Abstract
A million copies of the Alu short interspersed nuclear element (SINE) are scattered throughout the human genome, providing ∼11% of our total DNA. SINEs spread by retrotransposition, using a transcript generated by RNA polymerase (pol) III from an internal promoter. Levels of these pol III-dependent Alu transcripts are far lower than might be expected from the abundance of the template. This was believed to reflect transcriptional suppression through DNA methylation, denying pol III access to most SINEs through chromatin-mediated effects. Contrary to expectations, our recent study found no evidence that methylation of SINE DNA reduces its occupancy or expression by pol III. However, histone H3 associated with SINEs is prominently methylated on lysine 9, a mark that correlates with transcriptional silencing. The SUV39 methyltransferases that deposit this mark can be found at many SINEs. Furthermore, a selective inhibitor of SUV39 stimulates pol III recruitment to these loci, as well as SINE expression. These data suggest that methylation of histone H3 rather than DNA may mediate repression of SINE transcription by pol III, at least under the conditions we studied.
Keywords: Alu, H3K9, MeCP2, methylation, RNA polymerase III, SINE, SUV39
SINE Expression Can Have Detrimental Consequences and is Subject to Repression
SINEs are retrotransposons that evolved from transcripts made by pol III, most commonly tRNAs.1 For example, mouse B2 SINEs evolved from a tRNA,2 whereas human Alu and mouse B1 SINEs evolved from the 7SL RNA scaffold of the signal recognition particle.3 The principle pol III promoter elements of tRNA and 7SL genes are located within the transcribed region and are therefore propagated during retrotransposition, a feature that contributed substantially to the prolific spread of SINEs throughout mammalian genomes. Insertion of retrotransposed Alu SINEs has been implicated in many cases of human disease, a problem exacerbated by their concentration near protein-coding genes.4,5 Indeed, Alus are responsible for the majority of documented disease cases attributed to the insertion of retrotransposons.6 Comparison of 44 human genomes revealed an average of 791 Alu insertions in each that are not found in the reference assembly,7 that serves as a representative standard. The number of Alu insertions increases in cancer cells,7 perhaps reflecting elevated pol III activity.8 SINEs also provide hotspots for recombination between non-allelic loci, undermining genomic stability.5,9 In addition, overexpression of SINE transcripts can be cytotoxic and cause macular degeneration.10 Protective mechanisms have evolved to limit these deleterious effects. One such mechanism is transcriptional suppression and it was estimated that ∼99% of potentially active Alus may be subject to chromatin-mediated silencing.11 Genome-wide analyses suggest that only ∼0.1% of Alu loci are transcribed or occupied by pol III in the cell lines investigated.12-14 Other studies reported even lower pol III occupancy,15-17 but this may reflect technical differences, such as the choice of statistical threshold. Nevertheless, all agree that there is powerful suppression and the favored model for this has been transcriptional inhibition by DNA methylation.
DNA Methylation at SINEs
SINE DNA is heavily methylated in mouse and human cells.18-20 That this contributes to transcriptional repression was suggested by increased SINE expression induced by 5-azacytidine, an inhibitor of DNA methylation.21 However, the cells in these experiments had been treated for 8 days, which is more than long enough for indirect secondary effects. When we analyzed cells after 16 or 72 hrs exposure to 5-azacytidine, SINE expression was unchanged, despite robust induction of Apo-E, a pol II-transcribed gene shown previously to be silenced by DNA methylation.14 We confirmed that SINE DNA is demethylated in cells treated for 72 hrs with 5-azacytidine, without transcriptional induction.14 Similarly, SINE expression is unaltered in fibroblasts from mice with genetic knockout of Dnmt1, despite a 20-fold reduction in DNA methylation relative to wild-type.14 One possibility is that cells compensate for elevated SINE transcription by increasing transcript degradation, so that overall expression is unchanged. However, we saw no change in SINE occupancy by pol III or its associated factors TFIIIB and TFIIIC in Dnmt1 knockout or 5-azacytidine-treated cells.14 This argues strongly against the long-standing model that DNA methylation is responsible for denying accessibility of SINEs to pol III. Indeed, bisulphite sequencing after chromatin immunoprecipitation (ChIP-BS-Seq) of pol III-bound Alu DNA revealed no decrease in methylation relative to input genomic DNA.14 This proved directly that pol III engages methylated Alu DNA in vivo. A possibility remains that template methylation is unfavourable for passage of pol III along the DNA, but we found no evidence to support this conjecture. The data do, however, argue strongly against models in which SINE accessibility to the transcription apparatus is suppressed by DNA methylation.
