Abstract
Quantitative characterization of dynamic exchange between various conformational states provides essential insights into the molecular basis of many regulatory RNA functions. Here, we present an application of nucleic-acid-optimized carbon chemical exchange saturation transfer (CEST) and low spin-lock field R1ρ relaxation dispersion (RD) NMR experiments in characterizing slow chemical exchange in nucleic acids that is otherwise difficult if not impossible to be quantified by the ZZ-exchange NMR experiment. We demonstrated the application on a 47-nucleotide fluoride riboswitch in the ligand-free state, for which CEST and R1ρ RD profiles of base and sugar carbons revealed slow exchange dynamics involving a sparsely populated (p ~ 10%) and shortly lived (τ ~ 10 ms) NMR “invisible” state. The utility of CEST and low spin-lock field R1ρ RD experiments in studying slow exchange was further validated in characterizing an exchange as slow as ~60 s−1.
Many regulatory RNA functions depend on dynamic exchange between different conformations that can occur over a broad range of time scales from picosecond to second and longer.1 Conformational dynamics that involves formation of new distinct base pair interactions at either the secondary or tertiary structural level is a ubiquitous form of RNA dynamics and occurs on relatively slower microsecond to second time scales.1 NMR spectroscopy has been a powerful atomic-resolution tool for quantifying these conformational dynamics in nucleic acids.2,3 The imino/amino proton exchange experiment has a long history of characterizing base-pair-opening dynamics on time scales slower than a millisecond.4 ZZ-exchange5–8 and time-resolved9,10 NMR spectroscopy have allowed characterization of equilibrium and nonequilibrium to equilibrium base pair formation, respectively, provided that the state of interest is sufficiently populated and the rate of exchange falls within subsecond to second time scales. Fast microsecond base-pairing dynamics have been studied using conventional R1ρ relaxation dispersion (RD)11,12 and the development of low spin-lock field R1ρ RD13–15 has enabled discoveries of extensive micro-to-millisecond base pair reconfiguration in nucleic acids with exchange rate as slow as ~370 s−1 being successfully characterized.16–18 However, accurate quantification of functionally important slow millisecond dynamics in nucleic acids still remains elusive. While Carr–Purcell–Meiboom–Gill (CPMG) RD is widely applied in studying chemical exchange ranging from ~200 to 2000 s−1 in proteins,19,20 its application to nucleic acids can be complicated due to extensive carbon–carbon scalar couplings,21 unless employing site-specific labeling schemes.6,7,22 Here, we describe an application of carbon chemical exchange saturation transfer (CEST) and low spin-lock field R1ρ RD experiments, which provided accurate characterization of slow chemical exchange in a fluoride riboswitch, occurring on the time scale that, as demonstrated in proteins,23 is challenging to be accurately quantified by CPMG RD and is difficult if not impossible to be studied by ZZ-exchange NMR spectroscopy.
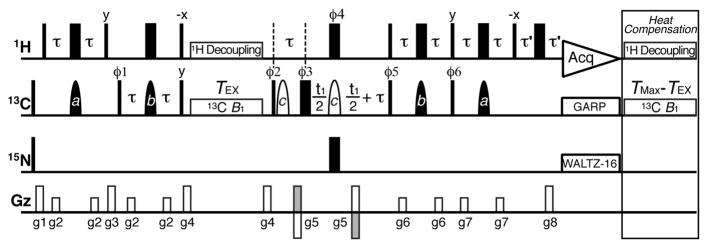
The saturation transfer type NMR experiment was originally developed by Forsen and Hoffman in the early 1960s.24 Recently, Clore and co-workers have developed a novel 2D 15N dark-state exchange saturation transfer (DEST) NMR experiment to study slow interconversion between peptide monomers and proto-fibrils.25 Kay and co-workers have subsequently developed a suite of 2D 1H, 13C, and 15N CEST NMR experiments, which have opened new routes to characterizing slow chemical exchange in proteins.23,26–29 Building upon the scheme by Kay and coworkers,23 we developed a nucleic-acid-optimized 2D 13C CEST experiment that uses a series of shaped pulses to selectively invert and refocus carbon magnetization of interest and to refocus carbon–carbon scalar coupling from neighboring carbons (Figure 1). A 90x240y90x composite pulse train,30 as previously described,23 is used for 1H decoupling to suppress C–H cross relaxation, dipolar–dipolar/carbon CSA cross-correlated relaxation, and the 13C multiplet structure in the CEST profile.23
Figure 1.
