Abstract
Background
CASR gene appears to be involved in cancer biology and physiology. However, a number of studies investigating CASR polymorphisms and cancer risks have presented inconclusive results. Thus, a systematic review and a meta-analysis of the effect of CASR polymorphisms on several cancer risks were performed to suggest a statistical evidence for the association of CASR polymorphisms with cancer risks.
Methods
MEDLINE, EMBASE, Web of Science, Scopus, and the HuGE databases were searched. Nineteen articles of case–control and cohort studies were included for the final analysis.
Results
The colorectal cancer risk was reduced in proximal (odds ratio [OR] =0.679, P=0.001) and distal (OR =0.753, P=0.026) colon sites with GG genotype of CASR rs1042636 and increased in distal colon site (OR =1.418, P=0.039) with GG genotype of rs1801726 by additive genetic model. The rs17251221 demonstrated noticeable associations that carrying a homozygote variant increases breast and prostate cancer risk considerably.
Conclusion
The significant association of CASR polymorphisms with several cancer risks was observed in this review. In particular, the act of CASR polymorphisms as a tumor suppressor or an oncogene differs by cancer site and can be the research target for tumorigenesis.
Keywords: rs1042636, rs1801725, rs1801726, systematic review, colorectal cancer
Introduction
The effect of calcium intake on various cancer risks is an ongoing topic of investigation. Besides the physiologic calcium level, the calcium-sensing receptor (CaSR), through which calcium balance is regulated, is thought to play an important role in the regulation of cancer expression. The activated CaSR can stimulate intracellular signal pathways including mitogen-activated protein kinase, phosphatidylinositol 3 kinase/protein kinase B, and cy-mic and cyclin D1 pathways; these processes are involved in cellular secretion, proliferation, differentiation, chemotaxis, and apoptosis.1 The CaSR expression is related to the CASR gene that seems to have a role in cancer cells, acting both as a tumor suppressor and an oncogene, depending on the cancer site and environmental condition. In colonic epithelial cells, high calcium intake could reduce the risk of colorectal cancer development.2 E-cadherin stimulated by CaSR can interact with β-catenin, an important protooncogene, contribute to reducing the cancer cell activity, and downregulate cell proliferation.3 Whereas, the increased expression of CaSR by high calcium levels promoted MCF-7, PC-3, and C4-2B breast and prostate cancer cells known to metastasize to the bone and the cancer cell proliferation process is linked to extracellular signal-regulated kinases 1 and 2 (ERK 1/2) phosphorylation.4
The CASR gene contains seven exons and is located on chromosome 3q13. Among the single-nucleotide polymorphisms (SNPs) in the CASR gene, rs1801725 (A986S, 2956G>T) causes an amino acid change from alanine (A) to serine (S), and the T allele is associated with higher levels of serum calcium.5 The rs1042636 (R990G, 2968A>G) polymorphism causes an amino acid change from arginine (A) to glycine (G) and induces a gain-of-function mutation associated with primary hyperparathy-roidism and calcium stone formation.6–8 The rs1801726 (Q1101E, 3403C>G) is a common polymorphism in African ethnicity whose functional characteristics need further investigation;9,10 glutamine (Q) to glutamic acid (A) change is observed.
The rs17251221 (1378–1412A>G) in introns, which is in high linkage disequilibrium with rs1801725,11 induces a gain-of-function mutation associated with total serum calcium concentration11 and stone multiplicity in patients with nephrolithiasis.12
Recently, many studies have focused on the association between CASR gene polymorphism and multiple cancer risks. Three common nonsynonymous SNPs (rs1801725, rs1042636, and rs1801726) have been the primary research targets for cancer risk, but inconsistent results have been reported. Dong et al13 reported that CASR variants are not associated with colorectal cancer risk, whereas Jenab et al14 suggested possible association between CASR rs1042636 variations with colorectal cancer risk. Additional genetic variants of the large CASR gene (102 kb), which cannot be sufficiently explained by the three nonsynonymous SNPs, are also the research targets of cancer risks. Thus, a systematic review on the effect of CASR polymorphisms with several cancer risks and a meta-analysis on colorectal cancer risk were performed to suggest statistical evidence for the clinical use of cancer markers.
Methods
Search strategy and eligibility criteria
The electronic databases of MEDLINE, EMBASE, Web of Science, Scopus, and the HuGE Published Literature database were searched with the following keywords: (“calcium sensing receptor” OR “casr protein” OR “CASR” OR “Calcium sensing receptor gene”) AND (“cancers” OR “neoplasia”). The references of included articles were checked to include any additional relevant articles.
