Abstract
Hexameric helicases control both the initiation and the elongation phase of DNA replication. The toroidal structure of these enzymes provides an inherent challenge in the opening and loading onto DNA at origins, as well as the conformational changes required to exclude one strand from the central channel and activate DNA unwinding. Recently, high-resolution structures have not only revealed the architecture of various hexameric helicases but also detailed the interactions of DNA within the central channel, as well as conformational changes that occur during loading. This structural information coupled with advanced biochemical reconstitutions and biophysical methods have transformed our understanding of the dynamics of both the helicase structure and the DNA interactions required for efficient unwinding at the replisome.
Keywords: Hexameric Helicase, DNA replication, DNA unwinding, Helicase Loading, Helicase Assembly
Introduction and context
Cell proliferation relies on the exact replication of an organism’s genetic material in a rapid but precisely controlled and efficient manner. The process and mechanism of DNA replication directs targeted and repetitive enzymatic activities towards long linear polymers of DNA. Interestingly, organisms have evolved a number of toroidal DNA replication and repair protein complexes that can maintain repetitive catalysis by encircling the DNA substrate. These protein-DNA rotaxane-like systems have the intrinsic ability to be processive enzymes due to their topological linkage with the substrate. As such, they provide inherent challenges to the loading and encircling of DNA. The steps required for the loading and encircling of circular protein complexes onto DNA provides for a higher level of regulation, which is required to restrict cell cycle progression and control DNA replication initiation. Because of this, the most highly regulated component within the DNA replisome is the loading and activation of the hexameric helicase, which dictates both the initiation steps and the elongation rate of DNA replication. Even though the general toroidal hexameric helicase structure has been known for more than two decades, the mechanisms for loading, encircling, activating, and unwinding are only just being discovered. These recent advances have been primarily aided by higher resolution structures that include DNA, better mechanistic descriptions of the interactions of the helicase with each separated strand of single-stranded DNA (ssDNA), and higher order in vitro reconstitution of DNA replication systems. It is an exciting time to be a part of the hexameric helicase field as big questions regarding dynamic structure-function relationships with DNA are poised to be revealed.
Hexameric helicase architectural conservation
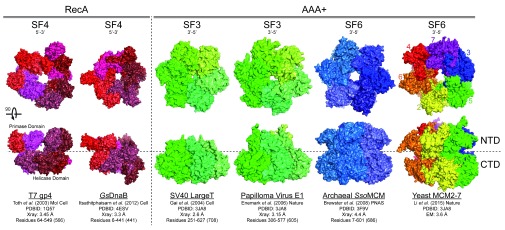
Although the general architecture of hexameric DNA replication helicases is shared across organismal domains, there is strong evidence that classes of these enzymes have evolved independently for a role in DNA replication 1. Although all hexameric helicases are members of the broader P-loop family of ATPases 2, individual evolution of RecA domains gave rise to the superfamily (SF) 4 helicases including T4 gp41, T7 gp4, bacterial DnaB, and mitochondrial Twinkle 3, while SF3, including SV40 Large T antigen (SV40 L-Tag) and papilloma virus E1, and SF6, including archaeal and eukaryotic minichromosome maintenance proteins (MCM), helicases came from an ATPases associated with a variety of cellular activities (AAA +) clade ( Figure 1 & Table 1) 4, 5. Regardless of the origin, these systems have all converged on a common ring-shaped architecture wherein a central channel is used to repetitively engage and translocate along ssDNA during unwinding. Bacterial and phage SF4 helicases are perhaps the best studied and have contributed most to our understanding of DNA unwinding, but more recent emphases on SF3 and SF6 helicases are providing insight into structure-function relationships across SFs.
Figure 1. Structural conservation of hexameric helicases.
Hexameric helicases are shown from different domains (RecA or AAA +) and superfamilies (SFs) with associated unwinding polarity and references. View from the N-terminal domain (NTD) rotated 90° to visualize the lateral length from the C-terminal domain (CTD) to the NTD.
Table 1. Model Replication Helicase Loading and Activation Components.
| Initiator | Helicase | SF
(Polarity) |
Loader | Activator | Accessory | |
|---|---|---|---|---|---|---|
| Eukaryotes | ||||||
| Sce/ Hsa | Orc1-6 | MCM2-7 | SF6 (3’-5’) | Cdt1/Cdc6 | CDK/DDK | GINS/Cdc45 |
| Archaea | ||||||
| Sso/ Mth | Orc1-2(3) | MCM | SF6 (3’-5’) | Cdt1 | - | GINS/RecJ |
| Bacteria | ||||||
| Eco/ Bsu | DnaA | DnaB | SF4 (5’-3’) | DnaC (DnaI) | - | Rep |
| Mitochondria | ||||||
| Hsa | - | Twinkle | SF4 (5’-3’) | - | - | - |
| Viral | ||||||
| papilloma E1 | - | E1 | SF3 (3’-5’) | - | - | - |
| polyoma SV40 | - | Large T-ag | SF3 (3’-5’) | - | - | - |
| Bacteriophage | ||||||
| T4 | - | gp41 | SF4 (5’-3’) | gp59 | - | - |
| T7 | - | gp4 | SF4 (5’-3’) | - | - | - |
Helicase activity requires the presence of both nucleotide triphosphates (NTPs) 6 and Mg 2+ for unwinding 7– 9. Although it is not known exactly how ATP hydrolysis directly drives DNA unwinding, it is likely to progress in a sequential manner, with each subunit driving conformational changes throughout the hexamer that contribute to unwinding polarity 4, 10– 14. RecA-like helicases (SF4) translocate along ssDNA in the 5’ → 3’ direction, while AAA + enzymes (SF3 and SF6) translocate in the 3’ → 5’ direction 13, 15– 18. The core ATP binding and hydrolysis domains consist of conserved RecA-like or AAA + folds that exist within a single subunit or between adjacent subunits that include “Walker A and B motifs” for ATP binding and hydrolysis and a basic “arginine finger” residue for nucleotide turnover and conformational coupling 3, 19– 22. Conserved β-hairpin structures in AAA + helicases contribute differentially to DNA binding and unwinding to direct DNA through the channel 11, 16, 22– 26, although additional interactions with DNA have also been detected on the exterior surface of helicases 15, 27, 28.
