Abstract
Two variants (c.[301_302delAG][301_302delAG] and c.[150delA][150delA]) in the PROP1 gene are the most common genetic causes of recessively inherited combined pituitary hormones deficiency (CPHD). Our objective was to analyze in detail the origin of the two most prevalent variants. In the multicentric study were included 237 patients with CPHD and their 15 relatives carrying c.[301_302delAG][301_302delAG] or c.[150delA][150delA] or c.[301_302delAG][ 150delA]. They originated from 21 different countries worldwide. We genotyped 21 single-nucleotide variant markers flanking the 9.6-Mb region around the PROP1 gene that are not in mutual linkage disequilibrium in the general populations – a finding of a common haplotype would be indicative of ancestral origin of the variant. Haplotypes were reconstructed by Phase and Haploview software, and the variant age was estimated using an allelic association method. We demonstrated the ancestral origin of both variants – c.[301_302delAG] was carried on 0.2 Mb-long haplotype in a majority of European patients arising ~101 generations ago (confidence interval 90.1–116.4). Patients from the Iberian Peninsula displayed a different haplotype, which was estimated to have emerged 23.3 (20.1–29.1) generations ago. Subsequently, the data indicated that both the haplotypes were transmitted to Latin American patients ~13.8 (12.2–17.0) and 16.4 (14.4–20.1) generations ago, respectively. The c.[150delA] variant that was carried on a haplotype spanning about 0.3 Mb was estimated to appear 43.7 (38.4–52.7) generations ago. We present strong evidence that the most frequent variants in the PROP1 gene are not a consequence of variant hot spots as previously assumed, but are founder variants.
Introduction
A cascade of transcription factors orchestrates the development of the anterior pituitary gland and lifelong maintenance of its proper function – production and secretion of growth hormone (GH), thyroid-stimulating hormone (TSH), prolactin, gonadotropins and adrenocorticotropic hormone (ACTH). A gene encoding one of these transcription factors, Prophet of Pit1 (prop1), was originally identified in the Ames dwarf mouse.1 In humans, several biallelic variants of the PROP1 gene located on the distal end of the short arm of chromosome 5 (5q35.3), lead to an insufficient expression of the downstream gene POU1F1 (PIT1). It results in the defective differentiation of at least three pituitary cell lines (somatotrophs, lactotrophs and thyrotrophs) manifested as an autosomal recessive disorder – combined pituitary hormones deficiency (CPHD [MIM262600]). Affected individuals present in early childhood with growth failure owing to a lack of GH and TSH. Later in life, deficiencies of gonadotropins and ACTH can also occur.2
Among 30 PROP1 variants causing CPHD reported so far3, two prevail – the deletion of two nucleotides (NM_0006261.4: c.[301_302delAG][301_302delAG], referred also as c.296delGA)2 and the deletion of one nucleotide (NM_0006261.4: c.[150delA][150delA]).4 Both represent frameshift variants and predict premature termination of the protein synthesis.
High prevalence of specific variants can be attributed to two causes – the presence of an ancestral variant or an alteration hot spot. In the first case, a founder (ancestral) variant was introduced into a population by a single individual in the past that transmitted it to the subsequent generations. As the founder variant was established on a particular chromosome, it is carried on a background of a unique combination of variants with probably no functional effect (markers), designated as a haplotype. The ancestral haplotype is shared initially by all variant carriers, but it will subsequently decrease over generations, as a result of random genetic recombination events. In the second case, the high prevalence of a particular variant occurs in a genetic region susceptible to variant events due to its nucleotide composition, that is, a so-called variant hot spot. In this case, variants arise independently in each individual carrying a different set of markers on the respective chromosome. Thus, no single common haplotype surrounding the variant would be expected.5
A study published in 1998 analyzed the c.[301_302delAG] in the PROP1 gene as putative founder variant in five families with CPHD from Brazil, Portugal, Austria and Russia. The variant was associated with a different microsatellite marker sequence for D5S408 on an affected individual in each family.6 This finding supported the suggestion that c.[301_302delAG] was a recurring variant.7 On the other hand, literature review summarized in Supplementary Table 1 shows that the distribution of reported patients with the variants c.[301_302delAG] and/or c.[150delA] in the PROP1 gene is restricted to a few regions, mainly Central and Eastern Europe and Latin America, suggesting ancestral origins of these variants.
