Abstract
Hantavirus Cardiopulmonary Syndrome is a severe disease caused by exposure to New World hantaviruses. Early diagnosis is difficult due to the lack of specific initial symptoms. Anti-hantavirus antibodies are usually negative until late in the febrile prodrome or the beginning of cardiopulmonary phase while Andes hantavirus (ANDV) RNA genome can be detected before symptoms onset. We analyzed the effectiveness of RTqPCR as a diagnostic tool detecting ANDV-Sout genome in peripheral blood cells from 78 confirmed hantavirus patients and 166 negative controls. Our results indicate that RTqPCR had a low detection limit (~10 copies), with a specificity of 100% and a sensitivity of 94.9%. This suggests the potential for establishing RT-qPCR as the assay of choice for early diagnosis, promoting early effective care of patients and improve other important aspects of ANDV infection management, such as compliance of biosafety recommendations for health personnel in order to avoid nosocomial transmission.
Keywords: hantavirus, Andes virus, diagnosis, RT-qPCR
1. Introduction
Hantaviruses are enveloped, segmented, negative strand-RNA viruses that belong to the Bunyaviridae family. The genome is composed by three segments: S, which encodes for the nucleocapsid (N) protein, M, which encodes for the membrane glycoproteins Gn and Gc, and L, which encodes for the RNA dependent RNA polymerase (Plyusnin et al. 1996; Meissner et al. 2002). There are two main categories for Hantavirus diseases, depending on organs involved and source of infection: hemorrhagic fever with renal syndrome (HFRS), mainly in Asia and Europe, are caused by Old World hantaviruses, while hantavirus cardiopulmonary syndrome (HCPS), in North and South America, are caused by New World hantaviruses (Pini 2004; Hjelle and Torres-pérez 2010; Manigold and Vial 2014). Specifically, in South America there are four different phylogenetic clades of hantaviruses corresponding to Andes, Laguna Negra, Rio Mamore and Jabora hantavirus (Firth et al. 2012; Souza and Figueiredo 2014). Several South American hantavirus have been associated to HCPS cases, including Andes, Anajatuba, Araraquara (ARQV), Paranoá, Bermejo, Castelo dos Sonhos, Juquitiba, Araucária, Laguna Negra, Lechiguanas, Maripa, Oran, Rio Mamore and Tunari; A total of approximately 4,000 cases associated to these hantavirus have been reported in South America (Figueiredo et al. 2014; Manigold and Vial 2014).
Andes hantavirus (ANDV) is endemic in Chile and Argentina. Specifically, ANDV-Sout strain is the variant circulating in Chile and South of Argentina. This is the only hantavirus known to be transmissible between humans (Padula et al. 1998; Martinez et al. 2005). According to reports from the Chilean Department of Health, through December 31st 2014, a total of 902 cases have been reported in Chile with a case fatality rate between 30 to 40% depending on the clinical center per year (Unidad de Vigilancia/Departamento de Epidemiologia/MINSAL 2012, 2014, 2015). The main reservoir for ANDV is the long-tailed pygmy rice rat (Oligoryzomys longicaudatus) (Palma et al. 2012) and transmission to humans occurs primarily by inhalation of the virus in aerosolized rodent excretions (Lee and van der Groen 1989; Wells et al. 1997). This mechanism suggests that people living in rural areas or routinely performing activities in these locations such as farmers, forest workers or people that engage in recreational activities in endemic places have more chance of becoming infected. In cases of ANDV infection, sexual partners and contacts who slept in the same bed during the prodromic period of the case have ten times more risk of becoming infected when compared to other household contacts (Ferres et al. 2007). The incubation period of ANDV varies from 7 to 39 days (Vial et al. 2006), and viral particles can be detected in blood as early as two weeks prior to symptoms or antibody response (Evander et al. 2007; Ferres et al. 2007).