2D 13C CEST pulse sequence for characterizing slow chemical exchange in nucleic acids. Narrow (wide) rectangles are 90° (180°) pulses and closed (open) shapes are selective on (off) resonance 180° pulses. Delays are τ = 1/4JCH and τ′ = g8. Phase cycle is ϕ1 = {x,−x}, ϕ2 = {y}, ϕ3 = {2x,2y,2(−x),2(−y)}, ϕ4 = {4x,4(−x)}, ϕ5 = {4x,4(−x)}, ϕ6 = {4y,4(−y)}, receiver = {x,2(−x),x,−x,2(x),−x}. Briefly, 1H magnetization is transferred to 13C longitudinal magnetization, which relaxes under a weak 13C B1 field during TEX. 13C transverse magnetization then evolves during t1 and is returned to 1H for detection. Peak intensities are monitored as a function of B1 offset and power. See details in SI.
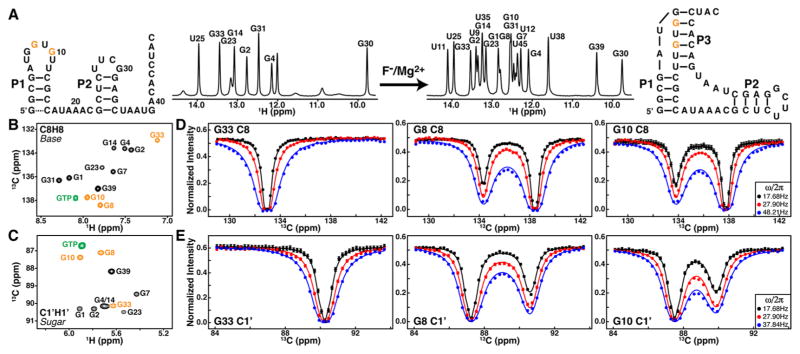
Riboswitches are an important class of noncoding RNAs that regulate gene expression by exposing or sequestering regulatory elements through base pairing in response to specific cellular cues.31 Tremendous progress in determining high-resolution ligand-bound structures has provided significant insights into the molecular basis of ligand recognition. However, high-resolution characterization of ligand-free riboswitches, which is essential for understanding the conformational landscape that underlies the “switching” process, is rather limited.8,32,33 Here, we applied the 13C-CEST experiment on a Bacillus cereus fluoride riboswitch in its ligand-free state (Figure 2). This recently discovered riboswitch regulates the transcription of putative fluoride transporters.34 The crystal structure of a ligand-bound fluoride riboswitch revealed a compacted pseudoknot that remarkably encapsulates a single fluoride ion in complex with three magnesium ions.35 However, this pseudoknot structure, which forms in solution in the presence of ligand, does not fold in the ligand-free state, as only signals from P1 and P2 stems are observed in the 1D imino 1H spectrum (Figure 2A). An individually guanosine 13C/15N labeled sample was prepared to provide well-distributed probes across all stem regions with greatly simplified NMR spectra. 2D 1H-13C HSQC spectra of nonexchangeable resonances from the ligand-free G-labeled sample further support the absence of pseudoknot. In particular, base (C8) and sugar (C1′) carbon chemical shifts of G8 and G10, which otherwise would be located in the center of the pseudoknot P3 stem, are significantly shifted toward those of a single GTP and far away from those of Gs residing within the P1 and P2 stems (Figure 2B,C). Thus, these conventional methods would strongly suggest that ligand binding induces pseudoknot formation.
Figure 2.
Quantification of an “invisible” ES in the ligand-free B. cereus fluoride riboswitch by 2D 13C CEST. (A) Secondary structures and 1D imino 1H spectra of the ligand-free and bound riboswitches. (B,C) 1H–13C HSQC spectra of base (C8) and sugar (C1′) region of a G 13C/15N labeled riboswitch. Overlaid on the spectra in green are resonances of a single GTP. Colored in orange are residues whose CEST profiles are shown. (D,E) 13C B1 field strength and carrier (in ppm) dependence of intensity profiles of base C8s and sugar C1’s for G33, G8, and G10. Solid lines represent the best fits to a two-state exchange process using the Bloch–McConnell equation.