The objective of the present multicenter study was to analyze in detail the origin of the two most prevalent variants in the PROP1 gene in an extensive cohort of patients with CPHD originating from 21 different countries worldwide.
Materials and methods
Patients
DNA samples from 252 individuals (237 patients and their 15 healthy family relatives) of 200 families were collected within a multinational, multicenter collaboration. The majority of patients with CPHD (153) were homozygous c.[301_302delAG][ 301_302delAG], 22 patients were homozygous c.[150delA][150delA], 52 patients were compound heterozygotes c.[301_302delAG][150delA], nine were compound heterozygotes for c.[301_302delAG] and other PROP1 variant and one patient was a compound heterozygote for c.[150delA] and other PROP1 variant. Regarding healthy family relatives, 11 were heterozygous for c.[301_302delAG], three carried c.[150delA] on one allele and in one family member neither c.[301_302delAG], nor c.[150delA] was detected because he/she carried another heterozygous variant in the PROP1 gene. More than 30% of samples were obtained from Eli Lilly's international post-marketing research project GeNeSIS (Genetics and Neuroendocrinology of Short Stature International Study). Families originated from 21 countries: Argentina, Austria, Belarus, Bosnia and Herzegovina, Brazil, Canada, Croatia, the Czech Republic, the Dominican Republic, Germany, Hungary, Jamaica, Lithuania, Panama, Poland, Portugal, Russia, Slovakia, Slovenia, Spain and USA (Tables 1 and 2).
Table 1. Patients with c.[301_302delAG] and their family relatives analyzed in the study according to their country of origin.
| c.[301_302delAG] [301_302delAG] | c.[301_302delAG][150delA] | c.[301_302delAG][?]a | c.[301_302delAG][=]b(healthy) | Total | |
|---|---|---|---|---|---|
| Argentina | 2 | 0 | 0 | 0 | 2 |
| Austria | 2 | 0 | 0 | 0 | 2 |
| Belarus | 4 | 6 | 0 | 0 | 10 |
| Bosnia and Hercegovina | 3 | 0 | 0 | 2 | 5 |
| Brazil | 14 | 0 | 0 | 0 | 14 |
| Canada | 0 | 0 | 2 | 0 | 2 |
| Croatia | 0 | 1 | 0 | 0 | 1 |
| Czech Republic | 7 | 10 | 1 | 2 | 20 |
| Dominican Republic | 10 | 0 | 0 | 0 | 10 |
| Germany | 12 | 7 | 0 | 3 | 22 |
| Hungary | 0 | 0 | 1 | 0 | 1 |
| Jamaica | 3 | 0 | 0 | 0 | 3 |
| Lithuania | 47 | 2 | 1 | 0 | 50 |
| Panama | 0 | 2 | 0 | 0 | 2 |
| Poland | 25 | 14 | 0 | 0 | 39 |
| Portugal | 10 | 0 | 0 | 0 | 10 |
| Russia | 7 | 7 | 3 | 4 | 21 |
| Slovakia | 0 | 1 | 0 | 0 | 1 |
| Spain | 1 | 0 | 0 | 0 | 1 |
| USA | 6 | 2 | 1 | 0 | 9 |
| Total | 153 | 52 | 9 | 11 | 225 |
Abbreviation: USA, United States of America.
? denoted that other variant than c.[301_302delAG] or c.[150delA] was detected in the PROP1 gene.
=denoted that no variant in the PROP1 gene was observed.
Reference sequence: NM_0006261.4.