There are four distinguishable stages in the clinical course of HCPS: Prodromic stage, which lasts between 2 and 8 days and that presents with unspecific symptoms that include but are not limited to, fever, myalgia, headache and gastrointestinal symptoms. Patients frequently seek attention during this stage but diagnosis is rarely made and serology, if requested, is usually negative. The Cardiopulmonary stage, which presents with development of dry cough, but can rapidly (within hours) set patients on the course for severe respiratory distress and shock, leading to death in one third of the cases. The Diuretic stage, characterized by full normalization of lung and cardiac functions in patients who survive the Cardiopulmonary stage. These patients are usually discharged without evident sequelae in one or two weeks. Finally, the Convalescent stage, in which some patients can still present with fatigue and malaise for weeks to months before fully recovering (Duchin et al. 1994; Hallin et al. 1996; Castillo 2001; Riquelme et al. 2003; Ferrés and Vial 2004; Sotomayor et al. 2009).
At present, reference centers in Chile and Argentina diagnose ANDV hantavirus infection through enzyme-linked immunosorbent assay (ELISA) for detection of IgM antibodies against hantavirus N protein (Padula et al. 2000). While these results are of high quality, frequently they are not available in the early stages of disease, where critical patient management decisions are required, leaving this process relying solely on clinical and epidemiological parameters. Other hematological parameters, such as thrombocytopenia, elevated white cell count, immature granulocytes and distinctive immunoblast cells are also supportive of a diagnosis of HCPS (Sotomayor et al. 2009). In many hospitals in Chile a commercially available, rapid test for IgM against Puumala hantavirus (Reagena POC® Puumala IgM) is also available, but its performance is not optimal. None of these techniques allow for an early diagnosis, as IgM may become detectable only during late stages of prodromic phase or at the beginning of the cardiopulmonary phase. HCPS patients have high levels of viremia (Terajima et al. 1999; Xiao et al. 2006) and we have previously found that RT-qPCR can detect the presence of ANDV in blood early in the disease, identifying viral RNA even before symptoms appear (Ferres et al. 2007). Despite this promising performance, RT-qPCR is still considered a research tool. In this study, we evaluate the performance of RT-qPCR as a diagnostic tool for ANDV infections and show that it complies with every requirement to become the assay of choice for early diagnosis of ANDV infections.
2. Materials and Methods
2.1 Samples
We analyzed blood samples from 244 anonymized patients that had been previously enrolled in different protocols from our Hantavirus Program between years 2001 and 2013: household contacts follow-up (Ferres et al. 2007); two treatment protocols: methylprednisolone (Vial et al. 2013) and immune plasma protocol (Vial et al. 2014) ; and a genetic study (a genome wide association study) (Fondecyt 1110397). Stored white blood cells (WBC) samples were obtained from whole blood drawn at the time of enrollment in the pertinent study, between days 0 and 3 of hospitalization. Peripheral blood was obtained by venipuncture, collected in EDTA blood tubes (4ml) and separated by centrifugation at low speed (1000 rpm for 15 minutes). After centrifugation, buffy coat containing WBC was removed with a small volume of plasma (200ul) and stored at −80°C until further use. Hantavirus positive patients were diagnosed on the basis of the presence of either a febrile prodrome followed by a cardiopulmonary phase with the development of bilateral pulmonary changes (determined by chest radiography) that may or may not trigger cardiogenic shock or by detection of anti-hantavirus antibodies by ELISA as discussed before. We also included samples from 166 negative household contacts of HCPS cases that were obtained between 1 to 5 weeks after HCPS index case detection. These contacts were followed during 40 days, period during which they did not develop any symptoms and remained serologically negative to hantavirus. All participants signed an informed consent, previously approved by institutional Ethics Committees.
2.2 Enzyme-Linked Immunosorbent Assay (serology)
ANDV-reactive antibodies were detected through an IgM capture enzyme-linked immunosorbent assay (ELISA) performed with patient sera and recombinant ANDV N protein following previously described procedures (Padula et al. 2000).
2.3 Viral RNA extraction and RT-qPCR
We extracted Viral RNA from peripheral WBC of patients from a starting volume of 200µl using High Pure Viral RNA Kit (Roche Appl Biosc, IN, USA) according to manufacturer instructions, obtaining approximately 50µL of RNA. For the reverse transcription reaction, 5µL of extracted RNA were used for cDNA synthesis (between 250–400 ng/ul RNA) with MMLV enzyme (Invitrogen, CA, USA) and ANDV-specific primer (forward strand) in a final reaction volume of 20 uL.
