Abstract
Lizards and snakes exhibit colour variation of adaptive value for thermoregulation, camouflage, predator avoidance, sexual selection and speciation. Furcifer pardalis, the panther chameleon, is one of the most spectacular reptilian endemic species in Madagascar, with pronounced sexual dimorphism and exceptionally large intraspecific variation in male coloration. We perform here an integrative analysis of molecular phylogeography and colour variation after collecting high‐resolution colour photographs and blood samples from 324 F. pardalis individuals in locations spanning the whole species distribution. First, mitochondrial and nuclear DNA sequence analyses uncover strong genetic structure among geographically restricted haplogroups, revealing limited gene flow among populations. Bayesian coalescent modelling suggests that most of the mitochondrial haplogroups could be considered as separate species. Second, using a supervised multiclass support vector machine approach on five anatomical components, we identify patterns in 3D colour space that efficiently predict assignment of male individuals to mitochondrial haplogroups. We converted the results of this analysis into a simple visual classification key that can assist trade managers to avoid local population overharvesting.
Keywords: colour patterns, Furcifer pardalis, panther chameleon, phylogeography, speciation, supervised learning, support vector machine classification
Introduction
Squamates (lizards and snakes) exhibit spectacular inter‐ and intraspecific colour variation generated by pigments (dark brown melanins, yellow/red pteridines and carotenoids) and structural elements (guanine nanocrystals causing interference of light waves) incorporated into various types of chromatophores (Kuriyama et al. 2006; Steffen & McGraw 2009; Cote et al. 2010; Weiss et al. 2012; Saenko et al. 2013; Teyssier et al. 2015). The adaptive value of colour variation in animals is associated with thermoregulation, camouflage, predator avoidance, sexual selection and speciation (Cooper & Greenberg 1992; Hoekstra 2006; Gray & McKinnon 2007; Magalhaes et al. 2010; Rosenblum et al. 2010; Brakefield & de Jong 2011; Olsson et al. 2013). In addition, intraspecific polymorphism in colour traits can also involve pleiotropy, epistatic interactions and stochastic processes (Steiner et al. 2007; Ducrest et al. 2008; McKinnon & Pierotti 2010; Sanchez‐Guillen et al. 2011).
Chameleons exhibit excellent cryptic coloration for predator‐specific camouflage (Stuart‐Fox et al. 2008), and several species have the striking ability to change colours (hue) for social signalling (Stuart‐Fox & Moussalli 2008; Ligon & McGraw 2013). We have recently shown that these colour changes are not due to dispersion/aggregation of intracellular pigment‐containing organelles, but are caused by the physical process of photonic interference involving the active and rapid tuning of a lattice of guanine nanocrystals within dermal iridophores (Teyssier et al. 2015). Some species exhibit significant sexual dimorphism in skin coloration, with males showing conspicuous red, blue, orange and yellow colours (Henkel & Schmidt 2000).
Chameleons form a highly derived clade of iguanian lizards that originated most likely in post‐Gondwanan Africa around 90 million years ago (Townsend et al. 2011; Tolley et al. 2013) and then dispersed to Madagascar, Europe and Asia. Madagascar shows exceptionally high levels of biodiversity and endemism (Myers et al. 2000; Ganzhorn et al. 2001) caused, among other phenomena, by vicariance events and the presence of locally steep environmental gradients (Bossuyt & Milinkovitch 2000; Bossuyt et al. 2006; Vences et al. 2009). This trend is particularly relevant for chameleons as the vast majority of the circa 160 extant species are found in Africa and Madagascar, with 62 species being endemic to the latter. A few additional recent lineages are distributed in the southern fringes of Europe and Asia as well as islands in the Indian Ocean.
Furcifer pardalis, the panther chameleon, is one of the most spectacular reptilian endemic species in Madagascar and is found in a wide range of humid, semi‐humid and transitional habitats from sea level to about 950 m of altitude along the northern and eastern coasts of the island (Fig. 1), as well as in Reunion and Mauritius islands where it has been recently introduced by humans (Ferguson et al. 2004). This diurnal species lives on trees and bushes both in relatively intact forests and in forest edges and degraded habitat and is frequently found around human dwellings and plantations. Among the most striking features of the panther chameleon are its sexual dimorphism and exceptionally large intraspecific variation in male coloration: females and juveniles are tan‐brown with hints of pink or orange, whereas adult males are much larger and have various combinations of bright red, green, blue and yellow. This extensive colour variation among individuals, which is independent from behavioural colour change abilities, made F. pardalis a popular species on the US, European and Asian pet markets where colour ‘morphs’ or ‘locales’ are often named after Malagasy villages and islands. Male coloration also varies with season and age, and local variation seems to exist within morphs (Ferguson et al. 2004).
Figure 1.
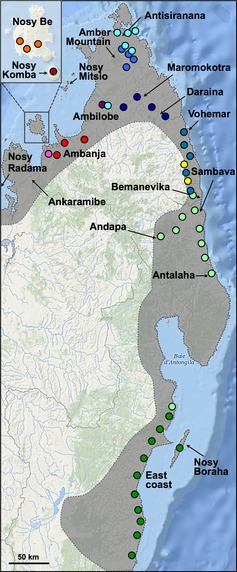
Sampling panther chameleons in Madagascar. Sampling locations are indicated with colours corresponding to mitochondrial haplogroups shown in Fig. 2. The location of a single individual from haplogroup 10 (light green) found on the east coast probably represents the southern limits of this haplogroup distribution. The most western locations of Nosy Radama and Ankaramibe were not sampled; instead, analysis was performed on captive individuals (haplotype L1 in Fig. 2). The shaded area represents the species entire distribution range.
We perform here molecular phylogeography and species delimitation analyses in F. pardalis and integrate these results with supervised learning classification of male colours for investigating the potential correlation between genetic and colour variations. First, we sampled 324 individuals in Madagascar across the species entire distribution and used mitochondrial DNA (mtDNA) and nuclear DNA (nuDNA) data to infer the genealogical relationships among populations and/or potential species. We also examine the genetic relatedness of individuals obtained on the pet market with those from the field. Second, using support vector machine (SVM; a family of supervised learning models) classification approaches, we test whether male colour pattern variation is correlated with molecular genetic population structure and could be used as a proxy for the management of local populations or potential species.
