Abstract
The cell cycle is integrated with many aspects of embryonic development. Not only is proper control over the pace of cell proliferation important, but also the timing of cell cycle progression is coordinated with transcription, cell migration, and cell differentiation. Due to the ease with which the embryos of aquatic organisms can be observed and manipulated, they have been a popular choice for embryologists throughout history. In the cell cycle field, aquatic organisms have been extremely important because they have played a major role in the discovery and analysis of key regulators of the cell cycle. In particular, the frog Xenopus laevis has been instrumental for understanding how the basic embryonic cell cycle is regulated. More recently, the zebrafish has been used to understand how the cell cycle is remodeled during vertebrate development and how it is regulated during morphogenesis. This review describes how some of the unique strengths of aquatic species have been leveraged for cell cycle research and suggests how species such as Xenopus and zebrafish will continue to reveal the roles of the cell cycle in human biology and disease.
Keywords: Differentiation, G1-phase, G2-phase, Gastrulation, Midblastula Transition, Sphase, Xenopus, Zebrafish
1. Introduction
Any biologist who has had the opportunity to watch and experiment with a developing fish or frog embryo would appreciate why embryologists have studied the development of aquatic animals for centuries. The embryos of many aquatic species are extraordinarily accessible for observation and manipulation. Aristotle (384 BC–322 BC) studied embryos from aquatic animals, comparing their development to other animals including humans and chickens. Even without the use of the microscope, Aristotle observed how species begin embryonic development by one of two major cell division patterns: holoblastic (in which the entire egg is divided into smaller cells, as in frogs) and meroblastic (in which the egg is separated into a yolk cell and the cells of the embryo proper, as in fish). The early cell divisions that Aristotle could observe in aquatic embryos are still the focus of many biologists today. In fact, studies of the easily observable cell divisions in the early embryos of sea urchins, frogs, and starfish, along with landmark studies in yeast and Drosophila melanogaster, have contributed greatly to our understanding of how the human cell cycle is controlled at the molecular level. We have much to learn about how the cell cycle is regulated during embryonic development and tissue homeostasis, and it is likely that aquatic model systems will continue to serve us well. The purpose of this review is to highlight how aquatic species have contributed to our understanding of the molecular mechanisms of cell cycle control and how aquatic models can still be used to address unanswered questions about how the cell cycle is coordinated with morphogenesis and cellular differentiation.
2. Fundamentals Revealed Through Studying the “Simple” Embryonic Cell Cycle
A major advantage of aquatic model organisms is that their oocytes and embryos develop externally and are relatively large, so they are particularly amenable to experimental manipulations such as microinjection. It is also extremely helpful that large numbers of oocytes, eggs or embryos can be collected at the same stage. This feature has enabled the biochemical analysis of the cell divisions occurring during oocyte maturation and early embryonic development. Such analyses have been important for the discovery of the fundamental mechanisms of the cell cycle.
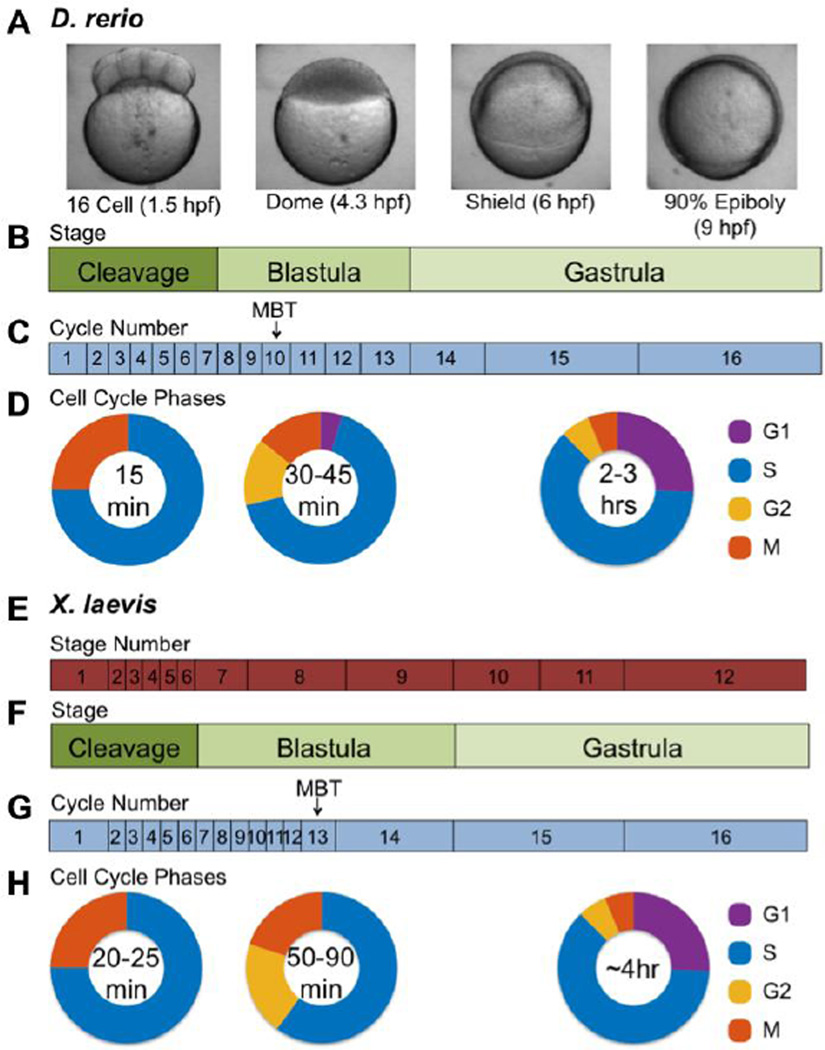
Many aquatic organisms begin development with a period of exponential cell expansion via a series of synchronous and rapid embryonic cell cycles. These early embryonic cell cycles of frogs and other aquatic organisms are simplified versions of the cell cycles of somatic cells (Evans et al., 1983; Kane and Kimmel, 1993; Newport and Kirschner, 1982a). These cycles differ from the canonical four-phase cell cycle in four important ways. First, an autonomous biochemical oscillator drives them, which is unaffected by developmental signals or checkpoints (Ikegami et al., 1997; Kane and Kimmel, 1993; Kimelman et al., 1987; Newport and Kirschner, 1982a; Newport and Kirschner, 1984). Second, they are extremely rapid and lack the gap phases (G1 and G2), which separate S-phase from mitosis (Figure 1). Third, they are characterized by a lack of growth in cytoplasmic volume, so the large egg cytoplasm is progressively cleaved into a large number of smaller nucleated cells. Finally, they occur without zygotic transcription, so they are entirely controlled by maternally provided mRNA and protein. For example, the frog Xenopus laevis undergoes twelve cleavage cycles to produce thousands of cells; and the zebrafish (Danio rerio) undergoes ten cleavage cycles (Kane and Kimmel, 1993; Newport and Kirschner, 1982a)(Figure 1). Thus, in these aquatic species, the egg cytoplasm contains all the protein and mRNA necessary for multiple complete embryonic cell cycles. The “loaded” egg cytoplasm of aquatic species, especially amphibians, has enabled cell biologists and biochemists to develop cell-free systems to study the embryonic cell cycle. These widely used systems were developed during the discovery of key regulators of the cell cycle, the Cyclins.
