fraction demonstrated autofluorescence of the VitA lipids in their cytoplasm at day 1 (Fig. 4B). Beginning at day 3, the HSCs became flat and increased their expression of activation markers, including Acta2, Col1a1, and Timp1 mRNAs (Fig. 4, B and C). FACS analysis validated the enrichment of the VitA+ HSCs (92%) in the 1.034 fraction compared with the 1.058 fraction (20%) (Fig. 4D). Although the VitA lipid size decreased in the day-21 HSCs (Fig. 4B), 88% of the HSCs still stored detectable amounts of VitA according to FACS (Fig. 4E), indicating that FACS is a sensitive method of detecting VitA lipids in HSCs.
Fig. 4.
FACS analysis of autofluorescence of vitamin A (VitA) in hepatic stellate cells (HSCs). A: nonparenchymal cells (NPCs) prepared from adult mouse livers were subjected to discontinuous density gradient ultracentrifugation and VitA-laden HSCs were enriched at interfaces of medium/1.034/1.043 densities (1.034). The 1.058 fraction containing VitA-poor HSCs and Kupffer was also used for analysis. B: morphology and VitA autofluorescence of cultured HSCs prepared from the 1.034 fraction. Top: morphology of cultured HSCs from day 1 to 21 (D1–D21, respectively) (n = 3). Bottom: autofluorescence of VitA lipids in HSCs captured by fluorescence microscopy. C: QPCR of cultured HSCs. The values were normalized against the Gapdh values. D: FACS analysis of autofluorescence of VitA lipids in the 1.034 and 1.058 fractions. E: FACS of cultured HSCs in B. Culture activated HSCs exhibit VitA autofluorescence even at day 21.
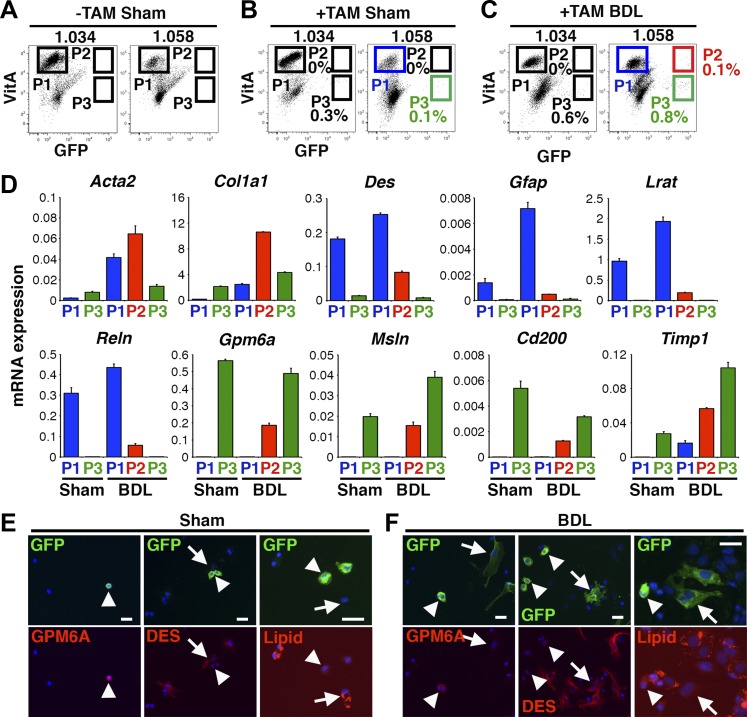
We prepared the 1.034 and 1.058 fractions from Wt1CreERT2/+;R26TGfl/fl mice and analyzed autofluorescence of VitA and GFP by FACS. Wt1CreERT2/+;R26TGfl/fl mice without tamoxifen treatment showed GFP− VitA+ HSCs in P1 in both 1.034 and 1.058 fractions (referred as 1.034-P1 and 1.058-P1 thereafter) and there were no GFP+ cells in P2 and P3 (Fig. 5A). As another negative control, Wt1CreERT2/+;R26TGfl/fl mice were treated with tamoxifen and then were subjected to sham operation. The sham control induced the presence of GFP+ VitA− cells in 1.034-P3 and 1.058-P3 (Fig. 5B). As shown in Fig. 2B, tamoxifen induced the GFP expression only in MCs in the Wt1CreERT2/+;R26TGfl/fl mouse liver. QPCR from the sham control revealed that 1.058-P1 expresses HSC markers including Des, Gfap, Lrat, and Reln (Fig. 5D, lane 1). On the other hand, 1.058-P3 expressed MC markers including Gpm6a, Msln, and Cd200 (Fig. 5D, lane 2), indicating that HSCs and MCs are enriched in P1 and P3, respectively. Moreover, FACS analyses revealed no GFP+ cells in both 1.034-P2 and 1.058-P2 (Fig. 5B), excluding a possibility of nonspecific induction of GFP in VitA+ HSCs by tamoxifen in this mouse model.
