Abstract
Background
Major depressive disorder (MDD) is a common and disabling condition with well-established heritability and environmental risk factors. Gene–environment interaction studies in MDD have typically investigated candidate genes, though the disorder is known to be highly polygenic. This study aims to test for interaction between polygenic risk and stressful life events (SLEs) or childhood trauma (CT) in the aetiology of MDD.
Method
The RADIANT UK sample consists of 1605 MDD cases and 1064 controls with SLE data, and a subset of 240 cases and 272 controls with CT data. Polygenic risk scores (PRS) were constructed using results from a mega-analysis on MDD by the Psychiatric Genomics Consortium. PRS and environmental factors were tested for association with case/control status and for interaction between them.
Results
PRS significantly predicted depression, explaining 1.1% of variance in phenotype (p = 1.9 × 10−6). SLEs and CT were also associated with MDD status (p = 2.19 × 10−4 and p = 5.12 × 10−20, respectively). No interactions were found between PRS and SLEs. Significant PRSxCT interactions were found (p = 0.002), but showed an inverse association with MDD status, as cases who experienced more severe CT tended to have a lower PRS than other cases or controls. This relationship between PRS and CT was not observed in independent replication samples.
Conclusions
CT is a strong risk factor for MDD but may have greater effect in individuals with lower genetic liability for the disorder. Including environmental risk along with genetics is important in studying the aetiology of MDD and PRS provide a useful approach to investigating gene–environment interactions in complex traits.
Key words: Depression, genetics, gene-environment interactions, polygenic risk scoring
Introduction
Major depressive disorder (MDD) is a global public health problem and the second leading cause of disability worldwide (Vos et al. 2012). The disorder has a well-established genetic contribution, with a heritability of 37% (Sullivan et al. 2000). Genome-wide association studies (GWAS) on depression have typically failed to identify the specific genetic variants involved (Ripke et al. 2013), although two loci have recently been implicated in the CONVERGE study of Chinese women with severe MDD (CONVERGE Consortium, 2015). Many environmental risk factors also increase the risk of depression, including unemployment, social isolation and relationship stressors (Brown & Harris, 1978). Stressful life events (SLEs) can trigger new depressive episodes and childhood trauma (CT) has been shown to double the risk for depression in adulthood (Kessler, 1997; Kendler et al. 1999; Nanni et al. 2012).
Gene–environment interactions (GxEs) whereby a person inherits sensitivity to environmental factors could also play an important role in MDD (Uher, 2014). GxEs have commonly been investigated using single loci in candidate genes, for example the serotonin transporter promoter polymorphism (5-HTTLPR) and its interaction with SLEs in major depression (Caspi et al. 2003). Despite this being the most widely investigated GxE in psychiatry, numerous studies including meta-analyses have produced discrepant results (Risch et al. 2009; Karg et al. 2011; Uher, 2014). There is more consistent evidence for an interaction between 5-HTTLPR and CT, conferring risk for persistent depression in adulthood (Karg et al. 2011; Uher et al. 2011; Brown et al. 2013; Fisher et al. 2013). Nevertheless, the conflicting results from GxE studies in psychiatry have made these findings controversial (Duncan & Keller, 2011).
Further analyses of the genetic effects on MDD have indicated that the disorder is likely to be highly polygenic, arising from the combined effect of many risk variants, each with small effect sizes (Wray et al. 2012; Ripke et al. 2013). Polygenic risk scoring can be used to test the predictive power of multiple genetic variants simultaneously. Subsets of single nucleotide polymorphisms (SNPs) from a discovery GWAS are selected according to their p value and weighted by their effect size to create a polygenic risk score (PRS) for each individual in an independent validation sample. The PRS can then be tested for its ability to differentiate between case and control status in the validation dataset (Purcell et al. 2009; Dudbridge, 2013). PRS derived using results from the largest GWAS on MDD by the Psychiatric Genomics Consortium have shown significant predictive ability for depression, explaining about 0.9% of variance in case–control samples (Ripke et al. 2013; Peyrot et al. 2014).
These findings have led to the hypothesis that GxEs in a highly polygenic trait such as MDD may involve multiple genetic variants rather than one specific locus. Indeed, interactions between polygenic scores for MDD and CT were found to increase risk for depression in the Netherlands Study of Depression and Anxiety (NESDA), accounting for 0.6% of variance in MDD status (Peyrot et al. 2014). The dearth of significant results in GWAS of MDD may be partially due to environmental influences not being accounted for and investigation of GxEs could provide important insights into the complex aetiology of the disorder. Here we test for interactions between polygenic risk for major depression and adult SLEs or CT in the RADIANT UK study of recurrent MDD.
Method
Clinical sample collection
Depression cases (n = 1605) were drawn from three studies previously described in the published literature. The RADIANT UK recurrent MDD sample is comprised of the Depression Case Control (DeCC) study and probands from the Depression Network (DeNT) study of affected sibling pairs (Farmer et al. 2004; Cohen-Woods et al. 2009). UK-ascertained cases from the Genome Based Therapeutic Drugs for Depression (GENDEP) study were also included. GENDEP is a prospective pharmacogenetic study of patients with unipolar depression of at least moderate severity, on a 12-week antidepressant treatment (Uher et al. 2010). Briefly, patients were diagnosed using the Schedules for Clinical Assessment in Neuropsychiatry Interview, according to standardized criteria (Wing et al. 1990). Information was recorded on patients’ worst and second worst episodes of depression in the DeCC and DeNT studies and on their current episode in the GENDEP study (Lewis et al. 2010). Exclusion criteria included personal or family history of other psychiatric diagnoses besides anxiety disorder (Farmer et al. 2004; Cohen-Woods et al. 2009; Uher et al. 2010).
Healthy controls (n = 1064) were available from the DeCC study and the London site of the Bipolar Affective Disorder Case–Control study (Gaysina et al. 2009; Lewis et al. 2010). Controls were screened for lifetime absence of all psychiatric disorders using the Past History Schedule (McGuffin et al. 1986). First-degree family history of any psychiatric disorder or a score of 10 or more on the Beck Depression Inventory at interview were further exclusion criteria (Beck et al. 1996; Cohen-Woods et al. 2009, 2010).