The opposite conclusion has been drawn in several previous studies, where transcription by pol III was found to be inhibited when the template DNA was methylated.19,22,23 Stimulation of transcription by Alu methylation has also been reported.24 A salient difference between our work and these contradictory studies was that the earlier analyses used plasmid DNA either in vitro or in transfected cells. This important difference might account, at least in part, for the discrepancies; perhaps pol III becomes more sensitive to DNA methylation when asked to perform outside the context of chromosomal DNA. It is also likely that transcriptional sensitivity to DNA methylation varies according to circumstances, even in the context of physiological chromatin.
MeCP2 at SINEs
In keeping with their strong methylation, ChIP assays demonstrated that SINEs are bound in vivo by proteins that recognize methylated DNA, such as MeCP2.14,25 MeCP2 interacts with histone deacetylases (HDACs)26 and has long been regarded as a transcriptional repressor.27 We had expected MeCP2 to have an inhibitory effect on SINE transcription, as active pol III promoters have acetylated histones16,28-31 and the HDAC inhibitor trichostatin A stimulates both pol III recruitment and SINE expression in vivo.28,32 However, HDACs are retained at SINEs in Dnmt1-knockout cells, where DNA is predominantly unmethylated, albeit at reduced levels.33 Sequential ChIP revealed that MeCP2 occupies SINEs simultaneously with pol III. Furthermore, release of MeCP2 following 5-azacytidine treatment was not accompanied by discernible effects on pol III binding or activity.14 Effects of MeCP2 on histone acetylation, chromatin structure and gene expression are seen predominantly in neurons, where it is far more abundant than in other cell types.34 This may explain its lack of apparent effect in the HeLa, ES cells and fibroblasts used in our study. Furthermore, repression by MeCP2 was recently reported to be proportional to gene length, it having minimal effect on genes shorter than ∼100kb;35 with lengths of ∼280bp or less, SINEs fall well below this threshold.
H3K9 Methylation at SINEs
Trimethylation of histone H3 on lysine 9 (H3K9me3) is a mark that is highly enriched at inactive tRNA genes, relative to genes that are pol III-occupied.16,29 H3K9me3 is also enriched at SINEs, even in the absence of DNA methylation.14 One of the enzymes that trimethylates H3K9 is SUV39H1, a methyltransferase that we detected at SINEs.14 SUV39H1 associates with HDACs.36 Furthermore, SUV39H1 and H3K9me3 together provide binding sites for HP1, a heterochromatin-associated protein that mediates transcriptional repression37 and was found at the same SINEs as SUV39H1.14 Significant decreases in H3K9me3, along with increased pol III occupancy and elevated SINE expression, followed treatment of cells with the fungal mycotoxin chaetocin,14 a selective inhibitor of SUV39H1 and related histone methyltransferases.38 This is consistent with a previous report that SINE expression can be stimulated by a dominant negative version of Suv39h.39 These data suggest that SUV39H1 contributes to the suppression of SINE transcription. Its involvement is likely to be substantial, as the increased pol III recruitment and expression were detected using consensus primer sets that recognize hundreds of members of the Alu, B1 and B2 SINE families.14 However, neither SUV39H1 nor HP1 were detected at a particular Alu on chromosome 6, despite enrichment for H3K9me3; in contrast to the other SINEs examined, this Alu showed no change in H3K9me3 or pol III binding following chaetocin treatment. Thus, other histone methyltransferases are also likely to modify H3K9 at some SINEs, presumably depending on local environment.