Several high-resolution X-ray and electron microscopy (EM) structures have been reported for the apo and nucleotide-bound forms of hexameric helicases (T7 gp4 29, 30, DnaB 31, 32, Mito Twinkle 33, SV40 Large-T 11, 34, Sulfolobus solfataricus MCM [ SsoMCM] 23, 35, and Saccharomyces cerevisiae MCM [ ScMCM2-7] 36). The global shared architecture of the ring-shaped helicases is generally composed of two tiers: an N-terminal DNA-binding domain (NTD) and a C-terminal AAA + or RecA motor domain (CTD) ( Figure 1). The orientation of most helicases on DNA places the CTD toward the duplex double-stranded DNA (dsDNA) region and the NTD outwards 22, 37. The exception is E1, where the orientation is reversed, placing the NTD toward the duplex 38. Thus, motor domains are often positioned close to the dsDNA duplex, which can leave the NTD regions free to bind, stabilize, or act on the resultant ssDNA.
Across species, hexameric helicase NTDs seem to have evolved differential functions. T7 gp4 can be expressed as either a 56 kDa helicase only form or as a full-length two-domain 65 kDa helicase-primase 39. The composition of the T7 gp4 helicase hexamer is thought to be a mixture of the two forms in vivo, controlling the number of primases present for faster replication and less pausing. Other SF4 helicases, including T4 gp41 and Escherichia coli DnaB, interact with a separately encoded primase at the NTD in an analogous configuration. In those cases, the composition and ratio of helicase to primase is <1:1, and more often recognized as 6:3 31, 40. For DnaB, ATP binding by the motor domain can induce conformational changes within the NTD collar that can regulate partner protein (i.e. DnaC or DnaG) selection 41. As can be seen in Figure 1, increasing organismal complexity through the SF3 and SF6 helicases (from left to right) generally increases the size of the NTD to where they have evolved additional β-hairpins and zinc-finger motifs for more stabilized binding of the encircled strand and double hexamer formation 42, 43. The expanded NTD also provides a platform for control of activity through helicase accessory protein binding (in the case of Cdc45 and GINS 44) or activation through phosphorylation by either cyclin-dependent kinase (CDK) or Dbf4-dependent Cdc7 kinase (DDK) 45– 47.
Helicase loading and the encircling of DNA
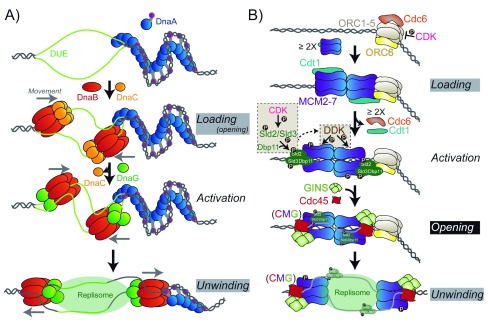
The loading of hexameric helicases at replication origins and the associated steps required for encircling only one strand have been the subject of much debate over the years. What is clear is that the loading of the hexameric helicase generally requires the concerted action of accessory initiator proteins to locally melt duplex DNA and facilitate encircling of DNA. However, phage T7, mitochondrial Twinkle, and SV40 L-Tag helicases can load onto circular dsDNA on their own 48– 50. Within the three domains of life, the core ATPase activity and ordered assembly of replication initiation factors seem to be preserved to control the start of DNA synthesis 51, 52 ( Table 1). In bacteria, the initiator, DnaA, forms a multimeric right-hand filament at the replication origin, oriC, to induce unwinding or melting at an A-T rich DNA unwinding element (DUE) ( Figure 2A) 53– 60. Afterwards, the DnaB helicase is loaded on the top and bottom strands by concerted activities of DnaC and DnaA 61– 66. Once loaded, the primase, DnaG, interacts with DnaB, displaces DnaC 40, 67, and activates unwinding 68. The association of an accessory helicase, Rep, with DnaB may aid in replication fork progression 69– 71.
Figure 2. Assembly of Hexameric Replication Helicase at Origins.
A) Loading of the bacterial DnaB helicase by the loader, DnaC, requires the destabilization of a DNA unwinding element (DUE) by the initiator protein DnaA. DnaB complexes with the primase, DnaG, to translocate along the lagging strand unwinding DNA ahead of the replication fork. B) Loading of the eukaryotic MCM2-7/Cdt1 complex requires the initial binding of the ORC complex (ORC1-6) and Cdc6. Interactions with Cdc6 and ORC with the CTD of MCM2-7 directs the adjacent loading of the first hexamer and dissociation of Cdc6 and Cdt1. Subsequent loading of the second hexamer is thought to proceed through direct interactions between the NTD of MCM2-7 to form the double hexamer. Activation of the helicase includes CDK phosphorylation of Sld2 and Sld3 to promote interaction with Dpb11 and stimulate DDK phosphorylation of MCM2/4/6 and recruitment of GINS and Cdc45 to form the CMG complex. Opening of the CMG complex and exclusion of the nontranslocating strand from the central channel activates unwinding and translocation on the leading strand. Gray and black boxes represent major and the foremost, respectively, queries remaining regarding structural conversions of helicases at origins.
In archaea and eukaryotes, the binding of the origin recognition complex (ORC1-6) and Cdc6 to origins of replication is necessary for loading MCM2-7/Cdt1 complexes onto dsDNA to generate a pre-replication complex (Pre-RC) ( Figure 2B) 72– 76. The precise structural conformations and dynamics of MCM loading are not fully known, but the steps and components for assembly and activation of the eukaryotic MCM2–7 complex have been recently biochemically reconstituted in vitro, providing significant insight into the process 77, 78. In archaea, the homohexameric MCM complex exists as a closed ring in solution and would require initiation factors to stimulate opening into a helical conformation onto DNA 79. Alternatively, increases in temperature for these model hyperthermophilic archaeal MCMs may provide the thermal energy required for the destabilization of a subunit interface required for loading. The eukaryotic MCM2-7 helicase appears to be naturally open, with a labile 2-5 interface that can be trapped by ORC/Cdc6 80– 82. The ORC1-6 complex is arranged in a two-layered cracked ring that encircles DNA and uses the helix-turn-helix domains to engage the MCM2-7 hexamer in a proposed ring-ring interaction 83, 84, in a manner similar to the loading mechanism of clamp/clamp-loader complexes onto dsDNA 85. The organization of the ORC complex also appears to be regulated and exists in either an autoinhibited ATP-bound form that precludes DNA binding or a proposed active form that requires a large conformational change in ORC1 that makes the complex competent for encircling DNA 83. Afterwards, the first MCM2-7 hexamer is loaded through direct interactions of MCM6-Cdt1 with the ORC1-6/Cdc6 complex 86, 87. The second MCM2-7 hexamer is loaded through contacts between the NTDs of the first loaded MCM2-7 hexamer, rather than through interactions with the ORC1-6/Cdc6 complex 78, 84, 88, 89. This generated the double hexamer complex, which has been known for years for MCM (and SV40 L-Tag) 42, 90– 92, but whether the double hexamer represents an active unwinding unit or an intermediate in the loading process was not known.
