Abstract
X-linked Alport syndrome (XLAS) is a progressive, hereditary nephropathy. Although men with XLAS usually develop end-stage renal disease before 30 years of age, some men show a milder phenotype and develop end-stage renal disease later in life. However, the molecular mechanisms associated with this milder phenotype have not been fully identified. We genetically diagnosed 186 patients with suspected XLAS between January 2006 and August 2014. Genetic examination involved: (1) extraction and analysis of genomic DNA using PCR and direct sequencing using Sanger's method and (2) next-generation sequencing to detect variant allele frequencies. We identified somatic mosaic variants in the type VI collagen, α5 gene (COL4A5) in four patients. Interestingly, two of these four patients with variant frequencies in kidney biopsies or urinary sediment cells of ≥50% showed hematuria and moderate proteinuria, whereas the other two with variant frequencies of <50% were asymptomatic or only had hematuria. De novo variants can occur even in asymptomatic male cases of XLAS resulting in mosaicism, with important implications for genetic counseling. This is the first study to show a tendency between the variant allele frequency and disease severity in male XLAS patients with somatic mosaic variants in COL4A5. Although this is a very rare status of somatic mosaicism, further analysis is needed to show this correlation in a larger population.
Introduction
Alport syndrome (AS) is a hereditary disorder of type IV collagen, characterized by chronic kidney disease progressing to end-stage renal disease (ESRD), sensorineural hearing loss, and ocular abnormalities. Approximately 85% of AS patients show X-linked inheritance (XLAS: OMIM301050) and variants in COL4A5, which encodes the type IV collagen α5 (α5(IV)) chain. COL4A5 variants result in abnormal α5(IV) expression, typically with complete absence of α5(IV) in the glomerular basement membrane (GBM) and Bowman's capsule in men, and a mosaic expression pattern in women.1
Male patients with XLAS can be classified as having either ‘adult type', associated with mild deafness and the development of ESRD >30 years of age, or ‘juvenile type', associated with hearing loss and often with lenticonus, and an onset of ESRD <30 years of age.2 These two phenotypes are partially related to the genotype; for example, missense variants or in-frame variants of COL4A5 were reported in cases of later-onset ESRD.3, 4, 5 We recently reported that 29% of male XLAS patients expressed the α5(IV) chain in the glomerulus and showed milder clinical manifestations.6 Interestingly, all α5(IV)-positive patients possessed non-truncating variants (n=13) or somatic mosaic variants (n=2) of COL4A5. One of these patients has been described in a previous case report.7 This implies that men with XLAS and somatic mosaic variants show milder phenotypes; however, no case series has reported the correlation between variant frequency and disease severity in patients with somatic mosaic variants. The present study, therefore, examined the correlation between variant frequency and phenotype in a case series of male XLAS patients with somatic mosaic variants using next-generation sequencing (NGS). We provide herein the first report of an asymptomatic male XLAS case and also describe the first cases of somatic and gonadal mosaic variants in COL4A5.
Materials and methods
Ethical considerations
All procedures were reviewed and approved by the Institutional Review Board of Kobe University School of Medicine. Informed consent was obtained from all patients or their parents.
Data collection
Clinical and laboratory findings of patients with XLAS were obtained from their medical records. Patients were referred to our hospital for clinical evaluation or genetic analysis. Most patients were followed in various local hospitals in Japan. DNA and data sheets were sent to our laboratory after acceptance of the request for mutational analysis.
Estimated glomerular filtration rates (eGFRs) were measured from the data in these data sheets. eGFRs were calculated using the Schwartz formula for patients aged ≤19 years, and the Cockcroft–Gault formula for patients aged ⩾20 years.8, 9, 10
Mutational analyses using Sanger sequencing
Mutational analyses of COL4A5 were carried out using the following methods: (1) PCR and direct sequencing of genomic DNA of all exons and exon–intron boundaries and (2) reverse-transcription PCR of mRNA and direct sequencing of abnormal mRNA products when a suspected splicing-site variant was detected.