Replication analysis was conducted using recurrent depression cases from The Genetics of Recurrent Early-Onset Depression (GenRED) 1 study (n = 260), the GenRED 2 study (n = 270) and the Depression Genes and Networks (DGN) study (n = 469) (Shi et al. 2011; Battle et al. 2014). Individuals in all analyses were of white European parentage and gave written informed consent to participate. Further information on clinical samples is provided in the online Supplementary material.
Measures
Recurrent MDD was defined according to standard criteria, as having at least two episodes of moderate severity, separated by two or more months of remission (World Health Organization, 1993; American Psychiatric Association, 1994). Number of episodes was not a requirement in the GENDEP study, although the majority of cases were recurrent (Lewis et al. 2010).
Whole-blood samples were collected in ethylene-diamine-tetra-acetic acid (EDTA) from depressed cases. DNA samples were collected from controls by taking blood or using buccal mucosa swabs returned via postal mail. DNA was extracted and samples of sufficient quantity and quality were genotyped on the Illumina Human610-Quad BeadChip (Illumina, Inc., USA) (Freeman et al. 2003; Lewis et al. 2010).
Adult SLEs were assessed using the Brief Life Event Questionnaire, which is a shortened version of the List of Threatening Experiences Questionnaire (LTE-Q) (Brugha et al. 1985). Childbirth was also included, giving a total of 12 items (Farmer et al. 2004) (online Supplementary material). Cases in the DeCC and DeNT studies were asked to report on whether or not they experienced each SLE in the 6 months prior to their worst episode of depression, while GENDEP cases were asked to report on the 6 months preceding the clinical trial (Keers et al. 2011; Fisher et al. 2012). Controls reported on the 6 months prior to their interview. The number of reported SLEs was summed for each individual and analysed as a quantitative variable with range 0–12. Following the LTE-Q categories, SLEs were split into those considered dependent on an individual's behaviour and those which seem independent (Brugha et al. 1985). Dependent SLEs included unemployment, separation, financial or legal difficulties and the birth of a baby. Independent events included personal illness, illness of a family member, death of a family member and being robbed. This gave a total of seven dependent and five independent SLEs (online Supplementary material). Mood at the time of interview was assessed using the self-report Beck Depression Inventory in the DeCC and GENDEP cases (Beck et al. 1996).
A subset of the sample (n = 240 cases, n = 272 controls) completed the self-report Childhood Trauma Questionnaire, which measures frequency and severity of sexual, physical and emotional abuse, physical and emotional neglect during childhood, using 25 Likert-type items (Bernstein et al. 2003). CT was firstly analysed as a quantitative score with range 25–125. To explore results, CT was divided into categories of none, mild and moderate/severe, according to a definition described previously in this sample (Fisher et al. 2013). The GenRED and DGN replication studies assessed CT with the self-report Childhood Events Questionnaire (E. Nelson and D. Levinson, unpublished observations), which is based on the US National Comorbidity Survey CT screening items (Kessler et al. 1997) and CT questionnaires from Washington University (Nelson et al. 2002). These items cover severity and frequency of sexual abuse, physical abuse and trauma (within and outside the family), and emotional neglect.
Quality control
Standard quality-control procedures were implemented to clean genetic data, leaving 471 747 SNPs (Lewis et al. 2010). Principal components (PCs) were calculated using EIGENSTRAT (Price et al. 2006). The first two PCs reduced the genomic control parameter (γ) to 1.02, indicating little difference between RADIANT UK cases and controls due to population stratification or other systematic genomic effects (Lewis et al. 2010). Missing information on age at worst episode of depression (233 cases) and age at interview (34 controls) was replaced with the mean age at worst episode or interview in males or females as appropriate. Number of SLEs was significantly associated with age (p = 3.64 × 10−8) and sex (p = 0.001), with younger individuals and females reporting more SLEs. Since depressed cases were younger than controls and contained a greater proportion of females, the number of SLEs was adjusted in cases to remove bias due to age and sex. Using controls as a proxy for the general population, a linear regression of SLEs on age and sex was used to estimate their association. These regression coefficients were then used to adjust the number of SLEs in depressed cases. Dependent and independent SLEs were adjusted separately in the same manner. CT score was not associated with age or sex, so no adjustment was performed.
Statistical analysis
Polygenic scores were constructed using summary results available online from the Psychiatric Genomics Consortium (https://pgc.unc.edu/) MDD GWAS (Ripke et al. 2013). The RADIANT UK sample was removed to provide an independent validation dataset and a meta-analysis of the remaining eight studies was conducted (7615 cases and 7931 controls). These discovery GWAS results were pruned for linkage disequilibrium (LD) using the p value informed clumping method in PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/), based on the LD structure from the RADIANT UK dataset (Purcell et al. 2007). Clumping preferentially retains SNPs with the strongest evidence of association and removes SNPs in high LD (r2 > 0.25 within a 300 kb window, filtering for significance, PLINK-command: --clump-p1 0.5 --clump-p2 0.1 --clump-r2 0.25 --clump-kb 300). Subsets of SNPs were selected from the results at nine increasingly liberal p value thresholds (pT < 0.0001, pT < 0.001, pT < 0.01, pT < 0.05, pT < 0.1, pT < 0.2, pT < 0.3, pT < 0.4, pT < 0.5). These sets of alleles, weighted by their log odds ratios (ORs) from the discovery study, were summarized into PRS for each individual in the validation sample using PLINK (Purcell et al. 2009). pT < 0.5 contained 87 737 SNPs.