Materials and methods
Sampling and molecular data production
Sampling and photographs on animals were approved by the Malagasy ethical regulation authority and performed according to Malagasy law. We collected blood samples from 324 F. pardalis individuals in Madagascar (see Fig. 1 and Table S1, Supporting Information) in November 2011 and November 2012, that is at the beginning of the breeding season. We travelled by car along paved and dirt roads and stopped each time an animal was spotted. On regular occasions (when proper habitat was identified), 3–5 people dispersed on foot and actively searched for animals during short periods of 15–45 min. In most cases, chameleons were spotted on the branches of trees and bushes along roads and rivers as well as around villages. The capture location was recorded using a global positioning system unit. Each individual was photographed, immediately after capture, alongside with a white balance card (X‐Rite ColorChecker) using a Nikon D700 (in 2011) or Nikon D800 (in 2012) digital single‐lens reflex camera before 0.2–1.0 mL of blood was collected from the caudal vein. Animals were then released at the point of capture. Blood samples were also taken from two individuals from Reunion Island and from 26 captive Malagasy individuals, all obtained from private breeders in 2010–2012. Blood was stored in lysis buffer (100 mm Tris, 100 mm EDTA and 2% SDS) at room temperature until DNA extraction.
Genomic DNA was extracted using the Promega Wizard SV Genomic DNA purification system for 96‐well plates, following the manufacturer's protocol. DNA was resuspended in water, treated with RNase and checked for quality and quantity on 2% agarose gels. Six DNA samples that appeared to be degraded were excluded from further analysis (see Table S1, Supporting Information). Primers 5′‐GACCCAAAACATCACCCACT and 5′‐GGCGGAATTTTATTGTTCGT were used to amplify and sequence a 1373‐bp mtDNA fragment consisting of 434 bp of the NADH dehydrogenase subunit 6 (ND6) gene, 870 bp of the cytochrome b gene (cytb) and 69 bp of intergenic sequence. Primers 5′‐GAGAACCAGCAGCCTCTCTG and 5′‐AACCTGGCAGAAGGGACTTT were used to amplify and sequence 1220 bp of the nuclear recombinase activating gene 1 (RAG1). PCRs were carried out in 25 μL volume reactions and consisted of initial denaturation for 5 min at 95 °C, followed by 40 cycles of 25s at 94 °C, 25 s at 57° C, 75 s at 72 °C and a final extension of 10 min at 72 °C. PCR products were purified and sequenced using capillary electrophoresis sequencing. DNA sequences were edited and aligned manually using BioEdit version 7.1.3.0 (Hall 1999) and deposited in GenBank under Accession nos. BankIt1741024: KM094213–KM094558 (mtDNA data) and BankIt1741148: KM094559–KM094904 (nuDNA data). All protein‐coding sequences were translated to confirm the absence of stop codons.
PHASE version 2.1 (Stephens et al. 2001) implemented in DnaSP v5.10.01 (Librado & Rozas 2009) was used to resolve RAG1 allele phase from individuals heterozygous for more than one nucleotide position. PHASE was run with default parameters (100 iterations, a thinning interval of one and a burn‐in of 100). To test PHASE results, RAG1 amplicons of six individuals, each having at least four polymorphic sites, were cloned into a pGEM‐Teasy vector (Promega). For each amplicon, five clones were sequenced in both directions with M13 primers and compared to the sequences of the two RAG1 alleles obtained from PHASE analysis. In each case, all five clones segregated into two and only two alleles whose sequences corresponded to those resolved by PHASE analysis. Hence, for all other sequenced individuals, the alleles resolved by PHASE were used for further analyses.
Network inference and diversity statistics
Genealogical relationships were investigated, separately for mtDNA (Fig. 2) and nuDNA haplotypes, using the union of maximum parsimonious trees (UMP) method (Cassens et al. 2005), implemented in CombineTrees v1.0 (available at http://applications.lanevol.org/combineTrees/), as it tends to perform better than algorithmic methods (Cassens et al. 2005; Woolley et al. 2008), and with the median‐joining network approach (Figs S1 and S2, Supporting Information) (Bandelt et al. 1999), as implemented in Network v4.611 (http://www.fluxus-engineering.com/sharenet.htm). Other statistical analyses were performed using Arlequin v3.5.1.3 (Excoffier & Lischer 2010). Genetic diversity was estimated by computing the number of haplotypes (k), haplotype/allele diversity (h), the number of polymorphic sites (PS) and nucleotide diversity (π) for mtDNA/nuDNA fragments. Diversity indices were estimated for the full sample (all Malagasy individuals) and for each mtDNA haplogroup. Differentiation among groups was investigated by computing (Table S2, Supporting Information) pairwise Φ ST (for mtDNA) and F ST (for nuDNA) values (Weir & Cockerham 1984) and performing analysis of molecular variance (AMOVA) with 1000 permutations (Excoffier et al. 1992).
Figure 2.
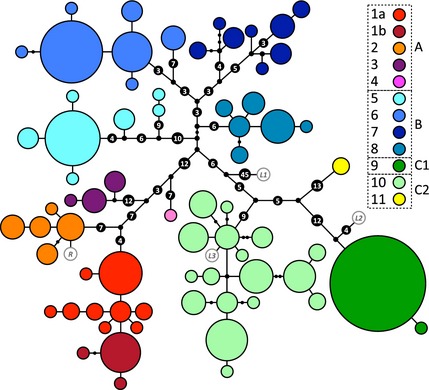
Panther chameleon mtDNA genealogical UMP (Cassens et al. 2005) network. Haplotypes are represented with coloured circles, the sizes of which are proportional to the number of individuals (smallest = 1, largest = 72). Inferred missing haplotypes are shown as small black dots, and each line indicates a mutational change (numbers of mutations >1 are shown inside larger black dots). Haplotypes are grouped into colour‐coded haplogroups; sub‐haplogroups 1a and 1b correspond to samples from the northwest coast and the island Nosy Komba, respectively. Haplotypes in white with grey border circles correspond to animals from Reunion (R) and the pet market (L1–L3).