Table 2. Patients with c.[150delA] and their family relatives analyzed in the study according to their country of origin.
| c.[150delA][150delA] | c.[301_302delAG][150delA] | c.[150delA][?]a | c.[150delA][=]b(healthy) | c.[=][=](healthy) | Total | |
|---|---|---|---|---|---|---|
| Belarus | 2 | 6 | 0 | 0 | 0 | 8 |
| Croatia | 3 | 1 | 0 | 0 | 0 | 4 |
| Czech Republic | 2 | 10 | 1 | 1 | 1 | 15 |
| Germany | 1 | 7 | 0 | 2 | 0 | 10 |
| Lithuania | 0 | 2 | 0 | 0 | 0 | 2 |
| Panama | 0 | 2 | 0 | 0 | 0 | 2 |
| Poland | 11 | 14 | 0 | 0 | 0 | 25 |
| Russia | 1 | 7 | 0 | 0 | 0 | 8 |
| Slovakia | 0 | 1 | 0 | 0 | 0 | 1 |
| Slovenia | 2 | 0 | 0 | 0 | 0 | 2 |
| USA | 0 | 2 | 0 | 0 | 0 | 2 |
| Total | 22 | 52 | 1 | 3 | 1 | 79 |
Abbreviation: USA, United States of America.
? denoted that other variant than c.[301_302delAG] or c.[150delA] was detected in the PROP1 gene.
= denoted that no variant in the PROP1 gene was observed.
Reference sequence: NM_0006261.4.
The genetic diagnosis of CPHD, that is, detection of causal variants in the PROP1 gene in all patients was performed in several centers by either denaturing high-pressure liquid chromatography followed by direct Sanger sequencing if a DNA sequence alteration was suspected,8 or by direct Sanger sequencing only,9, 10 or by DNA digestion with BcgI and Mnl, respectively and confirmation by direct Sanger sequencing.11 To avoid inconsistencies, randomly selected samples from all participating centers were retested by direct Sanger sequencing in the Prague laboratory with the same results as obtained from the original laboratories.
Clinical and hormonal characteristics of most of the patients included in this study have been reported previously.4, 8, 9, 10, 12, 13, 14 Participants gave written informed consent in their native language for genetic testing. This study was approved by the Institutional Ethics Committee of University Hospital Motol and 2nd Faculty of Medicine, Charles University, Prague, the Czech Republic.
Genetic investigation of variant origin
SNP typing
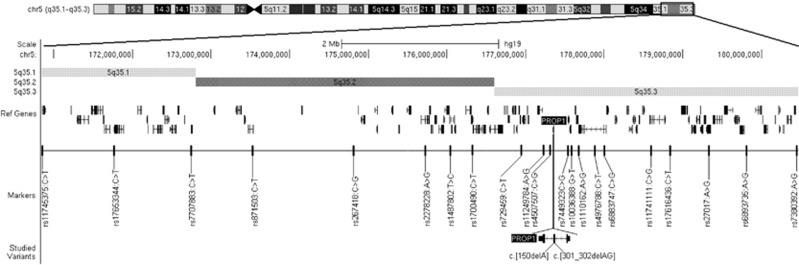
In order to assess the presence of eventual haplotype, we selected 21 single-nucleotide variant (SNP) markers surrounding the PROP1 gene (Figure 1) in length of 9.6 Mb (corresponding to ~21 cM in average based on the Rutgers Combined Map): rs11745375:C>T (chr5.hg19:g.170909160C>T), rs17653344:C>T, rs7707883:C>T, rs871503:C>T, rs267418:C>G, rs2278228:A>G, rs1487802:C>T, rs1700490:C>T, rs729459:C>T, rs11249784:A>G, rs4507507:C>G, rs7449323:C>G, rs10036388:G>T, rs1110162:A>G, rs4976788:C>T, rs6883747:C>G, rs11741111:C>G, rs17616436:C>T, rs27017:A>G, rs6893735:A>G and rs7380392:A>G. All markers were informative (the minor allele frequency in the general population ranged from 33 to 50% (ref. 15) and did not display mutual linkage disequilibrium according to the HapMap database15 (Supplementary Figure 1F) and also based on internal verification using 94 healthy Czech control samples (Pearson's r2≤0.1 for all pairs). The genotype of each marker in every subject was determined using the allelic discrimination method performed in TaqMan probes format (Applied Biosystems, Carlsbad, CA, USA) as described previously.16 The genotype as well as basic phenotype data have been submitted to the EGA database (https://www.ebi.ac.uk/ega/home), study number EGAS00001001165.
Figure 1.
Genetic region of interest with tested markers and variants.