Although there is no overlap between H3K9me3 and pol III binding in HeLa cells, there is a 24% overlap in human H1 ES cells.31 H3K9me3 may be less refractory to pol III recruitment in this context, because HP1 does not associate stably with chromatin in undifferentiated ES cells.40 This observation provides support for the contention that effects of chromatin modification vary according to context. Indeed, SINE expression varies substantially between tissues, with embryos showing notably higher levels of SINE-initiated transcripts.41 Responses to stimuli also depend on cell type. For example, acute (30 min) restraint of rats is a stress that stimulates SINE expression in most tissues analyzed, including frontal cortex and cerebellum, but has the opposite effect in the hippocampus, where a ∼5-fold decrease occurs.42 It is intriguing that this selective response correlates with induction of Suv39h1 and a localized increase in H3K9me3 at SINEs in the stressed hippocampus.42
As stated above, Dnmt1 knockout has minimal effect on SINE expression in fibroblasts, despite the loss of DNA methylation.14 However, we found that SINE induction by chaetocin is substantially enhanced in the absence of Dnmt1.33 This suggests the possibility that DNA methylation can assume importance for SINE repression under circumstances where SUV39H1 is inactive, potentially providing a back-up mechanism to protect against derepression. Further work will be needed to test this model, as indirect effects could also explain the increased SINE response to chaetocin in the absence of Dnmt1.
SINEs are Inefficient at Recruiting Pol III
The primate Alu family and the rodent B1 SINE family evolved from 7SL RNA, which provides the essential RNA scaffold for the signal recognition particle.3 7SL genes are highly active and every HeLa cell is estimated to carry ∼106 transcripts from the 3 7SL loci per human genome.21 In striking contrast, the same study estimated that Alu transcript abundance is only ∼103 transcripts per HeLa cell, despite the ∼106 templates dispersed across our genomes.21 The transcribed regions of 7SL genes contain an internal promoter sequence that provides the binding site for TFIIIC, the transcription factor responsible for recognizing most pol III templates.43-45 This internal promoter is propagated during retrotransposition, providing a TFIIIC-binding site within each new SINE. Although subsequent mutation has compromised these internal promoters in many SINEs, we detected TFIIIC at each of the 5 individual Alu loci examined.14 Indeed, TFIIIC occupancy of these SINEs in HeLa cells is comparable to that of 7SL, as judged by ChIP-qPCR.14 When consensus primers were used to amplify ∼103 members of one of the younger Alu subfamilies (PV), TFIIIC was detected at levels not far below its occupancy of 7SL genes.14 In contrast, pol III occupancy assessed in parallel was markedly higher on 7SL than on Alus amplified with either locus-specific or consensus primers.14 Inefficient recruitment of pol III was also observed on B1 and B2 SINE families in mouse cells.14 Quantification of multiple ChIP experiments revealed that the ratio of pol III to TFIIIC is significantly lower on Alu, B1 or B2 SINEs than it is on 7SL.14 Thus, SINEs are accessible to TFIIIC but then struggle to recruit pol III. When the H3K9me3 mark was removed in cells exposed to SUV39H1 inhibitor chaetocin, pol III occupancy increased significantly with minimal change in TFIIIC binding (Fig. 1). Thus, SUV39H1 appears to act at SINEs on a step after template recognition by TFIIIC. In contrast, pol III recruitment is highly efficient at 7SL, which shows only background levels of SUV39H1, H3K9me3 and HP1.14
Figure 1.
