This issue of Current Biology features five reviews covering various key aspects of the eukaryotic cell cycle. The topics include initiation of chromosome replication, assembly of the mitotic spindle, cytokinesis, the regulation of cell-cycle progression, and cell-cycle modeling, focusing mainly on budding yeast, fission yeast and animal cell model systems. The reviews underscore common themes as well as key differences in the way these processes are carried out and regulated among the different model organisms. Consequently, an important question is how cell-cycle mechanisms and controls have evolved, particularly in the broader perspective of the three domains of life.
Until recently, it appeared that prokaryotes and eukaryotes, with their different levels of complexity and cellular structure, did not share much kinship in cell-cycle mechanics. But with the rapid increase in the number of complete prokaryotic genome sequences, and advances in prokaryotic cell biology, surprising similarities have emerged that, for some organisms, suggest a direct evolutionary connection. Additional evidence indicates that, in other prokaryotes, the cell cycle is fundamentally different from the eukaryotic paradigm.
The initiation of chromosomal DNA replication in eukaryotes is characterized by the use of multiple replication origins, followed by the prevention of reinitiation until the next cell cycle. Origin recognition complex (ORC) proteins recognize the origins and direct the replication machinery to the correct locations in the chromosomes, thereby providing specificity to the process, and also directly participate in replication initiation. The archaea have a set of DNA replication proteins that is entirely homologous to that of eukaryotes, including one, or several, CDC6/ORC1-like proteins. Furthermore, several archaeal species have recently been shown to initiate replication in synchrony from multiple origins, providing yet another fundamental similarity to eukaryotes.
Key aspects of replication have thus been evolutionarily conserved across two kingdoms, and further study of the archaea will reveal to what extent cell-cycle control mechanisms also may be conserved. In the other major branch of prokaryotic life, the bacteria, the DnaA replication initiator protein is also structurally and functionally related to CDC6/ORC1, although more distantly, and ORC1 and DnaA also share the property of being involved in the transcriptional regulation of other genes. But in all bacterial species analyzed to date, chromosome replication is initiated at a single origin (oriC).
In eukaryotes, the reinitiation of replication is prevented by tight cell-cycle control over origin firing. This regulation requires a two-step system involving cyclin-dependent kinases (CDKs), which are regulated by cell-cycle dependent oscillation of cyclin levels, and the anaphase promoting complex/cyclosome (APC/C). High CDK activity prevents formation of the replication complex during S, G2 and M phases, and licensing of replication is renewed only in G1, prior to the next S phase. In archaea from the genus Sulfolobus, Orc1/Cdc6 protein levels vary dramatically over the cell cycle, similar to the variation in cyclin abundance over the eukaryotic cell cycle, although no cyclin homologues have yet been identified.
Bacteria such as Escherichia coli also prevent reinitiation at oriC until the next cell cycle, but their strategy is very different from that of eukaryotes. Instead of using a dedicated cell-cycle control system for replicative licensing, both the DnaA initiator and the oriC chromosome region are made transiently inactive in E. coli after initiation starts. This period of inactivity lasts a significant fraction of the cell cycle, thus accomplishing the same goals as in eukaryotes. Inactivation of DnaA occurs by conversion of the active initiator DnaA–ATP form to inactive DnaA–ADP, stimulated by direct contact with the assembled replisome. A second mechanism involves titration of DnaA by the large number of binding sites on the chromosome, which lowers the level of free DnaA immediately after this region is duplicated early in the replication process and keeps it low until a threshold level of DnaA–ATP is reached.
Transcriptional autoregulation of DnaA also plays a role, and transient inactivation of oriC itself occurs by physical sequestration for about one third of the cell cycle. This requires SeqA, a protein that binds to GATC sites adjacent to DnaA-binding sites, and the Dam methylase, which normally methylates the adenines in the GATC sequences on both strands. Immediately after replication, the new strand is not yet methylated at these sites, which results, by a mechanism that is still unclear, in oriC being unable to re-fire until the new strand is methylated.
So from the model species studied in detail so far, prokaryotes and eukaryotes seemingly use very different mechanisms to prevent the reinitiation of chromosome replication, which appear to have evolved independently. The phylogenetic range of E. coli-type methylation and sequestration proteins is limited, however, and it appears unlikely that the wellstudied E. coli strategy is a universal paradigm for prokaryotes.