Real Time PCR, was performed following a previously described method (Kramski et al. 2007), as detailed in Supplementary Table 1 and 2. All PCRs were done by an operator that was blinded to the clinical diagnosis of the samples, in a Light Cycler 2.0 thermal cycler (Roche Appl Biosc, IN, USA) except for the experiment comparing different thermal cyclers. PCRs were catalogued as positive when the crossing point (Cp) was less than 36. The whole process from RNA isolation to the RT-qPCR result takes around 3 hours and 30 minutes.
2.4 Plasmid preparation
We extracted RNA from in vitro cultured ANDV (Galeno et al. 2002) and RT-qPCR was performed using primers from S segment described in supplementary table 1 (Kramski et al. 2007). For the ligation step, we added adenine to the amplified product using Platinum®Taq DNA polymerase (Life Technologies, CA, USA). Briefly, 1U of polymerase and 10nM of dATP (Fermentas, MA, USA) were added to 10 µg of PCR product and incubated for 20 min at 72°C. We separated the product in a 1% agarose gel stained with SYBR® Safe DNA gel stain (Life Technologies, CA, USA) and the product was extracted from the gel using Wizard® SV gel (Promega Corporation, WI, USA) and PCR Clean-Up System (Promega Corporation, WI, USA). Plasmid pGemT-easy (Promega Corporation, WI,USA) and PCR fragments were ligated according to manufacturer instructions. E. coli (DH5α) competent bacteria were transformed with the plasmid, growth in LB medium (Life Technologies, CA, USA) and selected by Ampicillin (Life Technologies, CA, USA). The plasmid was purified, quantified and stored.
2.5 in vitro transcribed RNA
S segment RNA was kindly donated by Marcelo Lopez-Lastra, and produced by in vitro transcription using T7 RNA polymerase (Fermentas, NY, USA) as described (Vera-Otarola et al. 2012).
2.6 Statistical analysis
Concordance between diagnostic methods (RT-qPCR vs ELISA) was evaluated through calculation of the kappa coefficient (κ) using SPSS v20 (IBM, NY, USA). Diagnostic test parameters were calculated with Epidat v3.1 (Panamerican Health Association OPS).
3. Results
3.1 Analytical detection limit
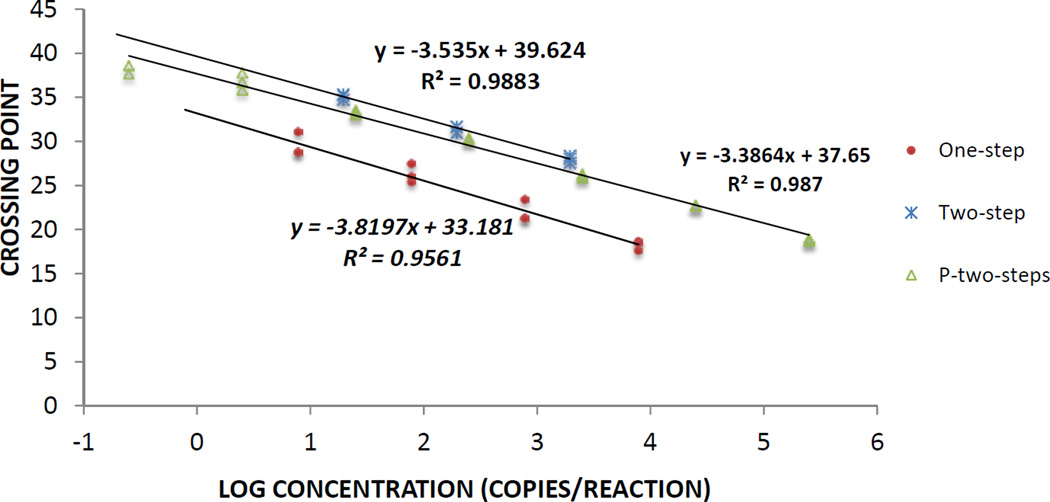
We performed RT-qPCR reactions for ANDV on serial dilutions of known copy number of either an in vitro transcribed RNA or a plasmid containing cDNA of the S segment. In Figure 1 we show the corresponding CP value for the logarithm of each dilution and compared these results between one-step and two-step assays done with an in vitro transcribed RNA. We estimated efficiency by calculating the slope of the trend line that groups all the data points, showing that the two-step assay has a better efficiency (91.8%) than the one-step assay (82.7%) (Supplementary Table 3). We also performed the two-step assay on a plasmid containing the S segment and this reaction had an efficiency of 97%. Since the efficiency was better in the two-step reaction, we performed all the experiments with patient samples using this method. Using this experimental data, we calculated that the two-step method has a limit of detection (LOD) of 11.96 RNA copies/reaction (Confidence Interval (CI) 95%: 0–11.96, logistic regression). The linear range of the RT-qPCR was between 107 and 10 copies/reaction.