Colour variation and support vector machine analyses
To analyse colour variation in vivo, images of 234 adult males photographed in the field were transformed from NEF to 16‐bit TIFF format using View NX 2 (http://nikonimglib.com/nvnx/). We then used Gimp 2.6.12 (http://www.gimp.org/) to adjust white balance and to transform the images to 8‐bit TIFF format. Five anatomical components (eyelid, lip, face, vertical bars and body background; see Fig. 3a) were manually cropped from the pictures of each individual using our home‐made tool CropIT v1.0 (script available on request). We then developed a program (ColourAnalysis v1.0, written in Python 2.7) that (i) converts cropped images into the HSV (hue/saturation/value) colour space and generates a corresponding colour histogram for each anatomical part of each individual and (ii) performs clustering and classification of individuals on the basis of these histograms. Briefly, Colour Analysis assigns each pixel of an image to a bin of a 3D histogram on the basis of its hue (represented in a circular space from 0°, i.e. red, to 180°, i.e. turquoise, to 359°, i.e. purple red), saturation (from 0 to 255) and value (from 0, i.e. black, to 255, i.e. white). The hue component is discretized into 16 bins, whereas the value and saturation components are discretized into 4 bins each, yielding a 3D histogram with 256 bins (Fig. 3b). We then computed, separately for each anatomical component, the relative distance L1 between each pair of individuals, using the equation:
| (1) |
where H 1[i] and H 2[i] are the values of the ith element of the histograms of the two individuals (Huang et al. 1997). We first used these L1 distances to hierarchically cluster the 234 individual panther chameleons using UPGMA (Sokal & Michener 1958). For each anatomical component, clustering trees identified six to seven groups separated by distance values >1.2 (Fig. S3, Supporting Information).
Figure 3.
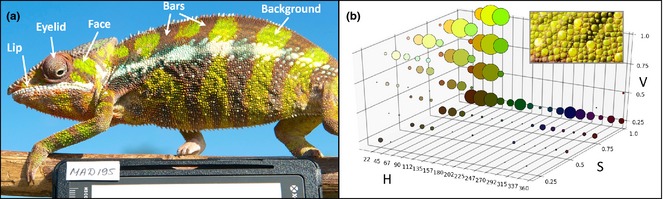
Colour variation in male panther chameleons. (a) Male panther chameleon (top left image) photographed with a white balance card; arrows indicate body parts used in colour analyses. (b) Examples of a cropped section [here background colour from the individual shown in (a)] and the corresponding 3D HSV histogram; the size of bubbles is shown on a logarithmic scale for ease of reading.
Second, we performed supervised machine learning analysis and classification on the basis of colour histograms and haplogroups: each data point consists of a pair (x,y), where x represents the 3D colour histogram of each anatomical part and y is a haplogroup. Individuals of each group were randomly placed into either the training set (containing 2/3 of individuals) or the evaluation set (1/3 of individuals). The former was used to train a multiclass SVM [support vector machine classifier (Cortes & Vapnik 1995)] with the chi‐squared nonlinear kernel (Chapelle et al. 1999) to find patterns in 3D colour space that predict assignment of individuals to groups predefined on the basis of the genealogical network shown in Fig. 2. To evaluate the quality of classification, we used the trained classifier to classify individuals of the evaluation set and calculated the precision value p (i.e. the ratio between number of individuals correctly assigned to a group and the number of all individuals assigned to that group), the recall value r (i.e. the number of individuals correctly assigned to a group divided by the number of all individuals that actually belong to that group), the F1 score (i.e. the harmonic mean of p and r) for each group and the total F1 score (i.e. arithmetic average of group F1 scores weighted by the number of individuals in each group). We performed classification for different haplogroup sets (Fig. 2): (i) all haplogroups separately, (ii) group A (haplogroups 1–3, haplotype 4 not included as it is a juvenile), haplogroups 6–7 combined, group C2 (haplogroups 10 and 11) and haplogroups 5, 8 and 9 in isolation and (iii) groups A, B, C1 and C2. We chose to split group C into monophyletic C1 and paraphyletic C2 because these correspond to two different geographical locations, namely east coast and northeastern coast. Note that haplogroup 11 (Fig. 2) was included in group C2 on the basis of its geographical proximity to haplogroup 10 (Fig. 1). As the first two sets produced substantially lower total F1 scores (0.50–0.66) than the third one (0.68–0.75), we performed subsequent analyses on the latter. We repeated the analyses 30 times with different training and evaluation sets and computed average p, r and F1 scores for each anatomical component (see Table S3, Supporting Information). We considered a group F1 score ≥0.8 to indicate good classification. As a control, the same analysis (repeated 30 times as well) was performed on four groups of the same size as the real ones, but with individuals randomly assigned to groups. Finally, we devised a classification key (Fig. 4) on the basis of the results from the SVM classifier.
Figure 4.
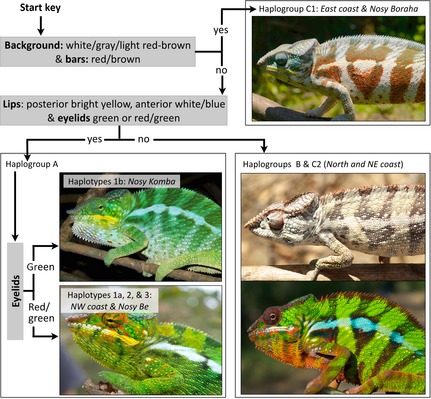
Classification key. Three questions pertaining to colour characters identified by supervised machine learning allow to partially identify which haplogroup individual male panther chameleons belong to.
Species delimitation analyses
Species delimitation analyses were performed with a Bayesian approach on the combined RAG1 and mtDNA data set (318 individuals) using the reversible‐jump Markov chain Monte Carlo (rjMCMC) algorithm (Yang & Rannala 2010) implemented in the bpp software version 3.1. The network among the 11 haplogroups (Fig. 2) was used as the reference fixed phylogeny (‘guide tree’). The priors were set to τ0 ~ G(26, 1000) with mean of 0.026 and θ ~ G(2, 699) with mean of 0.0029, and we tested each of the two proposal algorithms available in the software. We also performed unsupervised analyses, that is without providing a fixed phylogeny. Other settings were as follows: burn‐in, 3000 generations; sample frequency, 50 generations; and total number of samples, 1000.
Results and Discussion
Molecular phylogeography of Malagasy panther chameleons
We examined the phylogeography of the panther chameleon by sequencing and analysing mitochondrial (mtDNA) and nuclear (nuDNA) gene fragments (1373 bp from the ND6 and cytb genes and 1220 bp from the RAG1 gene) in 318 individuals (234 males, 55 females and 29 juveniles) sampled across nearly the entire species’ range (Fig. 1), including the islands of Nosy Be and Nosy Komba in the northwest, and Nosy Boraha in the east. We observed substantial variation in these molecular markers (Table 1): 178 (13.0%) polymorphic and 158 (11.5%) parsimony‐informative sites in the mtDNA and 75 (6.1%) polymorphic and 46 (3.8%) parsimony‐informative sites in the nuDNA fragments. Among 318 individuals, we resolved 69 mtDNA haplotypes (43 shared, 26 singletons) and 91 RAG1 alleles (41 shared, 50 singletons).
Table 1.