Figure 1. Illustration of important changes from fertilization through gastrulation in the zebrafish.
Images and timelines of the first (A) 10 hours of D. rerio development or (B) 14 hours of X. laevis development are shown above the corresponding cell cycle times and cell cycle phasing estimated from our unpublished data and published studies (Heasman, 2006; Kane and Kimmel, 1993; Kimmel et al., 1994; Nieuwkoop and Faber, 1994; Webb and Miller, 2006).
Studies of frog oocyte maturation laid the foundation for the discovery of Cyclins. Oocytes are arrested in prophase of meiosis I. The hormone progesterone triggers oocyte prophase exit and cell division (Masui, 1967). Using the frog Rana pipiens, Yoshio Masui showed that effects of progesterone could be mimicked by injecting cytoplasm from a maturing oocyte into a resting one, demonstrating that a soluble cytoplasmic factor, which was named maturation promoting factor (MPF), could induce meiotic progression (Masui and Markert, 1971). This result was repeated in the frog X. laevis, and through microinjection experiments in these two species, it was demonstrated that MPF activity is also present in the cytoplasm of mitotic cells, and it oscillates during the cell cycle, peaking in metaphase and disappearing with the completion of mitosis (Gerhart et al., 1984; Sunkara et al., 1979). MPF was shown to be an enzymatic activity that causes an increase in overall protein phosphorylation (Maller et al., 1977). We now know that MPF is a Cyclin-dependent kinase (CDK), which is composed of the Cdc2 catalytic subunit and Cyclin B regulatory subunit. Like the MPF, the discovery of Cyclin could be credited to the clever exploitation of highly accessible and manipulable oocytes and embryos of aquatic animals.
Tim Hunt and colleagues discovered Cyclin proteins when they noticed that a few proteins were preferentially translated upon sea urchin and clam egg fertilization, and those proteins were destroyed during the first mitosis and then resynthesized in the next interphase (Evans et al., 1983). The periodic expression of those proteins mirrored the periodic MPF activity that had been described earlier. Indeed, Cyclins activate MPF, but proving that fact involved the development of cell-free systems that recapitulate the embryonic cell cycle in frogs.
To study the MPF, Manfred Lohka and Yoshio Masui took advantage of the transformative activity of frog egg cytoplasm to develop a system in which a single cell cycle takes place in vitro (Lohka and Masui, 1983). Improvement of that system by Lohka and Maller enabled the first biochemical purification of MPF, which was comprised of two proteins (Lohka et al., 1988). One of the two proteins was demonstrated to be the Xenopus homolog of the S. pombe cell cycle kinase p34cdc2; the other protein was shown to be Cyclin B (Dunphy et al., 1988; Gautier et al., 1990; Gautier et al., 1988; Lohka et al., 1988). Together Cdc2 and Cyclin B form the prototypical Cyclin-dependent kinase (CDK) complex that drives mitosis. Cyclin B-Cdc2 is just one of several CDK complexes that promote cell cycle progression; other CDKs are critical for G1- and S-phase progression.
The discovery and molecular definition of MPF and Cyclin demonstrates the power of aquatic model systems for both embryology and biochemistry. Derivatives of the frog egg cytoplasmic extracts have continued to be extraordinarily productive in the cell cycle field. Blow and Laskey showed that the Xenopus derivative of the Lohka and Masui system could be used to study the initiation and completion of DNA replication in a cell-free system (Blow and Laskey, 1986). Murray and Kirschner developed the egg extract system even further, and showed that their improved system could drive multiple successive cell cycles (Murray and Kirschner, 1989). Variations in the Xenopus egg extract protocols have been optimized to study different aspects of cell biology including DNA replication licensing and initiation; sister chromatin cohesion; mitosis; the DNA damage and cell cycle checkpoints; and DNA repair (Dasso and Newport, 1990; Hekmat-Nejad et al., 2000; Hyrien et al., 1995; Kumagai et al., 1998; Lafont et al., 2010; Raschle et al., 2008; Sheehan et al., 1988; Walter et al., 1998). Just as aquatic model organisms have been used to study the fundamental mechanisms of cell cycle control, they have also been used to understand how the cell cycle changes during development. Organisms such as Xenopus and zebrafish have been useful for studying how the basic embryonic cell cycle is remodeled into the more complex somatic cell cycle.
3. Using Aquatic Model Organisms to Understand Cell Cycle Remodeling
Early development is associated with dramatic changes in cell cycle dynamics. The cell cycle is remodeled throughout the cleavage, blastula, gastrula, and segmentation stages, during which time critical changes in transcription, cell motility and cellular differentiation also occur. The early embryonic cell cycles of cleavage and early blastula embryos, which have been useful for elucidating the mechanisms of the “core” cell cycle engine, lack additional layers of regulation that are found in somatic proliferating cells. Ultimately the rapid cleavage embryo cell cycles with alternating S and M phases are transformed into mature cell cycles with G1 and G2 gap phases and extended S phases (Figure 1). This cell cycle remodeling begins halfway through the blastula stage of development, the midblastula transition (MBT). Cell cycle changes at the MBT are coincident with other important developmental changes including the onset of zygotic transcription and cell motility. After the MBT, the cell cycle continues to lengthen, and additional controls over cell cycle transitions are introduced. The mechanisms underlying cell cycle remodeling are not completely known, but its importance for proper embryonic development is clear. The addition of G1 and G2 allows for cell division to be coordinated with DNA repair, cell differentiation, and morphogenesis. There is also increasing evidence that S-phase lengthening represents a spatiotemporal reorganization of replication forks that is important for proper cellular differentiation and genome stability. Cell cycle remodeling during early vertebrate development has been studied extensively in frogs and fish, so we mainly describe what is known about remodeling of the X. laevis and D. rerio cell cycles.