Genomic DNA was isolated from peripheral blood leukocytes, urinary sediments, kidney biopsies, skin and/or hair roots from patients, and their parents using the Quick Gene Mini 80 System (Fujifilm Corporation, Tokyo, Japan) according to the manufacturer's instructions. For genomic DNA analysis, all 51 COL4A5 exons were amplified by PCR, as described previously.11 PCR-amplified products were then purified and subjected to direct sequencing using a Dye Terminator Cycle Sequencing Kit (Amersham Biosciences, Piscataway, NJ, USA) with an automatic DNA sequencer (ABI Prism 3130; Perkin Elmer Applied Biosystems, Foster City, CA, USA).
Mutational analysis data were submitted to the Alport syndrome and COL4A5 database (http://www.arup.utah.edu/database/ALPORT/ALPORT_welcome.php). For variant description, reference sequences were NC_000023.9 and NM_000495.3. Exons were numbered according to a previous report.12
Mutational analysis using NGS
A subset of exome-targeting genes with disease-causing variants were subjected to NGS using a commercially available kit (TruSight One, Illumina, San Diego, CA, USA) and targeted resequencing as a means of deep sequencing. Following the TruSight workflow, input genomic DNA was converted into adapter-tagged libraries by rapid Nextera (Nextera DNA Library Preparation Kit, Illumina)-based sample preparation. The libraries were then denatured into single-stranded DNA, and biotin-labeled probes specific to the targeted region were used for Rapid Capture hybridization. The pool was enriched for the desired regions by adding streptavidin beads that bound to the biotinylated probes. Biotinylated DNA fragments bound to the streptavidin beads were pulled down magnetically from the solution. The enriched DNA fragments were then eluted from the beads and hybridized for a second Rapid Capture. Sequence data generated from TruSight exome-enriched libraries were analyzed using the on-instrument MiSeq Reporter software (Illumina).
For deep sequencing of somatic mosaic variant analysis, 500-bp PCR products harboring each suspected mutation site were purified by gel extraction using the QIAquick gel extraction kit (Qiagen, Valencia, CA, USA). Each variant was then analyzed using the TruSeq PCR-free LT kit (Illumina). All procedures were conducted according to the manufacturers' instructions. The primer sequences were as follows:
COL4A5-exon25-F: 5′-CCCCAGTTGTATTCAGTA-3′ and COL4A5-exon25-R: 5′-GAGCAAAATTAACAGTAA-3′ COL4A5-exon28-F: 5′-AAAAGCATATGTTCCACA-3′ and COL4A5-exon28-R: 5′-GATGATTTGGGGTTAAAT-3′ COL4A5-exon44-F: 5′-ATTTATTCAGGGTAATCC-3′ and COL4A5-exon44-R: 5′-TAAAAGGTCTGCTATCAA-3′ and COL4A5-exon49-F: 5′-GGAGACAATACTTAGCAAATG-3′ and COL4A5-exon49-R: 5′-ACACCAAGGGTAGTCAAA-3′.
To determine the limit of variant frequency detection, we made test samples containing mixtures of DNA from an XLAS patient with a hemizygous COL4A5 c.1948+1G>A mutation and control DNA at variant frequencies of 0.5, 1, 2, 10, and 20%. Targeted resequencing was then conducted using the primer pair for COL4A5 exon25.
Results
Clinical, pathological, and mutational results are shown in Figures 1 and 2, Tables 1 and 2, and Supplementary Table 1. NGS analysis findings including the depth and forward/reverse reads are shown in Supplementary Table 1.
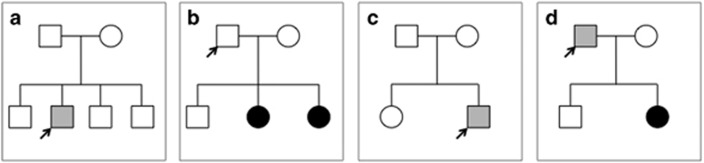
Figure 1.
Patient pedigrees. (a) Patient ID14 possessing COL4A5 mutation c.3998-2A>T in intron 43. This individual showed hematuria and moderate proteinuria. The parents are asymptomatic. (b) Patient ID28 possessing COL4A5 mutation c.2147-2A>G in intron 27. This individual is asymptomatic although possesses a somatic and gonadal mosaic variant. One daughter has hematuria and mild proteinuria, whereas the second daughter has hematuria. (c) Patient ID52 possessing COL4A5 mutation c.1912G>A in exon25. This individual has hematuria and moderate proteinuria. The parents are asymptomatic. (d) Patient ID 252 possessing COL4A5 mutation c.4787G>T. This individual has hematuria without proteinuria, and the daughter has hematuria and mild proteinuria.
