Abstract
Listeria monocytogenes is a dangerous bacterium that causes the food-borne disease listeriosis and accounts for nearly 20 % of food-borne deaths. This organism can survive the body's natural defences within the digestive tract, including acidic conditions and bile. Although the bile response has been analysed, limited information is available concerning the ability of L. monocytogenes to resist bile under anaerobic conditions, especially at acidic pH, which mimics conditions within the duodenum. Additionally, it is not known how the bile response varies between serotypes. In this study, the survival of strains representing six serotypes was analysed under aerobic and anaerobic conditions following exposure to bile. Exposure to bile salts at acidic pH increased toxicity of bile, resulting in a significant reduction in survival for all strains tested. However, following this initial reduction, no significant reduction was observed for an additional 2 h except for strain 10403S (P = 0.002). Anaerobic cultivation increased bile resistance, but a significant increase was only observed in virulent strains when exposed to bile at pH 5.5. Exposure to pH 3.0 prior to bile decreased viability amongst avirulent strains in bile in acidic conditions; oxygen availability did not influence viability. Together, the data suggested that being able to sense and respond to oxygen availability may influence the expression of stress response mechanisms, and this response may correspond to disease outcome. Further research is needed on additional strains to determine how L. monocytogenes senses and responds to oxygen and how this varies between invasive and non-invasive strains.
Introduction
Listeria monocytogenes is a Gram-positive, food-borne pathogen that is responsible for nearly 20 % of food-related deaths in the USA (Scallan et al., 2011). This microbe causes the opportunistic infection listeriosis and primarily affects pregnant women, the elderly and the immunocompromised (Farber & Peterkin, 1991). Upon ingestion, L. monocytogenes must survive a variety of stressors, including acidic conditions of the stomach, bile encountered within the gastrointestinal tract and hypoxic/anoxic conditions. Being able to efficiently respond to these stressors is key for survival within the host.
Following exposure to the acidic conditions within the stomach, L. monocytogenes is exposed to bile secreted into the duodenum by the liver during digestion (Monte et al., 2009). This complex fluid is composed primarily of bile acids, cholesterol, phospholipids and bilirubin (Coleman et al., 1979). Bile acids are the bactericidal component of bile, being able to disrupt the cell membrane and DNA (Bernstein et al., 1999; Coleman et al., 1979; Prieto et al., 2004, 2006). The primary bile acids (cholic acid and chenodeoxycholic acid) are formed from cholesterol in the liver and are subsequently conjugated with either a glycine or taurine prior to being secreted into the small intestine. Secondary bile acids (deoxycholic acid and lithocholic acid) can form due to dehydroxylation reactions performed by gut microbes (Merritt & Donaldson, 2009). Peptide linkages between secondary bile acids and glycine or taurine also form conjugated bile acids, such as glycodeoxycholic acid (GDCA) and taurodeoxycholic acid (TDCA). Human bile is primarily composed of these conjugated bile acids (Shioda et al., 1969).
The toxicity of bile is dependent upon pH (Begley et al., 2002). For example, the L. monocytogenes strain EGDe (serotype 1/2a) had decreased survival in ex vivo bile at pH 5.5 in comparison with pH 7. Survival under these acidic conditions was found to be dependent upon the sigma stress factor (sig B), principal virulence factor (prfA), bile salt hydrolase (bsh) and bile exclusion system (bilE). These resistance mechanisms, however, were not necessary for survival in bile at pH 7, which would be encountered within the gall bladder (Dowd et al., 2011). Hypoxic conditions have been linked to increased expression of bsh, which may influence bile resistance under anaerobic conditions (Dussurget et al., 2002). However, the L. monocytogenes strain LO28 (serotype 1/2c) had increased sensitivity to bile acids at pH 5.5 in comparison with pH 7.5 when cultured under anaerobic conditions (Begley et al., 2002). These data suggest that availability of oxygen may influence bile resistance, but the impact on survival may vary between strains.
Although much is known concerning the response of L. monocytogenes to stressors encountered within the gastrointestinal tract, little is understood in regard to these responses in anaerobic conditions, especially with respect to variations between serotypes. Therefore, the goal of this study was to examine the effect of pH on bile toxicity of different serotypes in the presence or absence of oxygen.
Methods
Bacterial strains
Bacterial strains used in this study are listed in Table 1. Frozen stocks were stored at − 80 °C and grown on Tryptic Soy Agar (TSA) prior to cultivation in Tryptic Soy Broth (TSB) at 37 °C.
Table 1. Strains used in this study.
| Strain | Serotype | Isolation source |
|---|---|---|
| F2365 | 4b | Human – epidemic |
| EGDe | 1/2a | Animal – sporadic |
| 10403S | 1/2a | Human – sporadic |
| 15313 | 1/2a | Animal – sporadic |
| HCC23 | 4a | Catfish |
| HCC7 | 1 | Catfish |
| 2011L-2663 | 1/2b | Human – cantaloupe outbreak |
| 2011L-2676 | 1/2a | Human – cantaloupe outbreak |
| ScottA | 4b | Human – epidemic |
| LO28 | 1/2c | Human – sporadic |
Survival analysis in bile
Overnight cultures were inoculated into fresh TSB and allowed to grow to mid exponential phase (OD600 0.35–0.4) at 37 °C, at which point cells were pelleted (8000 g for 5 min) and resuspended in fresh media at pH 7.5 or 5.5 supplemented with either 0 or 1 % porcine bile extract (Sigma). Aliquots (0.1 ml) were removed at 0, 1, 2 and 3 h post-exposure to bile extract, serially diluted in PBS, and plated onto TSA. Plates were incubated at 37 °C for 16 h prior to enumeration. A minimum of three independent replicates were performed. The log10c.f.u. ml− 1 values were calculated and the mean determined amongst the replicates. Percent viability was assessed based on non-bile-treated samples for each pH condition tested. GraphPad Prism was used to analyse the differences between pH on growth using an unpaired t-test, with significance declared at P < 0.05. For correlation analysis, survival was first normalized against the starting concentration at time 0, then compared with growth over time between strains using Pearson's correlation (xlstat version 2015 1.01; Addinsoft), with P < 0.05 indicating significance.