Population genetic statistics for mtDNA and nuclear RAG1 fragments. N – sample size, PS – number of polymorphic sites, k – number of haplotypes (for mtDNA)/alleles (for RAG1), h – haplotype diversity, π – nucleotide diversity, SD – standard deviation. Haplogroups 4 and 11 were not used for statistics as they include only one and two individuals, respectively
| Haplogroup | N | 1373‐bp mtDNA fragment | 1220‐bp nuclear RAG1 fragment | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PS | k | h (SD) | π (SD) | PS | k | h (SD) | π (SD) | ||
| 1 | 39 | 11 | 12 | 0.812 (0.044) | 0.00127 (0.00013) | 23 | 17 | 0.801 (0.039) | 0.00374 (0.00028) |
| 2 | 16 | 5 | 5 | 0.817 (0.051) | 0.00124 (0.00018) | 19 | 10 | 0.778 (0.064) | 0.00372 (0.00034) |
| 3 | 8 | 3 | 3 | 0.679 (0.122) | 0.00096 (0.00020) | 13 | 8 | 0.842 (0.075) | 0.00297 (0.00055) |
| 5 | 31 | 23 | 7 | 0.536 (0.103) | 0.00258 (0.00105) | 27 | 19 | 0.815 (0.041) | 0.00323 (0.00031) |
| 6 | 48 | 16 | 7 | 0.632 (0.063) | 0.00170 (0.00049) | 29 | 32 | 0.910 (0.019) | 0.00347 (0.00021) |
| 7 | 17 | 23 | 8 | 0.897 (0.042) | 0.00607 (0.00057) | 24 | 18 | 0.808 (0.070) | 0.00319 (0.00047) |
| 8 | 21 | 6 | 6 | 0.795 (0.059) | 0.00112 (0.00016) | 25 | 16 | 0.898 (0.030) | 0.00549 (0.00029) |
| 9 | 72 | 1 | 2 | 0.028 (0.027) | 0.00002 (0.00002) | 6 | 4 | 0.107 (0.035) | 0.00032 (0.00011) |
| 10 | 63 | 23 | 17 | 0.918 (0.016) | 0.00288 (0.00014) | 24 | 15 | 0.766 (0.027) | 0.00324 (0.00031) |
| Total | 318 | 178 | 69 | 0.929 (0.009) | 0.02223 (0.00025) | 75 | 91 | 0.898 (0.007) | 0.00520 (0.00007) |
The mtDNA haplotype network generated by the UMP approach [CombineTrees v1.0 (Cassens et al. 2005)] consists of eleven well‐differentiated haplogroups (colour‐coded in Fig. 2), separated from each other by at least six mutations. Note that haplogroups 4 and 11 consist of only one and two individuals, respectively, and probably represent undersampled populations. This network was chosen over the slightly different one (Fig. S1, Supporting Information) generated by median‐joining network (Bandelt et al. 1999) as the UMP approach generally outperforms algorithmic methods (Cassens et al. 2005; Woolley et al. 2008). This genealogy indicates that panther chameleon populations are strongly structured, with haplogroups restricted to geographical regions and overlapping merely in some locations (Fig. 1). The eleven haplogroups can be grouped into three major network partitions A, B and C (Fig. 2). Hierarchical amova for the mtDNA fragment shows that 38.5% of genetic variation is due to differentiation among these A, B and C partitions (Φ CT = 0.385, d.f. = 2, P < 0.001), 55.5% is explained by variance among haplogroups within the three partitions (Φ SC = 0.940, d.f. = 8, P < 0.001), and 6% is due to variation within haplogroups (Φ ST = 0.939, d.f. = 307, P < 0.001). Φ ST values are high (0.521–0.998) and significant (P < 0.05) for all pairwise haplogroup comparisons (Table S2, Supporting Information).
A similar, although less conspicuous, pattern was found for the nuDNA fragment (Fig. S2, Supporting Information), for which 45.2% of the genetic variance in a hierarchical amova test is associated with variance among the A, B and C mitochondrial groups (F CT = 0.452, d.f. = 2, P < 0.001), 9.8% with variance among haplogroups within groups (F SC = 0.179, d.f. = 8, P < 0.001) and 45.0% with variance within haplogroups (F ST = 0.551, d.f. = 625, P < 0.001). Nuclear genetic differentiation among mitochondrial partitions A, B and C is confirmed by significant (d.f. = 2, P < 0.001) F ST values: 0.209 (A vs. B), 0.575 (B vs. C) and 0.526 (A vs. C). F ST values for pairwise comparisons among haplogroups (Table S2, Supporting Information) generally increase with geographical distance, with highest levels of genetic differentiation between haplogroup 9 and all the others (F ST > 0.679, P < 0.05), whereas several F ST values are low and not significant among haplogroups within group B. Furthermore, a low but significant F ST value is observed between Nosy Komba (haplogroup 1b; Fig. 2) and the northwest coast (F ST = 0.091, P < 0.05), whereas the value for the comparison between Nosy Komba and Nosy Be (haplogroup 2; Fig. 2) is low and not significant (F ST = 0.049, P = 0.063). Lower F ST values for RAG1 than Φ ST values for the mtDNA are consistent with differences in nuclear vs. mitochondrial mutation rates, effective population sizes and, possibly, a higher dispersal rate of males than females. Moreover, slower fixation rates in nuclear vs. the mitochondrial genome make retention of nuclear ancestral polymorphisms more likely (Hare 2001), providing a possible explanation for the fact that 25 of the RAG1 alleles are shared among populations: for example, two alleles are present in 85 (groups A and B) and 88 (groups B and C) individuals, respectively (Table S1, Supporting Information). This also explains that, contrary to haplotypic networks reconstructed with mtDNA data, networks reconstructed with nuclear haplotypes include less well‐differentiated haplogroups (Fig. S2, Supporting Information).
Our phylogeography analyses indicate that Malagasy populations of the panther chameleon are characterized by strong genetic structure as assessed by mtDNA analyses and partially supported by the nuclear data. Indeed, mtDNA haplogroups are separated by a large number of mutations and the major lineages of F. pardalis are geographically restricted: only in a few geographical locations do mtDNA haplotypes from different haplogroups co‐occur (Fig. 1). Such a conspicuous genetic structure reveals restricted gene flow among populations that might be caused by low dispersal rates of individuals, pre‐ or postzygotic reproductive isolation (e.g. female preference for a particular male colour morph and/or reduced fitness of hybrids), geophysical features (e.g. river barriers, habitat fragmentation) or a combination of these factors.