After loading of the MCMs, a series of steps are required to form the active unwinding complex ( Figure 2B). ATP hydrolysis by Cdc6 and ORC1 causes the dissociation of Cdc6 and Cdt1 93. Subsequent phosphorylation of Sld2 and Sld3 by CDK promotes Dpb11 (DNA polymerase B-associated protein) to interact with MCM2-7 94– 96. Phosphorylation of Sld3, in particular, recruits Cdc45 and GINS (Sld5, Psf1, Psf2, Psf3) to MCM2-7 and stimulates the DDK-dependent phosphorylation of MCM2 97, as well as MCM4/6 98, 99. These phosphorylation events allow opening of the MCM2/5 interface to extrude ssDNA that remains bound to Sld2/Sld3/Dpb11 100, 101. The active unwinding CMG complex or “unwindosome” is formed through the association of Cdc45 and GINS with MCM2-7 at the labile MCM2-MCM5 interface 80, 81 along the waist between the NTD and the CTD 44, 80, 102. Cdc45 in particular blocks the MCM2-5 gate and prevents the loss of DNA from the central channel 103. In the reverse mechanism, Cdc45 may also be important for converting MCM2-7 encircled on dsDNA to encircling only a single DNA strand while excluding the other. Formation of the CMG complex widens the gap between the NTD and CTD at MCM2 and MCM5 while concomitantly narrowing the interface at the opposite MCM4 and MCM6 subunits. This induced spiral configuration may contribute to coupled ATP hydrolysis, propagating a conformational change through the MCM2-7 complex to translocate along and unwind duplex DNA 44, 103.
Interactions with DNA: views of the encircled strand
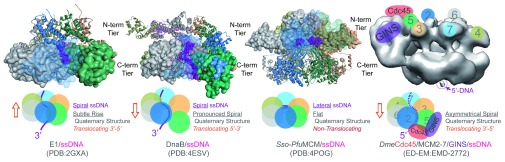
X-ray structures of hexameric helicases with oligonucleotides bound in the central channel (E1, E. coli Rho [ EcRho], Bacillus stearothermophilus [Bst] DnaB, and Pyrococcus furiosus MCM [ PfuMCM]) have informed our understanding of the contacts and conformations required for translocation along ssDNA. In these co-crystal structures, ssDNA is bound in a helical conformation in the central channel making direct contacts with each subunit ( Figure 3). For E1 and Rho, the hexameric ring is proposed to remain closed, but conformational changes between subunits, coupled with sequential ATP binding and hydrolysis around the ring, direct ssDNA through the central channel through contact with DNA binding loops in a staircase motion 13, 38, 104. Each hexamer subunit interacts with one nucleotide of the oligo, predicating a step-size of one nucleotide per ATP hydrolyzed. This is consistent with the measured step-size of T7 gp4 of one base-pair unwound per dTTP hydrolyzed 105. For DnaB, the crystal structure resembles more of a lock washer, where similar conformational changes throughout the quaternary structure facilitate movement, with a step-size of two nucleotides per ATP hydrolyzed, maintaining a cracked ring structure 106. The ssDNA bound to the archaeal MCM seems to be trapped in a lateral orientation around the interior of the NTD, possibly identifying specific contacts during activation or unwinding, implying a step-size greater than one nucleotide per ATP hydrolyzed during translocation 107. The EM structure of the intact eukaryotic CMG complex bound to DNA is in a spiral or lock washer conformation 44, more similar to the DnaB/ssDNA complex. The crack in the ring between the MCM2 and 5 subunits is again held in check by the Cdc45 and GINS subunits and helps to stabilize the spiral configuration ( Figure 3). Of course, the impact and absolute degree of spiraling, wrapping, or compaction of the encircled strand will need to be validated experimentally, most likely using single-molecule approaches to measure end-to-end distances during loading and unwinding. Almost certainly the flat ring, the asymmetrical spiral, and the cracked lock washer structures represent intermediates during helicase activation and unwinding, but both conformations will also need to be validated further with additional high-resolution structural studies or rigorous biophysical characterizations to monitor the changes in the conformations.
Figure 3. Conformational States of Hexameric Helicases Bound to the Encircled Strand.
Space-filling representation of the C-terminal domain (E1 [SF3, 3’-5’] and DnaB [SF4, 5’-3’]) or N-terminal domain ( Sso- Pfu hybrid [SF6, 3’-5’]) interacting with the encircled strand (purple). Also shown is the electron microscopy structure of the Drosophila Cdc45/MCM2-7/GINS complex (CMG) (SF6, 3’-5’) with color-coded subunits. The conformational states of the active translocating hexamers representing rings with subtle rises (E1) or obvious spirals (DnaB and CMG) in the structures as well as helical single-stranded DNA (ssDNA) are indicated in the schematics. The flat Sso- Pfu hybrid structure represents a nontranslocating state used to identify a novel lateral DNA binding site. The orange box arrow indicates the translocation direction of the hexamer relative to the encircled ssDNA.