pH dependency on bile extract, GDCA and TDCA toxicity
Isolates from freshly streaked cultures were used to inoculate 5 ml TSB for cultivation at 16 h at 37 °C. This culture was added to fresh media (1 % inoculum) with 1 % porcine bile extract (Sigma), 5 mM GDCA (Sigma) or 5 mM TDCA (Sigma) at pH 7.5, 6.5, 5.5 or 4.5, adjusted with 10 N HCl where needed. Cultures were incubated for 16 h at 37 °C, at which time aliquots (0.1 ml) were serially diluted in PBS, plated onto TSA and incubated for 16 h prior to enumeration. The log10c.f.u. ml− 1 values were calculated and the mean determined amongst three independent replicates. Percent survival was assessed based on non-bile-treated samples for each pH condition tested. Prism (GraphPad) was used to analyse the differences between pH on growth within each treatment using an unpaired t-test, with P < 0.05 declared as significant.
Anaerobic cultivation
Media to be used for anaerobic cultivation were prepared at least 2 days prior to use. Briefly, TSB was adjusted to pH 7.5, 6.5, 5.5 or 4.5, autoclaved and immediately placed in a Coy Anaerobic Airlock chamber (gas mix 95 % N2/5 % H2). Anaerobic conditions were monitored throughout the duration of the study with an oxygen monitor and also with a resazurin control sample.
Isolates from freshly streaked cultures were used to inoculate 5 ml TSB for cultivation at 16 h at 37 °C. Overnight cultures (1 %) were used to inoculate the TSB previously prepared and then were subsequently supplemented with either 0 or 1 % porcine bile extract (Sigma). Cultures were incubated at 37 °C within the anaerobic chamber for 16 h, after which aliquots (0.1 ml) were removed, serially diluted in PBS and plated onto TSA. Plates were incubated anaerobically for 24 h at 37 °C. Per cent survival was determined for each replication based on TSB media only controls. Prism (GraphPad) was used to analyse the differences between the survival of strains in bile under aerobic or anaerobic conditions using an unpaired t-test, with P < 0.05 declared as significant. At least three independent replicates were analysed.
Acid cross-protection to bile assay
Overnight cultures were diluted 1 : 100 in fresh TSB and allowed to incubate to late exponential phase (OD600 0.8), at which time media were exchanged for fresh TSB at pH 7.5 or 3.0. Cells were exposed to the acidic condition for 1 h, after which cells were pelleted, washed in PBS and resuspended in fresh TSB supplemented with either 0 or 1 % porcine bile extract at pH 7.5 or 5.5 for 16 h at 37 °C. Samples were serially diluted in PBS and plated onto TSA. Experiments analysed under anaerobic conditions were performed exactly as described for aerobic conditions, with the exception of incubation in a Coy anaerobic chamber (gas mix 95 % N2/5 % H2). Per cent survival was calculated based on the viability of controls not treated with bile extract, with respect to appropriate pH treatment. Prism (GraphPad) was used to analyse the impact of pretreatment with acid on bile survival using an unpaired t-test, with P < 0.05 declared as significant. A minimum of three independent replicates were analysed.
Results and Discussion
Survival of L. monocytogenes in bile
The toxicity of bile is directly related to pH (Begley et al., 2002). However, it is not known whether the severity of this bactericidal effect is similar amongst different serotypes of L. monocytogenes. Variation has been observed in bile (King et al., 2003; Merritt et al., 2010; Payne et al., 2013) and acid (King et al., 2003; Koutsoumanis et al., 2003; Liu et al., 2005; Melo et al., 2013; Olesen et al., 2009) resistance between different strains of L. monocytogenes. Due to this variation, it is imperative to analyse the biological response utilizing a multi-strain approach. Listeria monocytogenes consists of four genetic lineages (Orsi et al., 2011). Strains used in this study were chosen based on serotype and genetic lineage, as well as isolation source. Six of the 13 serotypes are represented in the current study. Genetic lineage I strains tested were F2365, ScottA and 2011L-2663. Genetic lineage II strains tested were EGDe, 10403S, 15313, 2011L-2676 and LO28. These two genetic lineages represent serotypes primarily associated with human cases of listeriosis and 95 % of the characterized isolates collected from recent outbreaks (1/2a, 1/2b and 4b), although variations exist in their pathogenic potential. For instance, F2365 (serotype 4b), 2011L-2663 (serotype 1/2b) and 2011L-2676 (serotype 1/2a) were isolated from two of the deadliest listeriosis outbreaks in history (Laksanalamai et al., 2012; Linnan et al., 1988). However, 15313 is avirulent in a mouse model, although it is a serotype 1/2a strain that contains genes related to virulence (Erdenlig et al., 2000; Kathariou & Pine, 1991; Liu et al., 2003). The 10403S strain also has reduced virulence in comparison with EGDe (Bécavin et al., 2014). Genetic lineage III and IV isolates are rare and primarily associated with animals; HCC23 and HCC7 were chosen to represent these lineages. The evolutionary divergence between these serotypes has been described by others (Ragon et al., 2008; Rychli et al., 2014). Although many genomic comparisons have been conducted to decipher the virulent genome, limited information is known about the physiological response of these different strains to stressors encountered within the human gastrointestinal tract. Even within lineages, some genes can be lost, modifying the ability of some strains to adapt to certain environments. For instance, proteomic analysis supports that bile salt tolerance varies amongst strains and regulation of stress response differs between serotypes (Payne et al., 2013). Additionally, genetic comparisons and evolution of the bacteria have been proposed to result in a loss of virulence (den Bakker et al., 2010; Ragon et al., 2008). Therefore, it is imperative to analyse multiple strains to gain a better understanding of how invasive strains and non-invasive strains respond to the host's environment and how this influences the outcome of disease. However, it is noted that this study is still very limited in terms of strain selection, as only 10 strains were analysed.
