Abstract
Background
Marine cold-temperature environments are an invaluable source of psychrophilic microbial life for new biodiscoveries. An Arctic marine bacterial strain collection was established consisting of 1448 individual isolates originating from biota, water and sediment samples taken at a various depth in the Barents Sea, North of mainland Norway, with an all year round seawater temperature of 4 °C. The entire collection was subjected to high-throughput screening for detection of extracellular laccase activity with guaiacol as a substrate.
Results
In total, 13 laccase-positive isolates were identified, all belonging to the Psychrobacter genus. From the most diverse four strains, based on 16S rRNA gene sequence analysis, all originating from the same Botryllus sp. colonial ascidian tunicate sample, genomic DNA was isolated and genome sequenced using a combined approach of whole genome shotgun and 8 kb mate-pair library sequencing on an Illumina MiSeq platform. The genomes were assembled and revealed genome sizes between 3.29 and 3.52 Mbp with an average G + C content of around 42 %, with one to seven plasmids present in the four strains. Bioinformatics based genome mining was performed to describe the metabolic potential of these four strains and to identify gene candidates potentially responsible for the observed laccase-positive phenotype. Up to two different laccase-like multicopper oxidase (LMCO) encoding gene candidates were identified in each of the four strains. Heterologous expression of P11F6-LMCO and P11G5-LMCO2 in Escherichia coli BL21 (DE3) resulted in recombinant proteins exhibiting 2,2'-azino-bis-3-ethylbenzothiazoline-6-sulphonic acid (ABTS) and guaiacol oxidizing activity.
Conclusions
Thirteen Psychrobacter species with laccase-positive phenotype were isolated from a collection of Arctic marine bacteria. Four of the isolates were genome sequenced. The overall genome features were similar to other publicly available Psychrobacter genome sequences except for P11G5 harboring seven plasmids. However, there were differences at the pathway level as genes associated with degradation of phenolic compounds, nicotine, phenylalanine, styrene, ethylbenzene, and ethanolamine were detected only in the Psychrobacter strains reported in this study while they were absent among the other publicly available Psychrobacter genomes. In addition, six gene candidates were identified by genome mining and shown to possess T1, T2 and T3 copper binding sites as the main signature of the three-domain laccases. P11F6-LMCO and P11G5-LMCO2 were recombinantly expressed and shown to be active when ABTS and guaiacol were used as substrates.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2445-4) contains supplementary material, which is available to authorized users.
Keywords: Psychrobacter, Multicopper oxidase, Laccase, High-throughput screening, Genome sequencing, Heterologous expression, Escherichia coli
Background
With 70 % of the Earth’s surface covered by oceans and 90 % of their water having temperatures of maximum 5 °C [1], the oceans represent a major resource for new bio-discoveries of psychrophilic and psychrotolerant species. Increasing efforts in recent years have aimed at describing microbial biodiversity in a variety of marine cold-temperature environments, including large-scale approaches. These studies led to unprecedented insight into the biodiversity of these environments, largely promoted by the significant technological developments in standardized sampling and next-generation sequencing of metagenome and metatranscriptome [2]. Psychrophilic microorganisms living at cold temperatures must cope with reduced rates of biochemical and physical processes in response to the low molecular kinetic energy of their environment. However, these organisms, through evolutionary processes, developed metabolic capabilities that enable them to survive and multiply at low temperatures [3, 4]. Cold-active biocatalysts from psychrophilic microbial strains are attractive with respect to applications that profit from low-temperature reaction conditions, for example due to thermal instability of products and reactants, like in the food processing industry, or the need to efficiently inactivate the biocatalysts during downstream processes, as exemplified by enzymes for bio-chemical synthesis or molecular research [5–8]. Advantages of such enzymes are their often high rates of catalytic activity at low temperature, low temperature optima and less resistance to thermal inactivation as compared to their mesophilic counterparts. Psychrophilic microbial strains with suitable biochemical capabilities may also be used in bio-remediation of cold-temperature environments polluted with recalcitrant chemicals, like aromatic or aliphatic hydrocarbons from crude oil [9].
Laccases (EC 1.10.3.2, p-diphenol:dioxygen oxidoreductase) are copper-containing oxidases that catalyze the monoelectronic oxidation of various aromatic substances at the expense of molecular oxygen, producing water as the sole by-product [10]. Belonging to the blue-copper family of oxidases, they have their main role in nature both in construction and de-construction of complex aromatic polymers, in particular lignin, and thus many different laccases have so far been described from plants, insects, fungi and bacteria [11]. In combination with electron transfer mediators, forming laccase-mediator systems (LMS), their oxidation capability can be even expanded beyond their natural phenolic substrates [12]. Due to considerable improvements in enzyme production and purification methods, today both laccases and LMS represent ideal biocatalysts for a plethora of biotechnological oxidation processes. At present, laccases or LMS are used as industrial enzymes in diverse applications: for delignification of lignocellulosic fibers and bleaching in pulp and paper industry [13, 14]; and in the de-colorization of dyes in the textile and printing industries [15]. They have also the potential to gain a broader role in chemical synthesis [10], as well as in bio-remediation of industrial waste waters and soils contaminated with e.g., dyes [16], polyaromatic hydrocarbons [17], or extra heavy crude oil [18]. Laccase functionality based on the conversion of classical laccase substrates such as 2,2'-azino-bis-3-ethylbenzothiazoline-6-sulphonic acid (ABTS), 2,6-dimethoxyphenol (2,6-DMP) and guaiacol has lately been shown to be exhibited by other proteins and enzymes not specifically assigned as laccases based on Enzyme Commission (EC) nomenclature, including copper resistance protein [19] and endospore coat protein [20]. As for the laccases themselves, the spectrum of substrates these enzymes convert varies [21], hence rendering the assignment of an enzyme as a laccase rather subjective.
Fungal laccases have been extensively studied [22–24]. Despite generally high level of enzymatic activity shown by fungal laccases they do not possess desired features exhibited by their bacterial counterparts such as higher stability over wide ranges of pH and temperature [25]. Therefore, there is a growing interest in the search of new bacterial laccases. Bacterial laccases are produced by phylogenetically diverse bacteria with various biological functions [25–27]. However, despite the recent advances in the discovery and characterization of laccases and laccase like proteins from bacterial sources [26–29], our knowledge is still limited regarding cold active bacterial laccases, while the importance of cold active enzymes both in academy and industry is well documented [6].
In this study, we describe the discovery and genome sequencing of four new Psychrobacter strains originating from the Barents Sea that exhibit laccase activity and heterologous expression of two of these enzymes in Escherichia coli BL21 (DE3). These strains and/or their aromatic compounds oxidizing enzymes may have broad application potentials as cold-active biocatalysts.
Results
Building and characterizing a culture collection of bacterial isolates from an Arctic marine environment
In collaboration with UiT, the Arctic University of Norway, Tromsø, Norway, biota, water and sediments were sampled between May 14th and 26th, 2009, on a research cruise of R/V Jan Mayen in North of Norway within the Arctic Circle. Sampling was done in ten different locations, spanning a region between and around the Svalbard archipelago and the Bear Island in the Barents Sea. The collection led to the establishment of a library of 1448 single bacterial isolates originating from biota (773), sediments (418), and water (257) samples. The strain library consists of, at least, 31 genera based on 16S rRNA gene sequences of 550 isolates, including Algibacter, Aliivibrio, Alteromonas, Bacillus, Bizionia, Cellulophaga, Cobetia, Colwellia, Cytophaga, Flavobacterium, Halomonas, Lacinutrix, Leeuwenhoekiella, Marinobacterium, Marinomonas, Moritella, Neptunomonas, Olleya, Photobacterium, Planococcus, Polaribacter, Pseudoalteromonas, Pseudomonas, Psychrobacter, Psychromonas, Psychroserpens, Shewanella, Sulfitobacter, Tenacibaculum, Thalassomonas, and Vibrio species.