Fig. 5.
FACS analysis of autofluorescence of VitA in HSCs and MC-derived HSCs in biliary fibrosis. Wt1CreERT2/+;R26TGfl/fl mice were treated with or without tamoxifen and were subjected to sham operation or BDL. A–C: FACS analysis of the 1.034 and 1.058 fractions prepared from sham or BDL models. Cells were analyzed based on the autofluorescence of VitA and GFP. GFP− VitA+, GFP+ VitA+, and GFP+ VitA− cells are gated as P1, P2, and P3, respectively. D: QPCR of 1.058-P1, -P2, and -P3 from the sham (B) and BDL (C) groups. P1 HSCs increase activation markers by BDL. P2 MC-derived HSCs express both HSC and MC markers. P3 MCs express MC markers. E and F: cultured 1.058 fractions prepared from the sham (B) or BDL (C) were stained with a lipid-specific fluorescent dye (red) and antibodies against GFP (green), GPM6A (red), or DES (red). E: arrowheads and arrows indicate GFP+ MCs and GFP− HSCs, respectively. F: arrowheads and arrows indicate GFP+ MCs and GFP+ MC-derived HSCs, respectively. Nuclei were counterstained with DAPI. Bar: 20 μm.
In the BDL model without tamoxifen treatment, we did not observe GFP+ cells in P2 and P3 (data not shown). As shown in Fig. 3B, BDL induces differentiation of GFP+ MCs to GFP+ HSCs in the tamoxifen-treated mice. If MC-derived HSCs stored VitA lipids, these cells would be fractionated in 1.034-P2 by FACS. However, mice treated with tamoxifen followed by BDL did not induce GFP+ cells in 1.034-P2 (Fig. 5C). Instead, GFP+ VitA+ cells were observed in 1.058-P2 (Fig. 5C, 0.1 ± 0.07%, n = 3). After sorting 1.058-P1, -P2, and -P3 from the BDL model, we analyzed gene expression by QPCR. As expected, 1.058-P1 and -P3 expressed HSC and MC markers, respectively (Fig. 5D, lanes 3 and 5). The 1.058-P1 from the BDL model increased expression of Acta2, Col1a1, and Timp1 mRNAs compared with that from the sham control (Fig. 5D, lanes 1 and 3), indicating the activation of HSCs in biliary fibrosis. The 1.058-P2 from the BDL model expressed Acta2, Col1a1, Des, and MC markers (Fig. 5D, lane 4), indicating the enrichment of MC-derived HSCs in 1.058-P2. Compared with P1 HSCs, P2 MC-derived HSCs expressed Gfap, Lrat, and Reln relatively low levels, implying that the phenotype of MC-derived HSCs is not identical to that of HSCs. The presence of GFP+ VitA+ cells in 1.058-P2, but not in 1.034-P2, suggests that MC-derived HSCs do not store VitA lipids at the same level as HSCs in biliary fibrosis. Compared with P1 HSCs in the BDL model, P2 MC-derived HSCs expressed more Acta2, Col1a1, and Timp1 mRNAs (Fig. 5D, lanes 3 and 4). P3 MCs also expressed these markers, indicating that MCs and MC-derived HSCs are fibrogenic cells in biliary fibrosis.