Panther chameleons from the Reunion Island and European pet market
We additionally sequenced the same mtDNA and nuclear gene fragments in two individuals from Reunion Island and in 26 captive individuals obtained on the European pet market. First, we found that Reunion Island individuals share RAG1 alleles with animals from clade A and have a mtDNA haplotype (labelled R in Fig. 2) closely related to haplogroup 2, suggesting that the introduced Reunion island population originated from Nosy Be in the northwest of Madagascar. Second, our analyses indicate that the geographical origin claimed by sellers of panther chameleons on the pet market is often unreliable. Indeed, only three among these 26 individuals show haplotype/alleles matching those of the presumed locality. In addition, 14 of 26 animals exhibit haplotype/alleles characteristic of haplogroup 9 (i.e. C1, east coast), even though their ‘morph’ was indicated as ‘Ambilobe’ or ‘Ankify’, that is northwestern locations that should correspond to group A or B. Such discrepancy can be explained by unsubstantiated commercial claims but also by undesired hybridization among captive bred individuals due to difficulties in identifying the geographical origin of females (because of their almost invariably dull coloration). For example, a female from the east coast (the location that is easiest to reach by car from the capital Antananarivo) could be sold as from a different locality and will produce offspring with the ‘eastern’ mtDNA haplotype.
Colour variation in Malagasy chameleons and support vector machine classification
To test for a possible predictive value of male colour characters for the assignment of individuals to genealogical groups (identified above through phylogeographic analyses), we analysed colour patterns in the 234 adult males among the 318 sampled individuals. We obtained colour composition information from high‐resolution photographs for five anatomical components (eyelid, lip, face, vertical bars and body background; Fig. 3A) that show the highest variation among individuals. We then performed, separately for each anatomical component, hierarchical clustering of individuals on the basis of pairwise distances (Equation (1) in Materials and methods) among 3D colour histograms (Fig. 3B). In general, clusters do not strictly correspond to haplogroups, and each haplogroup is represented in more than one cluster (Fig. S3, Supporting Information). However, these results suggest that some features could be used to distinguish individuals from specific haplogroups. For example, the ‘eyelid’ and ‘lip’ components seem to differentiate most group A individuals from individuals in other haplogroups.
Given the limitations of clustering approaches, we performed supervised machine learning analysis and classification (using a support vector machine classifier, SVM, see Materials and methods) on the basis of the colour histograms. Supervised learning tends to produce much better classifications than clustering methods when patterns in multidimensional space are not obvious. Individuals from groups A, B, C1 and C2 (Fig. 2) were randomly separated into a training and an evaluation set. The former was used to train a multiclass SVM (Cortes & Vapnik 1995) that was then used to classify individuals of the evaluation set. The classifier generated high‐quality classifications (F1 scores of 0.84–0.92) for the ‘lip’ and ‘eyelid’ components in group A, and for all anatomical parts but ‘lip’ in group C1 (Table S3, Supporting Information). Control analyses, that is classification of four randomized groups, resulted in substantially lower scores (total F1 = 0.24–0.28) than those of the classified A, B, C1 and C2 groups (total F1 = 0.68–0.75). These results indicate that, contrary to clustering approaches, the trained classifier allows for prediction of the position of many individuals in the mitochondrial genealogy (i.e. in either group A, B, C1 or C2) on the basis of their colour photographs. This result underlined specific characters that we used to devise a simple classification key (Fig. 4). First, males from group C1 (east coast and Nosy Boraha) can be distinguished by a very light (white or grey or light red‐brown, sometimes with red spots) background and red/brown (sometimes with white spots) bars. Note that they often additionally exhibit blue or white‐brown face and eyelids, and white or light yellow (occasionally light brown) lips. Second, males from group A (northwestern coast and islands) are characterized by conspicuous bright yellow posterior lips and the presence of green or red‐green eyelids. Additionally, they typically exhibit a blue‐green (sometimes with red speckles) or yellow‐green background, green or blue‐red bars, and bright green face. Nosy Komba individuals can be further identified (within group A) by the absence of red pigment in their eyelids. Third, males from the north and northeastern coast (groups B and C2) show a lot of variation in coloration, ranging from cryptic (brown‐yellow background and face, light brown or red‐brown bars, red‐brown or pink eyes) to much more vivid colours (bright green‐yellow background and face, blue or red bars, white‐blue‐brown or orange‐brown eyes). Their lips are brown‐yellow or bright white with various amounts of red and blue, but never as bright yellow as in males from the northwestern populations. Notably, males in northern haplogroups 5–7 are more often cryptic (~82.5%) than are those in northeastern coast haplogroups 8, 10 and 11 (~36.7%). The potential ecological adaptive value of these characters (see Supporting Information, Table S4 and Fig. S4 for climate and environmental variables across population range) warrants further investigation beyond the scope of the present work.
Species delimitation in panther chameleons
To investigate whether the restricted gene flow among populations, uncovered through haplotypic network analysis and trained classifier, would potentially imply species status for some of these populations, we performed species delimitation analyses using a Bayesian approach (Yang & Rannala 2010) on the combined RAG1 and mtDNA data set. Each of the mtDNA haplogroups (1–11, 1a and 1b being merged; Fig. 2) was considered as a separate population and the reversible‐jump Markov chain Monte Carlo (rjMCMC) algorithm generated maximum posterior probabilities (all values = 1.0) for all haplogroups, suggesting that they form eleven distinct species. The posterior means of the θs range from 0.0001 to 0.0209, whereas the posterior mean of the root τ is 0.0007. Algorithms 0 and 1 (for new state proposals in the MCMC implemented in the BPP software) do not generate significantly different results. Unsupervised BPP analyses (i.e. without a fixed guide tree) also suggested that the 11 haplogroups form 11 distinct species, albeit with lower probabilities ranging from 0.77 to 0.93, depending on the proposal algorithm. In essence, these results suggest species status of the various haplogroups because reproductive barriers generated through time increase genealogical depth and agreement among unlinked loci (Avise & Wollenberg 1997).