4. Timing the Midblastula Transition
The MBT represents a shift from a maternal developmental program to a program under zygotic control. Studies in frogs have contributed greatly to our understanding of how the MBT is timed so precisely during development. In a landmark study, Newport and Kirschner showed that transcription is unnecessary in X. laevis cell cycle lengthening at the MBT, so a maternal timer or counting mechanism triggers the first stages of cell cycle changes (Newport and Kirschner, 1982a). Remarkably, neither an endogenous clock nor mechanisms counting the rounds of cell division or DNA replication trigger the onset of cell cycle lengthening, cell motility, or transcription. Instead, all three events depend on the ratio of nuclear to cytoplasmic (N:C) volume (Newport and Kirschner, 1982a, f). This observation suggested that the MBT in X. laevis is triggered by the titration of one or more limiting factor(s) by nuclei. The MBT timing mechanism appears to be conserved in vertebrates, since transcription, cell motility, and cell cycle lengthening are also controlled by the N:C ratio in zebrafish (Kane and Kimmel, 1993). Recent work has revealed more detail about how the cell cycle changes after the MBT, and strong candidates for the proposed titrated limiting factors have been identified (Amodeo et al., 2015; Collart et al., 2013; Murphy and Michael, 2013).
Complete remodeling of the embryonic cell cycle involves the slowing and restructuring of S-phase and the addition of G1 and G2 (Figure 1). Earlier studies indicated that the cell cycle lengthens in X. laevis because of gradually increasing durations of all three of those cell cycle phases (Graham and Morgan, 1966). In a more detailed analysis, Masui et al. showed that the cell cycle changes in a more step-wise pattern; initially there is an expansion of S-phase, which is followed by the introduction and gradual lengthening of G2 (Iwao et al., 2005). The addition of G1 occurs during gastrulation in X. laevis, 2–3 hours after the introduction of G2 (Iwao et al., 2005). This pattern may also be conserved in zebrafish. The lengthening of the cell cycle at the zebrafish MBT is associated with the introduction of G2, and it does not require transcription (Dalle Nogare et al., 2009). In contrast, the G1 phase appears later and depends on zygotic transcription (Zamir et al., 1997)(Figure 1). The regulation of S-phase during zebrafish development has not been examined. Interestingly, the S-G2-G1 order of cell cycle remodeling is also conserved in the fruit fly (D. melanogaster), so it may be a common theme among metazoans (Farrell et al., 2012).
5. Remodeling S-Phase
5.1 Replication Fork Initiation
The S-phases of cleavage cycles are extremely short. In fact, S-phase in the early embryos represents the minimum S-phase length documented for any cell type of that species. To understand the changes that occur in S-phase during embryonic development, it is useful to explore which of the following factors influence the length of S-phase: fork rate, fork number, and replication timing.
The genomes of higher eukaryotes are replicated through the concerted activity of tens-of-thousands of replication forks. Altering the rate at which individual replication forks synthesize DNA and/or changing the total number of active forks in each S-phase could modulate the overall rate of DNA synthesis. Johannes Walter and John Newport used the cell-free X. laevis egg extract system to define the critical factors affecting S-phase length before and after the MBT (Walter and Newport, 1997). As the N:C ratio approaches the MBT level in the egg extract system, S-phase lengthens through an increase in the number of forks initiating in a given stretch of DNA (Walter and Newport, 1997). In contrast, S-phase elongation is not caused by a slowing of replication forks at the MBT (Walter and Newport, 1997). Thus, a decrease replication fork initiation is at least partly responsible for S-phase lengthening at the MBT in X. laevis.
There is now strong evidence that a reduction in replication fork initiation drives overall cell cycle lengthening at the MBT. As described above, it has been proposed that the titration of maternal cytoplasmic factors by nuclei times the MBT (Collart et al., 2013). Collart et al. tested whether proteins necessary for replication initiation were the titrated factors. The cellular levels of four replication initiation factors - Cut5, RecQL4, Ticrr/Treslin, and Drf1 (CRTD) - are reduced as Xenopus pre-MBT cells divide. Overexpression of the CRTD factors prevented the decrease in replication fork initiation that normally occurs at the MBT. Remarkably, CRTD overexpression also prevented the overall lengthening of the cell cycle, suggesting that a reduction in replication forks is a principal cause of cell cycle changes at the MBT. Proper control over replication initiation is critical for development, as embryos overexpressing the CRTD factors failed to properly express a fraction of zygotic genes and did not complete gastrulation (Collart et al., 2013).
5.2 Replication Timing
In somatic cells, the genome replicates according to a cell-type specific spatiotemporal pattern, in which different segments of the genome and compartments of the nucleus replicate during early or late S-phase (Taylor, 1960). The replication-timing program is largely a result of early or late initiation of replication forks in large chromosomal domains. Units of replication timing can be measured along linear chromosomal segments or in subnuclear replication foci (also called factories) (Hand, 1975; Jackson and Pombo, 1998; Leonhardt et al., 2000; Ma et al., 1998; O'Keefe et al., 1992). Chromosomal replication-timing domains of somatic cells hundreds-of-kilobases in length, and one or more clusters of synchronously firing forks replicate each domain (Hiratani et al., 2008; Karnani et al., 2007). At the level of the nucleus, timing domains are spatially organized into subnuclear foci (also called replication factories), and multitudes of simultaneously active replication factories create patterns of foci that can be used to classify early-, mid-, or late-S-phase cells (Leonhardt et al., 2000). Replication timing is developmentally regulated, and each cell type has a unique replication-timing profile (Rivera-Mulia et al., 2015). In mammalian cells, genome-wide replication-timing profiles strongly correlate with maps of three-dimensional chromatin interactions, so replication timing is highly related to higher order chromatin organization (Pope et al., 2014). Replication timing also has been linked to transcriptional control and genome stability (Donley and Thayer, 2013; Hiratani et al., 2009). It is unknown how the replication-timing program develops after the MBT in organisms such as frogs and fish. This is a fertile area of research because replication timing has been connected to many other cellular and developmental processes. Thus, understanding when and how the replication-timing program is formed will very likely clarify functional relationships between DNA replication and other aspects of cell and developmental biology.