To further analyze VitA lipids stored in HSCs, we cultured the 1.058 fraction and stained lipids with a lipid-specific fluorescent dye. In the sham control, GFP expression was observed in round GPM6A+ DES- MCs and these cells were negative for lipid staining (Fig. 5E). In the BDL model, GFP expression was observed in both GPM6A+ MCs and DES+ MC-derived HSCs, and these cells were negative for lipid staining (Fig. 5F). Collectively, these data indicate that MC-derived HSCs store little VitA lipids in biliary fibrosis and thus exhibit lower buoyancy compared with HSCs.
Fate tracing of MC-derived myofibroblasts during regression in CCl4-induced liver fibrosis.
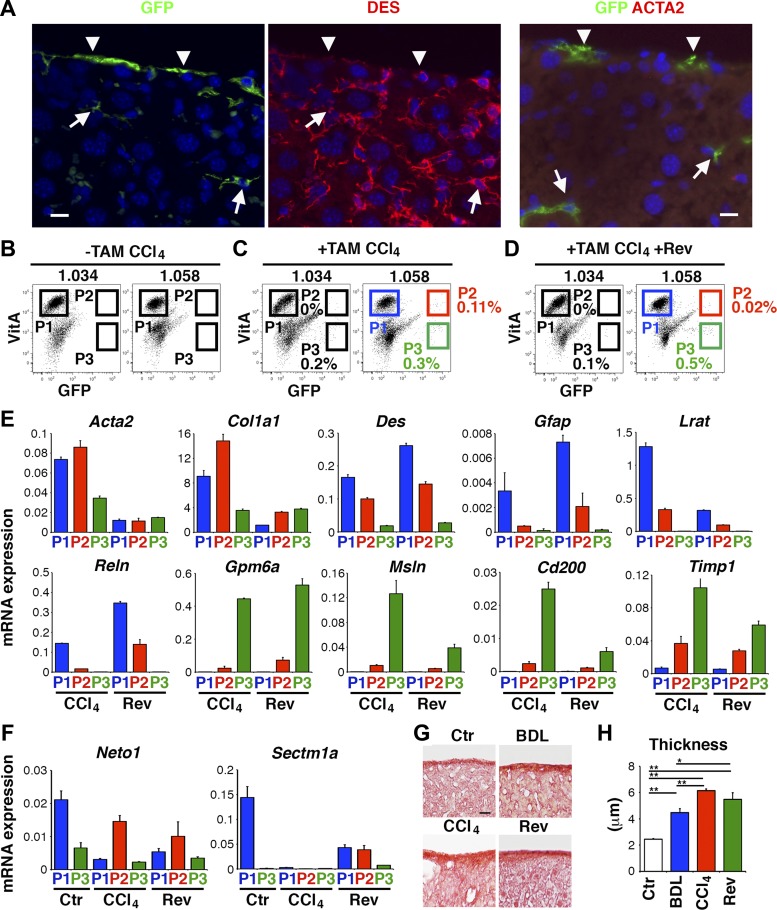
Recent studies indicate that activated HSCs revert to quiescent HSCs during regression in liver fibrosis (11, 29). To determine the fate of MC-derived myofibroblasts, we labeled MCs as GFP+ cells in Wt1CreERT2/+;R26TGfl/fl mouse livers by tamoxifen, induced liver fibrosis by CCl4 injections 12 times, and then kept mice for 1 mo without CCl4 treatment. One month after the last CCl4 injection, GFP+ MC-derived cells were observed beneath the liver surface and these cells expressed DES, but not ACTA2 (Fig. 6A), indicating that MC-derived myofibroblasts revert to DES+ HSCs during regression in CCl4-induced fibrosis.
Fig. 6.