Once chromosomal DNA synthesis is complete, eukaryotes assemble a mitotic spindle from microtubules so as to segregate their multiple chromosomes in a defined series of steps. As in eukaryotes, the termination of replication and initiation of mitosis are separated by a G2 phase in Sulfolobus species, and what may be a kind of pre-mitotic chromosome alignment has also been observed. In marked contrast to this archaeal system and eukaryotes, segregation of the bacterial chromosome occurs in parallel with its replication. Yet how is mitosis accomplished in prokaryotes, with no apparent spindle apparatus?
Chromosome segregation in these organisms is simpler, given that the cells are smaller, have no nucleus, and generally have just one or a few chromosomes. Elegant work with GFP fusion proteins has shown that replication origins move poleward much more rapidly than cell elongation in several bacterial model systems, ruling out mechanisms based on passive segregation. Consequently, it has been speculated that the force of extrusion of replicating DNA from a fixed central replication factory might be sufficient to provide the main force, and perhaps even the directionality, of movement necessary to partition bacterial chromosomes.
Recent evidence, however, indicates that bacterial mitosis may be a more active process, orchestrated by an actin-like cytoskeleton. MreB and its homologs in some bacteria and archaea are structurally and biochemically related to actin, and localize as helical filaments that extend from one cell pole to the other. Whereas complete deletion of MreB results in severe cell-shape abnormalities, milder disruption results in specific defects in chromosome segregation. ParM, a similar actin-like protein made by plasmid R1, localizes as a spindle-like track to push segregating R1 molecules poleward.
These examples suggest that bacterial chromosomes, and some plasmids, may segregate along an actin-like track, and that cables of bacterial actin might play roles analogous to the microtubules of the mitotic spindle. Further support comes from recent work on another bacterial model system, Caulobacter crescentus, in which disruption of MreB abolishes the polar localization of oriC as well as other proteins that normally localize specifically to the cell poles; this cellular polarity may stem from the inherent polarity of the MreB filaments themselves, not unlike the mitotic spindle. As centromere-like chromosome sequences have been identified in both bacteria and archaea, it is possible that kinetochore-like machines will be found that pull the chromosomes apart on the actin-like tracks. But many species of prokaryotes lack these actin homologues, suggesting that alternative mechanisms exist.
After the chromosomes are partitioned into future daughter cells, cytokinesis splits the cells in two. In eukaryotes, the mechanical process of cytokinesis is performed by the contraction of an actin-myosin ring, while cytokinesis in many prokaryotes, both archaea and bacteria, is orchestrated by a ring of FtsZ, which is a tubulin homolog. Although the septation proteins in both budding yeast and bacteria assemble in a defined order, in yeast, septins arrive first and the actomyosin ring assembles much later. In contrast, in bacteria, the FtsZ ring arrives first, analogous to the septin ring. Septins, which are found in most eukaryotes — though apparently not plants — are analogous to FtsZ, in that they bind GTP and assemble into filaments at the site of cytokinesis, but are not related structurally or by primary sequence.
The FtsZ ring, once assembled, recruits other division proteins and contracts at the leading edge of the invagination. It is intriguing that eukaryotes other than plants evolved to use mainly actin for cell division and tubulin for mitosis, as it appears that many prokaryotes do the reverse. Moreover, whereas archaeal organisms from the Euryarchaeota phylum contain an FtsZ-based cell division apparatus, species within the Crenarchaeota phylum, such as Sulfolobus, lack FtsZ, and no other cell division proteins have yet been identified in these organisms. Some groups of bacteria, such as the Planctomycetes and Chlamydia, also lack FtsZ homologs. It is completely unknown how these FtsZ-less species divide.
Proper spatial and temporal regulation of cytokinesis is crucial in order to produce viable daughter cells and for multicellular organisms to develop normally. As with other cell-cycle processes, eukaryotes regulate cytokinesis temporally such that only one division event will occur per cell cycle. In animal cells, for example, assembly of the actomyosin ring is inhibited at all times except during the metaphase-to-interphase transition, and its constriction depends on the degradation of cyclin B. In E. coli, the FtsZ ring assembles at about the time of replication termination, but it is not known whether termination specifically activates ring assembly, or if the timing of the two events is independently controlled. Multiple FtsZ rings can form in certain mutants but, interestingly, their constriction seems to be regulated to be activated only once per cell cycle, much like in eukaryotes. The mechanism behind this regulation is not understood.