Bile resistance is one of the key aspects to the infectious potential of L. monocytogenes (Gahan & Hill, 2014; Merritt & Donaldson, 2009). To determine the impact bile has on the growth of L. monocytogenes when in the more toxic environment of reduced pH, the survival of 10 different strains was determined hourly post-exposure to bile at pH 7.5 or 5.5 (Fig. 1). Viability was assessed at 1, 2 and 3 h post-exposure to bile; percent viability for each condition was determined based on the viable cell counts of non-bile-treated samples for individual time points. Exposure to 1 % bile at pH 7.5 decreased viability for 2011L-2676, 10403S, LO28 and 15313 during the 3 h time period analysed (P < 0.05). This was unexpected, as the other strains that represent genetic lineages I and II did not exhibit a significant decrease in survival under these conditions (P>0.05). In fact, F2365 showed a significant increase in viability within 3 h post-exposure to bile (P < 0.001).
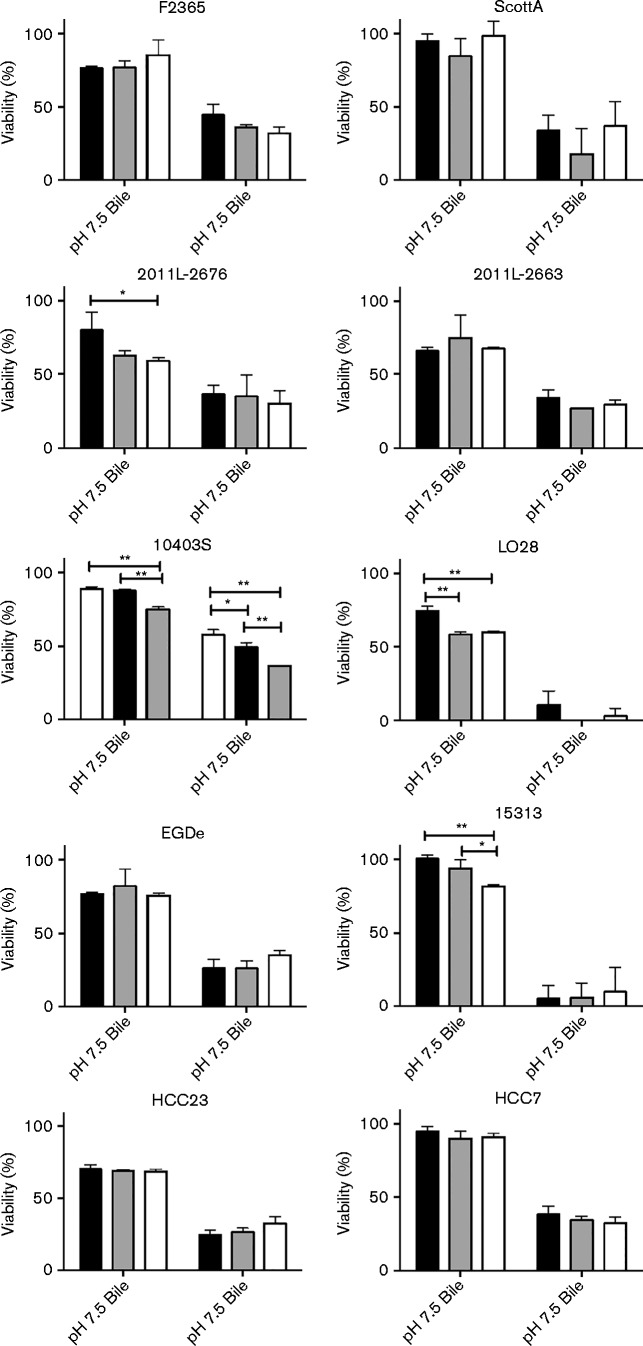
Fig. 1.
Viability of L. monocytogenes in porcine bile extract at pH 7.5 and 5.5. L. monocytogenes F2365, ScottA, 2011L-2636, 2011L-2676, 10403S, 15313, HCC23, HCC7, LO28 and EGDe were grown to mid exponential phase prior to treatment for 3 h in the following conditions: TSB pH 7.5 with either 0 or 1 % bile, or TSB pH 5.5 with either 0 or 1 % bile. Percent viability was calculated at 1 (black), 2 (grey) and 3 h (white) post-exposure to bile in relation to TSB-only controls at the respective pH. Results represent mean ± sd of three independent experiments. *P < 0.05; **P < 0.001.