Figure 1. RT-qPCR Reaction for ANDV.
Known amounts of in vitro transcribed S segment RNA was amplified by one-step reaction (circles) or two-step reaction (crosses). A plasmid (P) containing S Segment sequence (triangles) was assayed in a two-step RT-qPCR reaction. All experiments were done in LightCycler 2.0
3.2 Technical Reproducibility in two Real time Platforms
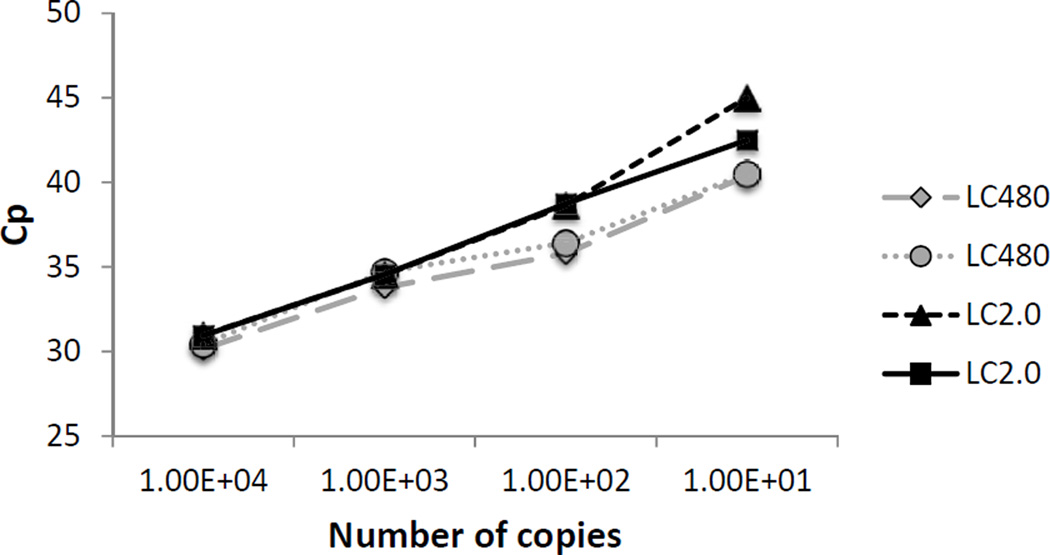
We run parallel serial dilutions of in vitro transcribed RNA to compare the assay performance in two different Light Cycler models (LighCycler 480® and LightCyler 2.0). As shown in Fig. 2, both platforms behaved similarly, indicating that the method can be extrapolated to other qPCR machines.
Figure 2.
Comparison of real-time ANDV assays performed in two platforms. Realtime PCR platforms LightCycler 480® (LC480), and LightCycler 2.0 (LC2.0), were used to assay the same amount of RNA copies. The graphic shows the comparison of the crossing point (Cp) for different amounts of in vitro transcribed RNA, and two technical duplicates for each lightCycler platform.
3.3 Patient Sample Analysis
We analyzed 244 human samples, 78 from positive hantavirus patients and 166 from controls, categorized as described in Methods. In Table 1 we show the two-step RT-qPCR results in which we found 240/244 concordant samples. Concordance between hantavirus cases and RT-qPCR had a kappa value of 0.96. To evaluate the performance of RT-qPCR as a diagnostic tool, we compared it to hantavirus positive and negative patients as a gold standard (Table 2) and found that RT-qPCR is very sensitive and highly specific, yielding high predictive positive and negative values, which converts this technique into an outstanding diagnostic test.