There is some evidence that DNA replicates in a temporal pattern in X. laevis cleavage cycles, but it is clear that the level of spatiotemporal organization of replication observed in somatic cells is absent in pre-MBT cells. In the Xenopus egg extract system, replication forks initiate synchronously within clusters to replicate 50–100 kilobase chromosomal segments, and different clusters initiate at different times during S-phase (Blow et al., 2001). Although the precise locations of the replication origin clusters are flexible, the replication timing of large chromosomal domains (1–5 megabases) is repeated in successive cell cycles (Labit et al., 2008). Thus, a rudimentary replication-timing pattern exists in the cleavage cell cycles of early embryos. The determinants of replication timing at this stage are unknown. The early timing program might represent some aspect of DNA sequence or basic chromatin structure that is already established before the MBT. Nonetheless, it is likely that the replication-timing program changes significantly after the MBT, as it is known that there are substantial changes in replication origin spacing, chromatin structure, and nuclear organization as cells progress past the MBT (Danis et al., 2004; Hyrien et al., 1995; Leibovici et al., 1992; Lemaitre et al., 1998; Vassetzky et al., 2000).
The first factor that might influence changes in replication timing at the MBT is the overall efficiency of replication fork initiation. The synchrony of origin firing is reduced, and the genomic positions of replication origins become more restricted as cells progress through MBT (Hyrien et al., 1995; Labit et al., 2008). These changes in the initiation of replication forks indicate that DNA replication begins to follow a somatic cell spatiotemporal pattern at the MBT. The emergence of this pattern could be caused by limiting levels of the CRTD initiation factors (Collart et al., 2013). This possibility would fit an attractive model in which timing patterns are a result of a competition between origins for limiting diffusible initiation factors (Bechhoefer and Rhind, 2012). Indeed, overexpression of the yeast CRTD homologs partially disrupts the normal temporal replication initiation pattern in Saccharomyces cerevisiae (Mantiero et al., 2011). Thus, the lengthening of S-phase at the MBT might result from both an overall reduction in replication fork number, and a delay in replication of regions of the genome that are outcompeted for the CRTD factors.
The idea that limiting replication factors could cause specific areas of the genome to become late replicating would need to invoke additional models to explain how early replicating segments of the genome compete better for the CRTD factors. This could be due to higher origin density or increased accessibility to initiation factors. Interestingly, recent mathematical models have shown that DNase hypersensitivity can accurately predict replication timing profiles, even when late origins are assigned the same efficiency as early origins, suggesting chromatin structure has a profound effect on origin initiation, replication timing, and S-phase remodeling (Gindin et al., 2014).
Changes in local chromatin structure at the MBT could contribute to the restructuring of S-phase at the MBT. Studies using zebrafish and Xenopus embryos have been especially useful for mapping the dynamics of epigenetic marks during vertebrate development. Several prominent histone modifications are not present at significant levels prior to zygotic genome activation but greatly increase after the MBT, when large-scale changes in chromatin modifications occur (Lindeman et al., 2011; Vastenhouw et al., 2010). Well-positioned nucleosome arrays are also established at the MBT (Zhang et al., 2014). Early zebrafish and Xenopus embryos express a maternal linker histone, which is substituted by somatic histone H1 variants starting at the MBT (Muller et al., 2002; Steinbach et al., 1997). Linker histones may be particularly important for controlling replication initiation, as studies in Xenopus extracts have shown that histone H1 represses initiation, resulting in reduced origin firing [35]. Chromatin associated proteins such as HP1, polycomb repressive complex (PRC), and chromatin remodeling complexes are known to be involved in regulating chromatin architecture, but the timing of their association with chromatin has not been assessed in aquatic embryos. In Drosophila, heterochromatin compaction appears to precede S-phase lengthening at the MBT, and HP1 accumulation does not occur until after late replication and S-phase lengthening (Shermoen et al., 2010). Whether chromatin modification or chromatin compaction influences the establishment of a DNA replication-timing program after the MBT in aquatic organisms is an unanswered question.
Large-scale nuclear reorganization after the MBT might influence the development of the timing program, as it has been demonstrated that higher-order chromatin organization significantly correlates with replication timing in many somatic cell types (Gilbert et al., 1995; Lawlis et al., 1996; Ryba et al., 2012; Ryba et al., 2010). Experiments with a variation of the X. laevis extract system have demonstrated that replication timing is established in G1, before S-phase begins. Xenopus egg extracts are capable of driving S- phase in Chinese hamster ovary (CHO) nuclei (Gilbert et al., 1995). Remarkably, when introduced into Xenopus egg extracts CHO nuclei replicate according to their natural spatiotemporal pattern, but only if they have passed beyond a specific time in G1 called the timing-decision point (TDP)(Dimitrova and Gilbert, 1999). The TDP coincides with a period in G1 when chromosomes are stably repositioned in the nucleus after mitosis (Dileep et al., 2015). Thus, the length of G1 might affect the higher order organization of the chromosomes, and thereby influence replication timing. Given that the pre-MBT cell cycles lack a G1 phase, it would be of great interest to examine whether the replication-timing program is established in aquatic species before the G1 phase is added to the cell cycle (Kane and Kimmel, 1993; Newport and Kirschner, 1982f).
Although limiting replication factors and changing chromatin architecture might be sufficient to establish the somatic replication-timing program in vertebrates, work from Drosophila suggests that more direct regulation of CDK might be involved. In the early Drosophila embryo, cycle 14 is significantly lengthened by prolongation of S-phase and a developmentally regulated pause in G2 (Edgar et al., 1994). The G2 arrest is caused by inhibition of Cdc2, the catalytic subunit of mitotic CDK (Edgar et al., 1994). Farrell et al recently examined the mechanism underlying the prolongation of the preceding S-phase (Farrell et al., 2012). Surprisingly, downregulation of Cdc2 was both necessary and sufficient for late replication and S-phase lengthening (Farrell et al., 2012). Preventing Cdc2 downregulation during cycle 14 effectively caused an additional cleavage cycle by keeping S-phase short and maintaining the mitotic entry time (Farrell et al., 2012). This demonstrated that the timing of origin firing is more influenced by active regulatory mechanisms than just the formation of compact heterochromatin in early Drosophila embryos. Interestingly, Cdc2 is also downregulated at the zebrafish and Xenopus MBT, but overriding this control over mitotic CDK has a different outcome in zebrafish; instead of promoting an additional cleavage cycle, it causes cells to enter mitosis before completing DNA replication (Dalle Nogare et al., 2009). This demonstrates that mitotic CDK downregulation is not necessary for S-phase lengthening in vertebrates.