Discussion
The large number of animals sampled across the Malagasy panther chameleon range allowed us to reveal and characterize a greater degree of intrapopulation variability than previously reported (Ferguson et al. 2004). Our phylogeography and trained multiclass SVM classification of colour variation revealed strong genetic structure, with distant geographically restricted lineages characterized by clade‐specific male colour features (e.g. bright yellow lips in northwestern haplogroups). Some of these lineages might warrant species status as suggested by our analyses based on a Bayesian approach to species delimitation. However, these results should be interpreted with caution, not only because of potential limitations of the approach (Olave et al. 2014; Zhang et al. 2014), but also because it is here dominated by the mitochondrial data, for which distinctness of populations tend to evolve rapidly (given that mtDNA markers are sensitive to genetic drift). Safe differentiation between two species (under the phylogenetic criterion) might require reciprocal monophyly of the two species (or paraphyly of one species with respect to a monophyletic other species), provided that this result is either observed in each of several genetically unlinked loci or involves the agreement between one or several gene tree(s) and a morphological/physiological designation, that is a disjunct state distribution in the corresponding nonmolecular character [e.g. (Milinkovitch et al. 2001)]. The first criterion is not fulfilled as none of the 11 haplogroups defined in the mtDNA haplotypic network (Fig. 2) form a clade in the nuDNA network (Fig. S2, Supporting Information). The second criterion is not fulfilled either as no mtDNA haplogroup exhibits a fixed colour character state. Still, the combination of our phylogeographic and supervised machine learning analyses suggests, but does not unambiguously indicate, that panther chameleons require being separated into multiple species, with a minimum of four corresponding to the bifurcation graph defined by the classification key shown in Fig. 4: the monophyletic haplogroup A1b, the paraphyletic group (A1a + A2 + A3 + A4), the monophyletic haplogroup C1 and the paraphyletic group (B + C2). In case our results are confirmed, taxonomic revision would be warranted.
Additional analyses are also warranted to determine whether the differences of colour characters among populations/haplogroups are associated with adaptation to local habitat and/or sexual selection. Note that, although panther chameleons from all geographical origin seem to successfully interbreed in captivity, anecdotal evidence (Ferguson et al. 2004) suggests that some of these hybrids present low reproductive performances, hinting at potential postzygotic isolation and, hence, at the existence of multiple species of panther chameleons as we suggested above.
High F1 scores generated by the multiclass SVM indicate substantial predictive efficiency of the supervised machine learning classification approach. We suggest that these methods should be more widely used in phylogeography and classification studies because they can produce useful predictions when clustering approaches fail. Note that the key we devised (Fig. 4) is a vast simplification of the multidimensional decision function generated by the SVM and is therefore likely to be less performant than a full SVM analysis. However, being much more practical than SVM analyses, the key could assist the Malagasy CITES Authority for managing the trade of wild‐caught panther chameleons and avoid local population overharvesting. Unreliable morph assignments on the pet market are not surprising as, even for males, general colour patterns are rarely exclusive to specific locations (Fig. S3, Supporting Information). Our classification key (Fig. 4) could provide an efficient means for inferring the origin of captive males, although hybridization in captive populations of animals from different haplogroups would have disrupted population structure. Note, however, that four animals sampled on the pet market exhibited a highly divergent haplotype (labelled L1 in Fig. 2) that we did not sample on the field. This suggests that the claimed geographical origin of these animals (‘Nosy Radama’ and ‘Ankaramibe’ in the northwest) might be correct as we did not sample these locations (Fig. 1).
One limitation of our SVM analyses is that males might fully develop their coloration at a specific period of the year (cf. our observations of captive males; Fig. S5, Supporting Information), suggesting that the SVM results, and the corresponding classification key, might be best valid during the breeding season and/or in natural conditions.
We do not expect the ability of panther chameleons to change colour [through tuning of a 3D lattice of intracellular guanine nanocrystals (Teyssier et al. 2015)] to have affected our analysis as (i) panther chameleons actively change skin hue only during social interactions (such as when encountering a male competitor or a potentially receptive female), (ii) other colour changes, which can be induced by stress during capture, mostly affect skin brightness (i.e. diffuse and/or specular reflectivity) through dispersion/aggregation of melanosomes within dermal melanophores (Taylor & Hadley 1970; Nilsson Skold et al. 2013) and are therefore unlikely to affect our analyses and (iii) colour photographs were taken on site immediately after capture, that is before potential reaction of the animal in terms of colour change. One potential improvement of our approach would be to incorporate skin reflectivity in the UV, highly relevant for colour perception in reptiles (Thorpe & Richard 2001; Fleishman et al. 2011) but not recorded by colour photographs. Measurements of skin reflectivity (including in the UV range) with portable spectrometers are unfortunately unreliable, mainly because chameleon skin darkens rapidly when it comes in contact with the optical probe.
Multiple pigments and structural elements are involved in colours and colour patterns in reptiles. Both carotenoid and pteridine pigments are common in reptiles’ skin in addition to melanin (Steffen & McGraw 2009; Cote et al. 2010; Weiss et al. 2012; Saenko et al. 2013) and can be identified by treating skin with solvents (Junqueira et al. 1978; Wijnen et al. 2007). Figure S6 shows that pigments in yellow and green skin of male panther chameleons are yellow carotenoids (dissolved in acetone) and those in red skin are red pteridines (dissolved in NH4OH). Hence, as in some other lizards [e.g. (Saenko et al. 2013; Teyssier et al. 2015)], green colour is caused by a yellow pigment layer on top of a blue/green structural background, and all skin samples contain various amounts of dark brown melanin. These findings indicate that colour variation in panther chameleons involves different genetic and physiological pathways. For example, while melanins, pteridines and structural colours are produced in the skin itself, carotenoids are obtained from the animal's diet (Fox 1976). Therefore, reduced yellow pigmentation can be attributed to scarcity of carotenoid‐rich food, but also to intrinsic physiological limitations in the animal's ability to absorb, metabolize and transport carotenoids to the skin (Parker 1996). Future work, investigating the physiology and genetic pathways involved in production of pigments and structural colours (Ziegler 2003; Higdon et al. 2013) is warranted in panther chameleons for a better understanding of the correlation between colour variation and the genealogical lineages (possibly species) we uncover here. In addition, the possibility to perform controlled captive breeding among individuals from different colour morphs, corresponding to different evolutionary lineages, opens the prospect of using gene mapping deep‐sequencing approaches (Lamichhaney et al. 2012) to identify the genetic basis of adaptive colour variation in this spectacular lineage.
Competing interests
The authors declare that they have no competing interests.
M.C.M. conceived and supervised the whole study. A.D., M.C.M., S.V.S., T.M.R. and A.P.R. performed field work and blood sampling. S.V.S. performed sequencing and pigment extraction. D.G. performed species delimitation MCMC searches, analysed the images, performed supervised learning and generated the classifier and all computer tools. S.V.S. and M.C.M. performed haplotype genealogical network analyses. M.C.M., S.V.S. and A.P.R. wrote the manuscript. All authors commented on the manuscript and approved the final version.