Table 1.
RT-qPCR results in hantavirus patients
| Hantavirus Positive |
Hantavirus Negative |
TOTAL | |
|---|---|---|---|
| PCR Positive | 74 | 0 | 74 |
| PCR Negative | 4 | 166 | 170 |
| TOTAL | 78 | 166 | 244* |
Measure of Agreement Kappa for the 244 samples=0.962
Table 2.
Clinical Diagnostic parameters of RT-qPCR
| Value | CI (95%)* | ||
|---|---|---|---|
| Sensitivity (%) | 94.9 | 89.3 | 100 |
| Specificity (%) | 100 | 99.7 | 100 |
| Positive Predictive Value (%) | 100 | 99.3 | 100 |
| Negative Predictive Value (%)) | 97.7 | 95.1 | 100 |
| Likelihood Ratio (+) | - | - | - |
| Likelihood Ratio (−) | 0.05 | 0.02 | 0.13 |
CI: confidence interval of 95%
4. Discussion
We have validated the use of RT-qPCR in white blood cells as a robust tool for the diagnosis of ANDV-Sout infection, displaying high sensitivity, specificity, positive and negative predictive values. Others have previously detected hantavirus by RT-qPCR but to our knowledge, this is the first effort to validate this technique as a diagnostic tool in a large number of patients (Evander et al. 2007; Ibrahim et al. 2011; Machado and Souza 2013).
Perhaps the characteristic of infection by ANDV that most increases mortality is that initial symptoms of hantavirus infections are highly unspecific and difficult to differentiate from other febrile illnesses. Because patients with hantavirus infection require urgent referral to a center with complex intensive care facilities, availability of the RT-qPCR assay is of tremendous relevance, since it has been proven as a reliable diagnostic test at early stages and could be used to direct the decision making process more efficiently. Also, an accurate and faster diagnosis is important in the reinforcement and adherence to isolation precautions in the health personnel to avoid nosocomial transmission (Martinez-Valdebenito et al. 2014). This method contrasts to hantavirus specific antibodies detection, which is usually not reliable until late into the febrile prodrome or beginning of the cardiopulmonary stage, making serologic testing not useful for early diagnosis. Ferres et al. reported that they can detect viral RNA in peripheral blood cells 5 to 15 days prior to the onset of symptoms and before the appearance of anti-hantavirus antibodies (Ferres et al. 2007), so RT-qPCR could prove a better tool than classic antibody detection by ELISA for diagnosis in an early phase of illness. This is also true for a few cases where even in the cardiopulmonary stage, serology by ELISA was negative, probably due to a slow immunological response of the patient (Martinez-Valdebenito et al. 2014). In addition, ANDV has been detected by RT-qPCR even 84 days after first day of hospitalization (Ferres et al. 2007; Vial et al. 2013), making PCR a good diagnostic tool at different stages of the disease, contrasting with infections such as dengue that have shorter viremia (Tang and Ooi 2012).
Because WBC is the compartment with the highest viral load when compared to respiratory secretions, saliva, urine and gingival crevicular fluid (Ferres et al. 2007; Godoy et al. 2009) it is probably the best sample for diagnosis. Despite the general good performance of RT-qPCR in WBC as a diagnostic tool, it failed to detect the viral RNA in four of the study samples (4 of 78, or 5%). We attribute this finding to the long storage time of these samples (3–10 years, at −80°C) causing viral RNA degradation, a hypothesis supported by the observation of a sensitivity of 100% when RT-qPCR was done in fresh samples (data not shown). Although we haven’t had false negative results when analyzing fresh samples, the negative result in the RT-qPCR test, should not be confirmatory. Overall, because HCPS has a high lethality, we think that the negative diagnosis should always be confirmed by serology, and if symptoms supportive of clinical diagnosis are maintained, a second PCR a day later should be performed.