6. Controlling Entry Into Mitosis
6.1 Checkpoint Control Over Mitotic Entry
The continuous oscillations in Cdk activity cause pre-MBT cells to synchronously enter mitoses with very precise timing. The oscillating timer of the early embryo lacks additional controls over the start of S-phase or mitosis, which are essential in somatic cells. For example, the timing of X. laevis and zebrafish cell divisions prior to the MBT is largely unaffected by inhibitors of DNA replication or DNA damaging agents (Ikegami et al., 1997; Kimelman et al., 1987). In contrast, somatic cells rely on evolutionarily conserved checkpoint pathways that block mitotic entry when DNA replication is inhibited or DNA is damaged. The DNA replication and DNA damage checkpoints are mechanistically related, and they both control mitotic entry through inhibitory phosphorylation of Cdc2 (Jin et al., 1996). The G2/M transition is also regulated through Cdc2 phosphorylation during morphogenesis, enabling the coordination of cell division with migration and differentiation (Bouldin et al., 2014).
Although Cdc2 phosphorylation is not influenced by checkpoints prior to the MBT, it does function as part of the autonomous Cdk oscillator in early X. laevis cell cycles. The inhibitory phosphorylation of Cdc2 is modulated by the opposing activities of the Wee1 kinases and Cdc25 phosphatases. Two members each of the Wee1 kinase family (Wee1A and Myt1) and the Cdc25 phosphatase family (Cdc25A and Cdc25C) are maternally provided in X. laevis (Okamoto et al., 2002). Cdc2 inactivates the Wee1 kinases to make a double negative feedback loop, while it activates the Cdc25 phosphatases to complete a positive feedback loop (Mueller et al., 1995). Computational modeling as well as experiments using the X. laevis egg extract system have demonstrated that the Cdc2/Cdc25 and Cdc2/Wee1 loops function in a bistable switch that sharpens the timing of mitotic entry once Cyclin B accumulates to a threshold level (Pomerening et al., 2005). Thus, the Cdc25-Cdc2-Wee1 axis is employed before the MBT in X. laevis even though it is not yet engaged by checkpoints or developmental signals. It is unclear whether this is true in other vertebrate species. In contrast to X. laevis, injection of non-phosphorylatable Cdc2 (Cdc2AF) or Cdc25 into zebrafish embryos does not accelerate pre-MBT cell cycles, suggesting that Cdc2 phosphorylation may not be important for the autonomous cell cycle oscillator in pre-MBT zebrafish (Dalle Nogare et al., 2009; Kim et al., 1999).
At the MBT, the role for Cdc2 phosphorylation changes significantly. The basal level of Cdc2 phosphorylation increases sharply, indicating that the Cdc25-Cdc2-Wee1 circuit is engaged to either introduce a G2 phase or to delay mitotic entry as part of the DNA damage or replication checkpoints (Hartley et al., 1996). In zebrafish, the increased phosphorylation of Cdc2 coincides with lengthening of the cell cycle (Dalle Nogare et al., 2009). Overriding the inhibition of Cdc2, by overexpressing Cdc25 or a nonphosphorylatable form of Cdc2, prevented the delay in mitotic entry that occurs normally at the zebrafish MBT (Dalle Nogare et al., 2009). Unlike Drosophila, in which Cdc25 overexpression shortened S-phase, deregulation of Cdc2 phosphorylation in zebrafish caused mitotic catastrophe, a sign that cells had entered mitosis with incompletely replicated DNA (Farrell et al., 2012). Thus, it seems that the critical function of Cdc2 phosphorylation at the zebrafish MBT is in the DNA replication checkpoint, which ensures that mitosis does not occur until after S-phase is complete. The need for the DNA replication checkpoint likely arises at the MBT because of S-phase lengthening, but changes in S-phase in zebrafish development have not been studied. A gap between S-phase and mitosis, a G2 phase, can be detected at the zebrafish MBT, but it is unknown whether this gap is functionally significant or is simply a result of a lag in checkpoint recovery (Dalle Nogare et al., 2009). In X. laevis, there is evidence that the delay in mitotic entry at the MBT is a result of S-phase lengthening rather than the addition of a G2 phase per se. As described above, overexpression of the limiting CRTD initiation factors prevented cell cycle lengthening by keeping S-phase short; thus, an independent mechanism delaying mitotic entry does not seem to exist at the MBT (Collart et al., 2013). Overall work from X. laevis and zebrafish indicates that checkpoint control over mitotic entry is added and is critical for preventing mitotic catastrophe at the MBT. A true G2 phase is added later, but the mechanism of G2 addition in vertebrates is still unknown.
6.2 Developmental Control Over Mitotic Entry
Recent work has shown that control over the G2/M transition is important in zebrafish tail morphogenesis. Bouldin et al. used a photo-convertible protein to track cell divisions in posterior progenitor cells (Bouldin et al., 2014). This system allowed them to observe how cell divisions are regulated during gastrulation, somitogenesis and at the onset of differentiation. By time-lapse imaging they determined that rapid cell divisions during gastrulation were followed by a quiescent period during which posterior progenitor cells took up to 8 hours to divide. Using a FUCCI (fluorescent ubiquitination-based cell cycle indicator) transgenic line, Bouldin et al. revealed that the quiescent cells were arrested in either S or G2, and they showed that the S/G2 arrest coincided with a downregulation of Cdc25. Forced mitotic entry caused by Cdc25 overexpression resulted in defects in cellular differentiation and morphogenesis. The work from Bouldin et al is another example of noncanonical developmental regulation of cell division that is outside of the G1 phase (Bouldin et al., 2014; Edgar and O'Farrell, 1989). This work also nicely demonstrates how using externally developing aquatic animals with state-of-the-art fluorescent reporters is a powerful way to gain insight into the cell cycle control during development.