Data accessibility
Sequence data are available from GenBank: BankIt1741024: KM094213–KM094558 (mtDNA data) and BankIt1741148: KM094559–KM094904 (nuDNA data). The image data and program code (written in Python 2.7) for clustering and SVM classification, as well as the input files for genealogical network inference and for the species delimitation analyses, have been deposited on Dryad (datadryad.org) with the Digital Object Identifier ‘doi:10.5061/dryad.74b7h’.
Supporting information
Data S1 Discussion on climate and environmental variables across population range.
Table S1 List of sampled individuals, including sample ID, region and GPS location of sampling, sex, date, mtDNA haplotype and RAG1 alleles IDs.
Table S2 mtDNA ΦST and nuclear FST values.
Table S3 SVM Classification statistics.
Table S4 Average temperature and precipitation variables.
Fig. S1 Panther chameleon MtDNA MJ network.
Fig. S2 Panther chameleon nuclear RAG1 MJ network.
Fig. S3 Hierarchical clustering trees for five anatomical components.
Fig. S4 Annual temperature and precipitation data.
Fig. S5 Morphological changes in coloration in two captive male panther chameleons.
Fig. S6 Results of chemical removal of pigments from skin.
Acknowledgements
We thank the ‘Direction du Système des Aires Protégées de Madagascar’ for their help in obtaining field work and sampling permits and Nicolas Salamin for advises on the use of Bayesian species delimitation methods. Sampling and photographs on animals were approved by the Malagasy ethical regulation authority and performed according to Malagasy law. We thank three reviewers for their constrictive comments that significantly improved the manuscript. This work was supported by grants from the University of Geneva (Switzerland), the Georges & Antoine Claraz Foundation, the Swiss National Science Foundation (FNSNF, grants 31003A_140785 and SINERGIA CRSII3_132430) and the SystemsX.ch initiative (project EpiPhysX).
References
- Avise JC, Wollenberg K (1997) Phylogenetics and the origin of species. Proceedings of the National Academy of Sciences of the United States of America, 94, 7748–7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A (1999) Median‐joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Bossuyt F, Milinkovitch MC (2000) Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proceedings of the National Academy of Sciences of the United States of America, 97, 6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt F, Brown RM, Hillis DM, Cannatella DC, Milinkovitch MC (2006) Phylogeny and biogeography of a cosmopolitan frog radiation: late cretaceous diversification resulted in continent‐scale endemism in the family ranidae. Systematic Biology, 55, 579–594. [DOI] [PubMed] [Google Scholar]
- Brakefield PM, de Jong PW (2011) A steep cline in ladybird melanism has decayed over 25 years: a genetic response to climate change? Heredity (Edinb), 107, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassens I, Mardulyn P, Milinkovitch MC (2005) Evaluating intraspecific “network” construction methods using simulated sequence data: do existing algorithms outperform the global maximum parsimony approach? Systematic Biology, 54, 363–372. [DOI] [PubMed] [Google Scholar]
- Chapelle O, Haffner P, Vapnik VN (1999) Support vector machines for histogram‐based image classification. Ieee Transactions on Neural Networks, 10, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Cooper W, Greenberg N (1992) Reptilian coloration and behavior In: Biology of the Reptilia. (eds Gans C, Crews DE.), pp. 298–422. Chicago University Press, Chicago. [Google Scholar]
- Cortes C, Vapnik V (1995) Support‐Vector Networks. Machine Learning, 20, 273–297. [Google Scholar]
- Cote J, Meylan S, Clobert J, Voituron Y (2010) Carotenoid‐based coloration, oxidative stress and corticosterone in common lizards. Journal of Experimental Biology, 213, 2116–2124. [DOI] [PubMed] [Google Scholar]
- Ducrest AL, Keller L, Roulin A (2008) Pleiotropy in the melanocortin system, coloration and behavioural syndromes. Trends in Ecology & Evolution, 23, 502–510. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson GW, Murphy JB, Ramanamanjato J‐B, Raselimanana AP (2004) The Panther Chameleon: Color Variation, Natural History, Conservation, and Captive Management. Krieger Publishing Company, Malabar, Florida. [Google Scholar]
- Fleishman LJ, Loew ER, Whiting MJ (2011) High sensitivity to short wavelengths in a lizard and implications for understanding the evolution of visual systems in lizards. Proceedings. Biological Sciences, 278, 2891–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DL (1976) Animal Biochromes and Structural Colours: Physical, Chemical, Distributional & Physiological Features of Coloured Bodies in the Animal World. University of California Press, Berkeley, Oakland. [Google Scholar]
- Ganzhorn JU, Lowry PP, Schatz GE, Sommer S (2001) The biodiversity of Madagascar: one of the world's hottest hotspots on its way out. Oryx, 35, 346–348. [Google Scholar]
- Gray SM, McKinnon JS (2007) Linking color polymorphism maintenance and speciation. Trends in Ecology & Evolution, 22, 71–79. [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium, 41, 95–98. [Google Scholar]
- Hare MP (2001) Prospects for nuclear gene phylogeography. Trends in Ecology & Evolution, 16, 700–706. [Google Scholar]
- Henkel F‐W, Schmidt W (2000) Amphibians and Reptiles of Madagascar and the Mascarene, Seychelles, and Comoro Islands. Krieger Publishing Co, Malabar, Florida. [Google Scholar]
- Higdon CW, Mitra RD, Johnson SL (2013) Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS One, 8, e67801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE (2006) Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity, 97, 222–234. [DOI] [PubMed] [Google Scholar]
- Huang J, Kumar SR, Mitra M, Zhu W‐J, Zabih R (1997) Image indexing using color correlograms. IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan.
- Junqueira LC, Lima MH, Farias EC (1978) Carotenoid and pterin pigment localization in fish chromatophores. Stain Technology, 53, 91–94. [DOI] [PubMed] [Google Scholar]
- Kuriyama T, Miyaji K, Sugimoto M, Hasegawa M (2006) Ultrastructure of the dermal chromatophores in a lizard (Scincidae: Plestiodon latiscutatus) with conspicuous body and tail coloration. Zoological Science, 23, 793–799. [DOI] [PubMed] [Google Scholar]
- Lamichhaney S, Martinez Barrio A, Rafati N et al (2012) Population‐scale sequencing reveals genetic differentiation due to local adaptation in Atlantic herring. Proceedings of the National Academy of Sciences of the United States of America, 109, 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Ligon RA, McGraw KJ (2013) Chameleons communicate with complex colour changes during contests: different body regions convey different information. Biology Letters, 9, 20130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes IS, Mwaiko S, Seehausen O (2010) Sympatric colour polymorphisms associated with nonrandom gene flow in cichlid fish of Lake Victoria. Molecular Ecology, 19, 3285–3300. [DOI] [PubMed] [Google Scholar]
- McKinnon JS, Pierotti ME (2010) Colour polymorphism and correlated characters: genetic mechanisms and evolution. Molecular Ecology, 19, 5101–5125. [DOI] [PubMed] [Google Scholar]
- Milinkovitch M, Leduc R, Tiedemann R, Dizon A (2001) Applications of molecular data in cetacean taxonomy and population genetics with special emphasis on defining species boundaries. Marine Mammals. P. H. Evans and J. Raga, Springer US: 325–359.