Growth at pH 5.5 was only impacted for 2011L-2676 (P < 0.05). Bile at pH 5.5 increased toxicity against all strains tested (P < 0.005; Fig. 1). However, the addition of bile did not impact survival significantly over the 3 h time course in comparison with TSB (pH 5.5)-only controls, indicating that the impact on viability was primarily due to the reduction in pH. In fact, only 10403S exhibited a decrease in survival following exposure to bile at pH 5.5 over the 3 h time course analysed (P < 0.05). Viability was not impacted for F2365, ScottA, 2011L-2676, 2011L-2663, LO28, EGDe, 15313, HCC23 and HCC7 (Fig. 1). Interestingly, ScottA, 15313, EGDe and HCC23 showed a slight increase in growth following exposure to bile, although this increase was not significant (Fig. 1). This suggests that 10403S has an impaired ability to efficiently respond to bile in acidic conditions.
The survival exhibited by certain strains of L. monocytogenes following exposure to bile at acidic pH could indicate the emergence of acid-resistant subpopulations that has been noted by others (Metselaar et al., 2013). Resistant subpopulations have been noted for LO28 following exposure to pH 3.5. Therefore, it is possible that the viability observed following exposure of mid-exponential-phase cells to bile at pH 5.5 was due to selected acid-resistant populations. Additional data are needed to analyse the impact of these subpopulations on extended exposure to bile.
Analysis of the viable bacterial populations following exposure to bile at reduced pH normalized against the concentration prior to exposure indicated that a correlation existed between the survival of ScottA with 2011L-2676 (P = 0.044) and HCC23 with EGDe (P = 0.049). Although these strains represent two different genetic lineages, this suggests that they may utilize a similar response mechanism in the presence of bile. As ScottA and EGDe have been found to have different glutamate decarboxylase activities, the impact of pH on bile resistance may be attributed to variations in acid resistance mechanisms (Olier et al., 2004).
Sensitivity of L. monocytogenes to TDCA and GDCA
Human bile is primarily composed of conjugated bile acids (Coleman et al., 1979; Shioda et al., 1969). Therefore, as variations were observed in survival in bile extract at reduced pH, the survival of these strains was analysed in media supplemented with the bile acids GDCA and TDCA at pH 7.5, 6.5, 5.5, or 4.5. GDCA at the concentration of 5 mM has been previously shown to be more toxic than 5 mM TDCA at reduced pH (Begley et al., 2002; De Smet et al., 1995). As expected, survival was severely impaired for all strains analysed after cultivation in media at pH 5.5 or 4.5 with GDCA for 16 h (Fig. 2); only F2365 showed detectable growth at pH 5.5 and 4.5. Bile acid toxicity under acidic conditions has been linked to cell death through the release of reactive oxygen species in eukaryotic cells (Jenkins et al., 2007), but has not been assessed under these conditions in bacteria. The impact of GDCA toxicity on energy demand by Listeria needs to be assessed.
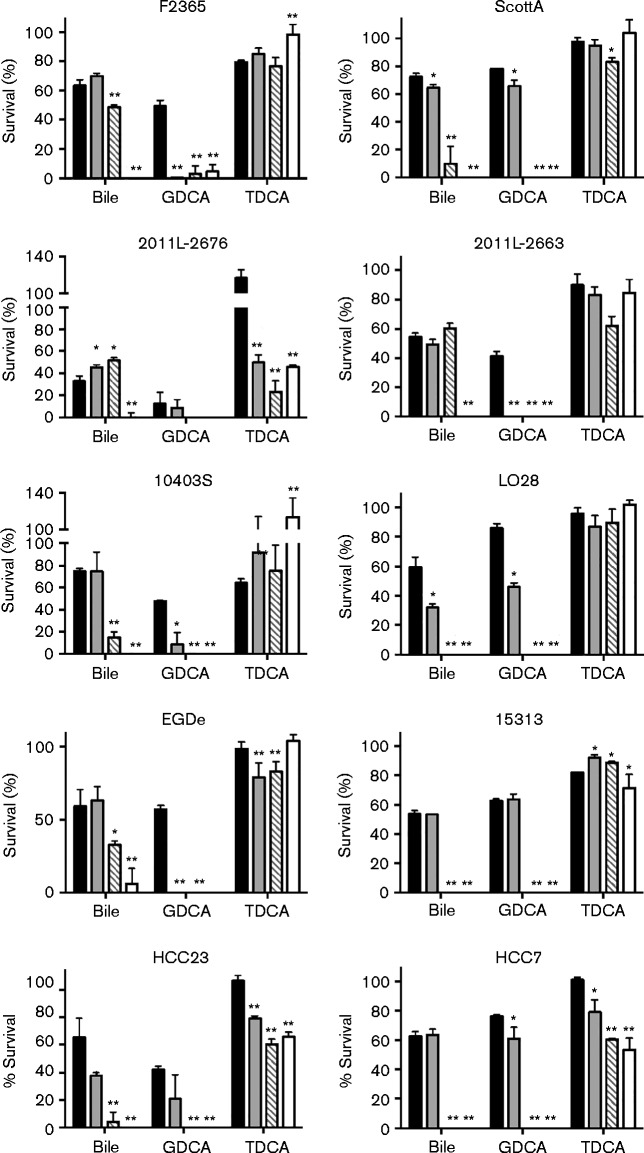
Fig. 2.
Influence of pH on toxicity of GDCA and TDCA. L. monocytogenes F2365, ScottA, 2011L-2636, 2011L-2676, 10403S, 15313, HCC23, HCC7, LO28 and EGDe were cultured in TSB at pH 7.5 (black), 6.5 (grey), 5.5 (hatched) or 4.5 (white). Cultures were grown for 16 h in media supplemented with 1 % porcine bile extract, 5 mM GDCA or 5 mM TDCA. Viability was assessed by plate counts and per cent survival was determined in relation to TSB-only controls at the respective pH. Results represent mean ± sd of three independent experiments. *P < 0.05; **P < 0.001.