Eukaryotes use diverse mechanisms for spatial regulation of cytokinesis. In animal cells, the position of the mitotic spindle is key, as is the RhoA protein, but the ultimate protein mediators of placement of the actomyosin ring are still unknown. Whereas budding yeast use the last bud site as a spatial cue, fission yeast find their midpoint via the position of the nucleus, which in turn is positioned medially by interphase microtubules. In plant cells, the preprophase band of microtubules is used as a spatial landmark for the future division site. The morphology of rod-shaped prokaryotes resembles that of fission yeast, and they, likewise, appear to use the chromosome as a spatial cue. But the unpartitioned chromosome is not a positive effector in bacteria; rather, it blocks assembly of the FtsZ ring. This effect is mediated at least in part by a recently discovered nucleoid binding protein, Noc, although the mechanism of inhibition is unknown. Local inhibition of division by an unpartitioned nucleoid makes sense, as its inappropriate scission by a division septum prior to partitioning would be a lethal event.
In addition to nucleoid-mediated division blockage, some bacteria also negatively regulate division with FtsZ inhibitors, the Min proteins, which localize to the cell poles either statically or dynamically depending on species. This polar localization appears to mediate inhibition of FtsZ ring assembly near the poles while allowing medial division. The combined negative spatial regulation by the nucleoid and Min proteins prevents FtsZ ring assembly until chromosome segregation has occurred. This global cellular masking system has no established counterpart in the eukaryotic cell cycle, although there is recent evidence that positioning of the cleavage furrow in animal cells may be negatively regulated by local microtubule density. In the archaea, which are morphologically more similar to bacteria than eukaryotes, virtually nothing is known about the molecular characteristics of the corresponding spatial regulatory systems.
In model bacterial systems such as E. coli and Bacillus subtilis, the cell-cycle timing of cytokinesis may be analogous to the timing of replication initiation; there is a dependence on other cell-cycle processes, but no dedicated cell-cycle control system has yet been discovered. Eukaryotic cell cycle checkpoints stop progression if there is a problem with a part of the cycle, but few such checkpoints in bacteria have been characterized at the molecular level. One, mediated by SulA, delays E. coli cell division when DNA is damaged, although cell growth continues unabated. Other, less well characterized, bacterial cell-cycle checkpoints include cell division arrest after inhibition of DNA replication initiation or chromosome segregation, which may involve the Noc system. There is also good evidence that cell-cycle checkpoints exist in Sulfolobus, and it will be interesting to see if archaeal checkpoints will reveal further cell cycle similarities to eukaryotes.
Furthermore, Caulobacter offers a bacterial cell cycle which, in some aspects, is intriguingly similar to those of eukaryotes. The organism spends its life alternating between a sessile stalked cell and a motile swarmer cell, in a sense hedging its nutritional bets. Every stalked cell eventually buds off a swarmer cell, and every swarmer cell loses its flagellum and grows a stalk. In so doing, Caulobacter cells undergo a differentiation program in every generation that is subject to strict cell-cycle control. DNA replication is shut off during the swarmer–stalk transition, equivalent to G1 phase, and is specifically activated in stalk cells (S phase). This is followed by partitioning of chromosomes (M phase) and asymmetric cell division to yield the two cell types.
Remarkably, just as the eukaryotic cell cycle is driven forward by timed protein phosphorylation and proteolysis, so is the Caulobacter cell cycle. Just as oscillation between two states of CDK activity is crucial to drive the eukaryotic cell cycle, timed proteolysis and transcription activation drive the reciprocal oscillation of two master transcriptional regulators in Caulobacter over the cell cycle. Once the story is more complete, Caulobacter should serve as a useful system in which to model a bacterial cell cycle and compare it to those of eukaryotes.
So despite their striking morphological differences, a number of similarities between prokaryotic and eukaryotic cell cycles have emerged, particularly evident in certain archaeal organisms, in terms of the molecular components, the regulatory principles, and the overall organization of the cell cycle. Still other prokaryotes display features that appear unrelated to those of eukaryotes. This is consistent with convergent evolution, and reflects the patchwork nature that is characteristic of the evolutionary process. There is clearly still much to learn.