Statistical analysis
Haplotype analysis
The Haploview software was used for detection of possible linkage disequilibrium between tested PROP1 gene variants and each of 21 SNP markers.17 Furthermore, the specific haplotypes based on population data (as family relatives were not available for all patients) have been reconstructed by Phase software.18 The analyses were performed separately for each of the studied variants with the exception of compound heterozygotes c.[301_302delAG][150delA] being included in both tests.
Variant age estimation
The history of given variants was estimated using a method based on the allelic association method.19 To avoid bias linked with relatedness among individuals, we performed the variant age estimation using only one patient from each family. The data used were the level of allelic association between variant alleles and surrounding markers obtained by haplotype analyses. The input parameters were the recombination rates derived from Rutgers Combined Map20, the estimated frequencies of the mutant alleles in the present populations (0.003 for c.[301_302delAG] and 0.001 for c.[150delA]) based on the prevalence of CPHD (1:8000 (ref. 21)), the current population sizes and the number of c.[301_302delAG] and c.[150delA] carriers derived from epidemiological studies10, 22 (see Supplementary Tables 3 and 4 for the values assumed in each population). We performed a joint maximum-likelihood estimation of the age of the studied variants as well as the population growth rates assuming neutrality. This maximum-likelihood estimation used a Mathematica Notebook available at https://sites.google.com/site/agegrowthestim/. This notebook implements equations (1) to (5) from Austerlitz et al19 (standard Luria–Debrück method).
Results
The origin of c.[301_302delAG]
The origin of the two-base deletion (c.[301_302delAG]) was studied in 225 subjects from all the countries mentioned above, with the exception of Slovenia. The majority of patients came from Central and Eastern Europe, less frequently from the Balkans, the Iberian Peninsula and Latin America (Table 1). Considering all c.[301_302delAG] carriers, the analysis showed that they shared a haplotype delimited by markers rs11249784:A>G and rs10036388:G>T (A-C-delAG-C-G) spanning ~175 kb around the PROP1 gene (Supplementary Figure 1A) confirming the ancestral origin of this variant. In total, 378 chromosomes carrying c.[301_302delAG] were investigated (Table 3) – the marker set-up A-C-delAG-C-G was observed on 252 chromosomes (67%) by Phase software, whereas 48 chromosomes (13%) carried A-G-delAG-C-G and other haplotypes were less frequent. In comparison, the respective most prevalent haplotype A-C-wt-C-G was observed only in 7.5% of healthy chromosomes of European ancestry.
Table 3. Ancestral haplotypes inferred by Phase software.
| Study group | Mutated chromosomes | Mutated chromosomes with ancestral haplotype | rs1700490:C>T | rs729459:C>T | rs11249784:A>G | rs4507507:C>G | VARIANT | rs7449323:C>G | rs10036388:G>T | rs1110162:A>G | rs4976788:C>T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Physical distance (Mb) | 1.02 | 0.39 | 0.04 | 0.01 | 0.00 | 0.08 | 0.14 | 0.34 | 0.54 | |||
| c.[301_302delAG] | Europea | 279 | 213 | C/T | C/T | A | C | delAG | C | G | A/G | C/T |
| Portugal and Spain | 22 | 15 | T | T | A | G | delAG | C | G | A/G | C/T | |
| Latin America 1b | 36 | 33 | C/T | T | A | G | delAG | C | G | A/G | C/T | |
| Latin America 2c | 24 | 23 | C/T | C/T | A | C | delAG | C | G | A | T | |
| North Americad | 17 | 16 | C/T | C/T | A | C | delAG | C | G | A/G | C/T | |
| c.[150delA] | Altogethere | 100 | 84 | C/T | C/T | A/G | C | delA | G | G | A | C/T |
Excluding patients from Iberian Peninsula (Portugal and Spain).
Argentina, Dominican Republic and partly Brazil.
Jamaica, Panama and partly Brazil.
USA and Canada.
Belarus, Croatia, Czech Republic, Germany, Lithuania, Panama, Poland, Russia, Slovakia, Slovenia, USA.
Ancestral haplotypes are indicated by bold. Reference sequence: NM_0006261.4.