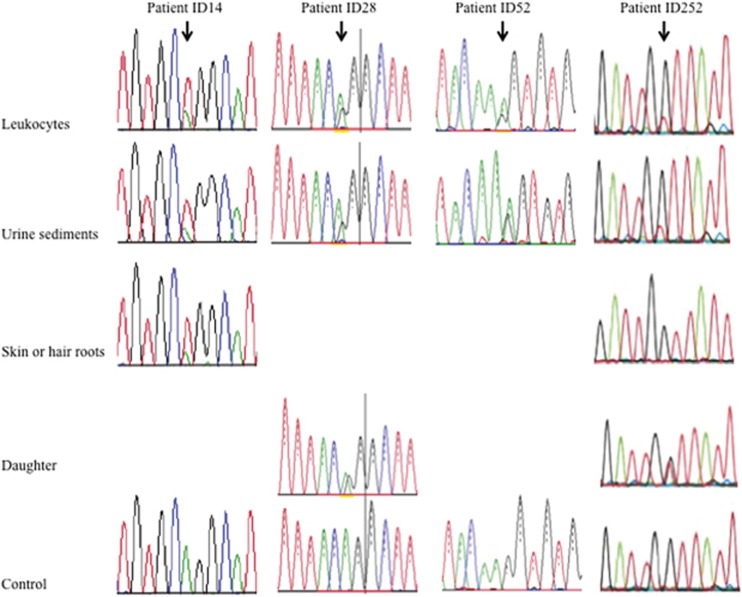
Figure 2.
Direct sequencing of patients with somatic mosaic variants. Patient ID14: c.3998-2A>T, IVS44-2A>T. NGS analysis revealed variant allele frequencies of 57.1% in leukocytes, 61.3% in urinary sediment, cells and 75.3% in skin. Patient ID28: c.2147-2A>G, IVS28-2A>G. NGS analysis revealed variant allele frequencies of 31.3% in leukocytes and 33.3% in urinary sediment cells. His daughter shows heterozygous variant. Patient ID52: c.1912G>A, p.(Gly638Ser). NGS analysis revealed variant allele frequencies of 60.9% in leukocytes and 68% in urinary sediment cells. Patient ID252: c.4787G>T, p.(Gly1596Val). NGS analysis revealed variant allele frequencies of 20.6% in leukocytes, 24.1% in urinary sediment cells, and 0% in hair roots. His daughter shows heterozygous variant.
Table 1. Clinical characteristics and laboratory data.
| Patient ID | Sex | Age (years) | ESRD (age) | Hearing loss (detected age) | sCr (mol/l) | eGFR (ml/min/1.73 m2) | Hematuria | U-P/Cr (g/g Cr) | EM | alpha-5 | Family history |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | M | 15–20 | — | — | 68.9 | 144.7 | 3+ | 0.74 | BWC | Mosaic | Sporadic |
| 28 | M | 35–40 | — | — | 68.9 | 87.1 | — | — | — | — | Two daughters pro/OB |
| 52 | M | 15–20 | — | — | 80.4 | 125 | 3+ | 0.83 | BWC | Mosaic | Sporadic |
| 252 | M | 40–45 | — | — | 86.6 | 104.1 | 3+ | — | — | — | Daughter pro/OB |
Abbreviations: BWC, basket-weave change; EASRD, end-stage renal disease; eGFR, estimated glomerular filtration rate; EM, electron microscopic findings; M, male; ND, not determined; OB, occult; pro, proteinuria; blood sCr, serum creatinine levels; U-P/Cr, urinary protein–creatinine ratio.
Table 2. COL4A5 variants and variant allele frequencies.