Screening and identification of strains showing laccase activity
The entire strain collection of 1448 individual bacterial isolates, arrayed in 96-well plates, was screened for isolates exhibiting extracellular laccase activity, and 13 of them scored positive based on the formation of brown zones around the colonies after 24 and 48 h of incubation at room temperature (20–23 °C). The color formation was observed after 24 h (Fig. 1a). However, the incubation time was prolonged to 48 h to capture the isolates that were slower in color formation due to either slower growth or reaction rates. Positive producers were re-streaked on modified marine agar plates from glycerol stocks and five well separated single colonies from each potential producer were picked and grown on screening plates (Fig. 1b). One colony from each producing isolate, confirmed for the laccase-positive phenotype, was chosen for further study.
Fig. 1.
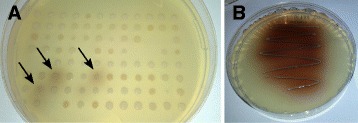
a A representative plate from primary high-throughput screening with three potentially laccase-positive hits indicated with arrows. b A representative plate from the laccase-activity confirmation step
All 13 laccase-positive strains were identified as members of the genus Psychrobacter based on the 16S rRNA gene sequence analysis. The aligned 16S rRNA gene sequences were used to generate a phylogenetic tree by the Weighbor weighted neighbor joining method (Fig. 2). The four distantly related strains P11F6, P2G3, P11G3 and P11G5 were selected for genome sequencing.
Fig. 2.
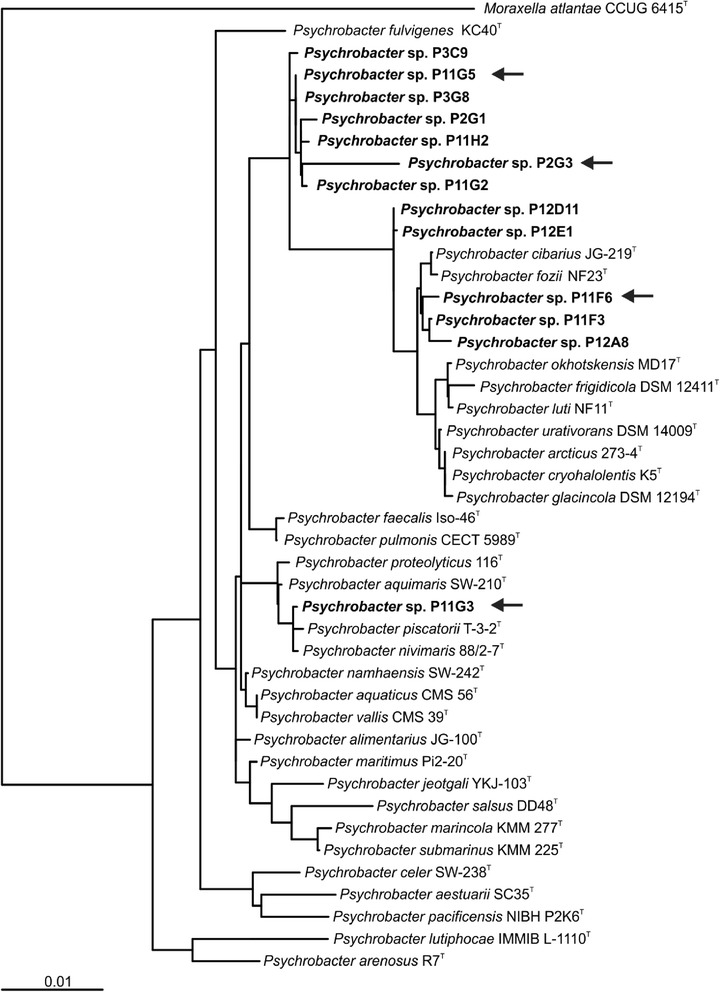
Phylogenetic tree of laccase-positive Psychrobacter species based on 16S rRNA gene sequence similarity. Psychrobacter strains with laccase-positive phenotype are indicated in bold font. Psychrobacter type strains were used as references, with Moraxella atlantae as an outgroup. The four strains selected for genome sequence determination are indicated with arrows
Genome features, annotation and comparative genomics
Manual in silico assembly of the initial Newbler assemblies resulted in one scaffold per replicon for all four genomes; P11G5 and P2G3 genomes completed, and two gaps in P11F6 and P11G3 chromosomes. The genomes of all four strains are almost identical in size and share a similar G + C content (Table 1), however display no significant average nucleotide identity (ANI) [30, 31], except for strains P2G3 and P11G5 which likely belong to the same novel species (97.22 % ANI). For the strain P11F6, high ANI values to Psychrobacter sp. JCM 18902 (95.77 % ANI) and Psychrobacter sp. JCM 18903 (95.65 % ANI) were found, while strain P11G3 shows significant similarity to Psychrobacter sp. JCM 18900 (94.09 % ANI) and Psychrobacter sp. JCM 18901 (93.30 % ANI). This fits well with P11F6 and P11G3 strains described as closely related to P. cibarius (strains JCM 18902 and JCM 18903) and P. nivimaris (strains JCM 18900 and JCM 18901) [32], respectively, thus validating the results for the 16S rRNA gene based classification.
Table 1.
Genome features of the four Psychrobacter strains and other publicly available Psychrobacter genomes
| Name | Ecosystem | Genome Size (bp) | GC (%) | CDSa Count | HTGb % | rDNA operon | tRNA Count | No. of Plasmid | Sequencing Method | Accession ID |
|---|---|---|---|---|---|---|---|---|---|---|
| Psychrobacter sp. 1501 | Host-associated, Circulatory system | 3,050,061 | 41 | 2660 | 6.45 | 1 | 46 | unknown | 454 | GenBank: AFHU01000000 |
| Psychrobacter sp. P11F6 | Host-associated, Tunicates ascidians, Marine, Arctic | 3,517,228 | 43 | 2871 | 2.58 | 6 | 53 | 1 | Illumina MiSeq | This study |
| Psychrobacter sp. P11G3 | Host-associated, Tunicates ascidians, Marine, Arctic | 3,285,555 | 43 | 2650 | 2.62 | 5 | 50 | 4 | Illumina MiSeq | This study |
| Psychrobacter sp. P11G5 | Host-associated, Tunicates ascidians, Marine, Arctic | 3,519,382 | 42 | 2859 | 3.66 | 4 | 48 | 7 | Illumina MiSeq | This study |
| Psychrobacter sp. P2G3 | Host-associated, Tunicates ascidians, Marine, Arctic | 3,366,800 | 42 | 2743 | 4.1 | 4 | 48 | 3 | Illumina MiSeq | This study |
| P. aquaticus CMS 56 | Antarctica | 3,216,409 | 43 | 2814 | 2.97 | 1 | 43 | unknown | 454 | IMG:37750 |
| P. pacificensis DSM 23406 | Seawater, Japan | 3,062,581 | 44 | 2666 | 1.69 | 2 | 40 | unknown | Illumina HiSeq 2500 | IMG:55227 |
| P. arcticus 273-4 | Terrestrial, Siberian permafrost | 2,650,701 | 43 | 2147 | 6.46 | 4 | 49 | unknown | Sanger | GenBank: CP000082 |
| P. cryohalolentis K5 | Aquatic, Siberian permafrost | 3,101,097 | 42 | 2515 | 8.83 | 4 | 48 | 1 | Sanger | GenBank: CP000323-4 |
| Psychrobacter sp. LV10R520-6 | Lake Visa, Antarctica | 3,218,993 | 43 | 2666 | 4.35 | 4 | 49 | unknown | PacBio RS | IMG:62179 |
| P. muriicola 2pS | Arctic cryopeg | 3,384,701 | 43 | 2820 | 1.9 | 6 | 53 | unknown | Illumina HiSeq 2000, HiSeq 2500, PacBio RS | IMG:43211 |
| P. lutiphocae DSM 21542 | Host-associated, digestive system | 3,176,011 | 41 | 2630 | 8.46 | 1 | 44 | unknown | Illumina HiSeq 2000, HiSeq 2500 | IMG:12623 |
| Psychrobacter sp. PRwf-1 | Host-associated, Skin, Off the coast, northeastern Puerto Rico. | 2,995,049 | 45 | 2402 | 4.64 | 5 | 57 | 2 | Sanger | GenBank: CP000713-5 |
| P. phenylpyruvicus DSM 7000 | Host-associated, Circulatory system | 3,099,946 | 42 | 2622 | - | 2 | 45 | unknown | Illumina HiSeq 2000, HiSeq 2500 | IMG:35831 |
| Psychrobacter sp. G | King george i land, Antarctica | 3,113,999 | 42 | 2622 | 1.64 | 4 | 48 | 3 | 454 GS-FLX, Illumina | IMG:26029 |
| Psychrobacter sp. UKMCC_SWTGB2 | Host-associated, Tinggi island, south China sea | 3,384,169 | 43 | 3054 | 14.6 | 1 | 31 | unknown | Solexa | IMG:744 |
| Psychrobacter sp. PAMC 21119 | Terrestrial,Antarctica | 3,510,716 | 43 | 2865 | 5.43 | 4 | 47 | unknown | 454-GS-FLX-Titanium, Illumina GAiix | IMG:24312 |
| Psychrobacter sp. TB67 | Host associated, Sponge, Antarctic | 3,585,631 | 43 | 3021 | - | 6 | 52 | unknown | Illumina HiSeq | IMG:43312 |
| Psychrobacter sp. AC24 | Host associated, Sponge, Antarctic | 3,574,524 | 43 | 2999 | - | 6 | 52 | unknown | Illumina HiSeq | IMG:39856 |