Interactions with DNA: Impact of the excluded strand
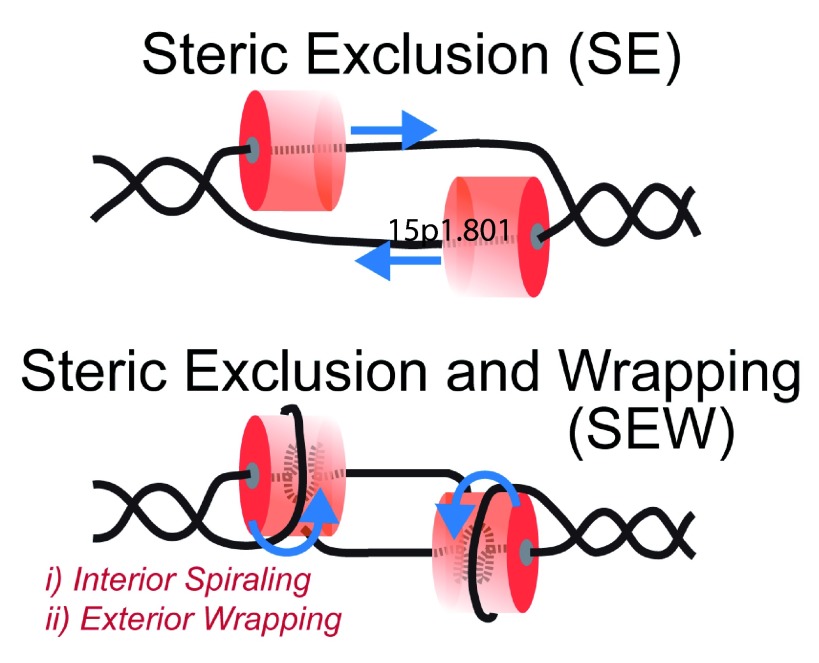
Based on the structures and associated biochemical data, the steric exclusion (SE) model, where one strand is encircled and the other is physically excluded, has become the consensus opinion for unwinding for hexameric replication helicases ( Figure 4) 108. One caveat to this model is that it generally ignores any contributions of the excluded strand to unwinding. Electrostatic interactions with the excluded strand on the external surface of hexameric helicases have been noted for archaeal MCM and shown to be important for unwinding, contributing to the development of the steric exclusion and wrapping (SEW) model ( Figure 4) 17, 109. Others have also noted that both ssDNA and dsDNA have a binding site on the external surface of other helicases 15, 27, 28. The dynamic and somewhat stochastic nature of unwinding has been attributed to interactions of ssDNA on the external surface of hexameric helicases E1 38, T4 gp41 110, and DnaB 111. In addition, subunits within the unwindosome complexes of SV40 L-Tag 112 and CMG 113 have been shown to interact with the excluded strand for loading and activation of unwinding. Intriguingly, DNA repair helicases have also been shown to sense damage or modifications on the excluded strand and stall unwinding 114– 117.
Figure 4. DNA Unwinding Models for Hexameric Replication Helicases.
Steric exclusion (SE) model encircles the translocating strand and physically separates the nontranslocating strand outside of the central channel. The steric exclusion and wrapping (SEW) model takes into account specific interactions within the central channel (compaction or spiraling) as well as external interactions (binding or wrapping) with the excluded strand. Blue arrows indicate the direction of movement of the DNA strands with respect to the hexamer.
For SV40 L-Tag, initial binding to the origin may be directed by internal β-hairpins making direct contacts with the minor groove 49 and specific contacts of the origin binding domain (OBD) to the major groove 118. It is not currently understood how this initial dimer contact nucleates assembly of a double hexamer around dsDNA. Once loaded, SV40 L-Tag is proposed to convert from encircling duplex DNA to encircling ssDNA by pumping and extruding one strand out through side-channels 11, 119. Conformational changes within internal β-hairpins may direct the translocating strand through the central channel, while extruding the opposing strand. Using single-molecule experiments, researchers have shown that DNA unwinding proceeds with a single hexamer of L-Tag in a steric exclusion mechanism that is somewhat conformational mobile and able to bypass bulky adducts during translocation 120. In comparison, a novel mechanism has recently been proposed for E1 where duplex DNA enters the hexamer before being separated internally and forcing individual strands out through separate exits channels 121. Of course, the steps and dynamics for how these hexameric helicases convert from encircling duplex DNA to single strand separases by pumping DNA out through side-channels, opening of a gate, or through another unknown mechanism need to be visualized directly with high resolution. A recent EM structure shows the leading strand Pol ε ahead of the yeast CMG complex (at the CTD) and suggests a possible model where the encircled leading strand bends back and threads through a side-channel via the MCM2-5/Cdc45/GINS gate to enter the polymerase active site 122. Alternative models of replisome-DNA interactions were also proposed in this study.
With this emerging information, excluded or opposing strand interactions shown in the SEW model ( Figure 4) are poised to play multifaceted roles in loading, encircling, unwinding, and sensing of DNA. In the case of archaeal MCM, the external ssDNA binding path in the SEW model serpentines along the lateral length of the homohexamer, spanning the CTD and NTD, and even crossing and wrapping across multiple subunits (Graham & Trakselis, unpublished data) 17. The SEW model for archaeal MCM is analogous to a socket wrench, whereby the encircling of one strand represents the socket and external interactions with the excluded strand represent the directional ratchet controlling the speed and stabilization of unwinding. Whether the SEW model is conserved in all or most hexameric helicases and/or at what stages of helicase assembly it may occur remains to be determined. Currently, we have found that external interactions and dynamics with the excluded strand in the E. coli DnaB helicase are practically identical to that of SsoMCM, despite their opposing polarities (Carney & Trakselis, unpublished). On the other hand, for T7 gp4 the excluded ssDNA interacts with T7 DNA polymerase to generate a replisome complex, where the helicase and polymerases are within one nucleotide of the fork junction and the helicase can make no external contact with the excluded strand 105, 123. Next, it will be important to determine which of the eukaryotic MCM subunits (MCM2–7) interact specifically with the excluded strand or whether uniform binding sites have evolved on all subunits. It is intriguing that this external contact may help fill in some of the missing steps highlighted in the gray or black boxes depicted in Figure 2.