Deactivation of MC-derived myofibroblasts during regression in CCl4-induced liver fibrosis. After labeling MCs as GFP+ cells in Wt1CreERT2/+;R26TGfl/fl mouse livers by tamoxifen, we induced liver fibrosis by CCl4 injections 12 times and then the regression of fibrosis was analyzed 1 mo after the cessation of CCl4. A: immunofluorescence staining of Wt1CreERT2/+;R26TGfl/fl mouse livers 1 mo after the last CCl4 injection. Arrowheads indicate GFP+ MCs on the liver surface. Arrows indicate ACTA2− DES+ GFP+ MC-derived HSCs remaining in the regenerated liver. Bar: 10 μm. B–D: FACS analysis of the 1.034 and 1.058 fractions prepared from CCl4 or CCl4 plus reversal (Rev) models with or without tamoxifen treatment. Cells were analyzed by the autofluorescence of VitA and GFP. GFP− VitA+, GFP+ VitA+, and GFP+ VitA− cells are gated as P1, P2, and P3, respectively. E: QPCR of 1.058-P1, -P2, and -P3 from the CCl4 (C) and CCl4 plus reversal (D) models. F: QPCR of 1.058-P1, -P2, and -P3 from the control, CCl4 and CCl4 plus reversal models. G: Sirius red staining. The deposition of collagen is detected in Glisson's capsule on the liver surface. H: quantification of the thickness of collagen in G. *P < 0.05, **P < 0.01 (n = 3 for each group).
We further characterize the storage of VitA and gene expression profile of MC-derived myofibroblasts and HSCs in CCl4-induced fibrosis and regenerated liver from fibrosis by FACS. Negative controls of CCl4 and its reversal models without tamoxifen treatment did not show GFP expression in P2 and P3 in both 1.034 and 1.058 fractions (Fig. 6B and data not shown). In the CCl4 model treated with tamoxifen, few GFP+ VitA+ MC-derived myofibroblasts were observed in 1.034-P2 (Fig. 6C). Similarly, no GFP+ VitA+ MC-derived HSCs were observed in 1.034-P2 in the reversal model (Fig. 6D). In contrast, GFP+ VitA+ cells were observed in 1.058-P2 in the CCl4 model (Fig. 6C, 0.11 ± 0.07%, n = 3), indicating that MC-derived myofibroblasts store little VitA lipids. In the reversal model, a few GFP+ VitA+ cells were detected in 1.058-P2 (Fig. 6D, 0.02 ± 0.01%, n = 3). We confirmed no lipid staining in GFP+ cells in cultured 1.058 fractions in both models (data not shown).
After sorting 1.058-P1, -P2, and -P3 from CCl4 and reversal models (Fig. 6, C and D), we analyzed gene expression by QPCR. In both models, P1 and P3 expressed HSC and MC markers, respectively (Fig. 6E). In the CCl4 model, P2 MC-derived myofibroblasts expressed both HSC and MC markers weakly and showed higher expression of Acta2 and Col1a1 mRNAs compared with P1 activated HSCs (Fig. 6E, lanes 1 and 2). P3 MCs expressed high Timp1 mRNA (Fig. 6E, lanes 3 and 6). One month after the last CCl4 injection, P2 downregulated the expression of Acta2 and Col1a1 mRNAs similar to P1 (Fig. 6E, lanes 2 and 5). During resolution of liver fibrosis, deactivated HSCs are known to downregulate several genes, including Neto1 and Sectm1a, compared with quiescent HSCs (29). Although the function of these genes remains elusive, we measured these gene's expression in MC-derived HSCs. Similar to the previous report (29), P1 deactivated HSCs in the reversal model downregulated the expression of Neto1 and Sectm1a low levels compared with P1 quiescent HSCs (Fig. 6F). The expression levels of these mRNAs were similarly low in P2 MC-derived HSCs in the reversal model (Fig. 6F). On the basis of these data, we concluded that MC-derived myofibroblasts revert to VitA-poor HSCs after reversal in CCl4-induced liver fibrosis.
Contribution of MC-derived myofibroblasts in capsular fibrosis.
In both BDL and CCl4 models, MC-derived myofibroblasts and HSCs are distributed near the liver surface (Figs. 2H and 3B). Sirius red staining revealed the deposition of collagen in Glisson's capsule of the normal adult mouse liver (Fig. 6G). After BDL, the thickness of collagen in Glisson's capsule was increased significantly from 2.5 to 4.5 μm (Fig. 6, G and H). In the CCl4 model, the thickness of collagen was increased to 6.1 μm and its thickness remained nearly similar (5.5 μm) 1 mo after the cessation of CCl4 (Fig. 6, G and H). These data suggest that MC-derived HSCs participate in capsular fibrosis beneath the liver surface.