7. Introducing the G1 Phase
The previous example not withstanding, control over the G1/S transition plays a very important role in the control of cell division (van den Heuvel and Dyson, 2008). The prevalence of human tumor suppressor mutations that disrupt control over the G1/S transition demonstrates the importance of G1 in somatic cells. In mammals, two groups of CDKs promote the G1/S transition. First, Cdks 4 and 6 with D-type Cyclins phosphorylate three members of the pocket protein family (pRb, p107 and p130). Pocket proteins regulate gene transcription by interacting with members of the E2F transcription factors. Cyclin D-Cdk4/6 phosphorylation of pocket proteins induces E2F-dependent transcription of key cell cycle genes such as E- and A-type Cyclins. The Cyclins E and A complex with Cdk2, and they stimulate the firing of replication forks through the phosphorylation of the Ticrr/Treslin protein during S-phase (Boos et al., 2011; Kumagai et al., 2011; Sansam et al., 2015). Somatic cells periodically express Cyclins E and A through a combination of transcriptional control and the cyclical activity of E3 ubiquitin ligases. In addition to the periodic expression of Cyclins, the G1/S CDKs are also regulated by inhibitory phosphorylation, through the activities of Wee1 and Cdc25 proteins, and by two families of CDK inhibitors.
Despite the importance of the G1/S transition, little is known about how the G1 phase is added to the embryonic cell cycles of aquatic species, nor is it known when it first becomes important for cellular differentiation or morphogenesis. Zebrafish develop a gap between mitosis and S-phase at the MBT, but the gap remains relatively short until gastrulation (Figure 1) (our unpublished data) (Zamir et al., 1997). Abrupt lengthening in G1 during gastrulation coincides with the determination of germ layer fates, so G1 expansion may represent an evolutionarily conserved role for G1 in cellular differentiation (Ho and Kimmel, 1993; Kimmel and Warga, 1986; Zamir et al., 1997). Gene expression changes also support that a prototypical vertebrate G1 phase is added during gastrulation in zebrafish. The mRNA levels of G1/S regulators such as Cyclin D1, Rb-like 1, and p21 increase at that time (Duffy et al., 2005). Knockdown of Cyclin D1 in zebrafish causes microphthalmia and microcephaly, phenotypes commonly associated with reduced proliferation (Amsterdam et al., 2004; Sansam et al., 2010; Sansam et al., 2006). Mutation of zebrafish pRb causes a delay in the differentiation of retinal ganglion cells, possibly because of a failure in cell cycle exit (Gyda et al., 2012).
Like in zebrafish, a gap between mitosis and S-phase develops shortly after the MBT, but a prominent G1 phase does not appear until gastrulation (Frederick and Andrews, 1994; Graham and Morgan, 1966; Iwao et al., 2005). Cyclin D1-Cdk4 is undetectable in X. laevis prior to the MBT (Hartley et al., 1996). E2F transcription factors are present even before the MBT, and they are already capable of binding DNA (Philpott and Friend, 1994). E2F-dependent transcription, however, is not activated until the MBT, at which time a dominant negative form of E2F blocks proliferation (Tanaka et al., 2003). pRb binding to E2F can be detected at very low levels prior to the MBT, but it does not bind to E2F significantly until much later than the first signs of a G1 phase (Philpott and Friend, 1994). In agreement with little or no binding of pRb to E2F, neither pRb knockdown nor overexpression has any affect on cellular proliferation during early X. laevis development (Cosgrove and Philpott, 2007). In fact, pRb is largely found in the hyperphosphorylated form in early X. laevis embryos, indicating that Cyclin D-Cdk4/6 effectively inactivates it (Cosgrove and Philpott, 2007). These results suggest that pocket protein inhibition of E2Fs may not play a role in G1 during early X. laevis development, but further analysis of the other pocket protein family members, such as p107 and p130, is needed. Multiple X. laevis CDK inhibitors are expressed in a developmentally regulated manner (Daniels et al., 2004; Shou and Dunphy, 1996; Su et al., 1995). While expression of CDK inhibitors inhibits the cell cycle and influences cellular differentiation later in development, it is unknown which of the CDK inhibitors might play a role in the introduction of the G1 phase after the MBT (Daniels et al., 2004; Ohnuma et al., 1999; Vernon and Philpott, 2003).
The mechanisms by which G1 is added to the cell cycle is a timely question, as the length of the G1 phase is now associated with pluripotency and cellular differentiation. The relationship between cell potency and G1 has been demonstrated in aquatic species and mammalian stem cells (ESCs), which have a relatively short G1 phase (Richard-Parpaillon et al., 2004; Roccio et al., 2013). Lengthening of G1 is associated with differentiation of ESCs, while a short G1 is associated with maintenance of pluripotency(Lange et al., 2009; Pauklin and Vallier, 2013; Roccio et al., 2013). The role for G1 is not entirely clear, but two G1-associated processes that are of great interest are the spatial reorganization of chromosomes after mitosis, and the establishment of the replication-timing program. Gastrulation is a time at which the lineage of many cell types is restricted (Ho and Kimmel, 1993; Kimmel and Warga, 1986). Intriguingly, the length of the G1 phase increases substantially during gastrulation in zebrafish (Figure 1) (our unpublished data)(Zamir et al., 1997). Aquatic models such as frogs and fish are excellent models to study the relationships between the G1 phase, cellular differentiation, nuclear organization, and DNA replication timing.
8. Conclusions
Embryologists were first attracted to aquatic species because of the ease with which they could observe the large externally developing embryos. The initial cleavage divisions are particularly striking, since they can occur rapidly and synchronously. These cleavage divisions are driven by maternally provided gene products, and they occur without zygotic transcription. The ability to collect large numbers of eggs, which are loaded with all of the necessary cell cycle factors, has enabled the development cell-free systems to study the fundamental mechanisms of the embryonic cell cycle in Xenopus. At the MBT, multiple cellular changes occur including the start of transcription and a remodeling of the embryonic cell cycle. One of the principle changes at the MBT is a lengthening of S-phase, which is caused by the titration of replication initiation factors and reduction in the density of replication forks. The chromatin and nuclear structure also change at the MBT, and those changes likely impact the spatiotemporal organization of replication forks during S-phase. The remodeling of S-phase necessitates checkpoints that delay mitosis until DNA replication is complete, and those checkpoints are someway “awakened” at the MBT. By the time Xenopus and zebrafish complete gastrulation, the cell cycle is much more complex, with the gap phases G1 and G2. Soon after gastrulation, during tail morphogenesis, developmental control over G2 is critical, as progenitor cells are paused in G2 to control the timing of their migration and differentiation. The mechanisms by which the G1 phase is added to the embryonic cell cycle are not entirely clear. The length of G1 increases sharply during gastrulation when cells lose pluripotency in zebrafish. Intriguingly, G1 is the time in the cell cycle that the nucleus is reorganized and the replication timing program is established. Recent work in mammalian stem cells has highlighted the importance of G1 in cellular potency. There is much to learn about the important ways that the cell cycle changes during development. Is G2 control important in other developmental contexts? How does G1 lengthening promote differentiation? What are the roles for changes in Sphase length and the replication-timing program in development? These are fundamental questions that highly accessible and manipulable model organisms such as zebrafish and Xenopus are well suited to answer.