| Variant frequency (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Variant position | Variants | Amino acid change | Methods | Leukocytes | Urine sediments | Kidney | Hair roots | Skin |
| 14 | intron43 | c.3998-2A>T | p.(Gly1333_Pro1356del) | Trusight One | 57.1 | 61.3 | — | — | 75.3 |
| c.3998-2A>T | Targeted resequencing | 60.8 | 63.0 | 62.9 | — | 64.8 | |||
| 28 | intron27 | c.2147-2A>G | p.(Gly716_Pro721del) | Trusight one | 31.3 | 33.3 | — | — | — |
| r.2147_2164del | Targeted resequencing | 43.7 | 45.5 | ||||||
| 52 | exon25 | c.1912G>A | p.(Gly638Ser) | Trusight one | 60.9 | 68.0 | — | — | — |
| r.(1912 g>a) | Targeted resequencing | 70.4 | — | 72.7 | — | — | |||
| 252 | exon49 | c.4787G>T | p.(Gly1596Val) | Trusight one | 20.6 | 24.1 | — | 0 | — |
| r.(4787 g>t) | Targeted resequencing | 24.9 | 19.4 | — | 1.5 | — | |||
Patient ID14
The pedigree of patient ID14 is shown in Figure 1a. The precise clinical course of this patient has been reported previously.7 At 16 years of age, the patient had microhematuria and moderate proteinuria with 0.74 g/g creatinine (Cr). Genetic analysis revealed the presence of an intron 43 splicing acceptor site variant (c.3998-2A>T, IVS44-2A>T). Transcriptional analysis showed that this variant caused skipping of exon44 (72 bp).
Patient ID28
A 38-year-old male was detected with microhematuria and proteinuria when he had a common cold; however, he had no urine abnormalities other than on that occasion. His pedigree is shown in Figure 1b. His older daughter also showed macrohematuria when she had a common cold at the age of 3 years, and subsequently demonstrated persistent microhematuria and mild proteinuria (0.2 g/g Cr). She underwent a kidney biopsy and was pathologically diagnosed with XLAS with a basket-weave change (BWC) on the GBM and mosaic α5(IV) expression. Genetic analysis revealed a COL4A5 heterozygous variant at the intron 27 splicing acceptor site (c.2147-2A>G, IVS28-2A>G), which has been reported previously without precise clinical information.13 Transcriptional analysis revealed this variant to cause skipping of part of exon28 (18 bp). Her mother was asymptomatic with no variants and her father was also asymptomatic, suggesting that she represents a sporadic case with a de novo COL4A5 variant. However, the second daughter was also detected with hematuria at a screening test at 3 years of age, and was genetically diagnosed with XLAS with the same variant (IVS28-2A>G). Subsequent genetic testing of the father revealed the same variant with somatic mosaicism in genomic DNA extracted from leukocytes and urine sediments (Figure 2). He was confirmed to have a normal karyotype (46,XY). Because both daughters carry the same heterozygous variant, this indicates that their father also has the same variant in a mosaic state that includes germinal cells. The father was subsequently diagnosed with asymptomatic XLAS with a somatic and gonadal mosaic variant in COL4A5.
Patient ID52
The pedigree of patient ID52 is shown in Figure 1c. He was an 18-year-old man who was first detected with hematuria and proteinuria by screening at the age of 3 years. Examination of a kidney biopsy taken at 10 years of age revealed AS with a BWC on the GBM. However, α5(IV) expression showed a mosaic pattern. His karyotype was 46,XY. Genetic analysis revealed an exon25 missense variant (c.1912G>A, p.(Gly638Ser)), which was reported previously without precise clinical information.14 At the age of 18 years, he had microhematuria and moderate proteinuria of 0.83 g/g Cr.
Patient ID252
Patient ID252 was a 42-year-old man in whom hematuria was first detected at 6 years of age. His pedigree is shown in Figure 1d. His daughter showed macrohematuria and mild proteinuria (0.2 g/g Cr) when she was 6 years old. She underwent kidney biopsy and was pathologically diagnosed with XLAS with a BWC on GBM and mosaic α5(IV) expression. Genetic analysis revealed a heterozygous missense variant at COL4A5 exon 49 (c.4787G>T, p.(Gly1596Val)). This amino-acid variant with a different amino-acid substitution was reported in a male patient who had not developed ESRD at the age of 19.15 Her mother was asymptomatic with no variants, but her father showed persistent microhematuria without proteinuria and had the same variant with somatic mosaicism in genomic DNA extracted from both leukocytes and urine sediments. We confirmed his karyotype to be normal (46,XY). His daughter had the same heterozygous variant, indicating that their father also had a germline variant. The father was diagnosed with XLAS with a somatic and gonadal mosaic variant in COL4A5.
Limit of variant frequency detection
Table 3 shows the results of our analysis to determine the limit of variant detection frequency. Targeted resequencing revealed that 1–2% was the lower limit of detection.