PRS for MDD were tested for ability to predict case/control status in RADIANT UK using logistic regression in R (http://www.r-project.org) to calculate the Nagelkerke's pseudo-R2 measure of variance explained, excluding the variance accounted for by two PCs (Nagelkerke, 1991). SLEs and CT score were also tested for association with case/control status. Interactions between PRS and SLEs or CT were investigated using two models. A multiplicative model tests interaction as departure from multiplicativity, meaning that the combined effect of PRS and environment differs from the product of their individual effects. This was tested using a logistic regression, co-varying for the main effects of PRS, environment and two PCs. Models were also adjusted for PC × environment and PC × PRS interactions (Keller, 2014). The interaction term was tested for its ability to differentiate between case and control status in the validation sample by calculating Nagelkerke's pseudo-R2. An additive interaction model tests whether the combined effect of PRS and environment differs from the sum of their individual effects. It has been suggested that this better captures the biological mechanism of GxEs (Rothman et al. 2008). Interaction as departure from additivity was tested using linear regression of MDD case/control status on the interaction term, with covariates as described previously. The multiple-R2 measure of variance explained by the interaction was calculated. To investigate gene–environment correlations, whereby genotypes may influence exposure to different environments, PRS for MDD were tested for association with SLEs or CT score using a linear regression, co-varying for two PCs. Empirical p values were calculated using permutation procedures for all analyses. Ten independent tests were conducted, giving a Bonferroni corrected significance threshold of 0.005. In the replication phase, gene–environment correlations between PRS and CT score in depressed subjects were tested in MDD cases from the independent GenRED and DGN samples.
Power calculations for our study were performed in QUANTO version 1.2.4 (Gauderman & Morrison, 2009), using ORs reported in the literature for the effects of PRS (OR = 1.22), 2+ SLEs (OR = 1.82) and CT (OR = 2.27) on MDD (Nanni et al. 2012; Motrico et al. 2013; Peyrot et al. 2014). The study had >80% power to detect an interaction between PRS and SLEs with an OR of 1.28 (at α = 0.05). In the subset with CT data, there was >80% power to detect an interaction between PRS and CT with an OR of 1.76 (at α = 0.05).
Ethical standards
All procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Results
Sample characteristics
Sample characteristics are shown in Table 1. Cases contained a significantly greater proportion of females than controls and their mean age at worst episode of depression was significantly younger than mean age at interview in controls. Cases had experienced significantly more SLEs and had higher CT scores than controls (Table 1). In the subset of the sample with CT data (240 cases and 272 controls), similar differences in sex and age at interview were found between cases and controls (p = 2.9 × 10−4 and p = 0.004, respectively).
Table 1.
Sample characteristics
| Cases (n = 1605) | Controls (n = 1064) | p a | |
|---|---|---|---|
| Sex, n (%) | 6.83 × 10−10 | ||
| Male | 471 (29.3) | 436 (41.0) | |
| Female | 1134 (70.7) | 628 (59.0) | |
| Age, years b | 36.7 (12.3) | 41.5 (13.2) | 4.16 × 10−19 |
| Age of onset, years | 23.1 (11.4) | ||
| Number of episodes | 2.48 (0.68) | ||
| Number of SLEs b | 1.57 (1.48) | 0.68 (0.87) | 1.67 × 10−67 |
| Number of dependent SLEs b | 0.88 (1.05) | 0.23 (0.51) | 3.05 × 10−79 |
| Number of independent SLEs b | 0.69 (0.89) | 0.44 (0.66) | 1.29 × 10−11 |
| Childhood trauma score c | 46.31 (16.25) | 32.75 (8.75) | 7.13 × 10−26 |
Data are given as mean (standard deviation) unless otherwise indicated.
SLEs, Stressful life events.
p Values were calculated using a non-parametric Mann–Whitney U test, with the exception of sex where a χ2 test was used.
For cases at worst episode of depression and for controls at interview.
Data were available on a subset of 240 cases and 272 controls. Statistics were calculated from individuals without missing data.
Interaction with SLEs
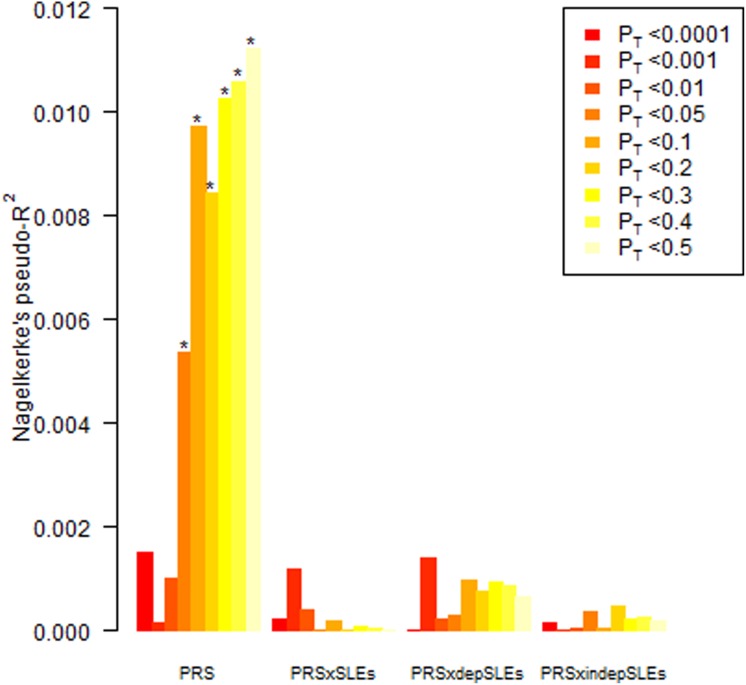
Polygenic scores derived from a meta-analysis of MDD using data from the Psychiatric Genomics Consortium showed significant predictive ability for depression in the RADIANT UK sample. As more SNPs were added to the PRS at increasingly liberal p value thresholds, the amount of variance explained increased. At pT < 0.5, the polygenic score explained 1.1% of variance in case/control status [p = 1.9 × 10−6, OR = 1.22, 95% confidence interval (CI) 1.12–1.32] (Fig. 1). After adjustment for age and sex, total SLEs were still significantly associated with MDD status (p = 2.19 × 10−4), explaining 0.7% of variance between cases and controls. A greater number of dependent SLEs was associated with case status and could predict 6.6% of variance in phenotype (p = 1.35 × 10−25). Independent SLEs showed significant but weaker predictive ability (p = 1.36 × 10−9, Nagelkerke's pseudo-R2 = 0.019) and in contrast to dependent SLEs, more independent events were found in controls v. cases, after correction for age and sex. Under a multiplicative model, there was no interaction between the PRS and total number of SLEs (Fig. 1). The largest R2 was 0.001 at pT < 0.001 (p = 0.12, OR = 1.05, 95% CI 0.98–1.12). Interactions between PRS and dependent or independent SLEs were also non-significant (Fig. 1). No interactions were found under additive models (online Supplementary material).