Acknowledgments
Grant support: NIH 5P20 GM103636-0
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This paper is based on work presented at the 7th Aquatic Animal Models of Human Disease Conference, hosted by Texas State University (Dec 13–Dec 19, 2014).
REFERENCES
- Amodeo AA, Jukam D, Straight AF, Skotheim JM. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1086–E1095. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhoefer J, Rhind N. Replication timing and its emergence from stochastic processes. Trends in genetics : TIG. 2012;28:374–381. doi: 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract Are 5–15 kilobases apart and are activated in clusters that fire at different times. The Journal of cell biology. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Current biology : CB. 2011;21:1152–1157. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- Bouldin CM, Snelson CD, Farr GH, 3rd, Kimelman D. Restricted expression of cdc25a in the tailbud is essential for formation of the zebrafish posterior body. Genes & development. 2014;28:384–395. doi: 10.1101/gad.233577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science (New York, N.Y.) 2013;341:893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove RA, Philpott A. Cell cycling and differentiation do not require the retinoblastoma protein during early Xenopus development. Developmental biology. 2007;303:311–324. doi: 10.1016/j.ydbio.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Dalle Nogare DE, Pauerstein PT, Lane ME. G2 acquisition by transcriptionindependent mechanism at the zebrafish midblastula transition. Developmental biology. 2009;326:131–142. doi: 10.1016/j.ydbio.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Daniels M, Dhokia V, Richard-Parpaillon L, Ohnuma S. Identification of Xenopus cyclin-dependent kinase inhibitors, p16Xic2 and p17Xic3. Gene. 2004;342:41–47. doi: 10.1016/j.gene.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nature cell biology. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome research. 2015 doi: 10.1101/gr.183699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Molecular cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Donley N, Thayer MJ. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Seminars in cancer biology. 2013;23:80–89. doi: 10.1016/j.semcancer.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy KT, McAleer MF, Davidson WR, Kari L, Kari C, Liu CG, Farber SA, Cheng KC, Mest JR, Wickstrom E, Dicker AP, Rodeck U. Coordinate control of cell cycle regulatory genes in zebrafish development tested by cyclin D1 knockdown with morpholino phosphorodiamidates and hydroxyprolyl-phosphono peptide nucleic acids. Nucleic acids research. 2005;33:4914–4921. doi: 10.1093/nar/gki799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes & development. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Farrell JA, Shermoen AW, Yuan K, O'Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes & development. 2012;26:714–725. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick DL, Andrews MT. Cell cycle remodeling requires cell-cell interactions in developing Xenopus embryos. The Journal of experimental zoology. 1994;270:410–416. doi: 10.1002/jez.1402700411. [DOI] [PubMed] [Google Scholar]
- Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller JL. Cyclin is a component of maturation-promoting factor from Xenopus. Cell. 1990;60:487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- Gautier J, Norbury C, Lohka M, Nurse P, Maller J. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+ Cell. 1988;54:433–439. doi: 10.1016/0092-8674(88)90206-1. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Wu M, Kirschner M. Cell cycle dynamics of an M-phase-specific cytoplasmic factor in Xenopus laevis oocytes and eggs. The Journal of cell biology. 1984;98:1247–1255. doi: 10.1083/jcb.98.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Miyazawa H, DePamphilis ML. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Molecular and cellular biology. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Molecular systems biology. 2014;10:722. doi: 10.1002/msb.134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CF, Morgan RW. Changes in the cell cycle during early amphibian development. Developmental biology. 1966;14:439–460. [Google Scholar]
- Gyda M, Wolman M, Lorent K, Granato M. The tumor suppressor gene retinoblastoma-1 is required for retinotectal development and visual function in zebrafish. PLoS genetics. 2012;8:e1003106. doi: 10.1371/journal.pgen.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R. Regulation of DNA replication on subchromosomal units of mammalian cells. The Journal of cell biology. 1975;64:89–97. doi: 10.1083/jcb.64.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley RS, Rempel RE, Maller JL. In vivo regulation of the early embryonic cell cycle in Xenopus. Developmental biology. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development (Cambridge, England) 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Current biology : CB. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS biology. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect--part II. Current opinion in genetics & development. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho RK, Kimmel CB. Commitment of cell fate in the early zebrafish embryo. Science (New York, N.Y.) 1993;261:109–111. doi: 10.1126/science.8316841. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science (New York, N.Y.) 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Rivera-Bennetts AK, Brooker DL, Yager TD. Effect of inhibitors of DNA replication on early zebrafish embryos: evidence for coordinate activation of multiple intrinsic cell-cycle checkpoints at the mid-blastula transition. Zygote (Cambridge, England) 1997;5:153–175. doi: 10.1017/s0967199400003828. [DOI] [PubMed] [Google Scholar]
- Iwao Y, Uchida Y, Ueno S, Yoshizaki N, Masui Y. Midblastula transition (MBT) of the cell cycles in the yolk and pigment granule-free translucent blastomeres obtained from centrifuged Xenopus embryos. Development, growth & differentiation. 2005;47:283–294. doi: 10.1111/j.1440-169X.2005.00802.x. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Gu Y, Morgan DO. Role of inhibitory CDC2 phosphorylation in radiationinduced G2 arrest in human cells. The Journal of cell biology. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development (Cambridge, England) 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome research. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Li C, Maller JL. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis. Developmental biology. 1999;212:381–391. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM. Tissue-specific cell lineages originate in the gastrula of the zebrafish. Science (New York, N.Y.) 1986;231:365–368. doi: 10.1126/science.231.4736.365. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development (Cambridge, England) 1994;120:265–276. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. The Journal of cell biology. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. The Journal of cell biology. 2011;193:995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Perewoska I, Germe T, Hyrien O, Marheineke K. DNA replication timing is deterministic at the level of chromosomal domains but stochastic at the level of replicons in Xenopus egg extracts. Nucleic acids research. 2008;36:5623–5634. doi: 10.1093/nar/gkn533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20364–20369. doi: 10.1073/pnas.1011069107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell stem cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Lawlis SJ, Keezer SM, Wu JR, Gilbert DM. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. The Journal of cell biology. 1996;135:1207–1218. doi: 10.1083/jcb.135.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovici M, Monod G, Geraudie J, Bravo R, Mechali M. Nuclear distribution of PCNA during embryonic development in Xenopus laevis: a reinvestigation of early cell cycles. Journal of cell science. 