Application of 13C-CEST to the ligand-free G-labeled riboswitch immediately revealed a hidden conformational state that is “invisible” to conventional NMR experiments. Shown in Figure 2D,E are representative 13C-CEST profiles of base and sugar carbons recorded at 30 °C with TEX = 0.3 s. While G33 from P2 displays single dips in peak intensity profiles that match peak positions in 1H-13C HSQC spectra, second intensity dips corresponding to an “invisible” excited state (ES) can be clearly seen for G8 and G10. Interestingly, except residues from P2, we observed either asymmetrically broadened intensity dips or more than one dip in intensity for all other guanosine residues. The nature of these is subject to further investigation. Here, we focus on G8 and G10, which not only have the most distinct carbon chemical shifts among all G residues in the free state but also display the most dramatic CEST profiles (Figure 2B–E). Base and sugar carbon CEST profiles of G8 and G10 were first fitted independently to a two-state model using the Bloch–McConnell equation36 to quantitatively extract ES carbon chemical shifts as well as kinetics and thermodynamics of the exchange between ES and ground state (GS) (see SI). Since carbon B1 fields from ~17 to ~48 Hz were used, small 1JC8-N7,9 (~15 Hz) couplings cannot be well-resolved and therefore were not included in the fitting of base (C8) CEST profiles. On the other hand, large 1JC1′-C2′ (~45 Hz) couplings have been taken into account in the data analysis of sugar (C1′) CEST profiles, as described previously.29
Excellent fits were obtained with the simple two-state model (Figure 2D,E and Table S1). Initial inspection of the ES carbon chemical shifts (ϖES = ϖGS + Δϖ), where Δϖ is the extracted chemical shift difference between ES and GS, immediately provides structural insights into the “invisible” ES. Base carbons (C8) of G8 and G10 are shifted −4.09 and −3.95 ppm to 134.30 and 133.80 ppm in the ES, respectively, which reside among resonances of residues within P1 and P2 stems with an average chemical shift of 133.47 ppm. Similarly, the ES sugar carbon (C1′) resonances are also among those from helical residues (Table S1). Thus, these carbon chemical shifts strongly indicate that both G8 and G10 are located within helical environments in the ES. The obtained exchange parameters from individual fits are also very similar, the rate of exchange (kex = kGE + kEG) ranges between 102 and 122 s−1 and the population of ES (pES) ranges between 9.8 and 10.4%, strongly suggesting a global exchange process. Global fitting of all CEST profiles of G8 and G10 to a single two-state model resulted in kex = 112 ± 4 s−1 and pES = 10.1 ± 0.1%, with the lifetime of ES being τ = 9.9 ± 0.3 ms. Collectively, these results indicate that G8 and G10 move concertedly from unfolded to helical-like states. Such a helical conformation, comprising both G8 and G10, would be consistent with the formation of the P3 stem, suggesting a pseudoknot-like ES in the absence of ligand. Given the importance of the pseudoknot conformation in ligand recognition, as observed in the crystal structure, this putative pseudoknot may represent a state that is selected by the ligand. It is also worth noting that, despite the presence of small and large scalar couplings, accurate exchange parameters can be extracted from all CEST profiles, consistent with a recent study in proteins showing that exchange parameters can be reliably extracted from 13CO CEST even in the presence of 13CO-13Cα scalar couplings.29
Interestingly, the CEST experiment is reminiscent of the R1ρ experiment,37 except that the magnetization of interest is kept along Z instead of being rotated into its effective magnetic field prior to spin lock. The developments of low spin-lock field R1ρ RD have allowed access to an exchange regime that is typically measured by CPMG RD.13–15 Exchange as slow as ~280 s−1 and ~370 s−1 has been reported in proteins14 and nucleic acids,16 respectively. However, it is not clear if even slower processes can be measured by low spin-lock field R1ρ RD. Previously, low spin-lock fields of ~25 Hz and ~90 Hz were demonstrated to ensure single-exponential relaxation decays of amide 15N nuclei in proteins14 and isolated 13C nuclei in nucleic acids,15 respectively. Consistent with these previous studies, we found that the spin-lock field can be roughly as low as ~45 Hz for C8 and ~150 Hz for C1′ (Figure S1), which is about three times larger than 1JC8–N7,9 and 1JC1′-C2′ couplings, respectively, to sufficiently lock the 13C multiplet components along the effective field. With this guidance, we applied the 1D selective R1ρ RD,15 with minor modifications, to measure dispersion of base and sugar carbons of G8 and G10 (see Supporting Information). We adapted a previously described constant-time approach14 where R1ρ values were obtained from a single delay period (TEX = 32 ms) and only the magnetization associated with the GS was prepared and rotated into the effective magnetic field prior to the spin-lock period. This initial magnetization condition was then taken into account in data analysis using the Bloch-McConnell equation36 (see Supporting Information).