The RT-qPCR tool has very low technical limit of detection and high technical reproducibility, thus it can be used in diverse platforms, having a high technical sensitivity and specificity for detecting ANDV. Although the one-step assay is widely used in virology for diagnosis, we found that for this particular RT-qPCR the two-step assay had better efficiency, so this should be the test of choice for early ANDV detection and decision-making, although confirmation of diagnosis by the national reference laboratories is still required.
In order to prove the reproducibility of this technique, we compared RT-qPCR results done in two different diagnosis centers, by 4 different technicians, finding that Cp results are similar (data not shown), showing this technique is highly reproducible. This should allow hospitals across the country equipped to perform RT-qPCR, the performance of this test that should be done in adherence to good laboratory quality control practices. This will allow the use of this technique on site allowing a fast delivery of results and improving the standard of care for patients with ANDV.
4.1 Conclusions
In this study, we have validated the use of two-step RT-qPCR as a diagnostic tool for ANDV-Sout. RT-qPCR can become an excellent tool for diagnosis allowing early and fast viral detection, positively affecting patient management.
Supplementary Material
Highlights.
We characterized a RT-qPCR protocol to detect ANDV from blood samples
We validated the RT-qPCR as a diagnostic tool for ANDV infection
This test behaves as a good diagnostic test with a sensibility of 94.9% and specificity of 100%
Acknowledgments
We thank Dr. Marcelo Lopez-Lastra for the donation of in vitro transcribed RNA for the S segment of ANDV. We thank Dr. Gregory J Mertz, Brian Hjelle, Francisco Calderón and Luis Leon for critical review of the manuscript. This work was partly supported by the National Institutes of Allergy and Infectious Diseases at the National Institutes of Health NIH 5UI9AI045452, Fondo Nacional de Investigación y Desarrollo en Salud FONIS SA07I20045, Fondo Nacional de Desarrollo de Cientifico y Tecnológico FONDECYT 11110397, Plan de Mejoramiento Institucional PMI UDD1204.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Castillo C. Hantavirus Pulmonary Syndrome Due to Andes Virus in Temuco, Chile *. CHEST J. American College of Chest Physicians. 2001 Aug 1;120(2):548. doi: 10.1378/chest.120.2.548. [DOI] [PubMed] [Google Scholar]
- Duchin J, Koster F, Peters C, Simpson G, Tempest B, Zaki SR, et al. Hantavirus Pulmonary Syndrome: a Clinical Description of 17 Patients with newly recognize disease. N Engl J Med. 1994;330(14):949–955. doi: 10.1056/NEJM199404073301401. [DOI] [PubMed] [Google Scholar]
- Evander M, Eriksson I, Pettersson L, Juto P, Ahlm C, Olsson GE, et al. Puumala Hantavirus Viremia Diagnosed by Real-Time Reverse Transcriptase PCR Using Samples from Patients with Hemorrhagic Fever and Renal Syndrome Puumala Hantavirus Viremia Diagnosed by Real-Time Reverse Transcriptase PCR Using Samples from Patients with. 2007 doi: 10.1128/JCM.01902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrés M, Vial P. Hantavirus infection in children. Curr Opin Pediatr. 2004 Feb;16(1):70–75. doi: 10.1097/00008480-200402000-00013. [DOI] [PubMed] [Google Scholar]
- Ferres M, Vial P, Marco C, Yanez L, Godoy P, Castillo C, et al. Prospective evaluation of household contacts of persons with hantavirus cardiopulmonary syndrome in chile. J Infect Dis. 2007 Jun 1;195(11):1563–1571. doi: 10.1086/516786. [DOI] [PubMed] [Google Scholar]