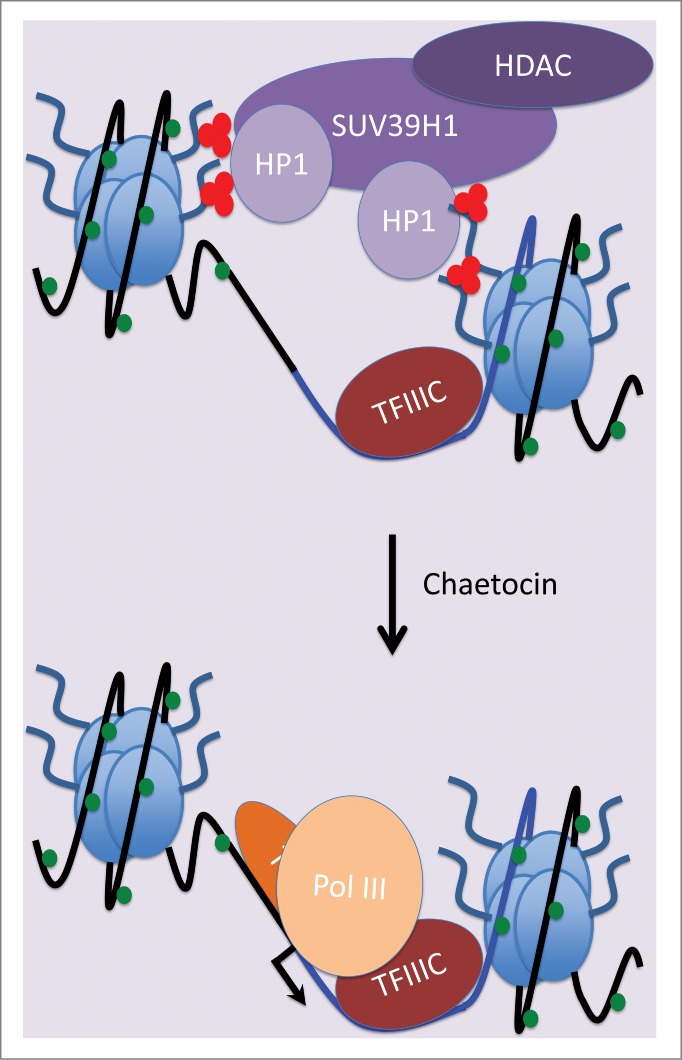
Model of SINE repression by SUV39H1. Lysine 9 of histone H3 is trimethylated in the vicinity of SINEs by SUV39H1, thereby attracting HP1. The resultant repressive chromatin structure impedes pol III recruitment to SINEs, while having much less effect on TFIIIC. The SUV39H1 inhibitor chaetocin can alleviate this repression and thereby stimulate transcription of SINEs by pol III. Blue line indicates SINE DNA; black line indicates flanking DNA. Red dots signify trimethylated H3K9. Green dots signify methylated DNA.
TFIIIC can be regarded as a “pioneer” factor that is good at accessing promoters in chromatin.46 Once it has initiated assembly of the transcription complex, recruitment of pol III depends on TFIIIB, a factor that is itself recruited by TFIIIC to DNA upstream of the transcription start site.47,48 (Fig. 2) For 7SL, the DNA immediately upstream of the start site, where TFIIIB binds, has a strong stimulatory effect on transcription.49 This upstream sequence from 7SL can also stimulate SINE expression in chimaeric constructs.50-52 However, it is not propagated during retrotransposition and is replaced in SINEs by random DNA flanking the site of insertion, which can influence transcription strongly but is unlikely to be optimal in most cases.13 TFIIIB can still be recruited to suboptimal upstream DNA via protein/protein interactions with TFIIIC bound to the internal promoter within the SINE, but this is probably less efficient in most cases than for the upstream sequence of 7SL. Indeed, we observed that TFIIIB occupancy on SINEs tends to be reduced in comparison to 7SL.14 However, the relative inefficiency of TFIIIB recruitment to SINEs is less marked than for pol III and did not reach statistical significance in our analyses.14 Thus, a trend toward diminished TFIIIB occupancy does not seem sufficient to explain the significant reduction in pol III that is consistently seen at SINEs.
Figure 2.