Table 3. Determining the limit of variant frequency detection.
| Variant frequency (%) | Wild type | Mutant | ||||
|---|---|---|---|---|---|---|
| Test sample | NGS result | Depth | Forward reads | Reverse reads | Forward reads | Reverse reads |
| 0.5 | 1.1 | 459 772 | 140 068 | 310 841 | 1665 | 3441 |
| 1 | 1.9 | 504 779 | 149 572 | 340 835 | 2954 | 6680 |
| 2 | 2.6 | 463 811 | 124 329 | 323 702 | 3158 | 8805 |
| 10 | 10.7 | 399 956 | 98 220 | 255 492 | 11 222 | 31 623 |
| 20 | 19.4 | 440 254 | 350 826 | 118 714 | 26 670 | 58 759 |
Comparison of variant frequencies between kidney biopsies and urinary sediments
We previously showed that urinary sediments can be used as an alternative cell source to kidney biopsies.16, 17 The present study compared the allele frequencies between DNA extracted from these two sources and obtained very similar findings (Table 2). Therefore, we compared the variant frequency of either kidney biopsies or urinary sediments with the phenotype in our analysis.
Discussion
Male patients with XLAS sometimes show a milder, ‘adult type' phenotype, with only mild deafness and an onset of renal failure >30 years old.2 This milder phenotype is associated with unique genotypes such as missense or in-frame variants in COL4A5.3, 4, 5 We previously reported a male XLAS patient with a missense COL4A5 variant who showed only hematuria without proteinuria at the age of 33.18 We also reported a male patient with a somatic COL4A5 variant who showed hematuria and mild proteinuria at the age of 8 years. His kidney biopsy expressed α5(IV) mosaicism in the glomerulus, which was associated with the somatic mosaic variant.7 To date, however, only six patients in four reports have been described with somatic mosaic variants in COL4A5, including our previous report (Table 4).7, 19, 20, 21 Although all six cases showed a milder phenotype and some of the female cases were asymptomatic, no asymptomatic male cases have previously been reported.
Table 4. Previously reported cases with COL4A5 mosaic variants.
| Mosaicism | Variants | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| First author | Sex | Age (years) | Somatic cells | Germline cells | ESRD (age) | Urinary exam | Hearing loss | Ocular lesion | Exon | Nucleotide | Amino acid |
| Plant KE | Female | ND | + | + | − | OB | − | − | 26 | c.2006G>C | p.Gly669Ala |
| Female | ND | + | + | − | − | − | − | IVS12-3 | c.848-3C>A | exon 12 skip | |
| Male | ND | + | + | 43 | ND | ND | − | 25 | c.1912G>A | p.Gly638Ser | |
| Bruttini M | Female | ND | − | + | − | − | − | − | IVS44+1 | c.4069+1G>C | exon44 skip |
| Krol RP | Male | 8 | + | ND | − | OB, mild pro | − | − | IVS44-2 | c.3998-2A>T | exon44 skip |
| Beicht S | Female | ND | + | + | − | OB | + | Myopia | IVS30-1 | c.2396-1G>A | exon 30 skip |
Abbreviations: ESRD, end-stage renal disease; ND, not determined; OB: occult blood; pro: proteinuria.
A recent publication by Beicht et al.19 described an asymptomatic female XLAS patient with a somatic mosaic variant who had variant allele frequencies of 14, 7, 4, and 7% in leukocytes, urine sediments, hair roots, and oral mucosa, respectively, as shown by NGS. However, it is difficult to evaluate variant allele frequencies and phenotypes in female XLAS patients because skewed X-inactivation might affect the phenotype. The present study examined the correlation between the percentage of variant alleles in genomic DNA extracted from kidney biopsies and/or urinary sediments and renal symptoms in men with XLAS and somatic mosaic variants for the first time, revealing a tendency for an association between lower variant allele frequency and milder phenotype. Interestingly, two patients with variant frequencies in kidney biopsies and/or urinary sediment cells of ≥50% showed hematuria and moderate proteinuria, whereas two patients with frequencies <50% were asymptomatic or only had hematuria.