Differing haplotypes in patients from various regions
On the basis of the obtained genetic data, we performed follow-up subgroup analyses, which indicated that there are different haplotype patterns among patients from the Iberian Peninsula as compared to the rest of Europe. The results are displayed in Table 3 and as Haploview diagrams in Supplementary Figures 1A–E. The linkage disequilibrium between surrounding markers and the variant was strengthened after including exclusively European subjects without Portugal and Spain (Supplementary Figure 1B). Among 301 affected chromosomes from European patients, 216 (72%) displayed the ancestral haplotype A-C-delAG-C-G. A different haplotype was observed in most patients from the Iberian Peninsula, (T-T-A-G-delAG-C-G, Supplementary Figure 1C). It reached a length of ~1.7 Mb and differed in the closest marker to c.[301_302delAG] (rs4507507:C>G), which was observed predominantly as nucleotide G in the Iberian samples versus nucleotide C in other European patients. Similarly, half of the chromosomes (55%) from patients referred from Latin America (Argentina, the Dominican Republic and partly Brazil) displayed a haplotype related to that detected in Portuguese and Spanish patients with c.[301_302delAG] (T-A-G-delAG-C-G, Supplementary Figure 1D). Other Latin American subjects with CPHD (from Jamaica, Panama and partly Brazil) displayed the ancestral European haplotype extended to 686 kb (A-C-delAG-C-G-A-T, Supplementary Figure 1E). Regarding the patients from North America (USA and Canada), 94% of their chromosomes with c.[301_302delAG] showed the same haplotype as the majority of European patients (A-C-delAG-C-G, Table 3).
Supplementary Table 2 summarizes the estimated age as the appearance of c.[301_302delAG] variant in the studied populations pinned together according to the results of haplotype analyses. We estimated that the ancestral haplotype prevailing in patients from Central and Eastern Europe as well as in USA and Canada was introduced ~101 generations ago corresponding to 2525 years with the assumption of 25 years per generation (confidence interval: 90.1–116.4 generations). The haplotype with c.[301_302delAG] observed in patients from the Iberian Peninsula seemed to be younger – its age was estimated to be 23.3 (20.1–29.1) generations, that is, 583 (503–728) years old. The estimated origin of the variant in patients from Argentina, the Dominican Republic and in Brazilian patients showing a similar haplotype as individuals from Portugal and Spain was 16.4 (14.4–20.1) generations ago, whereas for the haplotype in patients from Jamaica, Panama and the other Brazilian patients, the age of origin was estimated at 13.8 (12.2–17.0) generations ago. According to our statistical approach, which allowed us to estimate the variant age separately in the populations where more than one haplotype was observed, the c.[301_302delAG] occurred earliest in Lithuania (Supplementary Table 3).
The origin of c.[150delA]
Altogether, 79 patients from 11 countries (Belarus, Croatia, the Czech Republic, Germany, Lithuania, Panama, Poland, Russia, Slovakia, Slovenia and USA) carried the c.150delA and have been included in the study. Similar to the distribution of c.[301_302delAG] carriers, patients from Central Europe (Poland, Germany and the Czech Republic) prevailed (Table 2). An ancestral origin was documented also for the c.[150delA] that was transmitted on a haplotype spanning about 353 kb (Supplementary Figure 2). This common haplotype flanked by markers rs4507507:C>G and rs1110162:A>G (C-delA-G-G-A) was present in 84 of the 100 studied chromosomes (84%) carrying c.[150delA] (Table 3). Unlike variant c.[301_302delAG], the same ancestral haplotype found in European populations was observed also in patients from Panama and USA.
The c.[150delA] variant in the PROP1 gene emerged 43.7 (38.4–52.7) generations ago, corresponding to 1093 (960–1318) years with the assumption of 25 years per generation (Supplementary Table 2). Considering all studied populations with more than one detected haplotype independently, the c.[150delA] was estimated to have appeared earliest in the Belarus region (Supplementary Table 4).
Discussion
To our best knowledge, we have compiled the largest DNA collection of patients with CPHD caused by the most prevalent variants c.[301_302delAG] and/or c.[150delA] in the PROP1 gene. These patients originated from almost all countries with reported appearance of the respective variants. Our extended genetic investigation has proved that both c.[301_302delAG] and c.[150delA] variants in the PROP1 gene have been introduced to these populations by common ancestors.