| Psychrobacter sp. TB47 | Host associated, Sponge, Antarctic | 3,544,180 | 43 | 3134 | - | 2 | 53 | unknown | Illumina HiSeq | IMG:43267 |
| Psychrobacter sp. TB15 | Host associated, Sponge, Antarctic | 3,066,842 | 45 | 2574 | - | 2 | 45 | unknown | Illumina HiSeq | IMG:39779 |
| Psychrobacter sp. TB2 | Host associated, Sponge, Antarctic | 3,033,234 | 45 | 2544 | - | 2 | 45 | unknown | Illumina HiSeq | IMG:39800 |
| Psychrobacter sp. JCM 18900 | Host associated, Frozen marine animals | 3,272,645 | 43 | 2893 | 3.78 | 1 | 37 | unknown | Ion Torrent PGM | IMG:38002 |
| Psychrobacter sp. JCM 18903 | Host associated, Frozen marine animals | 3,427,960 | 43 | 3004 | 2 | 1 | 42 | unknown | Ion Torrent PGM | IMG:43060 |
| Psychrobacter sp. JCM 18901 | Host associated, Frozen marine animals | 3,145,827 | 43 | 2875 | 2.67 | 1 | 41 | unknown | Ion Torrent PGM | IMG:43027 |
| Psychrobacter sp. JCM 18902 | Host associated, Frozen marine animals | 3,274,327 | 43 | 2798 | 1.3 | 1 | 42 | unknown | Ion Torrent PGM | IMG:43050 |
aCDS: Coding DNA sequence; bHTG: Horizontally transferred gene
An interesting feature of the four genomes is the localization of the rRNA gene operons: all of the strains have two operons that are surrounded by CDS, while the remaining two, three and four operons (for strains P2G3-P11G5, P11G3 and P11F6, respectively) are clustered together. In all cases, two tRNAs are located between the 16S and 23S rRNA encoding genes.
Another point of variability among the four isolates is the number and the size of the plasmids they harbor: strain P11F6 contains one large plasmid (47.8 kbp); P2G3 contains three plasmids (23.9 kbp; 11.3 kbp, and 9.7 kbp); P11G3 has four small plasmids (8.5 kbp, 6.9 kbp, 5.6 kbp, and 5.5 kbp); and P11G5 harbors seven plasmids (41.0 kbp, 14.3 kbp, 13.6 kbp, 9.2 kbp, 6.0 kbp, 5.8 kbp, and 5.5 kbp).
The genome features were also compared to the other 22 Psychrobacter genomes that were publicly available as of December 2015 (Table 1), and shown to be similar to the other previously reported sequences except high number of plasmids in P11G5.
Pairwise genome alignments were performed for the four studied strains [see Additional file 1]. In agreement with ANI values for P2G3 and P11G5 (97.22 %), dot plot analysis for the two chromosomal DNA sequences [see Additional file 1: Figure S1C] confirm that they likely belong to the same species as they show very limited level of discontinuity in the main diagonal which could be explained by very few independent evolutionary events introducing insertions/deletions (indels). Also strains P11F6 and P11G3 show high level of similarity [see Additional file 1: Figure S1D]. In addition to indels observed throughout the sequences of the two strains, inverted sequence repeats were observed at the middle of the main diagonal which could be an indication of transposable elements. In other cases, significant variation was observed including indels, as well as transposable elements at the middle of the sequences, while the main diagonal remained relatively more consistent at both ends (see Additional file 1: Figure S1A, S1B, S1E and S1F).
In order to compare the metabolic profiles of the genomes, principal component analysis (PCA) and hierarchical clustering were performed based on KEGG pathways. According to hierarchical clustering, P11F6 and P11G3 were not clustered with any other species, while P11G5 and P2G3 were clustered together (Fig. 3). In PCA, however, P2G3 appeard as an outlier and clearly distinguished from P11G5 (Fig. 4). In order to get an insight as to which metabolic pathway distinguishes the studied genomes, not only from each other but also from the other previously reported Psychrobacter genomes, all genes were assigned to KEGG Orthology (KO) numbers and subsequently used to generate a heat map [see Additional file 2: Figure S2]. It should be noted that 36–50 % of the genes could not be connected to any KO. Interestingly, Psychrobacter sp. P11F6, P2G3 and P11G5 were the only strains possessing phenol hydroxylases (K16249, K16246, K16245, K16244, K16243 and K16242) that catalyzes the first step of phenol degradation pathway [33]. Psychrobacter sp. P11F6 and P2G3 were the only strains possessing maleate isomerase gene (K01799) which have been found in nicotine degrading bacteria catalyzing the conversion of maleate to fumarate which can subsequently be used to produce aspartic acid [34]. 4-hydroxy 2-oxovalerate aldolase (K01666), acetaldehyde dehydrogenase (K04073), and 3-phenylpropionate/trans-cinnamate dioxygenase ferredoxin reductase subunit (K00529, K05710) were detected only in Psychrobacter sp. P11F6. These enzymes are known to participate in the metabolism of aromatic amino acid phenylalanine along with other environmental pollutants with similar chemical structure such as styrene and ethylbenzene [35, 36]. Psychrobacter sp. P2G3 and P11G5 were the only strains possessing ethanolamine ammonia-lyase small subunit (K03736) and large subunit (K03735) which is an enzyme commonly found in Entrobacteriaceae family giving them the ability to utilize ethanolamine present in mammalian gastrointestinal tract as a source of carbone and nitrogen [37, 38]. Basic amino acid/polyamine antiporter-APA family (K03294) was only found in Psychrobacter sp. P11F6 which is an electron proton pump involved in extreme acidic resistance [39]. There were also certain genes that were found to be present in the four strains and only in a few of other previously reported genomes. For example, Psychrobacter sp. P2G3 and P11G5, together with Psychrobacter sp. UKMCC_SWTGB2 were the only strains with polysaccharide transporter (PST family; K03328) which potentially enables them to resist and survive in external environmental stress and host interaction settings [40]. Chemotaxis protein methyltransferase gene-cheR (K00575) was detected in Psychrobacter sp. P2G3, Psychrobacter sp. P11G5, P. pacificensis DSM 23406 and P. cryohalolentis K5. Interestingly, even though cheY and cheX were present in 16 of the analyzed strains, other chemotaxis and flagella-assembly associated genes were absent in all of the strains.
Fig. 3.