Although the structural features of the SEW model may be conserved with various hexameric helicases, both the mechanistic roles and molecular interaction sites may be different. In the case of SsoMCM, disruption of external interactions through mutagenesis reduced unwinding efficiency (3’-5’) 17, but analogous external mutations on DnaB show a stimulation in unwinding (5’-3’) (Carney & Trakselis, unpublished). Modification of the excluded strand to a morpholino oligo similarly stimulates the unwinding rate of T7 gp4 124. Whether these effects result from opposite unwinding polarities or finely tuned control of unwinding rates and maintenance of the excluded strand requires further testing. However, detection and identification of these novel external interactions may provide a unique opportunity to target specific helicases for inhibition. As none of the different hexameric helicase families exhibit significant sequence homology outside of the center P-loop NTPase fold, novel exterior patches (e.g. between the CTD and the NTD) may provide idealized locations for specific targeting of small molecules that perturb unwinding through disruption of excluded strand contacts and avoid direct inhibition of the internal conserved ATPase site.
Future directions
Although significant advances in our understanding of hexameric helicase assembly, loading, and unwinding have been made over the past few years from quantitative biophysical characterizations and various high-resolution structures, more work is required to reveal specific mechanistic steps and transitions. For example, the essential components for the initial loading of hexameric helicases onto DNA are well described, but the conformational changes that occur within the hexamers during the encircling of ssDNA are still unknown. After all these years, the black box in the whole mechanism is still the structural conversion of the helicase from encircling dsDNA to the encircled ssDNA directing the polarity of translocation and unwinding, primarily for SF3 and SF6 enzymes. Although much is known about the loading and activation mechanism in the Gram-negative E. coli system, far less is known about SF4 helicases in the Gram-positive organisms where DnaI acts as the loader 65, 125, 126 or in bacteria which lack DnaC/DnaI loader homologs altogether 127, 128.
Although there is a wealth of structural information on the static hexameric helicases themselves, there is still much debate on the mechanics of helicase action. No longer is the focus directly on the structure of the helicase protein itself. Instead, it has switched from identifying conformational changes, transacting proteins, and post-translational modifications that reveal how duplex DNA is destabilized and the path it takes to be excluded. Finally, although the unwinding mechanism of hexameric helicases was thought to be as simple as excluding one strand from the central channel, new information highlighting the specificity and importance of interactions with the nontranslocating strand have central implications on loading and unwinding mechanisms. It is these dynamic conformational steps from the viewpoint of both the helicase and the duplex DNA that will lead to the next transformational leap in replication helicase discovery.
Abbreviations
AAA +, ATPases associated with a variety of activities; ATP, adenosine triphosphate; Cdc6, cell division cycle 6; Cdc45, cell division cycle 45; CDK, cyclin-dependent kinase; Cdt1, chromatin licensing and DNA replication factor 1; CMG, Cdc45/MCM2-7/GINS complex; CTD, C-terminal domain; DDK, Dbf4-dependent Cdc7 kinase; Dpb11, DNA polymerase B-associated protein; dsDNA, double-stranded DNA; EM, electron microscopy; GINS, go-ichi-nii-san, Japanese for 5-1-2-3 for Sld5-Psf1-Psf2-Psf3; MCM, minichromosome maintenance proteins; NTD, N-terminal domain; NTPs, nucleotide triphosphates; ORC, origin recognition complex; Pre-RC, prereplication complex; Psf, partner with Sld5; SE, steric exclusion; SEW, steric exclusion and wrapping; SF, superfamily; Sld, synthetic lethal with dpb11; ssDNA, single-stranded DNA.
Acknowledgements
I am grateful to the broader helicase field as well as individual members of my laboratory (both past and present) for their inspiration, discussions, and collegiality. Thanks to Sean Carney and Brian Graham for critical reading of the manuscript.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Smita Patel, Department of Biochemistry and Molecular Biology, Robert Wood Johnson Medical School, Rutgers University, Piscataway, New Jersey, 08854, USA
James Berger, Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, Maryland, 21205, USA
Kevin Raney, Department of Biochemistry and Molecular Biology, University of Arkansas for Medical Sciences, Little Rock, Arkansas, 72205, USA
Funding Statement
Research in the Trakselis laboratory is sponsored by the American Cancer Society (RSG-11-049-01-DMC) and Baylor University.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
References
- 1. Leipe DD, Aravind L, Koonin EV: Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27(17):3389–3401. 10.1093/nar/27.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorbalenya AE, Koonin EV: Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17(21):8413–8440. 10.1093/nar/17.21.8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leipe DD, Aravind L, Grishin NV, et al. : The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10(1):5–16. [PubMed] [Google Scholar]
- 4. Erzberger JP, Berger JM: Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. 10.1146/annurev.biophys.35.040405.101933 [DOI] [PubMed] [Google Scholar]
- 5. Neuwald AF, Aravind L, Spouge JL, et al. : AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. [PubMed] [Google Scholar]
- 6. Matson SW, Richardson CC: DNA-dependent nucleoside 5'-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983;258(22):14009–14016. [PubMed] [Google Scholar]
- 7. Patel SS, Hingorani MM: Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J Biol Chem. 1993;268(14):10668–10675. [PubMed] [Google Scholar]
- 8. Dong F, Gogol EP, von Hippel PH: The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate. J Biol Chem. 1995;270(13):7462–7473. [DOI] [PubMed] [Google Scholar]
- 9. Bujalowski W, Klonowska MM, Jezewska MJ: Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1994;269(50):31350–31358. [PubMed] [Google Scholar]
- 10. Patel SS, Picha KM: Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. 10.1146/annurev.biochem.69.1.651 [DOI] [PubMed] [Google Scholar]
- 11. Gai D, Zhao R, Li D, et al. : Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119(1):47–60. 10.1016/j.cell.2004.09.017 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Enemark EJ, Joshua-Tor L: On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18(2):243–257. 10.1016/j.sbi.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomsen ND, Berger JM: Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139(3):523–534. 10.1016/j.cell.2009.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Liao JC, Jeong YJ, Kim DE, et al. : Mechanochemistry of t7 DNA helicase. J Mol Biol. 2005;350(3):452–475. 10.1016/j.jmb.2005.04.051 [DOI] [PubMed] [Google Scholar]
- 15. Hingorani MM, Patel SS: Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32(46):12478–12487. 10.1021/bi00097a028 [DOI] [PubMed] [Google Scholar]