Origin and dispersion of c.[301_302delAG]
We observed more than one common haplotype on which c.[301_302delAG] was transmitted. The results of the haplotype analyses as well as the lengths of the detected ancestral haplotypes indicate that a c.[301_302delAG] variant occurred probably in a Portuguese heterozygous carrier on a different haplotype (changed nucleotide C to G of rs4507507:C>G, Table 3) and this modified haplotype was subsequently transmitted not only to other subjects on the Iberian Peninsula but also to Latin America. The results from variant age estimation showed that the European ancestral haplotype is the oldest compared with the haplotypes detected either in patients from the Iberian Peninsula or Latin America (Supplementary Table 2).
Several approaches for ancestral variant age estimation from molecular data have been introduced so far.23, 24, 25 Because our data set comprised individuals from many different countries that could not be considered as a single homogenous population, we used a method that has been successfully applied to similar data sets from European subjects and which furthermore correlated well with other approaches to variant age inference.19 Splitting studied populations of patients with CPHD according to their shared, ancestral haplotype showed that the c.[301_302delAG] variant appeared in Europe in the first millennium BC. We might only speculate that the variant has been subsequently transmitted to the Iberian Peninsula or that it originated independently there on a modified haplotype in the fifteenth century AD. At that time, Portugal and Spain started to explore sea routes and other continents including Latin America. The plausible transmission of the c.[301_302delAG] variant from the Iberian Peninsula to regions of Latin America is supported by the fact that ancestral haplotypes related to the European ones were observed in Latin American patients and that the variant has been estimated to appear in Latin America in the seventeenth century, a time of exploration, colonization and conquest by Europeans.
Among all input parameters for variant age estimation described in the Materials and Methods section, the ratio of most frequently observed haplotypes out of all haplotypes and recombination rate are crucial. Therefore, for populations where only one haplotype has been detected (eg, Bosnia and Herzegovina, the Dominican Republic in case of c.[301_302delAG] and Slovenia in case of c.[150delA], respectively) the variant age could not be estimated. Moreover, the data for each population (Supplementary Table 3) could be biased as not all subjects with CPHD caused by the respective variant have been included in the study. Nevertheless, the country with the estimated first appearance of c.[301_302delAG] (third millennium BC) was Lithuania in the Baltic sea region with the highest reported proportion of c.[301_302delAG] variant carriers worldwide10, followed by variant occurrence in the neighboring or close-by countries including the Eastern part of Germany, Belarus, Poland, the Czech Republic and Russia.
History of c.[150delA]
Variant c.[150delA] in the PROP1 gene was originally described in patients with hereditary dwarfism originating exclusively from two adjacent villages, Baščanska Draga and Jurandvor, of the Island of Krk, Croatia.4 DNA samples of two patients from Krk (one from Baščanska Draga and one from Jurandvor village)26 were analyzed in the present study. Both patients displayed the same ancestral haplotype as the other patients with the c.[150delA] variant. The variant c.[150delA] was estimated to have appeared in Croatia in the seventeenth century AD, whereas in other European countries it emerged earlier (Supplementary Table 4). One may speculate that the variant was brought to the Island of Krk where it spread in this isolated population owing to the genetic drift phenomenon of changing allelic frequencies.5 Nevertheless, the overall variant age of c.[150delA] including all studied mutated chromosomes dated the respective DNA change back to the tenth century AD.
Revised prior assumptions of c.[301_302delAG] genesis
The dramatic geographic clustering of the respective subjects in the selected parts of the world (Supplementary Table 1), illustrated by 70% prevalence of c.[301_302delAG] in Lithuanian patients with CPHD10 and no patient with the PROP1 anomaly from Netherlands,27 suggests a founder effect. On the other hand, the c.[301_302delAG] variant is located in three tandem GA repeats and such genetic regions are prone to polymerase slippage during DNA replication.28 These genetic regions are therefore considered as variant hot spots. Independent origin of c.[301_302delAG] in each patient has also been suggested in a microsatellite study of one marker (D5S408) showing its different alleles in unrelated patients with the studied variant.6 However, the distance between the marker and the PROP1 gene was about 2.5 Mb, well beyond the distance of the common haplotypes identified in our study.