We recently reported a male XLAS patient with a mild phenotype caused by a unique intronic splicing variant, causing a cryptic exon in the transcript; however, mRNA extracted from the kidney showed both normal and abnormal transcripts, the former rescuing him from having the severe phenotype.22
The milder phenotype in men with XLAS is currently defined by the following five patterns: (1) missense variants in COL4A5;4, 5, 6 (2); in-frame variants in COL4A5;4, 6 (3) somatic mosaic variants in COL4A5;7, 19, 20, 21 (4) α5(IV)-positive expression in the glomerulus;6 and (5) aberrant splicing variants in COL4A5, leading to both normal and abnormal mRNAs.22 In this study, we reported four cases with milder phenotypes: two with splice site variants (ID14 and 28) and two with missense variants (ID52 and 252). These variant types could contribute to a modulation of the phenotype. However, among these four patients, the influence of somatic mosaicism appears to be stronger because, of the two patients with missense mutations, ID252 with a lower variant frequency showed a much milder phenotype.
We previously used the techniques of semi-quantitative PCR analysis, restriction enzyme digestion, and electrophoresis to report variant frequencies for patient ID14 of 37% in leukocytes, 71% in urine sediments, and 32% in the skin.7 Although at the time of this study (2008), we thought that our methods were highly efficient, it now appears that they were not reliable because the two techniques used in the current study (TruSight One and targeted resequencing) achieved almost identical frequencies, which differed from our previous data.
Patients ID28 and 252 of the present study also showed mosaic variants in germline cells. In these cases, we were unable to conduct an analysis of sperm cells because we were not given consent to do so. However, determining the mutation allele frequency in these cells would provide additional information about the genetic risk facing offspring inheriting the mutated allele, which would be invaluable for genetic counseling.
NGS is a highly relevant tool for use in the diagnosis of AS. Moreover, early diagnosis of this disease is becoming increasingly important because AS is now a treatable disease.23, 24 The targeted resequencing technique that we used in the present study is both efficient and cost effective, and we propose that it should be adopted worldwide for the use in disease diagnosis.
The present study reports a tendency between variant allele frequency and the severity of renal symptoms in four men with XLAS with somatic mosaic variants. Although asymptomatic female cases with mosaic variants have been reported previously, the current study provides the first report of an asymptomatic male XLAS patient with a mosaic variant in COL4A5. We also describe the first male XLAS cases with somatic and gonadal mosaic variants in COL4A5. These results indicate that de novo variants can occur even in asymptomatic men with XLAS, and that the variant frequency may influence the severity of XLAS in patients with somatic mosaic variants. These cases highlight the fact that genetic counseling for asymptomatic parents of a child with AS should consider the possibility that one of the parents may carry a variant and show somatic and gonadal mosaicism.
Acknowledgments
This study was supported by a grant from the Ministry of Health, Labour, and Welfare of Japan for Research on Rare Intractable Diseases in Kidney and Urinary Tract (H24-nanchitou (nan)-ippan-041 to KI) in the ‘Research on Measures for Intractable Diseases' Project; a Grant-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Subject ID: 25893131 to KN); and a grant from the Mother and Child Health Foundation (Subject ID: 25-7 to KN).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
KI received grants from Novartis Pharma K.K., Japan Blood Product Organization, Kyowa Hakko Kirion Co., Ltd, JCR Pharmaceuticals Co., Ltd, AbbVie Inc., Genzyme Japan K.K., Teijin Pharma Ltd, Daiichi Sankyo Co., Ltd, and Miyarisan Pharmaceutical Co., Ltd, and lecture fees from Kyowa Hakko Kirin Co., Ltd, Astellas Pharma Inc., Pfizer Japan Inc., Asahi Kasei Pharma Corp., Kowa Pharmaceutical Co., Ltd, Merck Sharp & Dohme Corp., Alexion, Meiji Seika Pharma Co., Ltd, and Novartis Pharma K.K.. KI is also an advisor for Zenyaku Kogyo Co., Ltd.