DISCUSSION
Primary role of the mesothelium is considered to be a barrier of the liver (18). Intriguingly, recent studies indicate that MCs act as mesenchymal progenitor cells in organogenesis including the liver, heart, lung, and intestine (3, 23, 30, 35). Furthermore, in liver fibrosis, MCs were shown to migrate inward from the liver surface and give rise to ACTA2+ myofibroblasts in the CCl4 model or DES+ HSCs in the BDL model (15). The present study further analyzed the mechanism of MMT in liver fibrosis. TGF-β1 treatment induced the expression of ACTA2 and the morphological change of MCs to myofibroblasts in culture. Upon the deletion of Tgfbr2 gene, TGF-β1 failed to induce the nuclear localization of P-SMAD3 and the expression of ACTA2 in MCs. In addition, the loss of TGF-β signaling blocked the downregulation of Gpm6a, an MC marker, by TGF-β in MCs. Thus our data indicate that TGF-β signaling is responsible to the conversion of MCs to myofibroblasts. The deletion of Tgfbr2 reduced the mRNA expression of Ctgf, which is a known fibrogenic factor regulated by TGF-β (6, 26). Interestingly, Tgfbr2-wild-type GFP− MCs also decreased Acta2 and Ctgf mRNA expression by TGF-β1 in the presence of Tgfbr2-null GFP+ MCs in culture (Fig. 1F). Reduced induction of Ctgf mRNA in Tgfbr2-null GFP+ MCs may be responsible for less induction of MMT in Tgfbr2-wild-type MCs in the presence of TGF-β1.
Using Wt1CreERT2;R26TGfl;Tgfbr2fl mice, we conditionally deleted Tgfbr2 gene in MCs. After tamoxifen injection, only MCs on the liver surface started to express GFP. Immunohistochemistry and FACS analyses validated no induction of GFP in VitA+ HSCs. Two weeks after tamoxifen injection, GFP+ Tgfbr2-null MCs remained on the liver surface without noticeable morphological changes. After CCl4 injections, GFP+ MCs migrated inward and differentiated into ACTA2+ myofibroblasts. The migrated myofibroblasts expressed DES, but not GPM6A, indicating that MCs lose GPM6A expression upon differentiation to myofibroblasts. After deletion of Tgfbr2 gene in MCs, the migration and differentiation of MCs to myofibroblasts was significantly reduced in CCl4-induced liver fibrosis. Similar to the CCl4 model, the deletion of Tgfbr2 gene in MCs resulted in less differentiation and migration of MCs to HSCs in the BDL model. These data suggest that TGF-β signaling plays an important role in the migration and differentiation of MCs to myofibroblasts or HSCs.
Intriguingly, the conditional deletion of Tgfbr2 gene in GFP+ MCs did not completely block the differentiation and migration of MCs to myofibroblasts and HSCs in the CCl4 and BDL models, respectively. Even though the deletion of Tgfbr2 gene resulted in less differentiation of MCs, we still observed MC-derived myofibroblasts and HSCs in the fibrotic liver, implying that other factors may also be involved in MMT in liver fibrosis. During mouse liver development, MCs on the liver surface give rise to HSCs (3). We also examined whether the loss of Tgfbr2 in MCs suppresses the differentiation and migration of MCs to HSCs in developing liver using the same Wt1CreERT2/+;R26TGfl/fl;Tgfbr2fl/fl mouse model. However, GFP+ Tgfbr2-null MCs differentiated into HSCs same as wild-type MCs in E13.5 embryos (data not shown). Thus the mechanism underlying MMT is not identical in liver development and fibrosis. During lung development, MCs give rise to mesenchymal cells and Sonic hedgehog signaling is involved in this process (7). Further studies are necessary to elucidate whether Sonic hedgehog signaling regulates MMT in liver development and fibrosis.