- 16. Bujalowski W, Jezewska MJ: Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry. 1995;34(27):8513–8519. 10.1021/bi00027a001 [DOI] [PubMed] [Google Scholar]
- 17. Graham BW, Schauer GD, Leuba SH, et al. : Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 2011;39(15):6585–6595. 10.1093/nar/gkr345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaplan DL, O'Donnell M: Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol Cell. 2004;15(3):453–465. 10.1016/j.molcel.2004.06.039 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Ye J, Osborne AR, Groll M, et al. : RecA-like motor ATPases--lessons from structures. Biochim Biophys Acta. 2004;1659(1):1–18. 10.1016/j.bbabio.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 20. Singleton MR, Dillingham MS, Wigley DB: Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- 21. Moreau MJ, McGeoch AT, Lowe AR, et al. : ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28(2):304–314. 10.1016/j.molcel.2007.08.013 [DOI] [PubMed] [Google Scholar]
- 22. McGeoch AT, Trakselis MA, Laskey RA, et al. : Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12(9):756–762. 10.1038/nsmb974 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Brewster AS, Wang G, Yu X, et al. : Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105(51):20191–20196. 10.1073/pnas.0808037105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu X, Hingorani MM, Patel SS, et al. : DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat Struct Biol. 1996;3(9):740–743. 10.1038/nsb0996-740 [DOI] [PubMed] [Google Scholar]
- 25. Egelman EH, Yu X, Wild R, et al. : Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci U S A. 1995;92(9):3869–3873. 10.1073/pnas.92.9.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morris PD, Raney KD: DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38(16):5164–5171. 10.1021/bi9822269 [DOI] [PubMed] [Google Scholar]
- 27. Jezewska MJ, Rajendran S, Bujalowski W: Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1998;273(15):9058–9069. 10.1074/jbc.273.15.9058 [DOI] [PubMed] [Google Scholar]
- 28. Costa A, van Duinen G, Medagli B, et al. : Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J. 2008;27(16):2250–2258. 10.1038/emboj.2008.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singleton MR, Sawaya MR, Ellenberger T, et al. : Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101(6):589–600. 10.1016/S0092-8674(00)80871-5 [DOI] [PubMed] [Google Scholar]
- 30. Toth EA, Li Y, Sawaya MR, et al. : The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12(5):1113–1123. 10.1016/S1097-2765(03)00442-8 [DOI] [PubMed] [Google Scholar]
- 31. Bailey S, Eliason WK, Steitz TA: Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318(5849):459–463. 10.1126/science.1147353 [DOI] [PubMed] [Google Scholar]
- 32. Wang G, Klein MG, Tokonzaba E, et al. : The structure of a DnaB-family replicative helicase and its interactions with primase. Nat Struct Mol Biol. 2008;15(1):94–100. 10.1038/nsmb1356 [DOI] [PubMed] [Google Scholar]
- 33. Fernández-Millán P, Lázaro M, Cansız-Arda Ş, et al. : The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res. 2015;43(8):4284–4295. 10.1093/nar/gkv189 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Li D, Zhao R, Lilyestrom W, et al. : Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423(6939):512–518. 10.1038/nature01691 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Fletcher RJ, Bishop BE, Leon RP, et al. : The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–167. 10.1038/nsb893 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Li N, Zhai Y, Zhang Y, et al. : Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524(7564):186–191. 10.1038/nature14685 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Jezewska MJ, Rajendran S, Bujalowski W: Complex of Escherichia coli primary replicative helicase DnaB protein with a replication fork: recognition and structure. Biochemistry. 1998;37(9):3116–3136. 10.1021/bi972564u [DOI] [PubMed] [Google Scholar]
- 38. Lee SJ, Syed S, Enemark EJ, et al. : Dynamic look at DNA unwinding by a replicative helicase. Proc Natl Acad Sci U S A. 2014;111(9):E827–35. 10.1073/pnas.1322254111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Zhang H, Lee SJ, Kulczyk AW, et al. : Heterohexamer of 56- and 63-kDa Gene 4 Helicase-Primase of Bacteriophage T7 in DNA Replication. J Biol Chem. 2012;287(41):34273–34287. 10.1074/jbc.M112.401158 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Mitkova AV, Khopde SM, Biswas SB: Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J Biol Chem. 2003;278(52):52253–52261. 10.1074/jbc.M308956200 [DOI] [PubMed] [Google Scholar]
- 41. Strycharska MS, Arias-Palomo E, Lyubimov AY, et al. : Nucleotide and partner-protein control of bacterial replicative helicase structure and function. Mol Cell. 2013;52(6):844–854. 10.1016/j.molcel.2013.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Fletcher RJ, Shen J, Gómez-Llorente Y, et al. : Double hexamer disruption and biochemical activities of Methanobacterium thermoautotrophicum MCM. J Biol Chem. 2005;280(51):42405–42410. 10.1074/jbc.M509773200 [DOI] [PubMed] [Google Scholar]
- 43. Liu W, Pucci B, Rossi M, et al. : Structural analysis of the Sulfolobus solfataricus MCM protein N-terminal domain. Nucleic Acids Res. 2008;36(10):3235–3243. 10.1093/nar/gkn183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Costa A, Renault L, Swuec P, et al. : DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife. 2014;3:e03273. 10.7554/eLife.03273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sheu YJ, Kinney JB, Lengronne A, et al. : Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proc Natl Acad Sci U S A. 2014;111(18):E1899–908. 10.1073/pnas.1404063111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Labib K: How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24(12):1208–1219. 10.1101/gad.1933010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Devault A, Gueydon E, Schwob E: Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol Biol Cell. 2008;19(5):2267–2277. 10.1091/mbc.E07-06-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jemt E, Farge G, Bäckström S, et al. : The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 2011;39(21):9238–9249. 10.1093/nar/gkr653 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Chang YP, Xu M, Machado AC, et al. : Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen. Cell Rep. 2013;3(4):1117–1127. 10.1016/j.celrep.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Ahnert P, Picha KM, Patel SS: A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19(13):3418–3427. 10.1093/emboj/19.13.3418 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. Duderstadt KE, Berger JM: A structural framework for replication origin opening by AAA+ initiation factors. Curr Opin Struct Biol. 2013;23(1):144–153. 10.1016/j.sbi.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bell SP, Kaguni JM: Helicase loading at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5(6): pii: a010124. 10.1101/cshperspect.a010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ozaki S, Kawakami H, Nakamura K, et al. : A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283(13):8351–8362. 10.1074/jbc.M708684200 [DOI] [PubMed] [Google Scholar]