- Figueiredo LTM, Souza WM De, Ferrés M, Enria DA. Hantaviruses and cardiopulmonary syndrome in South America. Virus Res. Elsevier B.V. 2014;187:43–54. doi: 10.1016/j.virusres.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Firth C, Tokarz R, Simith DB, Nunes MRT, Bhat M, Rosa EST, et al. Diversity and distribution of hantaviruses in South America. J Virol. 2012 Dec;86(24):13756–13766. doi: 10.1128/JVI.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeno H, Mora J, Villagra E, Fernandez J, Hernandez J, Mertz GJ, et al. First Human Isolate of Hantavirus (Andes virus) in the Americas. 2002;8(7):657–661. doi: 10.3201/eid0807.010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy P, Marsac D, Stefas E, Ferrer P, Tischler ND, Pino K, et al. Andes Virus Antigens Are Shed in Urine of Patients with Acute Hantavirus Cardiopulmonary Syndrome. 2009;83(10):5046–5055. doi: 10.1128/JVI.02409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin GW, Simpson SQ, Crowell RE, James DS, Koster FT, Mertz GJ, et al. Cardiopulmonary manifestations of hantavirus pulmonary syndrome. Crit Care Med. 1996 Feb;24(2):252–258. doi: 10.1097/00003246-199602000-00012. [DOI] [PubMed] [Google Scholar]
- Hjelle B, Torres-pérez F. Hantaviruses in the Americas and Their Role as Emerging Pathogens. 2010:2559–2586. doi: 10.3390/v2122559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SM, Aitichou M, Hardick J, Blow J, O’Guinn ML, Schmaljohn C. Detection of Crimean-Congo hemorrhagic fever, Hanta, and sandfly fever viruses by real-time RT-PCR. Methods Mol Biol. 2011 Jan;665:357–368. doi: 10.1007/978-1-60761-817-1_19. [DOI] [PubMed] [Google Scholar]
- Kramski M, Meisel H, Klempa B, Kru DH, Pauli G, Nitsche A. Detection and Typing of Human Pathogenic Hantaviruses by Real-Time Reverse Transcription-PCR and Pyrosequencing. 2007;1905:1899–1905. doi: 10.1373/clinchem.2007.093245. [DOI] [PubMed] [Google Scholar]
- Lee HW, van der Groen G. Hemorrhagic fever with renal syndrome. Prog Med Virol. 1989 Jan 1;36:62–102. [PubMed] [Google Scholar]
- Machado A, Souza W de. Development of a one-step SYBR Green I real-time RT-PCR assay for the detection and quantitation of Araraquara and Rio Mamore hantavirus. Viruses. 2013 doi: 10.3390/v5092272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manigold T, Vial P. Human hantavirus infections: Epidemiology, clinical features, pathogenesis and immunology. Swiss Med Wkly. 2014;144(March):1–10. doi: 10.4414/smw.2014.13937. [DOI] [PubMed] [Google Scholar]
- Martinez VP, Bellomo C, San Juan J, Pinna D, Forlenza R, Elder M, et al. Person-to-person transmission of Andes virus. Emerg Infect Dis. 2005;11(12):1848–1853. doi: 10.3201/eid1112.050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Valdebenito C, Calvo M, Vial C, Mansilla R, Marco C, Palma RE, et al. Person-to-person household and nosocomial transmission of andes hantavirus, Southern Chile, 2011. Emerg Infect Dis. 2014 Oct;20(10):1629–1636. doi: 10.3201/eid2010.140353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner JD, Rowe JE, Borucki MK, Jeor SCS. Complete nucleotide sequence of a Chilean hantavirus. 2002;89:131–143. doi: 10.1016/s0168-1702(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Padula PJ, Edelstein A, Miguel SDL, Lo NM, Rossi CM, Rabinovich RD. Hantavirus Pulmonary Syndrome Outbreak in Argentina : Molecular Evidence for Person-to-Person Transmission of Andes Virus. 1998;330 doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- Padula PJ, Rossi CM, Della Valle MO, Martinez PV, Colavecchia SB, Edelstein A, et al. Development and evaluation of a solid-phase enzyme immunoassay based on Andes hantavirus recombinant nucleoprotein. J Med Microbiol. 2000 Feb 1;49(2):149–155. doi: 10.1099/0022-1317-49-2-149. [DOI] [PubMed] [Google Scholar]