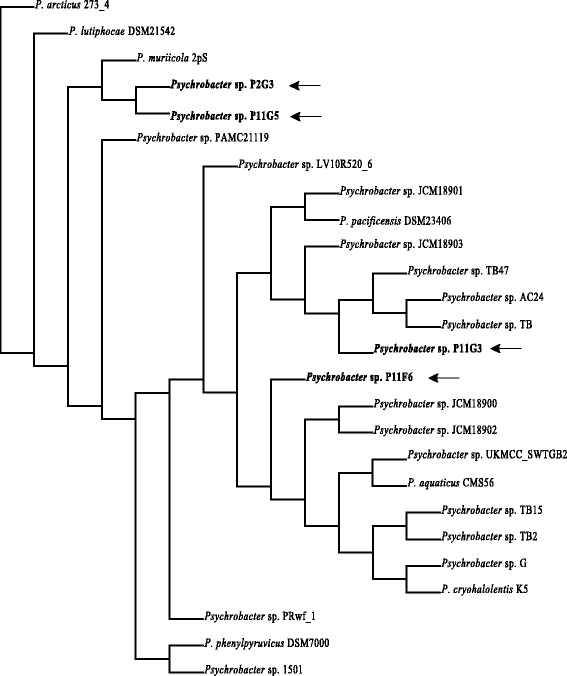
Hierarchical genome clustering of Psychrobacter species based on KEGG pathways. The four genome sequenced Psychrobacter species in this study are indicated in bold font and with arrows
Fig. 4.
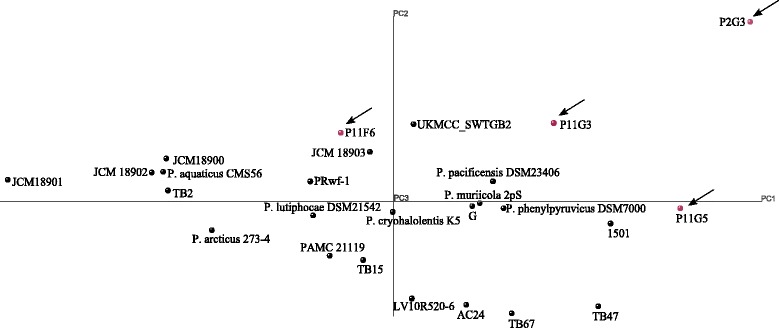
Genome clustering of Psychrobacter species based on KEGG pathways by principal component analysis (PCA). The four genome sequenced Psychrobacter species in this study are indicated with red spheres and arrows
Laccase-like gene candidates and sequence analysis
In total six laccase like multicopper oxidase (LMCO) genes were identified by genome mining (two each in P11G3 and P11G5, and one each in P11F6 and P2G3) possibly involved in the observed phenotype of laccase activity. These genes were annotated as copper-resistance protein (copA). The LMCO gene is located on a plasmid in the strain P11F6, while it is located on the chromosome of the other three strains. The two genes detected in P11G3 share 95 % similarity and located 1,000,347 bp apart from each other and oriented in opposite directions (Fig. 5). Similarly, the LMCO genes from P11G5 share 94 % similarity and are located 1,734,160 bp apart and also in opposite direction. In all six cases, the LMCO genes are followed by copB encoding a putative copper-resistance protein.
Fig. 5.
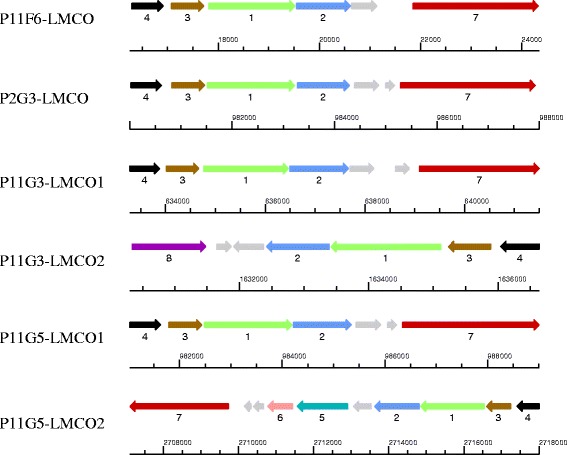
Gene organizations of gene candidates and their positions. 1: LMCO; 2: copB; 3: ATP-binding protein; 4: Cytochrome C; 5: amino acid permease; 6: Isoprenylcysteine carboxyl methyltransferase; 7: Copper-transporting P-type ATPase; 8: Oxidoreductase; Grey arrows represents hypothetical proteins
Table 2 summarizes the lengths, similarity analysis of the predicted candidate genes, and predicted residues forming the type 1 copper binding site. A complete list of amino acid residues predicted to form the substrate binding pocket is provided [see Additional file 3: Table S1]. The canonical three cupredoxin domains forming type 1 (T1), type 2 (T2) and type 3 (T3) copper binding sites of laccases were also detected in structural modeling analysis using Phyre2 [41]. In all cases, the T1 copper binding site was found to be composed of two histidines, one cysteine and one methionine residues (Table 2). The deduced amino acid sequence of all six LMCOs share 85–99 % similarity (Table 3). The signal sequences and the respective cleavage sites were detected in all sequences using the web based applications (see Methods for details).
Table 2.
Sequence analysis of LMCO genes. Length of the genes (bp) and the encoded proteins (aa), amino acid residues forming T1 copper binding site, and the level of similarity to the already known proteins as analyzed by BLAST search are listed
| Name | ORF (bp/aa) | T1 copper site | Amino acid similarity analysis | |
|---|---|---|---|---|
| Description | Coverage/Similarity (%) | |||
| P11F6-LMCO | 1707/568 | H503 C551 H556 M561 | multicopper oxidase [Psychrobacter sp. JCM 18902] | 100/100 |
| P2G3-LMCO | 1704/567 | H502 C550 H555 M560 | CopA family copper resistance protein [Psychrobacter sp. PRwf-1] | 100/95 |
| P11G3-LMCO1 | 1701/566 | H501 C549 H554 M559 | copper resistance protein CopA [Psychrobacter sp. PAMC 21119] | 100/96 |
| P11G3-LMCO2 | 1701/566 | H501 C549 H554 M559 | copper resistance protein CopA [Psychrobacter sp. PAMC 21119] | 100/94 |
| P11G5-LMCO1 | 1704/567 | H502 C550 H555 M560 | CopA family copper resistance protein [Psychrobacter sp. PRwf-1] | 100/95 |
| P11G5-LMCO2 | 1698/565 | H500 C548 H553 M558 | MULTISPECIES: copper resistance protein CopA [Psychrobacter] | 100/99 |
Table 3.
The percentage of sequence similarity between amino acid sequences of the six discovered LMCOs
| P11F6-LMCO | P2G3-LMCO | P11G3-LMCO1 | P11G3-LMCO2 | P11G5-LMCO1 | P11G5-LMCO2 | |
|---|---|---|---|---|---|---|
| P11F6-LMCO | N/A | 86 | 88 | 87 | 87 | 85 |
| P2G3-LMCO | 86 | N/A | 90 | 91 | 99 | 94 |
| P11G3-LMCO1 | 88 | 90 | N/A | 95 | 91 | 90 |
| P11G3-LMCO2 | 87 | 91 | 95 | N/A | 92 | 91 |
| P11G5-LMCO1 | 87 | 99 | 91 | 92 | N/A | 94 |
| P11G5-LMCO2 | 85 | 94 | 90 | 91 | 94 | N/A |
Enzymatic assay of P11F6-LMCO and P11G5-LMCO2
The reaction mixture was prepared and absorbance was monitored at A420 and A468 for ABTS and guaiacol, respectively. The increase in absorbance was an indication of enzymatic activity (Fig. 6a and b). Moreover, the oxidized product of ABTS and guaiacol are green and brown, respectively, which for both enzymes could be confirmed visually (Fig. 6c).
Fig. 6.