Fig. 1.
Polygenic risk scores (PRS) for major depressive disorder and multiplicative interactions with stressful life events (SLEs) used to predict depression in the RADIANT UK sample. The y-axis indicates Nagelkerke's pseudo-R2, a measure of the variance explained. On the x-axis the nine p value thresholds used to select single nucleotide polymorphisms in the discovery phase are plotted left to right. depSLEs, Dependent SLEs; indepSLEs, independent SLEs; pT, p value threshold. * p < 0.005. For a colour figure, see the online version.
Interaction with CT
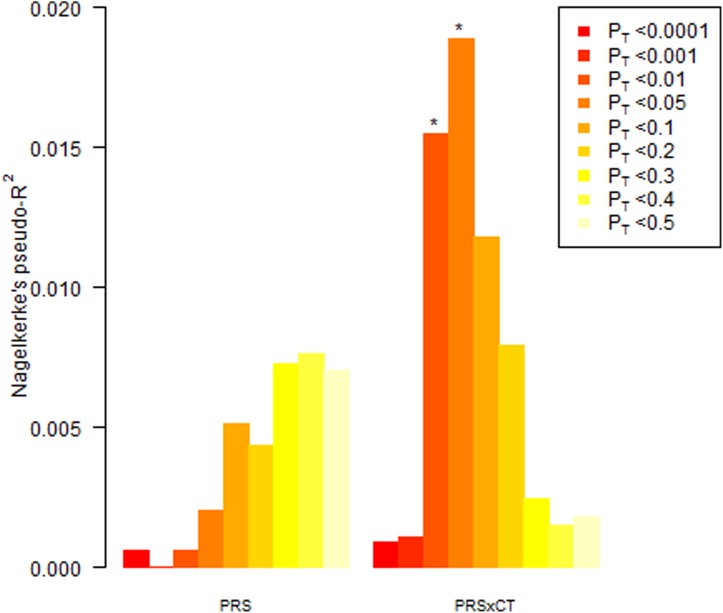
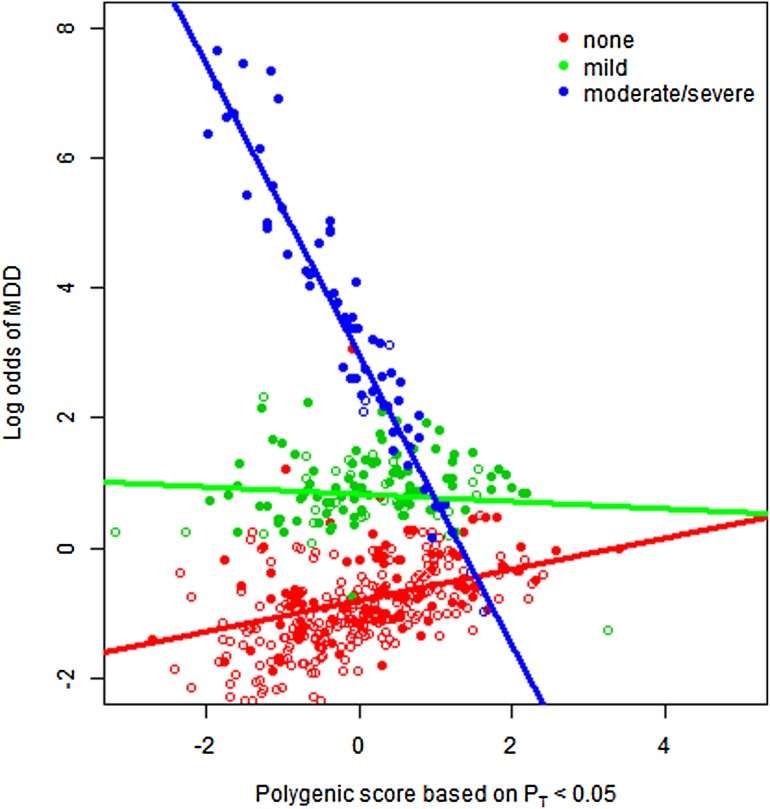
In the subset of the sample with CT data, the PRS did not show significant predictive ability for MDD (p = 0.078, Nagelkerke's pseudo-R2 = 0.007, pT < 0.4), though effects were in the expected direction (OR = 1.18, 95% CI 0.98–1.42) (Fig. 2). A higher CT score was significantly associated with depression status, explaining 30.2% of variance (p = 5.12 × 10−20). Multiplicative interactions were found between polygenic scores for MDD and CT (Fig. 2). The interaction at pT < 0.05 explained 1.9% of variance in the phenotype and was significant after multiple testing correction (p = 0.002). There was an inverse association between the interaction and MDD status (OR = 0.96, 95% CI 0.94–0.98). To visualize these results, interactions were plotted between categories of CT (none, mild, moderate/severe) and PRS standardized to mean 0 and s.d. 1. Plotting log odds of depression by polygenic score for each CT category at the p value threshold with most significant interaction (pT < 0.05; p = 0.002) allows visualization of the results (Fig. 3). For individuals who had not experienced CT, a higher PRS for MDD was associated with a higher risk of the disorder (Fig. 3; black line). Individuals in the mild CT category were at an increased risk of depression but this appeared to act independently of their genetic liability (mid-grey line). Those who had experienced moderate/severe CT were mostly depressed cases but interestingly the individuals at highest risk in this category had a lower PRS than average (Fig. 3; light grey line). There were no interactions between PRS and CT under additive models (online Supplementary material).
Fig. 2.
Polygenic risk scores (PRS) for major depressive disorder and multiplicative interaction with childhood trauma (CT) used to predict depression in the RADIANT UK sample. The y-axis indicates Nagelkerke's pseudo-R2, a measure of the variance explained. On the x-axis the nine p value thresholds used to select single nucleotide polymorphisms in the discovery phase are plotted left to right. pT, p value threshold. * p < 0.005. For a colour figure, see the online version.
Fig. 3.
Multiplicative interaction between standardized polygenic risk score for major depressive disorder (MDD) based on pT < 0.05 and categories of childhood trauma. Shaded circles are cases and open circles are controls. pT, p value threshold. For a colour figure, see the online version.