A systematic search for relevant literature was performed to include studies published up to July 26, 2014, by two independent reviewers (JS and KJ) without language restrictions. Any disagreement was resolved by discussion between the authors. Inclusion criteria for article selection were as follows: 1) case–control studies or cohort studies and 2) sufficient data reporting odds ratio (OR) with 95% confidence interval (CI) or sample frequency with which the appropriate calculations could be done. Studies were excluded if they were 1) duplicate or previously published, 2) letters, reviews, or editorials, and 3) CASR gene studies on cell lines or animals by PRISMA flow diagram.
Data extraction
The following information was extracted from included studies: first author, year of publication, country of study site, ethnic group, genotyping method, number of genotyped cases and controls, genotype frequencies for cases and controls, selection pool of control population (population-based controls and hospital-based controls) and Hardy–Weinberg equilibrium (HWE) in any population, tumor type and site, OR, and corresponding 95% CI. Ethnicity was classified as Caucasian, Asian, or African. When the study did not specify the ethnicity, the term “mixed ethnicity” was used. Any discrepancies in the extracted information were resolved by discussion among the authors.
Quality score assessment
Two reviewers (JS and KJ) independently evaluated the quality of the selected studies using the quality assessment scoring tool developed for genetic association studies by Thakkinstian et al,15 which was modified from previous meta-analyses of observational studies16–19 considering traditional epidemiologic and genetic issues20,21 (Table S1).
Statistical analysis
The association of three nonsynonymous CASR SNPs with colorectal cancer risk was examined by unconditional logistic regression to obtain ORs with 95% CIs in additive, dominant, and recessive genetic models and represented by forest plot. The pooled ORs were calculated for each genetic model and different cancer sites (eg, proximal colon, distal colon). Whenever ORs and 95% CIs were not reported, appropriate data were selected and calculated to produce OR with 95% CI. Between-study heterogeneity was assessed by the Q-statistic (heterogeneity was considered statistically significant if P<0.1)22 and quantified by the I2 value. Both fixed- and random-effects models were used to combine the aggregate data determined by the I2 value. When I2 was >50%, the random-effects model was used for analysis. Potential publication bias was assessed with the linear regression method of Egger’s test23 and funnel plot.24 Statistical analyses were performed using Comprehensive Meta-Analysis (Version 2; Biostat, Inc., Engelwood, NJ, USA) and PASW (Version 21; IBM Corporation, Armonk, NY, USA). All tests were two-sided, and P<0.05 was considered significant unless otherwise specified.
Results
Study selection
Twenty out of 1,309 publications were found to be eligible for systematic review as shown in Figure S1.
Among eligible publications, the study by Speer et al25 was excluded due to an overlapping population with another study by the same author.26 Also, a study for esophageal cancer27 was excluded due to insufficient SNP information. By hand search, a study by Mahmoudi et al28 was added, and the final number of studies included for systematic review was 19 (Table 1).
Table 1.
Main characteristics of included studies of CASR associated with cancer risks
| Cancer type | Reference | Country (ethnicity) | Study design | Cases (n) | Controls (n) | Genotyping method (HWE) | SNP | Tumor site |
|---|---|---|---|---|---|---|---|---|
| Colorectal | Speer et al26 | Hungary (Caucasian) | Hospital-based case–control | 56 | 112 | PCR (HWE: N/A) | rs1801725 (A986S) | Rectum |
| Peters et al33 | USA (94% Caucasian) | Population-based nested case–control | 716 | 729 | Taqman (HWE: A986S [P=0.92], A990G [P=0.69] Q1101E [P=0.62]) | rs1801725 (A986S), rs1042636 (A990G), rs1801726 (Q1101E) | Distal colorectum | |
| Fuszek et al81 | Hungary (Caucasian) | Population-based case–control | 70 | 201 | PCR (HWE: N/A) | rs1801725 | Colorectum | |
| Fuszek et al81 | Hungary (Caucasian) | Population-based case–control | 70 | 201 | PCR (HWE: N/A) | rs1801725 | Colorectum | |
| Bácsi et al76 | Hungary (Caucasian) | Population-based case–control | 278 | 260 | Taqman (HWE: N/A) | rs1801725 | Colorectum | |