The toxic effect that TDCA had on survival of L. monocytogenes varied between strains (Fig. 2). A reduction in survival was only evident for 2011L-2676, 15313, HCC23 and HCC7 (P < 0.05). Although two of the four 1/2a strains tested showed a significant decrease in survival at pH 4.5 in comparison with the media-only control (2011L-2676 and 15313), all other strains without a significant difference belonged to genetic lineages I and II.
Extended exposure to bile extract at reduced pH severely impacted survival. All strains tested showed a significant decrease in viability at pH 4.5. Strains that were isolated from the 2011 cantaloupe outbreak actually showed an increase in bile resistance at pH 5.5 in comparison with neutral pH. Together, these data suggest that an additional component to the bile may influence the survival of L. monocytogenes or that the influence on survival is strain specific. It is possible that the mechanism utilized may differ between strains and that this response may influence the outcome of disease. Further research is needed to dissect these two possibilities.
Anaerobic conditions increase survival of L. monocytogenes against bile salts at reduced pH
A previous study indicated that the activity of bile salt hydrolase increases under anaerobic conditions (Dussurget et al., 2002). As variations were observed in the survival of L. monocytogenes when exposed to bile at reduced pH, the influence of anaerobic conditions was tested on bile resistance. Strains representing various serotypes were analysed following 16 h cultivation in media supplemented with 1 % porcine bile extract at pH 7.5, 6.5, 5.5 or 4.5 (Fig. 3). At pH 7.5 under anaerobic conditions, F2365, 2011L-2663, 2011L-2676, LO28, 15313 and HCC7 showed a significant increase in survival in comparison with growth under aerobic conditions (P < 0.05). Contrary to previous studies, increased sensitivity to bile at pH 7.5 was not observed in EGDe in relation to other Listeria strains, particularly LO28 and ScottA (Begley et al., 2002). This difference is most likely due to variations in the cultivation methods used in this study in comparison with the previous study.
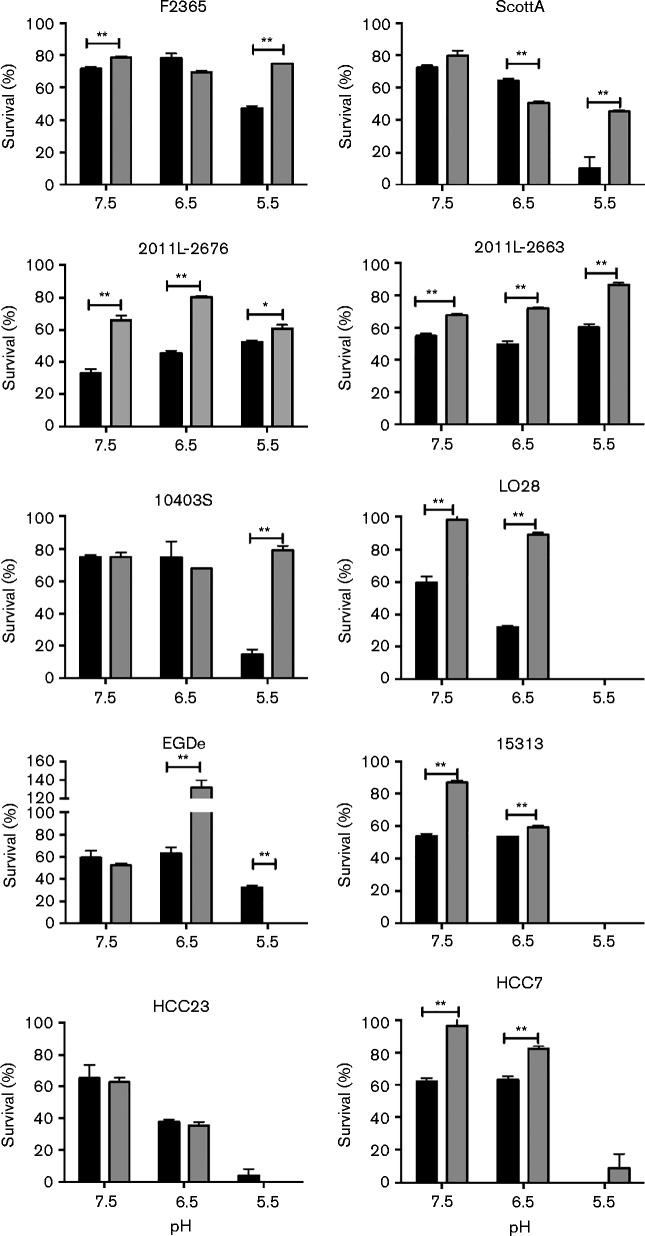
Fig. 3.
Influence of anaerobic cultivation on bile resistance of L. monocytogenes. L. monocytogenes F2365, ScottA, 2011L-2636, 2011L-2676, 10403S, 15313, HCC23, HCC7, LO28 and EGDe were cultured in media supplemented with either 0 or 1 % bile under either aerobic (black) or anaerobic (grey) conditions. Percent survivals were determined in relation to media-only controls at the respective pH. Results represent mean ± sd of three independent experiments. *P < 0.05; **P < 0.001.