Gene–environment correlations
Gene–environment correlations were explored between PRS for MDD and number of SLEs. Significant correlations were found within the MDD cases, specifically with dependent (pT < 0.001; p = 0.002) and not independent SLEs (online Supplementary material). No significant corrections were found between polygenic score and CT score in the RADIANT UK sample (online Supplementary material). As the interaction between PRS and CT showed an inverse association with MDD status, the relationship between PRS and CT score was tested in the independent GenRED and DGN depression cases. No significant correlations between PRS and CT score were found.
Mood at interview
To investigate whether low mood at the time of interview may result in a recall bias for negative events, cases severely depressed at interview were removed in a sensitivity analysis. Gene–environment correlations between PRS and dependent SLEs in cases were no longer significant, excluding those who were severely depressed at interview (online Supplementary material). Interactions between PRS and CT score remained significant after severely depressed cases were excluded (online Supplementary material).
Discussion
Polygenic scores derived from the Psychiatric Genomics Consortium MDD mega-analysis predicted depression in the RADIANT UK sample, explaining 1.1% of variance in case/control status. This modest figure is in line with previous estimates from this mega-analysis and confirms the presence of associated variants that the original GWAS was underpowered to detect (Ripke et al. 2013; Peyrot et al. 2014). SLEs were also significant predictors of case/control status. We hypothesized that given the polygenicity of MDD, testing interactions between polygenic scores and environmental adversity could be a more powerful approach than using single genetic variants in a candidate gene. No interactions were found between PRS for MDD and total, dependent or independent SLEs, which is in agreement with previous findings by Musliner et al. (2015).
In the subset of the sample with CT data, the PRS failed to show significant predictive ability for depression which probably reflects the restricted sample size. Consistent with previous reports, CT was a strong risk factor for recurrent MDD in adulthood (Nanni et al. 2012). Significant interactions were found between PRS and CT; however, there was an inverse association with depression status. This appeared to be driven by individuals who had experienced moderate/severe CT, as those at highest risk in this category tended to have a lower PRS than other cases or controls (Fig. 3). One possible explanation is that CT may be more important in the development of MDD for individuals who have a low genetic risk than for individuals who have a high genetic risk. This would be consistent with the liability threshold model for MDD, where a combined effect of many genetic risk variants together with an environmental contribution causes an individual to cross the liability threshold and become affected. Alternatively, the experience of CT may be such a strong risk factor for depression that genetics has a negligible effect.
In contrast to our results, the NESDA study found a significant PRS x CT interaction, whereby higher PRS and severe CT increased the risk for MDD (Peyrot et al. 2014). The conflicting results of these studies may be due to differences in design – for example, NESDA is a population-based study, includes single-episode, recurrent and chronic depression, used a different instrument for assessing CT and had a larger sample size (1645 MDD cases, 340 controls). It has been reported that interaction between 5-HTTLPR and CT specifically increases risk for chronic depression in adulthood (Brown et al. 2013). This suggests that GxEs may be more specific than anticipated and subtle differences between our study and NESDA could have contributed to the discrepant results. Similarly, our SLE assessment was for the 6 months prior to the worst episode of depression in recurrent depression cases. Testing SLEs preceding the initial onset of depression or in single-episode depression may identify different components of the gene–environment aetiology of MDD.
Evidence of gene–environment correlation was found, as polygenic scores for MDD increased exposure to (or reporting of) dependent SLEs in MDD cases (online Supplementary material). This suggests that depressed individuals may select themselves into environmental adversity by creating stressful life events due to their own behaviour, which is known as active gene–environment correlation. SLEs or the reporting of SLEs is heritable (Power et al. 2013) and twin studies have shown pleiotropy between the genetic contribution to SLEs and genetic liability to depression (Kendler & Karkowski-Shuman, 1997; Silberg et al. 1999).
There are several strengths of this study. SLEs were adjusted for age and sex prior to the analyses. The amount of variance explained by the SLEs decreased dramatically after adjustment (online Supplementary material), which demonstrates the importance of accounting for age and sex. It has been suggested that low mood at the time of interview may cause recall bias for negative events; however, we found no evidence that this influenced our results, consistent with two previous analyses in the RADIANT UK sample (Fisher et al. 2012, 2013).
A number of limitations also warrant noting. The discovery GWAS by the Psychiatric Genomics Consortium was underpowered to detect the likely effect sizes in MDD, which reduces the ability to separate modest signals from noise and achieve accuracy in estimation of the PRS (Dudbridge, 2013; Ripke et al. 2013). The PRS used in these analyses consists of SNPs selected from a study of their main effect on MDD, which may not be the same genetic variants that are involved in GxEs. This could explain the non-standard shape of the PRS histograms for the interactions, in contrast to the usual pattern where variance explained increases across the p value thresholds (Figs 1 and 2). Our study design relied on retrospective self-reports of depression and environment, which may be less accurate if the events occurred a long time ago. However, the worst episode of depression is arguably the most memorable, and retrospective self-reports of depressive episodes agree well with hospital records (McGuffin et al. 1986; Kendler et al. 1993).
The detection of GxEs has implications for future research strategies. Analysis of cohorts with heterogeneous environmental exposures may partially explain the lack of success in detecting genetic associations with MDD. Our results suggest that more power could be leveraged from GWAS by focusing only on individuals not exposed to CT as this might identify ‘more genetic’ cases of MDD. However, results of the NESDA study suggest that focusing on exposed individuals could render genetic effects larger, more homogeneous and easier to detect (Flint & Kendler, 2014; Peyrot et al. 2014). Polygenic interactions in MDD require further investigation in larger, similarly well-characterized samples and could provide important insights into the complex aetiology of depression.