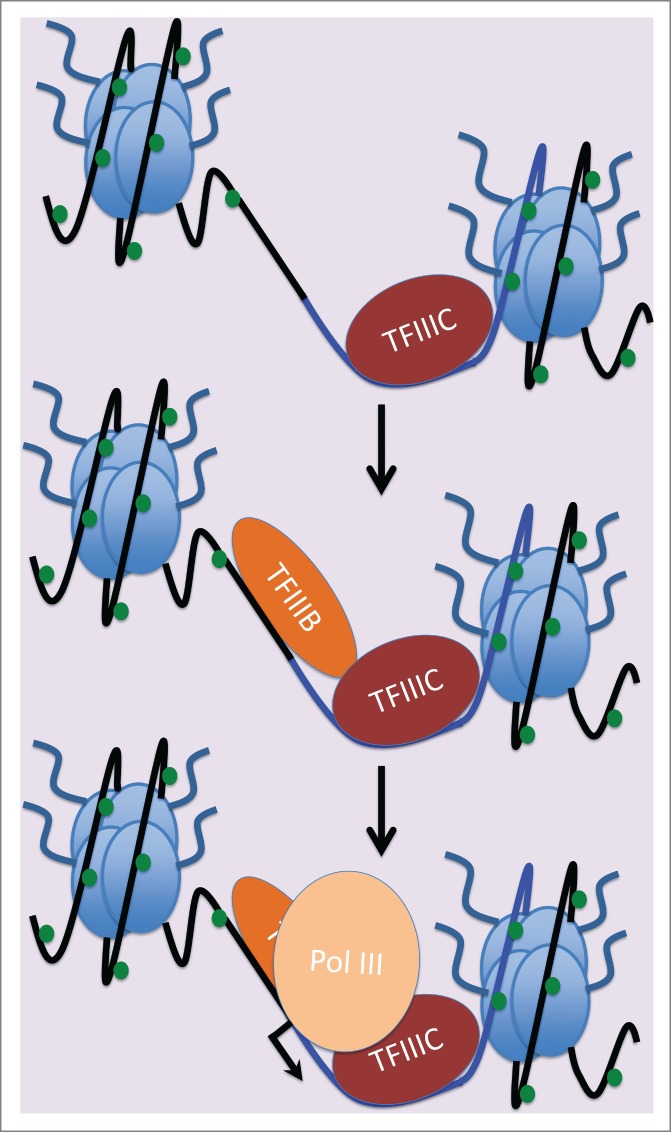
Model of pol III recruitment. Most pol III templates, including 7SL, SINEs and tRNA genes, have promoter sequences within the transcribed region that are recognized by TFIIIC. Protein/protein interactions with TFIIIC recruit TFIIIB and position it on flanking DNA immediately upstream of the transcription start site. TFIIIB occupancy can be strongly influenced by upstream DNA sequences (e.g. TATA), but does not depend on a specific DNA recognition motif. Recruitment of pol III is mediated by protein/protein interactions with TFIIIB. Blue line indicates SINE DNA; black line indicates flanking DNA. Green dots signify methylated DNA.
As mentioned above, expression of individual SINEs varies markedly between tissues.13,41 Thus, when 7 human cell types were compared, including HeLa and K562, the majority of Alu loci that were expressed in any one cell line appeared to be silent in the other 6.13 We detected pol III at ∼1,400 Alu SINEs in HeLa cells, but only ∼2% of these overlapped with the ∼1,000 Alu loci found in a different study to be occupied by pol III in K562 cells.12,14 Even within any given cell line, occupancy may vary according to growth conditions.53 Since all Alu SINEs are expected to share the same core machinery (TFIIIB, TFIIIC, pol III), the variability is most likely caused by factors that bind to flanking regions that are characteristic of each locus and may be tissue-specific and/or sensitive to external parameters. In a particular cell under particular conditions, a tiny fraction of Alu loci may come under the influence of activating factors that promote pol III recruitment. In the absence of such factors, the SINE will probably remain silent, like the majority of its siblings. Such silencing appears to involve histone modification.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet 2002; 3:370-9; PMID:11988762; http://dx.doi.org/ 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- 2.Daniels GR, Deininger PL. Repeat sequence families derived from mammalian tRNA genes. Nature 1985; 317:819-22; PMID:3851163; http://dx.doi.org/ 10.1038/317819a0 [DOI] [PubMed] [Google Scholar]
- 3.Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature 1984; 312:171-2; PMID:6209580; http://dx.doi.org/ 10.1038/312171a0 [DOI] [PubMed] [Google Scholar]
- 4.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab 1999; 67:183-93; PMID:10381326; http://dx.doi.org/ 10.1006/mgme.1999.2864 [DOI] [PubMed] [Google Scholar]
- 5.Burns KH, Boeke JD. Human transposon tectonics. Cell 2012; 149:740-52; PMID:22579280; http://dx.doi.org/ 10.1016/j.cell.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belancio VP, Hedges DJ, Deininger PL. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res 2008; 18:343-58; PMID:18256243; http://dx.doi.org/ 10.1101/gr.5558208 [DOI] [PubMed] [Google Scholar]
- 7.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al.. Landscape of somatic retrotransposition in human cancers. Science 2012; 337:967-71; PMID:22745252; http://dx.doi.org/ 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White RJ. RNA polymerase III transcription and cancer. Oncogene 2004; 23:3208-16; PMID:15094770; http://dx.doi.org/ 10.1038/sj.onc.1207547 [DOI] [PubMed] [Google Scholar]
- 9.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 2002; 35:97-112; PMID:12203773; http://dx.doi.org/ 10.1002/gcc.10111 [DOI] [PubMed] [Google Scholar]