Interestingly, strains isolated from epidemics of listeriosis all showed a significant increase in resistance at pH 5.5 under anaerobic conditions. F2365, ScottA, 2011L-2676, 2011L-2663 and 10403S showed an increase in survival at pH 5.5 in the presence of bile under anaerobic conditions (P < 0.05). This is very interesting to note, as this mimics the environment that L. monocytogenes would be exposed to when entering the duodenum. Survival in this part of the small intestine may allow for the upregulation of stress response mechanisms that will enhance invasiveness at deeper parts of the intestinal tract. This is in agreement with studies that have indicated that anaerobic cultivation increases the invasion potential of L. monocytogenes (Bo Andersen et al., 2007). The only strains that showed growth following 16 h incubation in media at pH 4.5 with bile were 2011L-2676 and EGDe. Anaerobic conditions did not influence the survival of these strains at pH 4.5 (P>0.05; data not shown).
Pre-exposure to acid does not improve bile survival of L. monocytogenes
The first stressor that L. monocytogenes encounters following ingestion is the acidic condition of the stomach. In addition to the alteration in pH, this also marks the first point of transition to a hypoxic environment (He et al., 1999). Although L. monocytogenes possesses acid tolerance mechanisms, the impact that exposure to pH 3.0 has on survival following exposure to additional stressors encountered within the gastrointestinal tract is not known, especially with regard to reduced oxygen availability. To determine if exposure to acidic pH 3.0, which closely mimics that of the stomach, can enhance survival under conditions encountered within the duodenum, various L. monocytogenes strains were exposed to media at pH 3.0 for 1 h, followed by exposure to media at pH 7.5 or 5.5 supplemented with bile extract. However, it should be noted that this analysis was performed with TSB at reduced pH, rather than in simulated gastric fluid. Fig. 4 represents the impact that pre-exposure to pH 3.0 has on survival of L. monocytogenes following exposure to media supplemented with bile. Survival was determined based on viability following exposure to bile in comparison with identical treatment without bile. Pretreatment to acidic conditions only increased the survival of HCC23 following exposure to bile at pH 7.5 under aerobic conditions (P < 0.001). For all other strains tested, exposure to the acidic conditions decreased the survival following exposure to bile at pH 7.5. Exposure to acidic conditions under anaerobic conditions did, ironically, stabilize viability of F2365, ScottA, 2011-2676, LO28, EGDe and HCC23 following exposure to bile at pH 7.5.
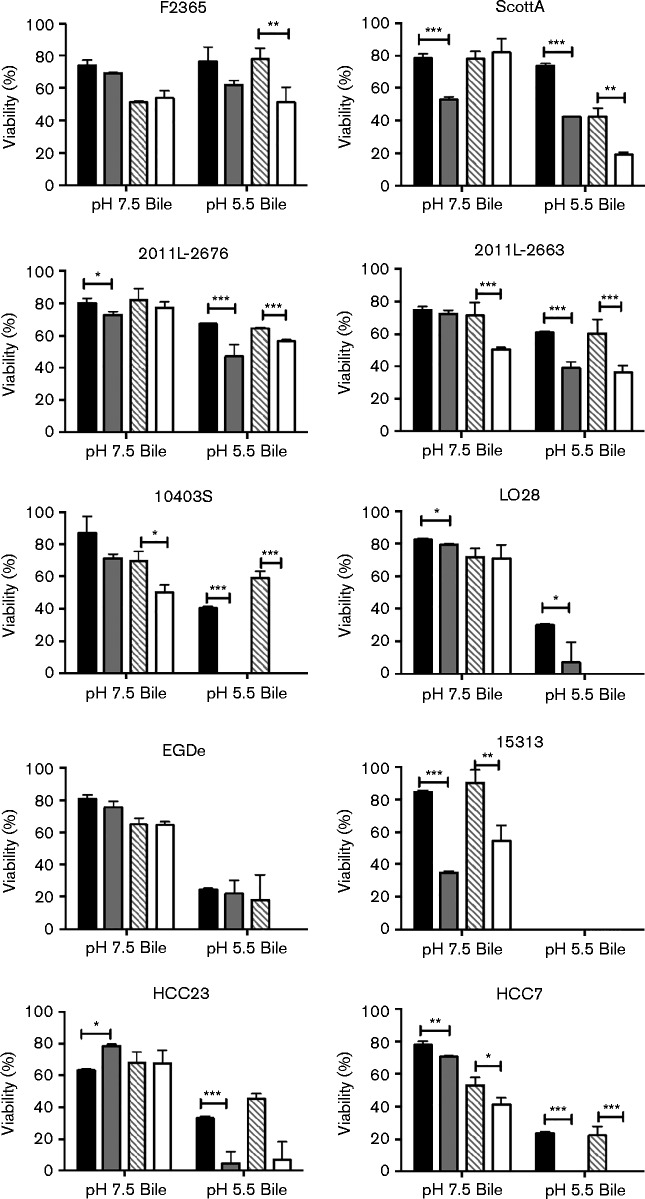
Fig. 4.
Viability of L. monocytogenes in bile when pre-exposed to acidic conditions. L. monocytogenes F2365, ScottA, 2011L-2636, 2011L-2676, 10403S, 15313, HCC23, HCC7, LO28 and EGDe were grown to late exponential phase, exposed to TSB at pH 7.5 or 3.0 for 1 h and then cultured in media at pH 7.5 or 5.5 supplemented with either 0 or 1 % bile for 16 h. Per cent survivals were determined in relation to media-only controls at the respective pH. Aerobic cultures exposed to pH 7.5 media, followed by exposure to bile (black); aerobic cultures exposed to pH 3.0 media, followed by exposure to bile (grey); anaerobic cultures exposed to pH 7.5 media, followed by exposure to bile (hatched); anaerobic cultures exposed to pH 3.0 media, followed by exposure to bile (white). Results represent mean ± sd of three independent experiments. *P < 0.05; **P < 0.001; ***P < 0.0001.