Enzymatic assay of P11F6-LMCO and P11G5-LMCO2 using ABTS and guaiacol as substrates. Increase in A 420 and A 468 when ABTS (a) and guaiacol (b) were used as the substrates, respectively. c Chromogenic products of ABTS and guaiacol oxidation by the studied LMCOs
Discussion
Psychrophilic microorganisms are found in all three domains of life: bacteria, archaea, and eukarya [42]. However, in deep sea and cold temperature marine environments bacteria, and in particular Gamma-proteobacteria, are dominant. Many novel cultivable species of the genera Colwellia, Moritella, Photobacterium, Psychromonas, Marinomonas and Shewanella have been first described from cold environments [42]. It is therefore not surprising that the screening of the Arctic marine bacterial collection led to identification of strains belonging to the Gamma-proteobacteria exhibiting laccase activity.
Comparative genomics were performed on KEGG-based metabolic profile of the studied four strains against the other publicly available Psychrobacter genomes. This analysis revealed noteworthy differences as well as novel genes not being present in the previously reported Psychrobacter genomes. For instance, genes associated with metabolic pathways of phenol (in P11F6, P2G3 and P11G5), nicotine (in P11F6 and P2G3), phenylalanine/styrene/ethylbenzene (in P11F6), ethanolamine (in P2G3 and P11G5) degradation are detected only in the Psychrobacter species that are reported in this study. In addition, P11F6 is the only strain with basic amino acid/polyamine antiporter gene responsible for extreme acidic resistance. Although not unique to our strains, several other features relevant to survival in extreme conditions are also detected such as resistance to heavy metals, regulation of osmotic pressure and the ability to utilize different carbon, nitrogen, and sulfur sources. All of these features potentially enable the species to survive in harsh conditions. Noteworthy, neither flagella assembly genes nor complete set of genes encoding chemotaxis components could be found based on KO analysis, in agreement that members of the Psychrobacter genus are generally known to be non-motile [43].
The discovery of the first bacterial laccase [44] motivated many researchers to explore the bacterial domain for laccases. It is now well established that those from bacterial sources exhibit higher thermal- and pH stability compared to their fungal counterparts [28]. In this study, we sought to find LMCOs from marine cold adapted bacteria. There are a limited number of reports on laccase activity from bacteria living in cold environments, while the importance of cold-active enzymes is well known [6]. Recent reports support the extracellular laccase like activity from Psychrobacter genus [45], as well as psychrophilic/psychrotolerant bacterial species [46].
The bacterial collection used in this study was screened for extracellular laccase activity without supplementing the screening medium with additional copper. This suggests that the identified isolates produce enzyme(s) with laccase-like activity, most likely, extracellularly, if not taking up the substrate and pumping out the chromogenic product back to the extracellular medium. We tested enzymatic activity of cell free supernatant from cultures grown in the absence of substrate which led to no measurable laccase activity (data not shown). This observation may suggest that the production of LMCOs by the studied isolates is inducible. The enzymes did not rely solely on copper for the activity as there were other metal ions such as Mg2+ and Ca2+ present in the medium that could possibly serve as cofactors for laccases [47]. However, we observed more intense brown zone formations in the presence of 0.25 mM CuCl2 (data not shown).
The laccase-positive isolates were all identified as members of the genus Psychrobacter, while members belonging to known laccase-active genera such as Shewanella and Pseudomonas [27, 48] were also present in our collection. This may be explained by our screening system being designed to target only species producing LMCOs extracellularly and only those able to oxidize guaiacol as substrate. Bacterial LMCOs from Gramella forsetii KT0803, Marivirga tractuosa DSM4126 were shown to not accept guaiacol although they oxidize ABTS and syringaldazine [21].
Similar to the present study, the majority of known bacterial genes encoding proteins with laccase-like activity have been annotated as multicopper oxidase (MCO) or copper-resistance proteins (copA) [25]. The amino acid sequence analysis of the studied LMCOs revealed the presence of conserved T1, T2 and T3 copper binding sites of 3-domain laccases. The T1 site is typically composed of two histidines, one cysteine and one position which is usually variable [49]. The variable residue in the studied sequences was methionine which is in agreement with known bacterial LMCOs [49]. This position is replaced by leucine or phenylalanine in fungal laccases and has been argued to influence the oxidation potential [49]. Therefore, the studied LMCOs are likely among the low redox potential laccases with redox potential below 500 mV [50]. Notably, however, a bacterial laccase with redox potential as high as 638 mV was recently reported [51] which falls into the middle range of redox potentials (up to 700 mV) [50]. Although the redox potential of the T1 Cu site (where the substrate oxidation takes place) is the key parameter in the oxidation capacity of laccases, changes in amino acid residues constituting the substrate binding pocket can considerably change the kcat and KM values of laccases while maintaining the same redox potential of T1 Cu site [50, 52]. In spite of sharing high level of similarity at the amino acid sequence level, the studied LMCOs showed considerable differences in the amino acid residues forming their predicted substrate binding pocket [see Additional file 3: Table S1]. This suggests that the studied enzymes may exhibit different substrate preferences.
Detection of single and multiple laccase genes, both on plasmid and chromosomes, in this study is in agreement with the earlier report by Ausec and coworkers [27] in which the authors showed that some members of certain bacterial genera (e.g., Rhodococcus) possess multiple laccase genes in their genome and that the genes can be present both on a plasmid or a chromosome. The presence of multiple laccase genes may confer adaptive benefits with respect to certain life styles and environmental factors [53]. Prediction of signal peptides encoded by the studied gene candidates supports the extracellular laccase activity observed during high-throughput screening (HTS). The majority of putative laccase genes found from publicly available finished/draft genomes and metagenomic datasets encode signal peptide sequences suggesting that the gene products are exported from the cytoplasm [27].
Heterologous expression of P11F6-LMCO and P11G5-LMCO2, in E.coli¸ confirmed the in silico based selection of the target enzymes showing laccase activity towards the phenolic substrate guaiacol, which was used during the initial HTS, as well as the bulky non-phenolic substrate ABTS. However, LMCO gene knock-out studies in the native strains are needed to confirm whether the identified candidate genes actually are responsible for the observed phenotype.
Conclusions
The screening of a bacterial collection from marine Arctic environments was performed that led to identification of 13 Psychrobacter strains exhibiting extracellular laccase activity. Four of these strains were genome sequenced. Based on genome scale metabolic profile analysis, genes associated with degradation of phenolic compounds, nicotine, phenylalanine, styrene, ethylbenzene, and ethanolamine were detected only in the Psychrobacter strains reported in this study while they were absent in the other publicly available Psychrobacter genomes. In addition, six LMCO gene candidates were identified by genome mining, and were located both in plasmid and chromosomal DNA. Conserved domains for copper binding sites for three-domain laccases were also detected in all six sequences. The activity of heterologously expressed P11F6-LMCO and P11G5-LMCO2 were validated using both ABTS and guaiacol as substrate. Further studies are underway to evaluate the role of the identified LMCOs in the four laccase-positive Psychrobacter strains, as well as their full biochemical characterization with respect to their potential in various biotechnological applications.
Methods
Bacterial culture collection
Details about the generation of the Arctic marine strain collection, including in total 1448 bacterial isolates, are presented in the Results section. Of particular relevance for this study, a Botryllus sp. (colonial ascidian tunicate) sample was taken from a depth of about 20 m, with seawater temperature of 4.1 °C (69° 44,98704 N, 30° 25,21654 E). The specimen was washed with sterile sea water, and was homogenized, which was then used as inoculum. The homogenate was prepared and diluted in sterile sea water, and aliquots of the diluted homogenate were spread on IM8 agar plates (1 g/L malt extract, 1 g/L glycerol, 1 g/L glucose, 1 g/L peptone, 1 g/L yeast extract, 18 g/L agar, 16 g/L sea salt, [54]) and incubated at 4 °C for two weeks, until visible colonies appeared. Single colonies were picked and grown in liquid IM8 with 225 rpm agitation at 4 °C until enough cell growth had occurred. Samples were frozen in individual vials at −80 °C. Later on, single colonies were transferred to 96 deep-well plates for screening purposes.