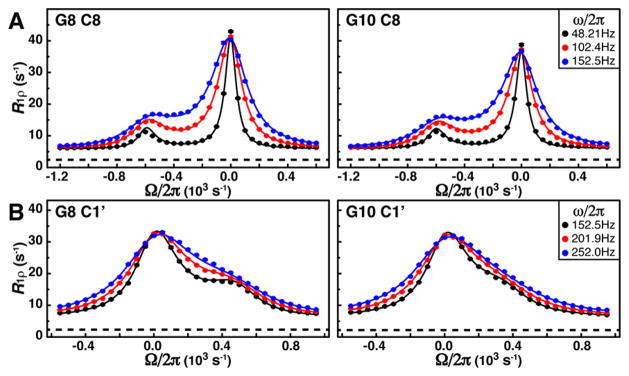
Shown in Figure 3 are R1ρ RD profiles of base and sugar carbons from G8 and G10. Clearly, the ES can be identified as either a second peak or a shoulder to the on-resonance peak in the profiles. We also noticed that the baselines of these RD profiles are much higher than the expected intrinsic longitudinal relaxation rates (R1) (Figure 3), while those from G33, which does not exhibit exchange, agree well (Figure S2). Simulation establishes that, if only GS magnetization is present prior to spin lock, such an elevation is an indication of a relatively highly populated ES (Figure S3). Independent fitting of each RD profile to a two-state model gave similar exchange parameters (kex ~ 110–120 s−1 and pES ~10–12%) (Table S1). Global fitting of all RD profiles gave values of kex = 121 ± 2 s−1 and pES = 10.8 ± 0.1%, which are essentially identical to those obtained from CEST. Extracted chemical shift differences for both base and sugar carbons are also in great agreement between the two methods (Table S1). For example, for C8 of G10, ΔϖR1ρ = −4.03 ± 0.02 ppm agrees very well with ΔϖCEST = −3.96 ± 0.01 ppm. Therefore, these two methods cross-validated each other as means to accurately study slow exchange. Moreover, to the best of our knowledge, this is also one of the slowest exchange processes that have been reported using R1ρ RD experiment.
Figure 3.
Quantification of slow exchange in the ligand-free fluoride riboswitch by low spin-lock field R1ρ RD. Shown are spin-lock power (ω/2π) and offset (Ω/2π) dependence of R1ρ for (A) base C8s and (B) sugar C1’s of G8 and G10, where Ω= ωrf − Ωobs is the frequency difference between spin-lock carrier (ωrf) and observed peak (Ωobs). Dashed lines are measured R1 rates. Solid lines represent the best fits to a two-state exchange process using the Bloch–McConnell equation.
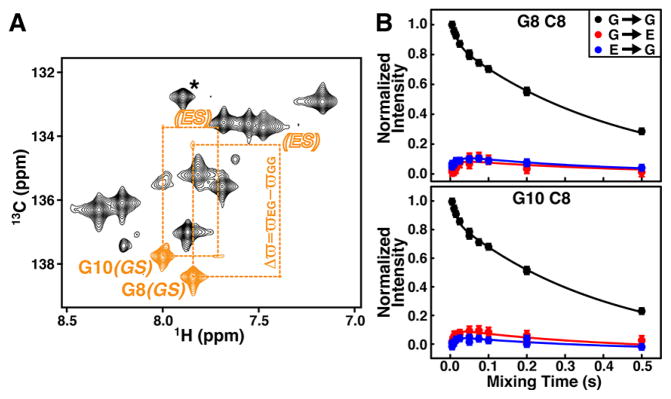
It is of interest to compare the advantage of CEST and low spin-lock field R1ρ RD in studying slow exchange involving lowly populated ES to ZZ-exchange (Figure S4), which is traditionally used to probe slow exchange, provided that exchanging states are sufficiently populated.38 Despite ES diagonal peaks being difficult to identify, very weak GS/ES cross-peaks can be observed for G8 and G10, allowing direct Δϖ measurement (Figure 4A). The obtained values are accurate and agree very well with those from CEST and R1ρ RD (i.e., ΔϖC8 (G10) = −4.03 ± 0.01 ppm, Table S1), providing independent validation of the latter methods in extracting ES chemical shifts. However, the obtained exchange parameters, such as kex(G8) = 65.4 ± 14.7 s−1 and kex(G10) = 90.0 ± 20.5 s−1, are much less accurate. Such deviations are likely due to the absence of ES peaks as well as weak cross-peak intensities, where, as shown in simulations, small measurement errors can result in large deviations in the extracted parameters (Figure S5), reflecting the limitation of ZZ-exchange in studying exchange involving lowly populated states.