Acknowledgements
The RADIANT studies were funded by a joint grant from the UK Medical Research Council, GlaxoSmithKline (G0701420) and by the National Institute for Health Research (NIHR) Biomedical Research Centre for Mental Health at South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, Psychology and Neuroscience, King's College London. The GENDEP study was funded by a European Commission Framework 6 grant (EC contract ref.: LSHB-CT-2003-503428). The GenRED project was supported by National Institute of Mental Health (NIMH) R01 grants MH061686 (D.F.L.), MH059542 (WH Coryell), MH075131 (WB Lawson), MH059552 (J.B.P.), MH059541 (WA Scheftner) and MH060912 (M.M.W.). We acknowledge the contributions of Dr George S. Zubenko and Dr Wendy N. Zubenko, Department of Psychiatry, University of Pittsburgh School of Medicine to the GenRED 1 project. The DGN study was supported by the NIMH (grant 5RC2MH089916). N.M. and C.M.L. have received funding from the European Community's Seventh Framework Programme under the Marie Curie Industry-Academia Partnership and Pathways (grant 286213). H.L.F. is supported by an MQ Fellows Award (MQ14F40). R.U. is supported by the Canada Research Chairs programme (http://www.chairs-chaires.gc.ca/).
We thank all individuals who participated in the RADIANT and GENDEP studies and all involved with data collection and management.
Declaration of Interest
A.E.F. and P.M.G. have received consultancy fees and honoraria for participating in expert panels from pharmaceutical companies including Lundbeck and GlaxoSmithKline. All other authors declare that they have no conflicts of interest.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0033291715002172.
click here to view supplementary material
References
- American Psychiatric Association (1994). Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV). American Psychiatric Association: Washington, DC. [Google Scholar]
- Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, Haudenschild CD, Beckman KB, Shi J, Mei R, Urban AE, Montgomery SB, Levinson DF, Koller D (2014). Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Research 24, 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AT, Steer RA, Brown GK (1996). Beck Depression Inventory – Second Edition Manual. The Psychological Corporation: San Antonio, TX. [Google Scholar]
- Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T, Stokes J, Handelsman L, Medrano M, Desmond D, Zule W (2003). Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse and Neglect 27, 169–190. [DOI] [PubMed] [Google Scholar]
- Brown GW, Ban M, Craig TK, Harris TO, Herbert J, Uher R (2013). Serotonin transporter length polymorphism, childhood maltreatment, and chronic depression: a specific gene–environment interaction. Depression and Anxiety 30, 5–13. [DOI] [PubMed] [Google Scholar]
- Brown GW, Harris TO (1978). Social Origins of Depression: A Study of Psychiatric Disorder in Women. Tavistock: London. [Google Scholar]
- Brugha T, Bebbington B, Tennant C, Hurry J (1985). The List of Threatening Experiences: a subset of 12 life event categories with considerable long-term contextual threat. Psychological Medicine 15, 189–194. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389. [DOI] [PubMed] [Google Scholar]
- Cohen-Woods S, Craig I, Gaysina D, Gray J, Gunasinghe C, Craddock N, Elkin A, Jones L, Kennedy J, King N, Korszun A, Knight J, Owen M, Parikh S, Strauss J, Sterne A, Tozzi F, Perry J, Muglia P, Vincent J, McGuffin P, Farmer A (2010). The Bipolar Association Case–Control Study (BACCS) and meta-analysis: no association with the 5,10-methylenetetrahydrofolate reductase gene and bipolar disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics 153B, 1298–1304. [DOI] [PubMed] [Google Scholar]
- Cohen-Woods S, Gaysina D, Craddock N, Farmer A, Gray J, Gunasinghe C, Hoda F, Jones L, Knight J, Korszun A, Owen MJ, Sterne A, Craig IW, McGuffin P (2009). Depression Case Control (DeCC) Study fails to support involvement of the muscarinic acetylcholine receptor M2 (CHRM2) gene in recurrent major depressive disorder. Human Molecular Genetics 18, 1504–1509. [DOI] [PubMed] [Google Scholar]
- CONVERGE Consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F (2013). Power and predictive accuracy of polygenic risk scores. PLoS Genetics 9, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC (2011). A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. American Journal of Psychiatry 168, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer A, Breen G, Brewster S, Craddock N, Gill M, Korszun A, Maier W, Middleton L, Mors O, Owen M, Perry J, Preisig M, Rietschel M, Reich T, Jones L, Jones I, McGuffin P (2004). The Depression Network (DeNT) Study: methodology and sociodemographic characteristics of the first 470 affected sibling pairs from a large multi-site linkage genetic study. BMC Psychiatry 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher HL, Cohen-Woods S, Hosang GM, Korszun A, Owen M, Craddock N, Craig IW, Farmer AE, McGuffin P, Uher R (2013). Interaction between specific forms of childhood maltreatment and the serotonin transporter gene (5-HTT) in recurrent depressive disorder. Journal of Affective Disorders 145, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher HL, Cohen-Woods S, Hosang GM, Uher R, Powell-Smith G, Keers R, Tropeano M, Korszun A, Jones L, Jones I, Owen M, Craddock N, Craig IW, Farmer AE, McGuffin P (2012). Stressful life events and the serotonin transporter gene (5-HTT) in recurrent clinical depression. Journal of Affective Disorders 136, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Kendler KS (2014). The genetics of major depression. Neuron 81, 484–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig IW (2003). DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behavior Genetics 33, 67–72. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ, Morrison JM (2009). QUANTO 1.2.4: A computer program for power and sample size calculations for genetic–epidemiology studies. University of Southern California: Los Angeles, CA: (http://biostats.usc.edu/Quanto.html). [Google Scholar]