High-throughput screening for laccase activity
The bacterial collection in 96-well plate format was replicated in modified marine broth medium [15 g/L Difco marine broth, 5 g/L peptone, 150 mL/L artificial sea water (425 mM NaCl, 9 mM KCl, 9 mM CaCl2.2H2O, 26 mM MgSO4.7H2O, 23 mM MgCl2.6 H2O, pH 7.8)] grown overnight at 20 °C with 800 rpm orbital agitation. Cultures were then transferred and stamped to modified marine agar (modified marine broth containing 15 g/L bacteriological agar) plates (14 cm) supplemented with 0.01 % guaiacol (Sigma-Aldrich) by a 96-pin replicator. The plates were covered to protect from light and incubated at room temperature (20–23 °C) for two days. The plates were checked after 24 and 48 h. The colonies with visible brown color zones around were selected for further characterization.
Isolation of total DNA and 16S rRNA gene sequence analysis
Total DNA from the producing isolates was purified using Wizard® Genomic DNA Purification Kit (Promega). 16S rRNA genes were amplified by PCR using primers 27f (5' AGAGTTTGATCMTGGCTCAG 3') and 1492r (5' TACGGYTACTTGTTACGACTT 3') [55]. The same primers were also used for direct sequencing of the PCR products. The similarity search of 16S rRNA gene sequences was performed by BLASTN [56]. A phylogenetic tree was then generated using the RDP database [57] with the 16S sequences of all Psychrobacter type strains as references and Moraxella atlantae as an outgroup.
Genome sequencing, annotation and comparative genomics
Genomic DNA was sequenced in a combined approach using a whole genome shotgun and a mate pair library for each strain to be sequenced. The whole genome shotgun libraries were constructed with the TruSeq DNA PCR-Free library preparation kit (Illumina). 8 kb mate pair libraries were prepared with the Nextera mate pair sample preparation kit (Illumina) according to the gel-plus protocol. All libraries were sequenced in a paired-end run using the MiSeq reagent kit v3 (600 cycles) and the MiSeq desktop sequencer (Illumina). Reads from the WGS libraries were quality trimmed (at least 5 nt with > = Q30 at the 3' end). Reads from both libraries were assembled with the Roche GS de novo Assembler software (Newbler; release 2.8). The results for the initial assemblies are listed in Table 4. The gap closure step was facilitated by the Consed software (version 26) [58]. Gene prediction was performed with Prodigal [59], tRNAscan-SE [60], and RNAmmer [61] for CDS, tRNAs and rRNAs in general. Functional annotation was performed using the GenDB annotation platform [62]. ANI values were calculated using the implementation in EzBioCloud (http://www.ezbiocloud.net/).
Table 4.
Statistics of the initial, automated assembly
| P2G3 | P11F6 | P11G3 | P11G5 | ||
|---|---|---|---|---|---|
| Aligned Reads | 2,096,689 | 2,858,651 | 1,705,396 | 1,811,833 | |
| Assembled Bases | 533,309,075 | 722,997,409 | 422,716,068 | 458,257,731 | |
| Contigsa | 28/30/45 | 32/39/51 | 28/32/51 | 39/46/68 | |
| Scaffolds | 6 | 6 | 8 | 8 | |
| Bases in scaffolds | 3,345,653 | 3,468,794 | 3,246,680 | 3,495,250 | |
| Coverage | 159.4 | 208.43 | 130.2 | 131.11 | |
| Gaps/MBase | 8.4 | 9.2 | 8.6 | 11.2 | |
| G + C content [%] | 41.79 | 42.73 | 42.76 | 41.81 | |
| Scaffold Size | Average | 557,608 | 578,132 | 405,835 | 436,906 |
| Largest | 3,289,362 | 2,649,068 | 2,388,248 | 3,395,360 | |
| Contig Size | Average | 111,293 | 88,690 | 63,489 | 75,633 |
| Largest | 378,865 | 441,017 | 655,512 | 487,469 | |
ain scaffolds/large/all
The embedded comparative tools in RAST version 2.0 [63] and integrated microbial genome (IMG) [64] were used for comparative genome analyses using publicly available genome sequences of Psychrobacter strains listed in Table 1.
Nucleotide sequence accession numbers
The Whole Genome Shotgun projects have been deposited at DDBJ/EMBL/GenBank under the accessions LJCE00000000 (strain P11F6) and LJCF00000000 (strain P11G3). The versions described in this paper are version LJCE01000000 and LJCF01000000, respectively. The complete annotated genome sequences (chromosomes and plasmids) have been deposited at DDBJ/EMBL/GenBank under the accession numbers CP012529-CP012532 (strain P2G3) and CP012533-CP012540 (strain P11G5).
Genome mining and sequence analysis of gene candidates
Genome mining, to identify genes encoding for enzymes potentially involved in the observed laccase-positive phenotype of the four strains, was performed using amino acid sequences of putative laccases, identified by Ausec and colleagues [27], against the Psychrobacter genome sequences obtained in the present study by using BLAST [65]. In addition, Psychrobacter genome annotations were browsed for genes annotated as laccases, multicopper oxidases (MCOs) and copper-resistance proteins. The DNA sequences mutually detected by both approaches were chosen for further study. Structure modeling and analysis of conserved domains of the studied laccase like multicopper oxidases (LMCOs) was performed using Phyre2 [41] and the laccase from Botrytis aclada (3SQR) as the template. The deduced amino acid sequence encoded by the gene candidates were further analyzed for the presence of a signal peptide using the web based applications SignalP 4.0 server [66], PRED-TAT [67], Phobius [68], and Phyre2 [41]. The level of similarity shared between the sequences was evaluated using the multiple sequence alignment function of the software CloneManager 9 Professional edition (Scientific & Educational Software).
Heterologous expression and activity validation of P11F6-LMCO and P11G5-LMCO2
In order to study the laccase activity of the LMCOs, P11F6-LMCO and P11G5-LMCO2 sharing the minimum homology (Table 3) were chosen as representatives. The region encoding mature protein, excluding the signal peptide predicted by SignalP, was heterologously expressed in Expresso™ T7 cloning and expression system (Lucigen). Cloning procedure was carried out according to manufacturer’s instructions. Expression was performed by inoculating 1 % of overnight-grown culture into 50 mL terrific broth medium [69] supplemented with 0.25 mM CuCl2 followed by incubation at 37 °C and 200 rpm until it reached OD600: ~1.0. The cultures were then induced by 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated overnight at 10 °C at 200 rpm. The cells were harvested by centrifugation at 4000 × g for 15 min, and were resuspended in 5 mL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) followed by the addition of 1 mg/mL lysozyme and incubation on ice for 30 min. Cells were then disrupted by sonication. The lysate centrifuged at 10000 × g for 20 min. Crude extract was prepared by filtering supernatants through 0.22 μM sterile filters and transferring into a new tube. Crude extract obtained from untransformed E. coli BL21 (DE3) was used as a negative control.
The reaction mixture was prepared in a total volume of 200 μL containing 50 mM Tris–HCl pH 7.0, 0.25 mM CuCl2, and 10 μL of crude extract. Commonly used nonphenolic and phenolic laccase substrates ABTS (1 mM) and guaiacol (10 mM), respectively, were added into separate reaction mixture and the reactions were then monitored at 420 nm and 468 nm, respectively.
Availability of supporting data
The datasets supporting the results of this article are included within the article and its Additional files.
Phylogenetic tree is available in the TreeBASE repository, [http://purl.org/phylo/treebase/phylows/study/TB2:S18786?x-accesscode=fa3db0ef767f25ea6936f6b6651efbd3&format=html].
Acknowledgements
We are thankful to Bjørn Altermark and Nils Peder Willassen for their efforts in establishing the Arctic bacterial collection, and initial 16S rDNA sequencing. The authors thank the anonymous reviewers for their guidance on improving the quality of this article.
Funding
This study was funded by the Norwegian Research Council (grant 192123) to establish the culture collection and genome sequencing of the Psychrobacter strains; and VISTA program (grant 6508) supporting high-throughput screening and heterologous expression studies.