Supplementary Material
References
- 1Kashtan CE: Alport syndrome and thin glomerular basement membrane disease. J Am Soc Nephrol 1998; 9: 1736–1750. [DOI] [PubMed] [Google Scholar]
- 2Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG: Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N Engl J Med 2003; 348: 2543–2556. [DOI] [PubMed] [Google Scholar]
- 3Bekheirnia MR, Reed B, Gregory MC et al: Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol 2010; 21: 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Gross O, Netzer KO, Lambrecht R, Seibold S, Weber M: Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant 2002; 17: 1218–1227. [DOI] [PubMed] [Google Scholar]
- 5Jais JP, Knebelmann B, Giatras I et al: X-linked Alport syndrome: natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol 2000; 11: 649–657. [DOI] [PubMed] [Google Scholar]
- 6Hashimura Y, Nozu K, Kaito H et al: Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV alpha5 chain. Kidney Int 2014; 85: 1208–1213. [DOI] [PubMed] [Google Scholar]
- 7Krol RP, Nozu K, Nakanishi K et al: Somatic mosaicism for a mutation of the COL4A5 gene is a cause of mild phenotype male Alport syndrome. Nephrol Dial Transplant 2008; 23: 2525–2530. [DOI] [PubMed] [Google Scholar]
- 8Cockcroft DW, Gault MH: Prediction of creatinine clearance from serum creatinine. Nephron 1976; 16: 31–41. [DOI] [PubMed] [Google Scholar]
- 9Schwartz GJ, Gauthier B: A simple estimate of glomerular filtration rate in adolescent boys. J Pediatr 1985; 106: 522–526. [DOI] [PubMed] [Google Scholar]
- 10Schwartz GJ, Haycock GB, Edelmann CM Jr, Spitzer A: A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics 1976; 58: 259–263. [PubMed] [Google Scholar]
- 11Martin P, Heiskari N, Zhou J et al: High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J Am Soc Nephrol 1998; 9: 2291–2301. [DOI] [PubMed] [Google Scholar]
- 12International Human Genome Sequencing Consortium: Finishing the euchromatic sequence of the human genome. Nature 2004; 431: 931–945. [DOI] [PubMed] [Google Scholar]
- 13Nagel M, Nagorka S, Gross O: Novel COL4A5, COL4A4, and COL4A3 mutations in Alport syndrome. Hum Mutat 2005; 26: 60. [DOI] [PubMed] [Google Scholar]
- 14Plant KE, Green PM, Vetrie D, Flinter FA: Detection of mutations in COL4A5 in patients with Alport syndrome. Hum Mutat 1999; 13: 124–132. [DOI] [PubMed] [Google Scholar]
- 15Renieri A, Bruttini M, Galli L et al: X-linked Alport syndrome: an SSCP-based mutation survey over all 51 exons of the COL4A5 gene. Am J Hum Genet 1996; 58: 1192–1204. [PMC free article] [PubMed] [Google Scholar]
- 16Kaito H, Nozu K, Fu XJ et al: Detection of a transcript abnormality in mRNA of the SLC12A3 gene extracted from urinary sediment cells of a patient with Gitelman's syndrome. Pediatr Res 2007; 61: 502–505. [DOI] [PubMed] [Google Scholar]
- 17Nozu K, Iijima K, Kawai K et al: In vivo and in vitro splicing assay of SLC12A1 in an antenatal salt-losing tubulopathy patient with an intronic mutation. Hum Genet 2009; 126: 533–538. [DOI] [PubMed] [Google Scholar]
- 18Kaneko K, Tanaka S, Hasui M et al: A family with X-linked benign familial hematuria. Pediatr Nephrol 2010; 25: 545–548. [DOI] [PubMed] [Google Scholar]
- 19Beicht S, Strobl-Wildemann G, Rath S et al: Next generation sequencing as a useful tool in the diagnostics of mosaicism in Alport syndrome. Gene 2013; 526: 474–477. [DOI] [PubMed] [Google Scholar]
- 20Bruttini M, Vitelli F, Meloni I et al: Mosaicism in Alport syndrome with genetic counselling. J Med Genet 2000; 37: 717–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Plant KE, Boye E, Green PM, Vetrie D, Flinter FA: Somatic mosaicism associated with a mild Alport syndrome phenotype. J Med Genet 2000; 37: 238–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Nozu K VI, Kaito H, Fu XJ et al. X-linked Alport syndrome caused by splicing mutations in COL4A5. Clin J Ame Soc Nephrol 2014; 9: 1958–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Kashtan CE, Ding J, Gregory M et al: Clinical practice recommendations for the treatment of Alport syndrome: a statement of the Alport Syndrome Research Collaborative. Pediatr Nephrol 2013; 28: 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Savige J, Gregory M, Gross O, Kashtan C, Ding J, Flinter F: Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol 2013; 24: 364–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.