- 54. Kaguni JM: DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. 10.1146/annurev.micro.60.080805.142111 [DOI] [PubMed] [Google Scholar]
- 55. Messer W: The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26(4):355–374. 10.1111/j.1574-6976.2002.tb00620.x [DOI] [PubMed] [Google Scholar]
- 56. Mott ML, Berger JM: DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5(5):343–354. 10.1038/nrmicro1640 [DOI] [PubMed] [Google Scholar]
- 57. Ozaki S, Katayama T: Highly organized DnaA- oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40(4):1648–1665. 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Duderstadt KE, Chuang K, Berger JM: DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478(7368):209–213. 10.1038/nature10455 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Ozaki S, Noguchi Y, Hayashi Y, et al. : Differentiation of the DnaA- oriC subcomplex for DNA unwinding in a replication initiation complex. J Biol Chem. 2012;287(44):37458–37471. 10.1074/jbc.M112.372052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duderstadt KE, Mott ML, Crisona NJ, et al. : Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem. 2010;285(36):28229–28239. 10.1074/jbc.M110.147975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Soultanas P: Loading mechanisms of ring helicases at replication origins. Mol Microbiol. 2012;84(1):6–16. 10.1111/j.1365-2958.2012.08012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arias-Palomo E, O'Shea VL, Hood IV, et al. : The bacterial DnaC helicase loader is a DnaB ring breaker. Cell. 2013;153(2):438–448. 10.1016/j.cell.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Marszalek J, Kaguni JM: DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269(7):4883–4890. [PubMed] [Google Scholar]
- 64. Liu B, Eliason WK, Steitz TA: Structure of a helicase-helicase loader complex reveals insights into the mechanism of bacterial primosome assembly. Nat Commun. 2013;4: 2495. 10.1038/ncomms3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abe Y, Jo T, Matsuda Y, et al. : Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem. 2007;282(24):17816–17827. 10.1074/jbc.M701841200 [DOI] [PubMed] [Google Scholar]
- 66. Mott ML, Erzberger JP, Coons MM, et al. : Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135(4):623–634. 10.1016/j.cell.2008.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Makowska-Grzyska M, Kaguni JM: Primase directs the release of DnaC from DnaB. Mol Cell. 2010;37(1):90–101. 10.1016/j.molcel.2009.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tougu K, Marians KJ: The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996;271(35):21398–21405. 10.1074/jbc.271.35.21398 [DOI] [PubMed] [Google Scholar]
- 69. Brüning JG, Howard JL, McGlynn P: Accessory replicative helicases and the replication of protein-bound DNA. J Mol Biol. 2014;426(24):3917–3928. 10.1016/j.jmb.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 70. Atkinson J, Gupta MK, McGlynn P: Interaction of Rep and DnaB on DNA. Nucleic Acids Res. 2011;39(4):1351–1359. 10.1093/nar/gkq975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guy CP, Atkinson J, Gupta MK, et al. : Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36(4):654–666. 10.1016/j.molcel.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Remus D, Diffley JF: Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21(6):771–777. 10.1016/j.ceb.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 73. Bell SP, Dutta A: DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. 10.1146/annurev.biochem.71.110601.135425 [DOI] [PubMed] [Google Scholar]
- 74. Barry ER, Bell SD: DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70(4):876–887. 10.1128/MMBR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wigley DB: ORC proteins: marking the start. Curr Opin Struct Biol. 2009;19(1):72–78. 10.1016/j.sbi.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 76. Erzberger JP, Pirruccello MM, Berger JM: The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21(18):4763–4773. 10.1093/emboj/cdf496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yeeles JT, Deegan TD, Janska A, et al. : Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519(7554):431–435. 10.1038/nature14285 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Ticau S, Friedman LJ, Ivica NA, et al. : Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161(3):513–525. 10.1016/j.cell.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Fu Y, Slaymaker IM, Wang J, et al. : The 1.8-Å crystal structure of the N-terminal domain of an archaeal MCM as a right-handed filament. J Mol Biol. 2014;426(7):1512–1523. 10.1016/j.jmb.2013.12.025 [DOI] [PubMed] [Google Scholar]
- 80. Costa A, Ilves I, Tamberg N, et al. : The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18(4):471–477. 10.1038/nsmb.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Bochman ML, Schwacha A: The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 'gate'. Nucleic Acids Res. 2010;38(18):6078–6088. 10.1093/nar/gkq422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lyubimov AY, Costa A, Bleichert F, et al. : ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci U S A. 2012;109(30):11999–12004. 10.1073/pnas.1209406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bleichert F, Botchan MR, Berger JM: Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519(7543):321–326. 10.1038/nature14239 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Sun J, Evrin C, Samel SA, et al. : Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20(8):944–951. 10.1038/nsmb.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bloom LB: Loading clamps for DNA replication and repair. DNA Repair (Amst). 2009;8(5):570–578. 10.1016/j.dnarep.2008.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu C, Wu R, Zhou B, et al. : Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res. 2012;40(7):3208–3217. 10.1093/nar/gkr1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fernández-Cid A, Riera A, Tognetti S, et al. : An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell. 2013;50(4):577–588. 10.1016/j.molcel.2013.03.026 [DOI] [PubMed] [Google Scholar]
- 88. Riera A, Tognetti S, Speck C: Helicase loading: how to build a MCM2-7 double-hexamer. Semin Cell Dev Biol. 2014;30:104–109. 10.1016/j.semcdb.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 89. Remus D, Beuron F, Tolun G, et al. : Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139(4):719–730. 10.1016/j.cell.2009.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 90. Wessel R, Schweizer J, Stahl H: Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66(2):804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Valle M, Chen XS, Donate LE, et al. : Structural basis for the cooperative assembly of large T antigen on the origin of replication. J Mol Biol. 2006;357(4):1295–1305. 10.1016/j.jmb.2006.01.021 [DOI] [PubMed] [Google Scholar]
- 92. Gómez-Llorente Y, Fletcher RJ, Chen XS, et al. : Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J Biol Chem. 2005;280(49):40909–40915. 10.1074/jbc.M509760200 [DOI] [PubMed] [Google Scholar]
- 93. Randell JC, Bowers JL, Rodríguez HK, et al. : Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21(1):29–39. 10.1016/j.molcel.2005.11.023 [DOI] [PubMed] [Google Scholar]