Abbreviations
- 2,6-DMP
2,6-Dimethoxyphenol
- ABTS
2,2'-azino-bis-3-ethylbenzothiazoline-6-sulphonic acid
- ANI
Average Nucleotide Identity
- BLAST
Basic Local Alignment Search Tool
- CDS
Coding DNA Sequence
- DDBJ
DNA Data Bank of Japan
- EC
Enzyme Commission
- EMBL
European Molecular Biology Laboratory
- HTS
High Throughput Screening
- HTG
Horizontally Transferred Genes
- IMG
Integrated Microbial Genomes
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- kbp
Kilobase pair
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG Orthology
- LMS
Laccase Mediator System
- LMCO
Laccase-like Multicopper Oxidase
- PCA
Principal Component Analysis
- RAST
Rapid Annotation Using Subsystem Technology
- RDP
Ribosomal Database Project
- rpm
Revolutions Per Minute
- WGS
Whole Genome Shotgun
Additional files
Dot plot analysis of chromosomal DNA sequences. A) P2G3 vs P11G3, B) P11F6 vs P2G3, C) P2G3 vs P11G5, D) P11F6 vs P11G3, E) P11F6 vs P11G5, F) P11G3 vs P11G5. (PDF 125 kb)
Heat map of selected genes (mentioned in the text) in the genomes of Psychrobacter species based on KEGG Orthology (KO) gene counts. Figure S3. Complete heat map of genes with KO number in the genomes of Psychrobacter species. (PDF 8085 kb)
Predicted amino acid residues forming substrate binding pocket. (PDF 87 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MSM and RL conceived and designed the study; MSM carried out all laboratory work; AA and AWi contributed to genome sequencing; LC conducted high-throughput screening and identification of the candidate isolates; KR involved in bioinformatics analysis; JK and CR carried out genome sequence assembly, annotation and comparative genomics analysis. MSM and RL wrote the paper with contributions from AWe and MFHM. All authors read and approved the manuscript in its final version.
Contributor Information
Morteza Shojaei Moghadam, Email: morteza.s.moghadam@ntnu.no.
Andreas Albersmeier, Email: aalbersm@cebitec.uni-bielefeld.de.
Anika Winkler, Email: awinkler@cebitec.uni-bielefeld.de.
Lorenzo Cimmino, Email: lore.cimmino@gmail.com.
Kjersti Rise, Email: kjersti.rise@ntnu.no.
Martin Frank Hohmann-Marriott, Email: martin.hohmann-marriott@ntnu.no.
Jörn Kalinowski, Email: joern@cebitec.uni-bielefeld.de.
Christian Rückert, Email: cruecker@mit.edu.
Alexander Wentzel, Email: Alexander.Wentzel@sintef.no.
Rahmi Lale, Email: rahmi.lale@ntnu.no.
References
- 1.Junge DK, Christner B, Staley DJT. Diversity of Psychrophilic Bacteria from Sea Ice - and Glacial Ice Communities. In: Horikoshi K, editor. Extremophiles Handbook. Japan: Springer; 2011. pp. 793–815. [Google Scholar]
- 2.Kopf A, Bicak M, Kottmann R, Schnetzer J, Kostadinov I, Lehmann K, Fernandez-Guerra A, Jeanthon C, Rahav E, Ullrich M, Wichels A, Gerdts G, Polymenakou P, Kotoulas G, Siam R, Abdallah RZ, Sonnenschein EC, Cariou T, O’Gara F, Jackson S, Orlic S, Steinke M, Busch J, Duarte B, Caçador I, Canning-Clode J, Bobrova O, Marteinsson V, Reynisson E, Loureiro CM, et al. The ocean sampling day consortium. GigaScience. 2015;4:27. [DOI] [PMC free article] [PubMed]
- 3.Pace NR. The universal nature of biochemistry. Proc Natl Acad Sci. 2001;98:805–808. doi: 10.1073/pnas.98.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sengupta D, Chattopadhyay MK. Metabolism in bacteria at low temperature: A recent report. J Biosci. 2013;38:409–412. doi: 10.1007/s12038-013-9306-y. [DOI] [PubMed] [Google Scholar]
- 5.Adapa V, Ramya LN, Pulicherla KK, Rao KRSS. Cold active pectinases: advancing the food industry to the next generation. Appl Biochem Biotechnol. 2014;172:2324–2337. doi: 10.1007/s12010-013-0685-1. [DOI] [PubMed] [Google Scholar]
- 6.Cavicchioli R, Charlton T, Ertan H, Omar SM, Siddiqui KS, Williams TJ. Biotechnological uses of enzymes from psychrophiles. Microb Biotechnol. 2011;4:449–460. doi: 10.1111/j.1751-7915.2011.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalmaso GZL, Ferreira D, Vermelho AB. Marine extremophiles: a source of hydrolases for biotechnological applications. Mar Drugs. 2015;13:1925–1965. doi: 10.3390/md13041925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiangwa J, Ali MSM, Salleh AB, Rahman RNZRA, Shariff FM, Leow TC. Adaptational properties and applications of cold-active lipases from psychrophilic bacteria. Extrem Life Extreme Cond. 2015;19:235–247. doi: 10.1007/s00792-014-0710-5. [DOI] [PubMed] [Google Scholar]
- 9.Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ Microbiol. 2007;9:177–186. doi: 10.1111/j.1462-2920.2006.01126.x. [DOI] [PubMed] [Google Scholar]
- 10.Riva S. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 2006;24:219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60:551–565. doi: 10.1016/S0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- 12.Bourbonnais R, Paice MG. Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267:99–102. doi: 10.1016/0014-5793(90)80298-W. [DOI] [PubMed] [Google Scholar]
- 13.Bajpai P. Application of enzymes in the pulp and paper industry. Biotechnol Prog. 1999;15:147–157. doi: 10.1021/bp990013k. [DOI] [PubMed] [Google Scholar]
- 14.Rochefort D, Leech D, Bourbonnais R. Electron transfer mediator systems for bleaching of paper pulp. Green Chem. 2004;6:14. doi: 10.1039/b311898n. [DOI] [Google Scholar]
- 15.Claus H, Faber G, König H. Redox-mediated decolorization of synthetic dyes by fungal laccases. Appl Microbiol Biotechnol. 2002;59:672–678. doi: 10.1007/s00253-002-1047-z. [DOI] [PubMed] [Google Scholar]
- 16.Faraco V, Pezzella C, Miele A, Giardina P, Sannia G. Bio-remediation of colored industrial wastewaters by the white-rot fungi Phanerochaete chrysosporium and Pleurotus ostreatus and their enzymes. Biodegradation. 2009;20:209–220. doi: 10.1007/s10532-008-9214-2. [DOI] [PubMed] [Google Scholar]
- 17.Niu J, Dai Y, Guo H, Xu J, Shen Z. Adsorption and transformation of PAHs from water by a laccase-loading spider-type reactor. J Hazard Mater. 2013;248–249:254–260. doi: 10.1016/j.jhazmat.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Naranjo-Briceño L, Pernía B, Guerra M, Demey JR, De Sisto A, Inojosa Y, González M, Fusella E, Freites M, Yegres F. Potential role of oxidative exoenzymes of the extremophilic fungus Pestalotiopsis palmarum BM-04 in biotransformation of extra-heavy crude oil. Microb Biotechnol. 2013;6:720–30. [DOI] [PMC free article] [PubMed]
- 19.Galai S, Lucas-Elio P, Marzouki MN, Sanchez-Amat A. Molecular cloning of a copper-dependent laccase from the dye-decolorizing strain Stenotrophomonas maltophilia AAP56. J Appl Microbiol. 2011;111:1394–1405. doi: 10.1111/j.1365-2672.2011.05164.x. [DOI] [PubMed] [Google Scholar]
- 20.Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis Endospore Coat. J Biol Chem. 2002;277:18849–59. [DOI] [PubMed]
- 21.Reiss R, Ihssen J, Richter M, Eichhorn E, Schilling B, Thöny-Meyer L. Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PLoS ONE. 2013;8:e65633. doi: 10.1371/journal.pone.0065633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldrian P. Fungal laccases – occurrence and properties. FEMS Microbiol Rev. 2006;30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- 23.Brijwani K, Rigdon A, Vadlani PV, Brijwani K, Rigdon A, Vadlani PV. Fungal laccases: production, function, and applications in food processing, fungal laccases: production, function, and applications in food processing. Enzyme Res Enzyme Res. 2010;2010:e149748. doi: 10.4061/2010/149748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera-Hoyos CM, Morales-Álvarez ED, Poutou-Piñales RA, Pedroza-Rodríguez AM, RodrÍguez-Vázquez R, Delgado-Boada JM. Fungal laccases. Fungal Biol Rev. 2013;27:67–82. doi: 10.1016/j.fbr.2013.07.001. [DOI] [Google Scholar]