Strengths and limitations of the study
The main strength of the study is its international extent covering a significant number of all patients with c.[301_302delAG] and/or c.[150delA] variants in the PROP1 gene. Moreover, the carefully selected set of 21 tested markers surrounding the PROP1 gene within a total length of 9.6 Mb and with the closest markers 12.3 kb upstream and 84.7 kb downstream from the gene represented a robust approach for both haplotype analyses and variant age estimation. The present study has also limitations. First, the PROP1 gene is located at the end of human chromosome five where the number of available informative SNP markers is limited. Therefore, the density of coverage was a compromise not allowing exact determination of the boundaries of the ancestral haplotypes and a mixture of ancestral and independently generated variants could not be implicitly excluded. Second, the ancestral variant age estimation is dependent on several parameters (frequency of mutated alleles in the populations, CPHD prevalence), which are difficult to estimate because of a lack of epidemiological and population data. Finally, it needs to be noted that the epidemiological data concerning CPHD patients are not available from all countries worldwide and although our data set comprises of a significant number of samples, it still does not cover all reported patients. This could influence the observed origin of the variants.
Conclusion
We present strong evidence that the recurrent variants in the PROP1 gene (c.[301_302delAG] and c.[150delA]), previously assumed to have arisen as a consequence of variant hot spots, are preferably founder variants. Results of this work have contributed to the population history of different nations. Moreover, in patients with clinically defined CPHD from Central and Eastern Europe, the Balkan, the Iberian Peninsula and Latin America genotyping of these two variants should be performed as a first step in the genetic investigation prior to entire PROP1 gene sequencing and may bring along significant time and costs savings.
Acknowledgments
We are grateful to attending endocrinologists and Professor Szalecki (Faculty of Health Sciences Jan Kochanowski University, Kielce, Poland) who provided the samples of their patients for the study. We also thank the participants (patients and their relatives from all countries). The study was supported by Eli Lilly IIT (grant number B9R-CY-O057), the Internal Grant Agency of the Czech Ministry of Health (grant number NT13692) and by the project for conceptual development of research organization 00064203/6001 (Ministry of Health, the Czech Republic).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
P.D., M.R.B., R.W.P., J.S.P. and J.L. have previously consulted for and received grant support from Eli Lilly and Company. W.F.B. was previously employed by and has equity interests in Eli Lilly and Company. Other authors declare no potential conflict of interest.
Supplementary Material
References
- 1Sornson MW, Wu W, Dasen JS et al: Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature 1996; 384: 327–333. [DOI] [PubMed] [Google Scholar]
- 2Wu W, Cogan JD, Pfaffle RW et al: Mutations in PROP1 cause familial combined pituitary hormone deficiency. Nat Genet 1998; 18: 147–149. [DOI] [PubMed] [Google Scholar]
- 3Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN: The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014; 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Krzisnik C, Kolacio Z, Battelino T, Brown M, Parks JS, Laron Z: The 'little people' of the island of Krk - revisited. Etiology of hypopituitarism revealed. J Endocr Genet 1999; 1: 9–19. [Google Scholar]
- 5Strachan T, Read A: Human Molecular Genetics. 4th edn. Garland Science: New York, NY, USA, 2010. [Google Scholar]
- 6Cogan JD, Wu W, Phillips JA 3rd et al: The PROP1 2-base pair deletion is a common cause of combined pituitary hormone deficiency. J Clin Endocrinol Metab 1998; 83: 3346–3349. [DOI] [PubMed] [Google Scholar]
- 7Fofanova OV, Takamura N, Kinoshita E et al: A mutational hot spot in the Prop-1 gene in Russian children with combined pituitary hormone deficiency. Pituitary 1998; 1: 45–49. [DOI] [PubMed] [Google Scholar]