Combining with the detection of autofluorescence of VitA and GFP by FACS, we separated HSCs, MCs, and MC-derived HSCs in the BDL model using Wt1CreERT2;R26TGfl mice. We prepared the 1.034 and 1.058 fractions by discontinuous gradient ultracentrifugation. By FACS, we could detect GFP+ VitA− cells in both fractions. QPCR revealed that GFP+ VitA− cells expressed MC markers, but not HSC markers. These data imply that during preparation of NPCs from the liver by enzymatic digestion, MCs are also digested and fractionated in the 1.034 and 1.058 fractions. Interestingly, MC-derived HSCs defined as GFP+ VitA+ were observed in the 1.058 fraction, but not in the 1.034 fraction, in the BDL model. Furthermore, cultured NPCs prepared from the 1.058 fraction showed no lipid staining in GFP+ cells including MCs and MC-derived HSCs. These results suggest that MC-derived HSCs in the BDL model store little VitA lipids that can be detected by FACS. However, VitA lipids in MC-derived HSCs may not be sufficient to be detected by lipid staining or enriched in the 1.034 fraction by their buoyancy in discontinuous gradient ultracentrifugation. Although MC-derived HSCs were negative for immunohistochemistry of ACTA2 in the BDL model, they increased the expression of Acta2 mRNA similar to activated HSCs. In addition, MC-derived HSCs expressed more activation markers (Col1a1, Timp1) and fewer HSC markers (Des, Gfap, Lrat) compared with HSCs. These data indicate that MC-derived HSCs do not fully acquire the HSC phenotype after MMT in biliary fibrosis and MC-derived HSCs have fibrogenic phenotype.
Liver fibrosis models in rodents reveal that liver fibrosis is a reversible process (8, 22). During regression in fibrosis, activated HSCs/myofibroblasts are eliminated by apoptosis or killing by natural killer cells (12, 22, 24). In addition, recent studies in mouse fibrosis models indicate that 40–50% of activated HSCs deactivate during resolution in liver fibrosis (11, 29). To test whether MC-derived myofibroblasts supply new quiescent HSCs during a process of fibrosis and its regression, we traced MC lineages in Wt1CreERT2;R26TGfl mice treated with CCl4 and followed by 1 mo recovery without CCl4 treatment. By CCl4 injections, GFP+ MCs differentiated into ACTA2+ DES+ GFP+ myofibroblasts. After cessation of CCl4 treatment, MC-derived DES+ GFP+ cells remained in the regenerating liver without expression of ACTA2. By FACS, we purified P2 GFP+ VitA+ MC-derived myofibroblasts and HSCs in the CCl4 and reversal models, respectively, and found that MC-derived myofibroblasts highly express Acta2, Col1a1, and Timp1 in CCl4-induced fibrosis. After regression in fibrosis, P2 MC-derived HSCs downregulated these activation markers similar to quiescent HSCs. Different from HSCs, P2 MC-derived HSCs were enriched in the 1.058 fraction in the reversal model and expressed less HSC markers including Gfap, Lrat, and Reln mRNAs. These data indicate that MC-derived myofibroblasts revert to HSCs during regression in liver fibrosis. However, these MC-derived HSCs do not fully acquire quiescent HSC phenotype.
FACS analysis revealed that P2 GFP+ VitA+ MC-derived HSCs occupy 0.1% of NPCs in the 1.058 fraction from the BDL model. Using the FACS data, we estimated the ratio of MC-derived HSCs against VitA+ HSCs is 0.28% in the 1.058 fraction. The average yield of VitA+ HSCs was 2.3 × 106 and 7 × 106 cells in the 1.034 and 1.058 fractions, respectively. If we calculated the occupancy of MC-derived HSCs in all VitA+ cells containing in both factions, the occupancy was 0.21% in all VitA+ HSCs. Given that the labeling efficiency of tamoxifen to MCs is ∼15%, contribution of MCs to all VitA+ HSCs is estimated as 1.4% in BDL-induced biliary fibrosis. In the CCl4 and reversal models, we detected 0.11 and 0.02%, respectively, of P2 GFP+ VitA+ cells in the 1.058 fractions by FACS. Similar to the BDL, we calculated that MCs contribute to 2.0% activated HSCs in CCl4-induced fibrosis and to 0.4% HSCs after regression in fibrosis. On the basis of these values, we estimated that ∼20% of MC-derived myofibroblasts survive and revert to HSCs during regression in CCl4-induced fibrosis.