- 25.Sharma P, Goel R, Capalash N. Bacterial laccases. World J Microbiol Biotechnol. 2006;23:823–832. doi: 10.1007/s11274-006-9305-3. [DOI] [Google Scholar]
- 26.Santhanam N, Vivanco JM, Decker SR, Reardon KF. Expression of industrially relevant laccases: prokaryotic style. Trends Biotechnol. 2011;29:480–489. doi: 10.1016/j.tibtech.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Ausec L, Zakrzewski M, Goesmann A, Schlüter A, Mandic-Mulec I. Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes. PLoS ONE. 2011;6:e25724. doi: 10.1371/journal.pone.0025724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh G, Bhalla A, Kaur P, Capalash N, Sharma P. Laccase from prokaryotes: a new source for an old enzyme. Rev Environ Sci Biotechnol. 2011;10:309–326. doi: 10.1007/s11157-011-9257-4. [DOI] [Google Scholar]
- 29.Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G. Laccases: a never-ending story. Cell Mol Life Sci. 2009;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 31.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudo T, Kidera A, Kida M, Kawauchi A, Shimizu R, Nakahara T, Zhang X, Yamada A, Amano M, Hamada Y, Taniyama S, Arakawa O, Yoshida A, Oshima K, Suda W, Kuwahara H, Nogi Y, Kitamura K, Yuki M, Iida T, Moriya S, Inoue T, Hongoh Y, Hattori M, Ohkuma M. Draft genome sequences of Psychrobacter strains JCM 18900, JCM 18901, JCM 18902, and JCM 18903, isolated preferentially from frozen aquatic organisms. Genome Announc. 2014;2 [DOI] [PMC free article] [PubMed]
- 33.Krastanov A, Alexieva Z, Yemendzhiev H. Microbial degradation of phenol and phenolic derivatives. Eng Life Sci. 2013;13:76–87. doi: 10.1002/elsc.201100227. [DOI] [Google Scholar]
- 34.Chen D, Tang H, Lv Y, Zhang Z, Shen K, Lin K, Zhao Y-L, Wu G, Xu P. Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside. Mol Microbiol. 2013;87:1237–44. [DOI] [PubMed]
- 35.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A. 2010;107:14390–5. [DOI] [PMC free article] [PubMed]
- 36.Navarro-Llorens JM, Patrauchan MA, Stewart GR, Davies JE, Eltis LD, Mohn WW. Phenylacetate catabolism in Rhodococcus sp. Strain RHA1: a central pathway for degradation of aromatic compounds. J Bacteriol. 2005;187:4497–504. [DOI] [PMC free article] [PubMed]
- 37.Scarlett FA, Turner JM. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J Gen Microbiol. 1976;95:173–176. doi: 10.1099/00221287-95-1-173. [DOI] [PubMed] [Google Scholar]
- 38.Stojiljkovic I, Bäumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer R, Williams C, Miller C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol. 2003;185:6556–6561. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam ST, Lam JS. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ Microbiol. 2013;15:1001–1015. doi: 10.1111/j.1462-2920.2012.02890.x. [DOI] [PubMed] [Google Scholar]
- 41.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 42.Margesin R, Miteva V. Diversity and ecology of psychrophilic microorganisms. Res Microbiol. 2011;162:346–361. doi: 10.1016/j.resmic.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Maruyama A, Honda D, Yamamoto H, Kitamura K, Higashihara T. Phylogenetic analysis of psychrophilic bacteria isolated from the Japan Trench, including a description of the deep-sea species Psychrobacter pacificensis sp. nov. Int J Syst Evol Microbiol. 2000;50(Pt 2):835–846. doi: 10.1099/00207713-50-2-835. [DOI] [PubMed] [Google Scholar]
- 44.Givaudan A, Effosse A, Faure D, Potier P, Bouillant M-L, Bally R. Polyphenol oxidase in Azospirillum lipoferum isolated from rice rhizosphere: Evidence for laccase activity in non-motile strains of Azospirillum lipoferum. FEMS Microbiol Lett. 1993;108:205–210. doi: 10.1111/j.1574-6968.1993.tb06100.x. [DOI] [Google Scholar]
- 45.Pangallo D, Šimonovičová A, Chovanová K, Ferianc P. Wooden art objects and the museum environment: identification and biodegradative characteristics of isolated microflora. Lett Appl Microbiol. 2007;45:87–94. doi: 10.1111/j.1472-765X.2007.02138.x. [DOI] [PubMed] [Google Scholar]
- 46.Jóelsson JP, Friðjónsdóttir H, Vilhelmsson O. Bioprospecting a glacial river in Iceland for bacterial biopolymer degraders. Cold Reg Sci Technol. 2013;96:86–95. doi: 10.1016/j.coldregions.2013.03.001. [DOI] [Google Scholar]
- 47.Ye M, Li G, Liang WQ, Liu YH. Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and highly soluble expression. Appl Microbiol Biotechnol. 2010;87:1023–1031. doi: 10.1007/s00253-010-2507-5. [DOI] [PubMed] [Google Scholar]
- 48.Kellner H, Luis P, Zimdars B, Kiesel B, Buscot F. Diversity of bacterial laccase-like multicopper oxidase genes in forest and grassland Cambisol soil samples. Soil Biol Biochem. 2008;40:638–648. doi: 10.1016/j.soilbio.2007.09.013. [DOI] [Google Scholar]
- 49.Claus H. Laccases: structure, reactions, distribution. Micron. 2004;35:93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 50.Pardo I, Camarero S. Laccase engineering by rational and evolutionary design. Cell Mol Life Sci. 2015;72:897–910. doi: 10.1007/s00018-014-1824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galai S, Korri-Youssoufi H, Marzouki MN. Characterization of yellow bacterial laccase SmLac/role of redox mediators in azo dye decolorization. J Chem Technol Biotechnol. 2014;89:1741–1750. doi: 10.1002/jctb.4254. [DOI] [Google Scholar]
- 52.Xu F, Berka RM, Wahleithner JA, Nelson BA, Shuster JR, Brown SH, Palmer AE, Solomon EI. Site-directed mutations in fungal laccase: effect on redox potential, activity and pH profile. Biochem J. 1998;334(Pt 1):63–70. [DOI] [PMC free article] [PubMed]
- 53.Mærk M, Johansen J, Ertesvåg H, Drabløs F, Valla S. Safety in numbers: multiple occurrences of highly similar homologs among Azotobacter vinelandii carbohydrate metabolism proteins probably confer adaptive benefits. BMC Genomics. 2014;15:192. doi: 10.1186/1471-2164-15-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bredholdt H, Galatenko OA, Engelhardt K, Fjaervik E, Terekhova LP, Zotchev SB. Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ Microbiol. 2007;9:2756–64. [DOI] [PubMed]
- 55.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 56.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 57.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(Database issue):D633–42. [DOI] [PMC free article] [PubMed]
- 58.Gordon D, Green P. Consed: a graphical editor for next-generation sequencing. Bioinformatics. 2013;29:2936–2937. doi: 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(Web Server issue):W686–9. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A. GenDB--an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–95. [DOI] [PMC free article] [PubMed]
- 63.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:75. [DOI] [PMC free article] [PubMed]
- 64.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D115–22. [DOI] [PMC free article] [PubMed]
- 65.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu Y-K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–9. [DOI] [PMC free article] [PubMed]
- 66.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 67.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics. 2010;26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 68.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35(Suppl 2):W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tartof KD, Hobbs CA. Improved media for growing plasmid and cosmid clones. Focus. 1987;9:12. [Google Scholar]