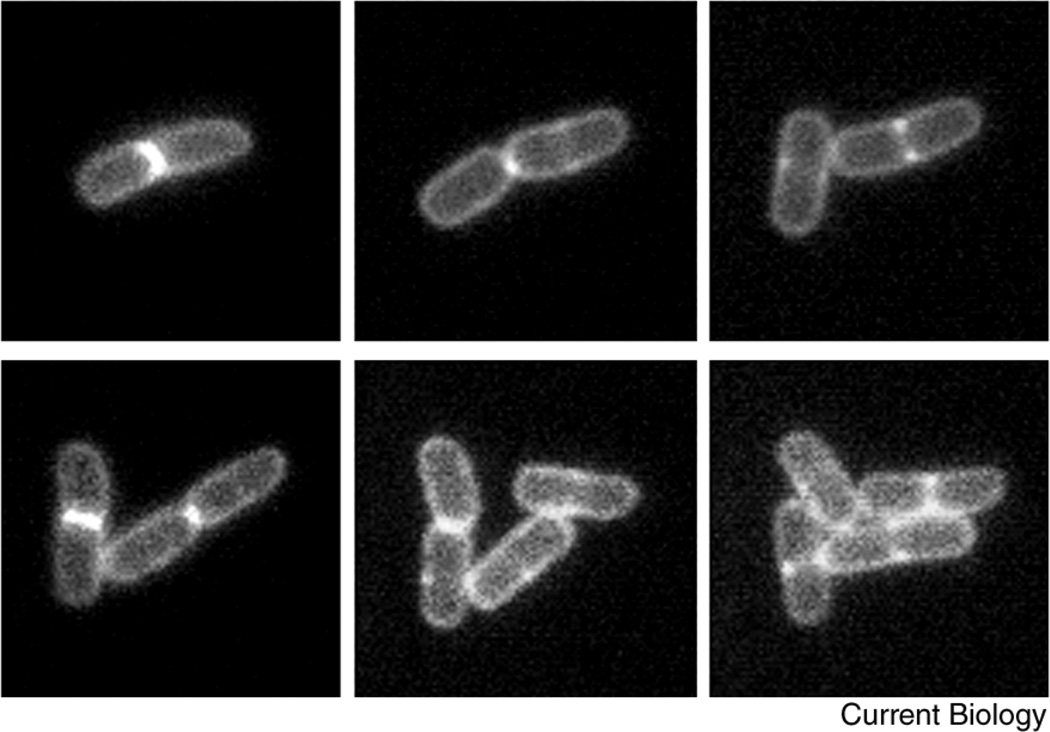
A time course of several cell division cycles of a microcolony of E. coli cells, fluorescently labeled at the cytoplasmic membrane and the division septum.
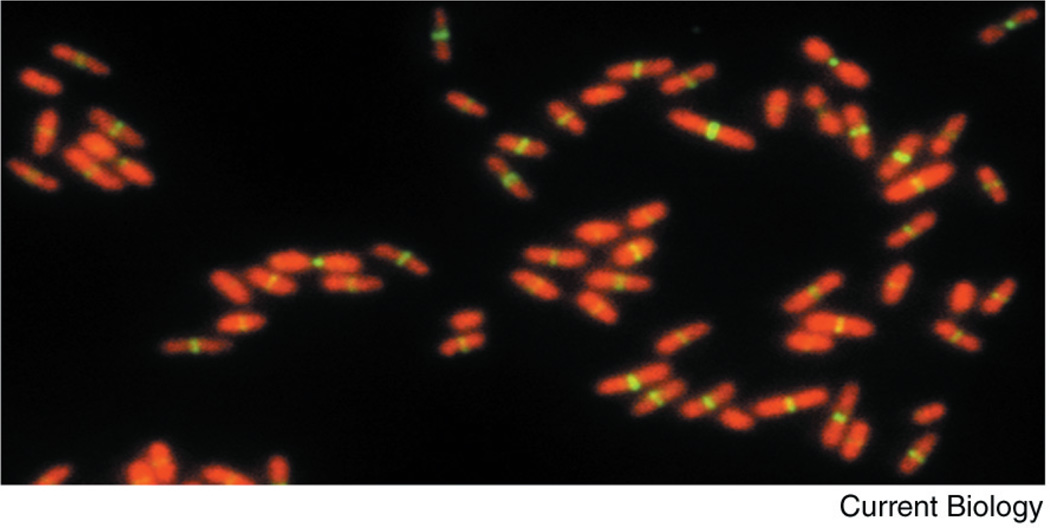
A field of E. coli cells at different stages of the cell cycle, labeled for chromosomal DNA (red) and FtsZ (green).