- 8Lebl J, Vosahlo J, Pfaeffle RW et al: Auxological and endocrine phenotype in a population-based cohort of patients with PROP1 gene defects. Eur J Endocrinol 2005; 153: 389–396. [DOI] [PubMed] [Google Scholar]
- 9Lemos MC, Gomes L, Bastos M et al: PROP1 gene analysis in Portuguese patients with combined pituitary hormone deficiency. Clin Endocrinol (Oxf) 2006; 65: 479–485. [DOI] [PubMed] [Google Scholar]
- 10Navardauskaite R, Dusatkova P, Obermannova B et al: High prevalence of PROP1 defects in Lithuania: phenotypic findings in an ethnically homogenous cohort of patients with multiple pituitary hormone deficiency. J Clin Endocrinol Metab 2014; 99: 299–306. [DOI] [PubMed] [Google Scholar]
- 11Deladoey J, Fluck C, Buyukgebiz A et al: 'Hot spot' in the PROP1 gene responsible for combined pituitary hormone deficiency. J Clin Endocrinol Metab 1999; 84: 1645–1650. [DOI] [PubMed] [Google Scholar]
- 12Mendonca BB, Osorio MG, Latronico AC, Estefan V, Lo LS, Arnhold IJ: Longitudinal hormonal and pituitary imaging changes in two females with combined pituitary hormone deficiency due to deletion of A301,G302 in the PROP1 gene. J Clin Endocrinol Metab 1999; 84: 942–945. [DOI] [PubMed] [Google Scholar]
- 13Mody S, Brown MR, Parks JS: The spectrum of hypopituitarism caused by PROP1 mutations. Best Pract Res Clin Endocrinol Metab 2002; 16: 421–431. [DOI] [PubMed] [Google Scholar]
- 14Osorio MG, Marui S, Jorge AA et al: Pituitary magnetic resonance imaging and function in patients with growth hormone deficiency with and without mutations in GHRH-R, GH-1, or PROP-1 genes. J Clin Endocrinol Metab 2002; 87: 5076–5084. [DOI] [PubMed] [Google Scholar]
- 15Altshuler DM, Gibbs RA, Peltonen L et al: Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Dusatkova P, Pruhova S, Borowiec M et al: Ancestral mutations may cause a significant proportion of GCK-MODY. Pediatr Diabetes 2012; 13: 489–498. [DOI] [PubMed] [Google Scholar]
- 17Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 18Stephens M, Smith NJ, Donnelly P: A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Austerlitz F, Kalaydjieva L, Heyer E: Detecting population growth, selection and inherited fertility from haplotypic data in humans. Genetics 2003; 165: 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Matise TC, Chen F, Chen W et al: A second-generation combined linkage physical map of the human genome. Genome Res 2007; 17: 1783–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21de Graaff LCG: PROP1-related combined pituitary hormone deficiency. in Pagon RA, Adam MP, Bird TD (eds): GeneReviews® [Internet]. University of Washington, Seattle, WA, USA, 2000. [Google Scholar]
- 22Halasz Z, Toke J, Patocs A et al: High prevalence of PROP1 gene mutations in Hungarian patients with childhood-onset combined anterior pituitary hormone deficiency. Endocrine 2006; 30: 255–260. [DOI] [PubMed] [Google Scholar]
- 23Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L: Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet 2004; 41: 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Reeve JP, Rannala B: DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 2002; 18: 894–895. [DOI] [PubMed] [Google Scholar]
- 25Slatkin M, Rannala B: Estimating the age of alleles by use of intraallelic variability. Am J Hum Genet 1997; 60: 447–458. [PMC free article] [PubMed] [Google Scholar]
- 26Krzisnik C, Grguric S, Cvijovic K, Laron Z: Longevity of the hypopituitary patients from the Island Krk: A follow-up study. Pediatr Endocrinol Rev 2010; 7: 357–361. [PubMed] [Google Scholar]
- 27de Graaff LC, Argente J, Veenma DC, Drent ML, Uitterlinden AG, Hokken-Koelega AC: PROP1, HESX1, POU1F1, LHX3 and LHX4 mutation and deletion screening and GH1 P89L and IVS3+1/+2 mutation screening in a Dutch nationwide cohort of patients with combined pituitary hormone deficiency. Horm Res Paediatr 2010; 73: 363–371. [DOI] [PubMed] [Google Scholar]
- 28Richards RI, Sutherland GR: Simple repeat DNA is not replicated simply. Nat Genet 1994; 6: 114–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.