Figure 4.
Characterizing slow exchange in the ligand-free fluoride riboswitch by ZZ-exchange. (A) 1H-13C spectrum of base C8s from ZZ-exchange at 75 ms mixing time. Colored in orange are diagonal peaks from GS and cross-peaks between GS and ES of G8 and G10. “Invisible” ES diagonal peaks are labeled as open letters. Asterisk is aliased G30 peak. (B) Mixing time dependence of diagonal and cross-peak intensities for G8 and G10 from ZZ-exchange. Solid lines represent the best fits to a two-state exchange process.
To this end, we wanted to explore the utility of CEST and low spin-lock field R1ρ RD in studying even slower exchange by carrying out measurements at a lower temperature (25 °C) and focusing on base carbons of G8 and G10. Despite only 5 °C difference, a more than 2-fold intensity decrease of these resonances made it prohibitively time-consuming to record 2D 13C-CEST with sufficient signal-to-noise, motivating us to develop a 1D 13C-CEST, which is built on the 1D 13C-R1ρ RD scheme15 (Figure S6). By focusing on one resonance at a time, the 1D scheme can provide 10–100 fold time savings compared with its 2D counterpart.14,15 We first validated the 1D scheme at 30 °C, where almost identical profiles were obtained between 1D and 2D CEST (Figure S7). We then performed 1D 13C CEST, R1ρ RD, and ZZ-exchange at 25 °C (Figure S8 and Table S2). The values of kex = 55.1 ± 3.3 s−1 and pES = 16.5 ± 0.5% were obtained from global fitting of CEST data. Remarkably, even at such slow exchange, R1ρ RD still gave quite accurate values of kex = 61.3 ± 0.8 s−1 and pES = 19.9 ± 0.2%. For ZZ-exchange, with a relatively higher ES population, the obtained kex = 44.0 ± 6.7 s−1 is in better agreement with CEST, yet, pES = 29.5 ± 7.2% still shows large deviation. Thus, these results further establish the robustness of CEST and low spin-lock field R1ρ RD experiments in quantifying slow exchange, especially when a sparsely populated state is involved in the exchange process.
In conclusion, we have presented NMR approaches for accurate quantification of slow chemical exchange in nucleic acids, which is difficult if not impossible for conventional methods. The versatility of employing very weak radiofrequency fields even in the presence of large C-C scalar couplings makes CEST particularly powerful in studying slow exchange in fully labeled nucleic acids. We further demonstrated that, despite the conventional R1ρ RD being widely considered as the method of choice for studying fast chemical exchange,37 the low spin-lock field R1ρ RD13–15 is indeed a powerful method in studying slow chemical exchange. From a practical point of view, since exchange processes in nucleic acids are often localized to confined regions,16,18 we suggest initial characterization using the 2D CEST scheme with a large offset step size (i.e., 50 Hz) at a relatively high B1 field (i.e., 25–50 Hz) to identify residues undergoing exchange. With this 2D blueprint, complete sets of selective 1D CEST and/or 1D R1ρ RD can be collected to thoroughly characterize the exchange process for key residues. The currently presented 2D/1D 13C CEST experiments, together with the 1D 13C R1ρ RD experiment,15 provide powerful tools to investigate slow chemical exchange. The robustness of these methods promises a unique opportunity to facilitate atomic understanding of slow conformational interconversion that is essential to many vital nucleic acid functions.
Supplementary Material
Acknowledgments
We thank Dr. Greg Young for maintenance of the NMR instruments. A.L.H. is supported by postdoctoral training grants from the National Science Foundation (OISE-0852964) and the Canadian Institutes of Health Research (CIHR). Q.Z. acknowledges start-up fund from the University of North Carolina at Chapel Hill and a grant from the March of Dimes Foundation.
Footnotes
The authors declare no competing financial interest.