- 10.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al.. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011; 471:325-30; PMID:21297615; http://dx.doi.org/ 10.1038/nature09830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russanova VR, Driscoll CT, Howard BH. Adenovirus type 2 preferentially stimulates polymerase III transcription of Alu elements by relieving repression: a potential role for chromatin. Mol Cell Biol 1995; 15:4282-90; PMID:7623822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 2010; 17, 635-40; http://dx.doi.org/ 10.1038/nsmb.1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conti A, Carnevali D, Bollati V, Fustinoni S, Pellegrini M, Dieci G. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res 2015; 43:817-35; PMID:25550429; http://dx.doi.org/ 10.1093/nar/gku1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun 2015; 6:6569; http://dx.doi.org/ 10.1038/ncomms7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res 2010; 20:710-21; PMID:20413673; http://dx.doi.org/ 10.1101/gr.101337.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al.. Human RNA polymerase III transcriptomes and relationships to pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 2010; 17:620-8; http://dx.doi.org/ 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci USA 2010; 107:3639-44; PMID:20139302; http://dx.doi.org/ 10.1073/pnas.0911315106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid CW. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res 1991; 19:5613-561; PMID:1945838; http://dx.doi.org/ 10.1093/nar/19.20.5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J 1993; 12:1141-51; PMID:8384552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al.. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454:766-70; PMID:18600261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu WM, Maraia RJ, Rubin CM, Schmid CW. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res 1994; 22:1087-95; PMID:7512262; http://dx.doi.org/ 10.1093/nar/22.6.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res 1993; 21:1351-9; PMID:8464725; http://dx.doi.org/ 10.1093/nar/21.6.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res 2001; 29:4493-501; PMID:11691937; http://dx.doi.org/ 10.1093/nar/29.21.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vorce RL, Lee B, Howard BH. Methylation- and mutation-dependent stimulation of Alu transcription in vitro. Biochem Biophys Res Commun 1994; 203:845-51; PMID:8093066; http://dx.doi.org/ 10.1006/bbrc.1994.2260 [DOI] [PubMed] [Google Scholar]
- 25.Koch C, Stratling WH. DNA binding of methyl-CpG-binding protein MeCP2 in human MCF7 cells. Biochemistry 2004; 43:5011-21; PMID:15109260; http://dx.doi.org/ 10.1021/bi0359271 [DOI] [PubMed] [Google Scholar]
- 26.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998; 393:386-9; PMID:9620804; http://dx.doi.org/ 10.1038/30764 [DOI] [PubMed] [Google Scholar]
- 27.Bird AP, Wolffe AP. Methylation-induced repression - belts, braces, and chromatin. Cell 1999; 99:451-4; PMID:10589672; http://dx.doi.org/ 10.1016/S0092-8674(00)81532-9 [DOI] [PubMed] [Google Scholar]
- 28.Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci USA 2007; 104:14917-22; PMID:17848523; http://dx.doi.org/ 10.1073/pnas.0702909104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 2010; 17:629-34; http://dx.doi.org/ 10.1038/nsmb.1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White RJ. Transcription by RNA polymerase III - more complex than we thought. Nat Rev Genet 2011; 12:459-63; PMID:21540878; http://dx.doi.org/ 10.1038/nrg3001 [DOI] [PubMed] [Google Scholar]
- 31.Alla RK, Cairns BR. RNA polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS ONE 2014; 9:e85648; http://dx.doi.org/ 10.1371/journal.pone.0085648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutcliffe JE, Brown TRP, Allison SJ, Scott PH, White RJ. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol Cell Biol 2000; 20:9192-202; PMID:11094071; http://dx.doi.org/ 10.1128/MCB.20.24.9192-9202.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varshney D. Regulation of RNA polymerase III transcription by DNA methylation and chromatin. PhD Thesis, University of Glasgow 2012. [Google Scholar]