Electron microscopy showed that the liver surface is covered by Glisson's capsule consisting of a single layer of mesothelium and underlying capsular fibroblasts (5). In BDL and CCl4 models, GFP+ MCs migrated inward from the liver surface and many of them remained in close proximity to the mesothelium. Sirius red staining revealed the deposition of collagen in Glisson's capsule in normal adult mice. After BDL or CCl4 treatment, the thickness of collagen was significantly increased. QPCR showed that MC-derived myofibroblasts and HSCs express high Col1a1 mRNA. We assume that these cells participate in capsular fibrosis. Interestingly, the thickness of collagen in Glisson's capsule remained the same between CCl4-induced fibrosis and 1 mo after the cessation of CCl4. High Timp1 mRNA expression in MCs and MC-derived HSCs may interfere the resolution of collagen fibers in capsular fibrosis beneath the liver surface. We also analyzed the effect of MC-specific deletion of Tgfbr2 gene on the thickness of collagen in Glisson's capsule. Although BDL and CCl4 treatment increased the thickness of collagen in the control Tgfbr2 fl/+ mice, the deletion of Tgfbr2 gene did not reduce the thickness significantly (data not shown). The labeling efficiency of GFP in Tgfbr2 fl/fl livers was ∼11% and the low deletion efficiency of Tgfbr2 gene in MCs may not be sufficient to change the development of capsular fibrosis in Glisson's capsule.
GRANTS
This work was supported by NIH grant R01AA020753 (to K. Asahina), pilot project funding (to K. Asahina) from P50AA011999, pilot project funding (to K. Asahina) from P30DK048522, training program (to I. Lua) from T32HD060549, and the Robert E. and May R. Wright foundation award (to K. Asahina).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Y.L. and K.A. conception and design of research; Y.L., I.L., S.W.F., and K.A. performed experiments; Y.L., I.L., and K.A. analyzed data; Y.L., I.L., S.W.F., and K.A. interpreted results of experiments; Y.L. and K.A. prepared figures; Y.L. and K.A. edited and revised manuscript; Y.L., I.L., S.W.F., and K.A. approved final version of manuscript; K.A. drafted manuscript.
REFERENCES
- 1.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat Cell Biol 4: 599–604, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahina K, Tsai SY, Li P, Ishii M, Maxson RE Jr, Sucov HM, Tsukamoto H. Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development. Hepatology 49: 998–1011, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahina K, Zhou B, Pu WT, Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology 53: 983–995, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borkham-Kamphorst E, Schaffrath C, Van de Leur E, Haas U, Tihaa L, Meurer SK, Nevzorova YA, Liedtke C, Weiskirchen R. The anti-fibrotic effects of CCN1/CYR61 in primary portal myofibroblasts are mediated through induction of reactive oxygen species resulting in cellular senescence, apoptosis and attenuated TGF-β signaling. Biochim Biophys Acta 1843: 902–914, 2014. [DOI] [PubMed] [Google Scholar]
- 5.Chapman GB, Eagles DA. Ultrastructural features of Glisson's capsule and the overlying mesothelium in rat, monkey and pike liver. Tissue Cell 39: 343–351, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Charrier AL, Leask A, French SW, Brigstock DR. Ethanol-stimulated differentiated functions of human or mouse hepatic stellate cells are mediated by connective tissue growth factor. J Hepatol 55: 399–406, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit R, Ai X, Fine A. Derivation of lung mesenchymal lineages from the fetal mesothelium requires hedgehog signaling for mesothelial cell entry. Development 140: 4398–4406, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis EL, Mann DA. Clinical evidence for the regression of liver fibrosis. J Hepatol 56: 1171–1180, 2012. [DOI] [PubMed] [Google Scholar]
- 9.Friedman SL. Evolving challenges in hepatic fibrosis. Nat Rev Gastroenterol Hepatol 7: 425–436, 2010. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol 6: 425–456, 2011. [DOI] [PubMed] [Google Scholar]