- Gaysina D, Cohen-Woods S, Chow PC, Martucci L, Schosser A, Ball HA, Tozzi F, Perry J, Muglia P, Craig IW, McGuffin P, Farmer A (2009). Association of the dystrobrevin binding protein 1 gene (DTNBP1) in a bipolar case–control study (BACCS). American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics 150B, 836–844. [DOI] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S (2011). The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Archives of General Psychiatry 68, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keers R, Uher R, Huezo-Diaz P, Smith R, Jaffee S, Rietschel M, Henigsberg N, Kozel D, Mors O, Maier W, Zobel A, Hauser J, Souery D, Placentino A, Larsen ER, Dmitrzak-Weglarz M, Gupta B, Hoda F, Craig I, McGuffin P, Farmer AE, Aitchison KJ (2011). Interaction between serotonin transporter gene variants and life events predicts response to antidepressants in the GENDEP project. Pharmacogenomics Journal 11, 138–145. [DOI] [PubMed] [Google Scholar]
- Keller MC (2014). Gene x environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biological Psychiatry 75, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Prescott CA (1999). Causal relationship between stressful life events and the onset of major depression. American Journal of Psychiatry 156, 837–841. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski-Shuman L (1997). Stressful life events and genetic liability to major depression: genetic control of exposure to the environment? Psychological Medicine 27, 539–547. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ (1993). The lifetime history of major depression in women. Reliability of diagnosis and heritability. Archives of General Psychiatry 50, 863–870. [DOI] [PubMed] [Google Scholar]
- Kessler RC (1997). The effects of stressful life events on depression. Annual Review of Psychology 48, 191–214. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Davis CG, Kendler KS (1997). Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychological Medicine 27, 1101–1119. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, Weale ME, Schosser A, Paredes UM, Rivera M, Craddock N, Owen MJ, Jones L, Jones I, Korszun A, Aitchison KJ, Shi J, Quinn JP, Mackenzie A, Vollenweider P, Waeber G, Heath S, Lathrop M, Muglia P, Barnes MR, Whittaker JC, Tozzi F, Holsboer F, Preisig M, Farmer AE, Breen G, Craig IW, McGuffin P (2010). Genome-wide association study of major recurrent depression in the U.K. population. American Journal of Psychiatry 167, 949–957. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Katz R, Aldrich J (1986). Past and Present State Examination: the assessment of ‘lifetime ever’ psychopathology. Psychological Medicine 16, 461–465. [DOI] [PubMed] [Google Scholar]
- Motrico E, Moreno-Kustner B, de Dios Luna J, Torres-Gonzalez F, King M, Nazareth I, Monton-Franco C, Gilde Gomez-Barragan MJ, Sanchez-Celaya M, Diaz-Barreiros MA, Vicens C, Moreno-Peral P, Bellon JA (2013). Psychometric properties of the List of Threatening Experiences – LTE and its association with psychosocial factors and mental disorders according to different scoring methods. Journal of Affective Disorders 150, 931–940. [DOI] [PubMed] [Google Scholar]
- Musliner KL, Seifuddin F, Judy JA, Pirooznia M, Goes FS, Zandi PP (2015). Polygenic risk, stressful life events and depressive symptoms in older adults: a polygenic score analysis. Psychological Medicine 45, 1709–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerke NJ (1991). A note on a general definition of the coefficient of determination. Biometrika 78, 691–692. [Google Scholar]
- Nanni V, Uher R, Danese A (2012). Childhood maltreatment predicts unfavorable course of illness and treatment outcome in depression: a meta-analysis. American Journal of Psychiatry 169, 141–151. [DOI] [PubMed] [Google Scholar]
- Nelson EC, Heath AC, Madden PA, Cooper ML, Dinwiddie SH, Bucholz KK, Glowinski A, McLaughlin T, Dunne MP, Statham DJ, Martin NG (2002). Association between self-reported childhood sexual abuse and adverse psychosocial outcomes: results from a twin study. Archives of General Psychiatry 59, 139–145. [DOI] [PubMed] [Google Scholar]
- Peyrot WJ, Milaneschi Y, Abdellaoui A, Sullivan PF, Hottenga JJ, Boomsma DI, Penninx BW (2014). Effect of polygenic risk scores on depression in childhood trauma. British Journal of Psychiatry 205, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power RA, Wingenbach T, Cohen-Woods S, Uher R, Ng MY, Butler AW, Ising M, Craddock N, Owen MJ, Korszun A, Jones L, Jones I, Gill M, Rice JP, Maier W, Zobel A, Mors O, Placentino A, Rietschel M, Lucae S, Holsboer F, Binder EB, Keers R, Tozzi F, Muglia P, Breen G, Craig IW, Muller-Myhsok B, Kennedy JL, Strauss J, Vincent JB, Lewis CM, Farmer AE, McGuffin P (2013). Estimating the heritability of reporting stressful life events captured by common genetic variants. Psychological Medicine 43, 1965–1971. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38, 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PA, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Muller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O'Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Volzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF (2013). A mega-analysis of genome-wide association studies for major depressive disorder. Molecular Psychiatry 18, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR (2009). Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. Journal of the American Medical Association 301, 2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman KJ, Greenland S, Lash TL (2008). Modern Epidemiology. Wolter Kluwer Health, Lippincott Williams and Wilkins: Philadelphia, PA. [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, Lawson WB, DePaulo JR Jr., Gejman PV, Sanders AR, Johnson JK, Adams P, Chaudhury S, Jancic D, Evgrafov O, Zvinyatskovskiy A, Ertman N, Gladis M, Neimanas K, Goodell M, Hale N, Ney N, Verma R, Mirel D, Holmans P, Levinson DF (2011). Genome-wide association study of recurrent early-onset major depressive disorder. Molecular Psychiatry 16, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg J, Pickles A, Rutter M, Hewitt J, Simonoff E, Maes H, Carbonneau R, Murrelle L, Foley D, Eaves L (1999). The influence of genetic factors and life stress on depression among adolescent girls. Archives of General Psychiatry 56, 225–232. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS (2000). Genetic epidemiology of major depression: review and meta-analysis. American Journal of Psychiatry 157, 1552–1562. [DOI] [PubMed] [Google Scholar]