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature, 403, 853–858. [DOI] [PubMed] [Google Scholar]
- Nilsson Skold H, Aspengren S, Wallin M (2013) Rapid color change in fish and amphibians ‐ function, regulation, and emerging applications. Pigment Cell Melanoma Research, 26, 29–38. [DOI] [PubMed] [Google Scholar]
- Olave M, Sola E, Knowles LL (2014) Upstream analyses create problems with DNA‐based species delimitation. Systematic Biology, 63, 263–271. [DOI] [PubMed] [Google Scholar]
- Olsson M, Stuart‐Fox D, Ballen C (2013) Genetics and evolution of colour patterns in reptiles. Seminars in Cell & Developmental Biology, 24, 529–541. [DOI] [PubMed] [Google Scholar]
- Parker RS (1996) Carotenoids. 4. Absorption, metabolism, and transport of carotenoids. FASEB Journal, 10, 542–551. [PubMed] [Google Scholar]
- Rosenblum EB, Rompler H, Schoneberg T, Hoekstra HE (2010) Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proceedings of the National Academy of Sciences of the United States of America, 107, 2113–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenko SV, Teyssier J, van der Marel D, Milinkovitch MC (2013) Precise colocalization of interacting structural and pigmentary elements generates extensive color pattern variation in Phelsuma lizards. BMC Biology, 11, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Guillen RA, Hansson B, Wellenreuther M, Svensson EI, Cordero‐Rivera A (2011) The influence of stochastic and selective forces in the population divergence of female colour polymorphism in damselflies of the genus Ischnura. Heredity (Edinb), 107, 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R, Michener C (1958) A statistical method for evaluating systematic relationships. University of Kansas Science Bulletin, 38, 1409–1438. [Google Scholar]
- Steffen JE, McGraw KJ (2009) How dewlap color reflects its carotenoid and pterin content in male and female brown anoles (Norops sagrei). Comparative Biochemistry and Physiology B‐Biochemistry & Molecular Biology, 154, 334–340. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE (2007) Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biology, 5, e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics, 68, 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart‐Fox D, Moussalli A (2008) Selection for social signalling drives the evolution of chameleon colour change. PLoS Biology, 6, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart‐Fox D, Moussalli A, Whiting MJ (2008) Predator‐specific camouflage in chameleons. Biology Letters, 4, 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JD, Hadley ME (1970) Chromatophores and color change in the lizard, Anolis carolinensis. Z Zellforsch Mikrosk Anat, 104, 282–294. [DOI] [PubMed] [Google Scholar]
- Teyssier J, Saenko SV, van der Marel D, Milinkovitch MC (2015) Photonic crystals cause active colour change in chameleons. Nature Communications, 6, 6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe RS, Richard M (2001) Evidence that ultraviolet markings are associated with patterns of molecular gene flow. Proceedings of the National Academy of Sciences of the United States of America, 98, 3929–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley KA, Townsend TM, Vences M (2013) Large‐scale phylogeny of chameleons suggests African origins and Eocene diversification. Proceedings. Biological Sciences, 280, 20130184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend TM, Mulcahy DG, Noonan BP et al (2011) Phylogeny of iguanian lizards inferred from 29 nuclear loci, and a comparison of concatenated and species‐tree approaches for an ancient, rapid radiation. Molecular Phylogenetics and Evolution, 61, 363–380. [DOI] [PubMed] [Google Scholar]
- Vences M, Wollenberg KC, Vieites DR, Lees DC (2009) Madagascar as a model region of species diversification. Trends in Ecology & Evolution, 24, 456–465. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC (1984) Estimating F‐Statistics for the analysis of population‐structure. Evolution, 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- Weiss SL, Foerster K, Hudon J (2012) Pteridine, not carotenoid, pigments underlie the female‐specific orange ornament of striped plateau lizards (Sceloporus virgatus). Comparative Biochemistry and Physiology B‐Biochemistry & Molecular Biology, 161, 117–123. [DOI] [PubMed] [Google Scholar]
- Wijnen B, Leertouwer HL, Stavenga DG (2007) Colors and pterin pigmentation of pierid butterfly wings. Journal of Insect Physiology, 53, 1206–1217. [DOI] [PubMed] [Google Scholar]
- Woolley SM, Posada D, Crandall KA (2008) A comparison of phylogenetic network methods using computer simulation. PLoS One, 3, e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B (2010) Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences of the United States of America, 107, 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rannala B, Yang Z (2014) Bayesian species delimitation can be robust to guide‐tree inference errors. Systematic Biology, 63, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler I (2003) The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell‐fate. Pigment Cell Research, 16, 172–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Discussion on climate and environmental variables across population range.
Table S1 List of sampled individuals, including sample ID, region and GPS location of sampling, sex, date, mtDNA haplotype and RAG1 alleles IDs.
Table S2 mtDNA ΦST and nuclear FST values.
Table S3 SVM Classification statistics.
Table S4 Average temperature and precipitation variables.
Fig. S1 Panther chameleon MtDNA MJ network.
Fig. S2 Panther chameleon nuclear RAG1 MJ network.
Fig. S3 Hierarchical clustering trees for five anatomical components.
Fig. S4 Annual temperature and precipitation data.
Fig. S5 Morphological changes in coloration in two captive male panther chameleons.
Fig. S6 Results of chemical removal of pigments from skin.
Data Availability Statement
Sequence data are available from GenBank: BankIt1741024: KM094213–KM094558 (mtDNA data) and BankIt1741148: KM094559–KM094904 (nuDNA data). The image data and program code (written in Python 2.7) for clustering and SVM classification, as well as the input files for genealogical network inference and for the species delimitation analyses, have been deposited on Dryad (datadryad.org) with the Digital Object Identifier ‘doi:10.5061/dryad.74b7h’.