- 11.Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, Evans SM, Dillmann W, Glass CK, Brenner DA. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci USA 109: 9448–9453, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell 134: 657–667, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15: 178–196, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leveen P, Larsson J, Ehinger M, Cilio CM, Sundler M, Sjostrand LJ, Holmdahl R, Karlsson S. Induced disruption of the transforming growth factor β type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood 100: 560–568, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci USA 110: 2324–2329, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lua I, James D, Wang J, Wang KS, Asahina K. Mesodermal mesenchymal cells give rise to myofibroblasts, but not epithelial cells, in mouse liver injury. Hepatology 60: 311–322, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lua I, Li Y, Pappoe LS, Asahina K. Myofibroblastic conversion and regeneration of mesothelial cells in peritoneal and liver fibrosis. Am J Pathol 185: 3258–3273, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutsaers SE. The mesothelial cell. Int J Biochem Cell Biol 36: 9–16, 2004. [DOI] [PubMed] [Google Scholar]
- 19.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605, 2007. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa T, Tateno C, Asahina K, Fujii H, Kawada N, Obara M, Yoshizato K. Identification of vitamin A-free cells in a stellate cell-enriched fraction of normal rat liver as myofibroblasts. Histochem Cell Biol 127: 161–174, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Onitsuka I, Tanaka M, Miyajima A. Characterization and functional analyses of hepatic mesothelial cells in mouse liver development. Gastroenterology 138: 1525–1535, 2010. [DOI] [PubMed] [Google Scholar]
- 22.Pellicoro A, Ramachandran P, Iredale JP. Reversibility of liver fibrosis. Fibrogenesis Tissue Repair 5: S26, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Que J, Wilm B, Hasegawa H, Wang F, Bader D, Hogan BL. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci USA 105: 16626–16630, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radaeva S, Sun R, Jaruga B, Nguyen VT, Tian Z, Gao B. Natural killer cells ameliorate liver fibrosis by killing activated stellate cells in NKG2D-dependent and tumor necrosis factor-related apoptosis-inducing ligand-dependent manners. Gastroenterology 130: 435–452, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Ramm GA, Britton RS, O'Neill R, Blaner WS, Bacon BR. Vitamin A-poor lipocytes: a novel desmin-negative lipocyte subpopulation, which can be activated to myofibroblasts. Am J Physiol Gastrointest Liver Physiol 269: G532–G541, 1995. [DOI] [PubMed] [Google Scholar]
- 26.Sakai N, Chun J, Duffield JS, Wada T, Luster AD, Tager AM. LPA1-induced cytoskeleton reorganization drives fibrosis through CTGF-dependent fibroblast proliferation. FASEB J 27: 1830–1846, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuppan D, Kim YO. Evolving therapies for liver fibrosis. J Clin Invest 123: 1887–1901, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Testerink N, Ajat M, Houweling M, Brouwers JF, Pully VV, van Manen HJ, Otto C, Helms JB, Vaandrager AB. Replacement of retinyl esters by polyunsaturated triacylglycerol species in lipid droplets of hepatic stellate cells during activation. PLoS One 7: e34945, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Troeger JS, Mederacke I, Gwak GY, Dapito DH, Mu X, Hsu CC, Pradere JP, Friedman RA, Schwabe RF. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 143: 1073–1083, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winters NI, Thomason RT, Bader DM. Identification of a novel developmental mechanism in the generation of mesothelia. Development 139: 2926–2934, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang MD, Chiang YM, Higashiyama R, Asahina K, Mann DA, Mann J, Wang CC, Tsukamoto H. Rosmarinic acid and baicalin epigenetically derepress peroxisomal proliferator-activated receptor γ in hepatic stellate cells for their antifibrotic effect. Hepatology 55: 1271–1281, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yata Y, Gotwals P, Koteliansky V, Rockey DC. Dose-dependent inhibition of hepatic fibrosis in mice by a TGF-β soluble receptor: implications for antifibrotic therapy. Hepatology 35: 1022–1030, 2002. [DOI] [PubMed] [Google Scholar]
- 33.Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, Brenner DA, Rippe RA. DNase I-hypersensitive sites enhance α1(I) collagen gene expression in hepatic stellate cells. Hepatology 37: 267–276, 2003. [DOI] [PubMed] [Google Scholar]
- 34.Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest 123: 1902–1910, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454: 109–113, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]