1992;102(Pt 1):63–69. doi: 10.1242/jcs.102.1.63. [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Geraud G, Mechali M. Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. The Journal of cell biology. 1998;142:1159–1166. doi: 10.1083/jcb.142.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. The Journal of cell biology. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Muller F, Alestrom P, Collas P. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Developmental cell. 2011;21:993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Hayes MK, Maller JL. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:3009–3013. doi: 10.1073/pnas.85.9.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science (New York, N.Y.) 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. The Journal of cell biology. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller J, Wu M, Gerhart JC. Changes in protein phosphorylation accompanying maturation of Xenopus laevis oocytes. Developmental biology. 1977;58:295–312. doi: 10.1016/0012-1606(77)90093-8. [DOI] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. The EMBO journal. 2011;30:4805–4814. doi: 10.1038/emboj.2011.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui Y. Relative roles of the pituitary, follicle cells, and progesterone in the induction of oocyte maturation in Rana pipiens. The Journal of experimental zoology. 1967;166:365–375. doi: 10.1002/jez.1401660309. [DOI] [PubMed] [Google Scholar]
- Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. The Journal of experimental zoology. 1971;177:129–145. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1- like kinase. Molecular biology of the cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, Thisse C, Thisse B, Raz E. Expression of a linker histone-like gene in the primordial germ cells in zebrafish. Mechanisms of development. 2002;117:253–257. doi: 10.1016/s0925-4773(02)00174-0. [DOI] [PubMed] [Google Scholar]
- Murphy CM, Michael WM. Control of DNA replication by the nucleus/cytoplasm ratio in Xenopus. The Journal of biological chemistry. 2013;288:29382–29393. doi: 10.1074/jbc.M113.499012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982f;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Newport JW, Kirschner MW. Regulation of the cell cycle during early Xenopus development. Cell. 1984;37:731–742. doi: 10.1016/0092-8674(84)90409-4. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Pub., New York. 1994 [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. The Journal of cell biology. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S, Philpott A, Wang K, Holt CE, Harris WA. p27Xic1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell. 1999;99:499–510. doi: 10.1016/s0092-8674(00)81538-x. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Nakajo N, Sagata N. The existence of two distinct Wee1 isoforms in Xenopus: implications for the developmental regulation of the cell cycle. The EMBO journal. 2002;21:2472–2484. doi: 10.1093/emboj/21.10.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A, Friend SH. E2F and its developmental regulation in Xenopus laevis. Molecular and cellular biology. 1994;14:5000–5009. doi: 10.1128/mcb.14.7.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerening JR, Kim SY, Ferrell JE., Jr Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–578. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, Thurman RE, Cheng Y, Gulsoy G, Dennis JH, Snyder MP, Stamatoyannopoulos JA, Taylor J, Hardison RC, Kahveci T, Ren B, Gilbert DM. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Parpaillon L, Cosgrove RA, Devine C, Vernon AE, Philpott A. G1/S phase cyclin-dependent kinase overexpression perturbs early development and delays tissuespecific differentiation in Xenopus. Development (Cambridge, England) 2004;131:2577–2586. doi: 10.1242/dev.01121. [DOI] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, Schulz TC, Menendez L, Kulik MJ, Dalton S, Gabr H, Kahveci T, Gilbert DM. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome research. 2015 doi: 10.1101/gr.187989.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccio M, Schmitter D, Knobloch M, Okawa Y, Sage D, Lutolf MP. Predicting stem cell fate changes by differential cell cycle progression patterns. Development (Cambridge, England) 2013;140:459–470. doi: 10.1242/dev.086215. [DOI] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, Devidas M, Druker BJ, Gilbert DM. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome research. 2012;22:1833–1844. doi: 10.1101/gr.138511.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome research. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CG, Goins D, Siefert JC, Clowdus EA, Sansam CL. Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes & development. 2015;29:555–566. doi: 10.1101/gad.246827.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes & development. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes & development. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. The Journal of cell biology. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O'Farrell PH. Developmental control of late replication and S phase length. Current biology : CB. 2010;20:2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Dunphy WG. Cell cycle control by Xenopus p28Kix1, a developmentally regulated inhibitor of cyclin-dependent kinases. Molecular biology of the cell. 1996;7:457–469. doi: 10.1091/mbc.7.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach OC, Wolffe AP, Rupp RA. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- Su JY, Rempel RE, Erikson E, Maller JL. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27XIC1. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10187–10191. doi: 10.1073/pnas.92.22.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkara PS, Wright DA, Rao PN. Mitotic factors from mammalian cells induce germinal vesicle breakdown and chromosome condensation in amphibian oocytes. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:2799–2802. doi: 10.1073/pnas.76.6.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ono T, Kitamura N, Kato JY. Dominant negative E2F inhibits progression of the cell cycle after the midblastula transition in Xenopus. Cell structure and function. 2003;28:515–522. doi: 10.1247/csf.28.515. [DOI] [PubMed] [Google Scholar]
- Taylor JH. Asynchronous duplication of chromosomes in cultured cells of Chinese hamster. The Journal of biophysical and biochemical cytology. 1960;7:455–464. doi: 10.1083/jcb.7.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nature reviews. Molecular cell biology. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Vassetzky Y, Hair A, Mechali M. Rearrangement of chromatin domains during development in Xenopus. Genes & development. 2000;14:1541–1552. [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AE, Philpott A. A single cdk inhibitor, p27Xic1, functions beyond cell cycle regulation to promote muscle differentiation in Xenopus. Development (Cambridge, England) 2003;130:71–83. doi: 10.1242/dev.00180. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport JW. Regulation of replicon size in Xenopus egg extracts. Science (New York, N.Y.) 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Molecular cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Webb SE, Miller AL. Ca2+ signaling and early embryonic patterning during the blastula and gastrula periods of zebrafish and Xenopus development. Biochimica et biophysica acta. 2006;1763:1192–1208. doi: 10.1016/j.bbamcr.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Molecular and cellular biology. 1997;17:529–536. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. Canonical nucleosome organization at promoters forms during genome activation. Genome research. 2014;24:260–266. doi: 10.1101/gr.157750.113. [DOI] [PMC free article] [PubMed] [Google Scholar]