To determine if exposure to acidic conditions encountered within the stomach (pH 3.0) increased the ability of L. monocytogenes to survive conditions encountered in the duodenum (pH 5.5 and bile), viability was assessed following pretreatment with either pH 7.5 or 3.0 prior to treatment with bile at pH 5.5 under aerobic or anaerobic conditions (Fig. 4). None of the strains tested showed an increase in viability in bile when pretreated with media at pH 3.0. This indicates that exposure to acidic conditions does not improve viability of L. monocytogenes against stressors encountered in the gastrointestinal tract. In fact, exposure to acid prior to bile exposure increased the sensitivity of most strains analysed. Additionally, as there was no impact on survival of acid pretreated samples and non-treated samples based on oxygen availability, this provides further support for the involvement of oxygen availability, not acid exposure, in increased resistance against stressors. Tolerance to acidic conditions encountered within the stomach is important for initial survival, but does not seem to provide cross-protection against conditions encountered in the small intestine. However, it is possible that exposure to conditions more closely simulating in vivo conditions, such as simulated gastric fluid, may result in different results. This will need to be further analysed in future studies.
Together, these data suggest that the ability of L. monocytogenes to sense oxygen may influence stress resistance. However, the impact of oxygen availability on bile resistance was not consistent amongst all strains analysed. This could indicate that the expression of stress response mechanisms differs between strains. Although studies have been conducted to analyse the genomic comparisons between serotypes, this study indicates that there is a necessity to decipher the transcriptome of multiple strains in response to stressors encountered under physiologically relevant conditions. It is also imperative that additional strains are analysed in future studies to determine correlations to resistance amongst L. monocytogenes.
Acknowledgements
We would like to thank Drs Frank Austin, Peter Gerner-Smidt, Mark Lawrence, Daniel Portnoy and Zuzana Zucerova for the bacterial strains used in this study. We would like to thank Drs Tjakko Abee and Justin Thornton for their helpful advice concerning this project. This project was funded through the National Institutes of Health (award number P20GM103646).
Abbreviations:
- GDCA
glycodeoxycholic acid
- TDCA
taurodeoxycholic acid
References
- Bécavin C., Bouchier C., Lechat P., Archambaud C., Creno S., Gouin E., Wu Z., Kühbacher A., Brisse S., other authors (2014). Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity MBio 5 e00969–e001014 10.1128/mBio.00969-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M., Gahan C. G., Hill C. (2002). Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance Appl Environ Microbiol 68 6005–6012 10.1128/AEM.68.12.6005-6012.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein C., Bernstein H., Payne C. M., Beard S. E., Schneider J. (1999). Bile salt activation of stress response promoters in Escherichia coli Curr Microbiol 39 68–72 10.1007/s002849900420 . [DOI] [PubMed] [Google Scholar]
- Bo Andersen J., Roldgaard B. B., Christensen B. B., Licht T. R. (2007). Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs BMC Microbiol 7 55 10.1186/1471-2180-7-55 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R., Iqbal S., Godfrey P. P., Billington D. (1979). Membranes and bile formation. Composition of several mammalian biles and their membrane-damaging properties Biochem J 178 201–208 10.1042/bj1780201 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Van Hoorde L., Vande Woestyne M., Christiaens H., Verstraete W. (1995). Significance of bile salt hydrolytic activities of lactobacilli J Appl Bacteriol 79 292–301 10.1111/j.1365-2672.1995.tb03140.x . [DOI] [PubMed] [Google Scholar]
- den Bakker H. C., Cummings C. A., Ferreira V., Vatta P., Orsi R. H., Degoricija L., Barker M., Petrauskene O., Furtado M. R., Wiedmann M. (2010). Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss BMC Genomics 11 688 10.1186/1471-2164-11-688 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd G. C., Joyce S. A., Hill C., Gahan C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile Infect Immun 79 369–379 10.1128/IAI.00330-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O., Cabanes D., Dehoux P., Lecuit M., Buchrieser C., Glaser P., Cossart P. (2002). European Listeria Genome Consortium Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis Mol Microbiol 45 1095–1106 10.1046/j.1365-2958.2002.03080.x . [DOI] [PubMed] [Google Scholar]
- Erdenlig S., Ainsworth A. J., Austin F. W. (2000). Pathogenicity and production of virulence factors by Listeria monocytogenes isolates from channel catfish J Food Prot 63 613–619 . [DOI] [PubMed] [Google Scholar]
- Farber J. M., Peterkin P. I. (1991). Listeria monocytogenes, a food-borne pathogen Microbiol Rev 55 476–511 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan C. G., Hill C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract Front Cell Infect Microbiol 4 9 10.3389/fcimb.2014.00009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Shankar R. A., Chzhan M., Samouilov A., Kuppusamy P., Zweier J. L. (1999). Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging Proc Natl Acad Sci U S A 96 4586–4591 10.1073/pnas.96.8.4586 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G. J., D'Souza F. R., Suzen S. H., Eltahir Z. S., James S. A., Parry J. M., Griffiths P. A., Baxter J. N. (2007). Deoxycholic acid at neutral and acid pH, is genotoxic to oesophageal cells through the induction of ROS: the potential role of anti-oxidants in Barrett's oesophagus Carcinogenesis 28 136–142 10.1093/carcin/bgl147 . [DOI] [PubMed] [Google Scholar]