Details of experiments and data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Nature. 2012;482:322. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinnenthal J, Buck J, Ferner J, Wacker A, Furtig B, Schwalbe H. Acc Chem Res. 2011;44:1292. doi: 10.1021/ar200137d. [DOI] [PubMed] [Google Scholar]
- 3.Bothe JR, Nikolova EN, Eichhorn CD, Chugh J, Hansen AL, Al-Hashimi HM. Nat Methods. 2011;8:919. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russu IM. Methods Enzymol. 2004;379:152. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- 5.Wenter P, Bodenhausen G, Dittmer J, Pitsch S. J Am Chem Soc. 2006;128:7579. doi: 10.1021/ja060344a. [DOI] [PubMed] [Google Scholar]
- 6.Kloiber K, Spitzer R, Tollinger M, Konrat R, Kreutz C. Nucleic Acids Res. 2011;39:4340. doi: 10.1093/nar/gkq1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wunderlich CH, Spitzer R, Santner T, Fauster K, Tollinger M, Kreutz C. J Am Chem Soc. 2012;134:7558. doi: 10.1021/ja302148g. [DOI] [PubMed] [Google Scholar]
- 8.Reining A, Nozinovic S, Schlepckow K, Buhr F, Furtig B, Schwalbe H. Nature. 2013;499:355. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- 9.Furtig B, Buck J, Manoharan V, Bermel W, Jaschke A, Wenter P, Pitsch S, Schwalbe H. Biopolymers. 2007;86:360. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 10.Lee MK, Gal M, Frydman L, Varani G. Proc Natl Acad Sci USA. 2010;107:9192. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoogstraten CG, Wank JR, Pardi A. Biochemistry. 2000;39:9951. doi: 10.1021/bi0007627. [DOI] [PubMed] [Google Scholar]
- 12.Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE. J Mol Biol. 2005;353:540. doi: 10.1016/j.jmb.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Massi F, Johnson E, Wang C, Rance M, Palmer AG., III J Am Chem Soc. 2004;126:2247. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 14.Korzhnev DM, Orekhov VY, Kay LE. J Am Chem Soc. 2005;127:713. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 15.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. J Am Chem Soc. 2009;131:3818. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 16.Nikolova EN, Kim E, Wise AA, O’Brien PJ, Andricioaei I, Al-Hashimi HM. Nature. 2011;470:498. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolova EN, Gottardo FL, Al-Hashimi HM. J Am Chem Soc. 2012;134:3667. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Nature. 2012;491:724. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer AG, 3rd, Kroenke CD, Loria JP. Methods Enzymol. 2001;339:204. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 20.Korzhnev DM, Kay LE. Acc Chem Res. 2008;41:442. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 21.Lundstrom P, Hansen DF, Kay LE. J Biomol NMR. 2008;42:35. doi: 10.1007/s10858-008-9260-4. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JE, Jr, Hoogstraten CG. J Am Chem Soc. 2008;130:16757. doi: 10.1021/ja805759z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallurupalli P, Bouvignies G, Kay LE. J Am Chem Soc. 2012;134:8148. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 24.Forsen S, Hoffman RA. J Chem Phys. 1963;39:2892. [Google Scholar]
- 25.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Nature. 2011;480:268. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouvignies G, Kay LE. J Phys Chem B. 2012;116:14311. doi: 10.1021/jp311109u. [DOI] [PubMed] [Google Scholar]
- 27.Bouvignies G, Kay LE. J Biomol NMR. 2012;53:303. doi: 10.1007/s10858-012-9640-7. [DOI] [PubMed] [Google Scholar]
- 28.Hansen AL, Bouvignies G, Kay LE. J Biomol NMR. 2013;55:279. doi: 10.1007/s10858-013-9711-4. [DOI] [PubMed] [Google Scholar]
- 29.Vallurupalli P, Kay LE. Angew Chem, Int Ed Engl. 2013;52:4156. doi: 10.1002/anie.201209118. [DOI] [PubMed] [Google Scholar]
- 30.Levitt MH. J Magn Reson. 1982;50:95. [Google Scholar]
- 31.Serganov A, Patel DJ. Annu Rev Biophys. 2012;41:343. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haller A, Souliere MF, Micura R. Acc Chem Res. 2011;44:1339. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 33.Liberman JA, Wedekind JE. Wiley Interdiscip Rev RNA. 2012;3:369. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Science. 2012;335:233. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren A, Rajashankar KR, Patel DJ. Nature. 2012;486:85. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McConnell HM. J Chem Phys. 1958;28:430. [Google Scholar]
- 37.Palmer AG, III, Massi F. Chem Rev. 2006;106:1700. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 38.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. J Biomol NMR. 1994;4:727. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.