- 34.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 2010; 37:457-68; PMID:20188665; http://dx.doi.org/ 10.1016/j.molcel.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015; 522:89-93; PMID:25762136; http://dx.doi.org/ 10.1038/nature14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res 2002; 30:475-81; PMID:11788710; http://dx.doi.org/ 10.1093/nar/30.2.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 2005; 25:2525-38; PMID:15767660; http://dx.doi.org/ 10.1128/MCB.25.7.2525-2538.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol 2005; 3:143-5; http://dx.doi.org/ 10.1038/nchembio721 [DOI] [PubMed] [Google Scholar]
- 39.Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein S. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 2005; 24:800-12; PMID:15678104; http://dx.doi.org/ 10.1038/sj.emboj.7600545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10:105-16; PMID:16399082; http://dx.doi.org/ 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al.. The regulated retrotransposon transcriptome of mammalian cells. Nature Genet 2009; 41:563-71; PMID:19377475; http://dx.doi.org/ 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- 42.Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker ME, Datson NA, McEwen BS, Pfaff DW. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA 2012; 109:17657-62; PMID:23043114; http://dx.doi.org/ 10.1073/pnas.1215810109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinert H, Gladen A, Geisler M, Benecke BJ. Differential regulation of transcription of human 7 S K and 7 S L RNA genes. J Biol Chem 1988; 263:11511-5; PMID:3403542 [PubMed] [Google Scholar]
- 44.Bredow S, Kleinert H, Benecke BJ. Sequence and factor requirements for faithful in vitro transcription of human 7SL DNA. Gene 1990; 86:217-25; PMID:2323574; http://dx.doi.org/ 10.1016/0378-1119(90)90282-V [DOI] [PubMed] [Google Scholar]
- 45.Muller J, Benecke BJ. Analysis of transcription factors binding to the human 7SL RNA gene promoter. Biochem Cell Biol 1999; 77:431-8; PMID:10593606; http://dx.doi.org/ 10.1139/o99-051 [DOI] [PubMed] [Google Scholar]
- 46.Burnol AF, Margottin F, Huet J, Almouzni G, Prioleau MN, Méchali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature 1993; 362:475-7; PMID:8464480; http://dx.doi.org/ 10.1038/362475a0 [DOI] [PubMed] [Google Scholar]
- 47.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 1990; 60:235-45; PMID:2404611; http://dx.doi.org/ 10.1016/0092-8674(90)90739-2 [DOI] [PubMed] [Google Scholar]
- 48.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev 2002; 16:2593-620; PMID:12381659; http://dx.doi.org/ 10.1101/gad.1018902 [DOI] [PubMed] [Google Scholar]
- 49.Ullu E, Weiner AM. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature 1985; 318:371-4; PMID:2415825; http://dx.doi.org/ 10.1038/318371a0 [DOI] [PubMed] [Google Scholar]
- 50.Chu WM, Liu WM, Schmid CW. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res 1995; 23:1750-7; PMID:7540287; http://dx.doi.org/ 10.1093/nar/23.10.1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chesnokov I, Schmid CW. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J Mol Evol 1996; 42:30-6; PMID:8576961; http://dx.doi.org/ 10.1007/BF00163208 [DOI] [PubMed] [Google Scholar]
- 52.Roy AM, West NC, Rao A, Adhikari P, Alemán C, Barnes AP, Deininger PL. Upstream flanking sequences and transcription of SINEs. J Mol Biol 2000; 302:17-25; PMID:10964558; http://dx.doi.org/ 10.1006/jmbi.2000.4027 [DOI] [PubMed] [Google Scholar]
- 53.Oler AJ, Traina-Dorge S, Derbes RS, Canella D, Cairns BR, Roy-Engel AM. Alu expression in human cell lines and their retrotranspositional potential. Mobile DNA 2012; 3:11; PMID:22716230; http://dx.doi.org/ 10.1186/1759-8753-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]