- Kathariou S., Pine L. (1991). The type strain(s) of Listeria monocytogenes: a source of continuing difficulties Int J Syst Bacteriol 41 328–330 10.1099/00207713-41-2-328 . [DOI] [PubMed] [Google Scholar]
- King T., Ferenci T., Szabo E. A. (2003). The effect of growth atmosphere on the ability of Listeria monocytogenes to survive exposure to acid, proteolytic enzymes and bile salts Int J Food Microbiol 84 133–143 10.1016/S0168-1605(02)00404-X . [DOI] [PubMed] [Google Scholar]
- Koutsoumanis K. P., Kendall P. A., Sofos J. N. (2003). Effect of food processing-related stresses on acid tolerance of Listeria monocytogenes Appl Environ Microbiol 69 7514–7516 10.1128/AEM.69.12.7514-7516.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laksanalamai P., Joseph L. A., Silk B. J., Burall L. S., Tarr C. L., Gerner-Smidt P., Datta A. R. (2012). Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US PLoS One 7 e42448 10.1371/journal.pone.0042448 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnan M. J., Mascola L., Lou X. D., Goulet V., May S., Salminen C., Hird D. W., Yonekura M. L., Hayes P., other authors (1988). Epidemic listeriosis associated with Mexican-style cheese N Engl J Med 319 823–828 10.1056/NEJM198809293191303 . [DOI] [PubMed] [Google Scholar]
- Liu D., Ainsworth A. J., Austin F. W., Lawrence M. L. (2003). Characterization of virulent and avirulent Listeria monocytogenes strains by PCR amplification of putative transcriptional regulator and internalin genes J Med Microbiol 52 1065–1070 10.1099/jmm.0.05358-0 . [DOI] [PubMed] [Google Scholar]
- Liu D., Lawrence M. L., Ainsworth A. J., Austin F. W. (2005). Comparative assessment of acid, alkali and salt tolerance in Listeria monocytogenes virulent and avirulent strains FEMS Microbiol Lett 243 373–378 10.1016/j.femsle.2004.12.025 . [DOI] [PubMed] [Google Scholar]
- Melo J., Schrama D., Andrew P. W., Faleiro M. L. (2013). Proteomic analysis shows that individual Listeria monocytogenes strains use different strategies in response to gastric stress Foodborne Pathog Dis 10 107–119 10.1089/fpd.2012.1297 . [DOI] [PubMed] [Google Scholar]
- Merritt M. E., Donaldson J. R. (2009). Effect of bile salts on the DNA and membrane integrity of enteric bacteria J Med Microbiol 58 1533–1541 10.1099/jmm.0.014092-0 . [DOI] [PubMed] [Google Scholar]
- Merritt M. E., Lawrence A. M., Donaldson J. R. (2010). Comparative study of the effect of bile on the Listeria monocytogenes virulent strain EGD-e and avirulent strain HCC23 Arch Clin Microbiol 1 4–9. [Google Scholar]
- Metselaar K. I., den Besten H. M., Abee T., Moezelaar R., Zwietering M. H. (2013). Isolation and quantification of highly acid resistant variants of Listeria monocytogenes Int J Food Microbiol 166 508–514 10.1016/j.ijfoodmicro.2013.08.011 . [DOI] [PubMed] [Google Scholar]
- Monte M. J., Marin J. J., Antelo A., Vazquez-Tato J. (2009). Bile acids: chemistry, physiology, and pathophysiology World J Gastroenterol 15 804–816 10.3748/wjg.15.804 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen I., Vogensen F. K., Jespersen L. (2009). Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress Foodborne Pathog Dis 6 669–680 10.1089/fpd.2008.0243 . [DOI] [PubMed] [Google Scholar]
- Olier M., Rousseaux S., Piveteau P., Lemaître J. P., Rousset A., Guzzo J. (2004). Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes Int J Food Microbiol 93 87–99 10.1016/j.ijfoodmicro.2003.10.010 . [DOI] [PubMed] [Google Scholar]
- Orsi R. H., den Bakker H. C., Wiedmann M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics Int J Med Microbiol 301 79–96 10.1016/j.ijmm.2010.05.002 . [DOI] [PubMed] [Google Scholar]
- Payne A., Schmidt T. B., Nanduri B., Pendarvis K., Pittman J. R., Thornton J. A., Grissett J., Donaldson J. R. (2013). Proteomic analysis of the response of Listeria monocytogenes to bile salts under anaerobic conditions J Med Microbiol 62 25–35 10.1099/jmm.0.049742-0 . [DOI] [PubMed] [Google Scholar]
- Prieto A. I., Ramos-Morales F., Casadesús J. (2004). Bile-induced DNA damage in Salmonella enterica Genetics 168 1787–1794 10.1534/genetics.104.031062 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto A. I., Ramos-Morales F., Casadesús J. (2006). Repair of DNA damage induced by bile salts in Salmonella enterica Genetics 174 575–584 10.1534/genetics.106.060889 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S. (2008). A new perspective on Listeria monocytogenes evolution PLoS Pathog 4 e1000146 10.1371/journal.ppat.1000146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychli K., Müller A., Zaiser A., Schoder D., Allerberger F., Wagner M., Schmitz-Esser S. (2014). Genome sequencing of Listeria monocytogenes Quargel listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential PLoS One 9 e89964 10.1371/journal.pone.0089964 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., Jones J. L., Griffin P. M. (2011). Foodborne illness acquired in the United States–major pathogens Emerg Infect Dis 17 7–15 10.3201/eid1701.P11101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda R., Wood P. D., Kinsell L. W. (1969). Determination of individual conjugated bile acids in human bile J Lipid Res 10 546–554 . [PubMed] [Google Scholar]