- Palma RE, Polop JJ, Owen RD, Mills JN. Ecology of rodent-associated hantaviruses in the Southern Cone of South America: Argentina, Chile, Paraguay, and Uruguay. J Wildl Dis. Wildlife Disease Association. 2012 Apr;48(2):267–281. doi: 10.7589/0090-3558-48.2.267. [DOI] [PubMed] [Google Scholar]
- Pini N. Hantavirus pulmonary syndrome in Latin America. Curr Opin Infect Dis. 2004;17(5):427–431. doi: 10.1097/00001432-200410000-00007. [DOI] [PubMed] [Google Scholar]
- Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: Genome structure, expression and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- Riquelme R, Riquelme M, Torres A, Rioseco ML, Vergara JA, Scholz L, et al. Hantavirus pulmonary syndrome, southern Chile. Emerg Infect Dis. 2003 Nov;9(11):1438–1443. doi: 10.3201/eid0911.020798. [DOI] [PubMed] [Google Scholar]
- Sotomayor V, Olea AM, Labraña M, Castillo C, Ortega C, Riquelme OR, et al. Diagnosis and treatment of cardiopulmonary hantavirus syndrome: Chile-2007. Rev Chilena Infectol. 2009 Feb;26(1):68–86. [PubMed] [Google Scholar]
- Souza W, Figueiredo L. Analysis of the nucleocapsid gene brings new insights to the classification of Sigmodontinae-borne hantaviruses. Arch Virol. 2014 doi: 10.1007/s00705-014-2088-1. [DOI] [PubMed] [Google Scholar]
- Tang KF, Ooi EE. Diagnosis of dengue: an update. Expert Rev Anti Infect Ther. 2012 Aug;10(8):895–907. doi: 10.1586/eri.12.76. [DOI] [PubMed] [Google Scholar]
- Terajima M, Hendershot JD, Kariwa H, Koster FT, Hjelle B, Goade D, et al. High levels of viremia in patients with the Hantavirus pulmonary syndrome. J Infect Dis. 1999 Dec 1;180(6):2030–2034. doi: 10.1086/315153. [DOI] [PubMed] [Google Scholar]
- Executive Summary of Hantavirus [in spanish] 2012. Unidad de Vigilancia/Departamento de Epidemiologia/MINSAL. [Google Scholar]
- Weekly Report [in spanish] Santiago: 2014. Unidad de Vigilancia/Departamento de Epidemiologia/MINSAL. [Google Scholar]
- Unidad de Vigilancia/Departamento de Epidemiologia/MINSAL. Informe Semanal Semana. 2015;4:1–6. 2015. [Google Scholar]
- Vera-Otarola J, Solis L, Soto-Rifo R, Ricci EP, Pino K, Tischler ND, et al. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J Virol. 2012 Feb;86(4):2176–2187. doi: 10.1128/JVI.06223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial Pa, Valdivieso F, Ferres M, Riquelme R, Rioseco ML, Calvo M, et al. High-dose intravenous methylprednisolone for hantavirus cardiopulmonary syndrome in Chile: A double-blind, randomized controlled clinical trial. Clin Infect Dis. 2013;57:943–951. doi: 10.1093/cid/cit394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial PA, Valdivieso F, Calvo M, Rioseco ML, Riquelme R, Araneda A, et al. A non-randomized multicentre trial of human immune plasma for treatment of hantavirus cardiopulmonary syndrome by ANDV. Antivir Ther. 2014 Oct 15; doi: 10.3851/IMP2875. [DOI] [PubMed] [Google Scholar]
- Vial PA, Valdivieso F, Mertz G, Castillo C, Belmar E, Delgado I, et al. Incubation period of hantavirus cardiopulmonary syndrome. Emerg Infect Dis. 2006 Aug;12(8):1271–1273. doi: 10.3201/eid1208.051127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RM, Sosa Estani S, Yadon ZE, Enria D, Padula P, Pini N, et al. An unusual hantavirus outbreak in southern Argentina: person-to-person transmission? Hantavirus Pulmonary Syndrome Study Group for Patagonia. Emerg Infect Dis. 1997 Jan;3(2):171–174. doi: 10.3201/eid0302.970210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Yang S, Koster F, Ye C, Stidley C, Hjelle B. Sin Nombre viral RNA load in patients with hantavirus cardiopulmonary syndrome. J Infect Dis. 2006 Nov 15;194(10):1403–1409. doi: 10.1086/508494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.