- 94. Tanaka S, Umemori T, Hirai K, et al. : CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445(7125):328–332. 10.1038/nature05465 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Zegerman P, Diffley JF: Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445(7125):281–285. 10.1038/nature05432 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Tanaka S, Komeda Y, Umemori T, et al. : Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1-GINS interaction. Mol Cell Biol. 2013;33(13):2614–2622. 10.1128/MCB.00431-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bruck I, Kaplan DL: Conserved mechanism for coordinating replication fork helicase assembly with phosphorylation of the helicase. Proc Natl Acad Sci U S A. 2015;112(36):11223–11228. 10.1073/pnas.1509608112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Randell JC, Fan A, Chan C, et al. : Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40(3):353–363. 10.1016/j.molcel.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sheu YJ, Stillman B: The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–117. 10.1038/nature08647 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 100. Bruck I, Kaplan DL: The Replication Initiation Protein Sld3/Treslin Orchestrates the Assembly of the Replication Fork Helicase during S Phase. J Biol Chem. 2015;290(45):27414–27424. 10.1074/jbc.M115.688424 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101. Bruck I, Kaplan DL: The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J Biol Chem. 2015;290(2):1210–1221. 10.1074/jbc.M114.608232 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102. Pacek M, Tutter AV, Kubota Y, et al. : Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21(4):581–587. 10.1016/j.molcel.2006.01.030 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 103. Petojevic T, Pesavento JJ, Costa A, et al. : Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc Natl Acad Sci U S A. 2015;112(3):E249–58. 10.1073/pnas.1422003112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 104. Enemark EJ, Joshua-Tor L: Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442(7100):270–275. 10.1038/nature04943 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. Pandey M, Patel SS: Helicase and polymerase move together close to the fork junction and copy DNA in one-nucleotide steps. Cell Rep. 2014;6(6):1129–1138. 10.1016/j.celrep.2014.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 106. Itsathitphaisarn O, Wing RA, Eliason WK, et al. : The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151(2):267–277. 10.1016/j.cell.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 107. Froelich CA, Kang S, Epling LB, et al. : A conserved MCM single-stranded DNA binding element is essential for replication initiation. eLife. 2014;3:e01993. 10.7554/eLife.01993 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Takahashi TS, Wigley DB, Walter JC: Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30(8):437–444. 10.1016/j.tibs.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 109. Rothenberg E, Trakselis MA, Bell SD, et al. : MCM forked substrate specificity involves dynamic interaction with the 5'-tail. J Biol Chem. 2007;282(47):34229–34234. 10.1074/jbc.M706300200 [DOI] [PubMed] [Google Scholar]
- 110. Ribeck N, Saleh OA: DNA unwinding by ring-shaped T4 helicase gp41 is hindered by tension on the occluded strand. PLoS One. 2013;8(11):e79237. 10.1371/journal.pone.0079237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ribeck N, Kaplan DL, Bruck I, et al. : DnaB helicase activity is modulated by DNA geometry and force. Biophys J. 2010;99(7):2170–2179. 10.1016/j.bpj.2010.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhou B, Arnett DR, Yu X, et al. : Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase α-primase. J Biol Chem. 2012;287(32):26854–26866. 10.1074/jbc.M112.363655 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 113. Ilves I, Petojevic T, Pesavento JJ, et al. : Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–258. 10.1016/j.molcel.2009.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Khan I, Suhasini AN, Banerjee T, et al. : Impact of age-associated cyclopurine lesions on DNA repair helicases. PLoS One. 2014;9(11):e113293. 10.1371/journal.pone.0113293 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 115. Suhasini AN, Brosh RM, Jr: Mechanistic and biological aspects of helicase action on damaged DNA. Cell Cycle. 2010;9(12):2317–2329. 10.4161/cc.9.12.11902 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 116. Buechner CN, Heil K, Michels G, et al. : Strand-specific recognition of DNA damages by XPD provides insights into nucleotide excision repair substrate versatility. J Biol Chem. 2014;289(6):3613–3624. 10.1074/jbc.M113.523001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 117. Suhasini AN, Sommers JA, Mason AC, et al. : FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J Biol Chem. 2009;284(27):18458–18470. 10.1074/jbc.M109.012229 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 118. Meinke G, Phelan P, Moine S, et al. : The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS Biol. 2007;5(2):e23. 10.1371/journal.pbio.0050023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cuesta I, Núñez-Ramírez R, Scheres SH, et al. : Conformational rearrangements of SV40 large T antigen during early replication events. J Mol Biol. 2010;397(5):1276–1286. 10.1016/j.jmb.2010.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yardimci H, Wang X, Loveland AB, et al. : Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492(7428):205–209. 10.1038/nature11730 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 121. Chaban Y, Stead JA, Ryzhenkova K, et al. : Structural basis for DNA strand separation by a hexameric replicative helicase. Nucleic Acids Res. 2015;43(17):8551–8563. 10.1093/nar/gkv778 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 122. Sun J, Shi Y, Georgescu RE, et al. : The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22(12):976–982. 10.1038/nsmb.3113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 123. Nandakumar D, Pandey M, Patel SS: Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. eLife. 2015;4:e06562. 10.7554/eLife.06562 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 124. Jeong YJ, Rajagopal V, Patel SS: Switching from single-stranded to double-stranded DNA limits the unwinding processivity of ring-shaped T7 DNA helicase. Nucleic Acids Res. 2013;41(7):4219–4229. 10.1093/nar/gkt133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Smits WK, Merrikh H, Bonilla CY, et al. : Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J Bacteriol. 2011;193(3):640–648. 10.1128/JB.01253-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tsai KL, Lo YH, Sun YJ, et al. : Molecular interplay between the replicative helicase DnaC and its loader protein DnaI from Geobacillus kaustophilus. J Mol Biol. 2009;393(5):1056–1069. 10.1016/j.jmb.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 127. Robinson A, Causer RJ, Dixon NE: Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr Drug Targets. 2012;13(3):352–372. 10.2174/138945012799424598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bazin A, Cherrier MV, Gutsche I, et al. : Structure and primase-mediated activation of a bacterial dodecameric replicative helicase. Nucleic Acids Res. 2015;43(17):8564–8576. 10.1093/nar/gkv792 [DOI] [PMC free article] [PubMed] [Google Scholar]