- Uher R (2014). Gene–environment interactions in common mental disorders: an update and strategy for a genome-wide search. Social Psychiatry and Psychiatric Epidemiology 49, 3–14. [DOI] [PubMed] [Google Scholar]
- Uher R, Caspi A, Houts R, Sugden K, Williams B, Poulton R, Moffitt TE (2011). Serotonin transporter gene moderates childhood maltreatment's effects on persistent but not single-episode depression: replications and implications for resolving inconsistent results. Journal of Affective Disorders 135, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, Mors O, Placentino A, Rietschel M, Souery D, Zagar T, Czerski PM, Jerman B, Larsen ER, Schulze TG, Zobel A, Cohen-Woods S, Pirlo K, Butler AW, Muglia P, Barnes MR, Lathrop M, Farmer A, Breen G, Aitchison KJ, Craig I, Lewis CM, McGuffin P (2010). Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. American Journal of Psychiatry 167, 555–564. [DOI] [PubMed] [Google Scholar]
- Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V, Abraham J, Ackerman I, Aggarwal R, Ahn SY, Ali MK, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Bahalim AN, Barker-Collo S, Barrero LH, Bartels DH, Basanez MG, Baxter A, Bell ML, Benjamin EJ, Bennett D, Bernabe E, Bhalla K, Bhandari B, Bikbov B, Bin Abdulhak A, Birbeck G, Black JA, Blencowe H, Blore JD, Blyth F, Bolliger I, Bonaventure A, Boufous S, Bourne R, Boussinesq M, Braithwaite T, Brayne C, Bridgett L, Brooker S, Brooks P, Brugha TS, Bryan-Hancock C, Bucello C, Buchbinder R, Buckle G, Budke CM, Burch M, Burney P, Burstein R, Calabria B, Campbell B, Canter CE, Carabin H, Carapetis J, Carmona L, Cella C, Charlson F, Chen H, Cheng AT, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahiya M, Dahodwala N, Damsere-Derry J, Danaei G, Davis A, De Leo D, Degenhardt L, Dellavalle R, Delossantos A, Denenberg J, Derrett S, Des Jarlais DC, Dharmaratne SD, Dherani M, Diaz-Torne C, Dolk H, Dorsey ER, Driscoll T, Duber H, Ebel B, Edmond K, Elbaz A, Ali SE, Erskine H, Erwin PJ, Espindola P, Ewoigbokhan SE, Farzadfar F, Feigin V, Felson DT, Ferrari A, Ferri CP, Fevre EM, Finucane MM, Flaxman S, Flood L, Foreman K, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabbe BJ, Gabriel SE, Gakidou E, Ganatra HA, Garcia B, Gaspari F, Gillum RF, Gmel G, Gosselin R, Grainger R, Groeger J, Guillemin F, Gunnell D, Gupta R, Haagsma J, Hagan H, Halasa YA, Hall W, Haring D, Haro JM, Harrison JE, Havmoeller R, Hay RJ, Higashi H, Hill C, Hoen B, Hoffman H, Hotez PJ, Hoy D, Huang JJ, Ibeanusi SE, Jacobsen KH, James SL, Jarvis D, Jasrasaria R, Jayaraman S, Johns N, Jonas JB, Karthikeyan G, Kassebaum N, Kawakami N, Keren A, Khoo JP, King CH, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lalloo R, Laslett LL, Lathlean T, Leasher JL, Lee YY, Leigh J, Lim SS, Limb E, Lin JK, Lipnick M, Lipshultz SE, Liu W, Loane M, Ohno SL, Lyons R, Ma J, Mabweijano J, MacIntyre MF, Malekzadeh R, Mallinger L, Manivannan S, Marcenes W, March L, Margolis DJ, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGill N, McGrath J, Medina-Mora ME, Meltzer M, Mensah GA, Merriman TR, Meyer AC, Miglioli V, Miller M, Miller TR, Mitchell PB, Mocumbi AO, Moffitt TE, Mokdad AA, Monasta L, Montico M, Moradi-Lakeh M, Moran A, Morawska L, Mori R, Murdoch ME, Mwaniki MK, Naidoo K, Nair MN, Naldi L, Narayan KM, Nelson PK, Nelson RG, Nevitt MC, Newton CR, Nolte S, Norman P, Norman R, O'Donnell M, O'Hanlon S, Olives C, Omer SB, Ortblad K, Osborne R, Ozgediz D, Page A, Pahari B, Pandian JD, Rivero AP, Patten SB, Pearce N, Padilla RP, Perez-Ruiz F, Perico N, Pesudovs K, Phillips D, Phillips MR, Pierce K, Pion S, Polanczyk GV, Polinder S, Pope CA III, Popova S, Porrini E, Pourmalek F, Prince M, Pullan RL, Ramaiah KD, Ranganathan D, Razavi H, Regan M, Rehm JT, Rein DB, Remuzzi G, Richardson K, Rivara FP, Roberts T, Robinson C, De Leon FR, Ronfani L, Room R, Rosenfeld LC, Rushton L, Sacco RL, Saha S, Sampson U, Sanchez-Riera L, Sanman E, Schwebel DC, Scott JG, Segui-Gomez M, Shahraz S, Shepard DS, Shin H, Shivakoti R, Singh D, Singh GM, Singh JA, Singleton J, Sleet DA, Sliwa K, Smith E, Smith JL, Stapelberg NJ, Steer A, Steiner T, Stolk WA, Stovner LJ, Sudfeld C, Syed S, Tamburlini G, Tavakkoli M, Taylor HR, Taylor JA, Taylor WJ, Thomas B, Thomson WM, Thurston GD, Tleyjeh IM, Tonelli M, Towbin JA, Truelsen T, Tsilimbaris MK, Ubeda C, Undurraga EA, van der Werf MJ, van Os J, Vavilala MS, Venketasubramanian N, Wang M, Wang W, Watt K, Weatherall DJ, Weinstock MA, Weintraub R, Weisskopf MG, Weissman MM, White RA, Whiteford H, Wiersma ST, Wilkinson JD, Williams HC, Williams SR, Witt E, Wolfe F, Woolf AD, Wulf S, Yeh PH, Zaidi AK, Zheng ZJ, Zonies D, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA (2012). Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2163–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing JK, Babor T, Brugha T, Burke J, Cooper JE, Giel R, Jablenski A, Regier D, Sartorius N (1990). SCAN. Schedules for Clinical Assessment in Neuropsychiatry. Archives of General Psychiatry 47, 589–593. [DOI] [PubMed] [Google Scholar]
- World Health Organization (1993). The ICD-10 Classification of Mental and Behavioural Disorders. Diagnostic Criteria for Research. World Health Organization: Geneva, Switzerland. [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, Ripke S, MacIntyre DJ, McGhee KA, Maclean AW, Smit JH, Hottenga JJ, Willemsen G, Middeldorp CM, de Geus EJ, Lewis CM, McGuffin P, Hickie IB, van den Oord EJ, Liu JZ, Macgregor S, McEvoy BP, Byrne EM, Medland SE, Statham DJ, Henders AK, Heath AC, Montgomery GW, Martin NG, Boomsma DI, Madden PA, Sullivan PF (2012). Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Molecular Psychiatry 17, 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0033291715002172.
click here to view supplementary material