| Dong et al13 | USA (Mixed, Caucasian predominant) | Population-based case–control | 1,600 | 1,949 | MALDI-TOF (HWE: P>0.01) | 17 SNPs | Proximal colon, distal colon | |
| Jenab et al14 | Europe (Caucasian) | Population-based nested case–control | 1,160 | 1,248 | Taqman (meet HWE) | rs1801725 | Colorectum, colon, rectum | |
| Jacobs et al79 | USA, Australia (mixed, Caucasian predominant) | Population-based discordant sibship case–control | 1,802 | 2,874 | Illumina Golden gate platform (HWE: N/A) | 36 SNPs | Proximal colon, distal colon, rectum | |
| Safaei et al77 | Iran (Caucasian) | Hospital-based case–control | 105 | 105 | PCR-RFLP (HWE: N/A) | rs1801725 | Colorectum | |
| Fedirko et al82 | Europe (Caucasian) | Population-based cohort | 1,137 | N/A | Taqman (HWE: N/A) | rs1801725 | Colorectum | |
| Hibler et al78 | USA Caucasian (white) | Population-based cohort | 1,439 | N/A | Illumina Golden gate platform (meet HWE) | 35 SNPs | Proximal colon, distal colon | |
| Kim et al34 | Korea (Asian) | Hospital-based case–control | 420 | 815 | Taqman (meet HWE) | rs10934578, rs12485716, rs4678174, rs2270916 | Proximal colon, distal colon, rectum | |
| Mahmoudi et al28 | Iran (Caucasian) | Hospital-based case–control | 350 | 510 | PCR-RFLP (HWE: N/A) | rs1801725 | Colorectum | |
| Prostate | Schwartz et al30 | USA (African–American) | Population-based case–control | 458 | 248 | Illumina Beadlab system: rs1042636, rs1801726; Taqman: rs1801725 (meet HWE) | rs1801725, rs1042636, rs1801726 | Prostate |
| Szendroi et al54 | Hungary (Caucasian) | Hospital-based case–control | 204 | 102 | PCR (HWE >0.05) | rs1801725 | Prostate | |
| Shui et al35 | USA (Caucasian with European decent) | Population-based nested case–control | 1,193 | 1,244 | Open-array SNP genotyping platform (HWE: P>0.01) | 18 SNPs | Prostate | |
| Jorde et al31 | Norway (Caucasian) | Population-based case–cohort | 370 | 1,647 | KBioscience competitive allele-specific PCR (meet HWE) | rs17251221, rs1801725 | Prostate, lung, breast, colorectum | |
| Breast | Li et al32 | People’s Republic of China (Asian) | Hospital-based case–control | 217 | 231 | Taqman (HWE: P>0.05) | rs17251221 | Breast |
| Pancreas | Anderson et al53 | Canada (Caucasian) | Population-based case–control | 628 | 1,193 | MassARRAY, iPLEX Gold sequenom Platform (meet HWE) | 13 SNPs | Pancreas |
| Neuroblastoma | Masvidal et al29 | Spain (Caucasian) | Cohort | 65 | N/A | RT-PCR (meet HWE) | Haplotype of rs1801725, rs1042636, rs1801726 | Nerve |
Abbreviations: HWE, Hardy–Weinberg equilibrium; PCR, polymerase chain reaction; N/A, not applicable; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight; SNP, single-nucleotide polymorphism; RFLP, restriction fragment length polymorphism; RT, reverse transcription.
In meta-analysis, two articles that reported colorectal cancer risk of rs1801725 were excluded because the reported frequency of homozygote variants was 0. Meta-analyses for colorectal cancer risk included 4,209 cases and 4,801 controls for rs1801725 and 5,557 cases and 5,552 controls for rs1042636 and rs1801726, respectively.
Synthesis of result by meta-analysis on the colorectal cancer risk
The association between rs1801725, rs1042636, rs1801726 and colorectal cancer risk, stratified by genetic model and cancer site, is presented in Table 2.
Table 2.
Stratified analysis of the three nonsynonymous SNPs (rs1801725, rs1042636, rs1801726) in CASR and colorectal cancer risk by three genetic models and cancer sites
| Variable
|
N* | n (case/control) | Association
|
Heterogeneity
|
Publication bias
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic model | Site | OR | 95% CI | P-value | I2 | P(Q)-value | Model | Funnel plot | Egger’s P-value | ||
| rs1801725 | |||||||||||
| TT vs GG | Colorectal | 6 | 4,209/4,801 | 1.152 | 0.859–1.543 | 0.379 | 25.769 | 0.345 | Fixed | None | 0.181 |
| rs1042636 | |||||||||||
| GG vs AA | Proximal | 3 | 4,841/4,823 | 0.679 | 0.536–0.859 | 0.001** | 42.519 | 0.176 | Fixed | None | 0.634 |
| Distal | 4 | 5,557/5,552 | 0.753 | 0.587–0.967 | 0.026** | 0 | 0.396 | Fixed | None | 0.957 | |
| AG + GG vs AA | Proximal | 3 | 4,841/4,823 | 0.797 | 0.505–1.260 | 0.332 | 83.839 | 0.002 | Random | None | 0.175 |
| Distal | 3 | 4,841/4,823 | 0.854 | 0.710–1.029 | 0.097 | 44.491 | 0.165 | Fixed | None | 0.451 | |
| rs1801726 | |||||||||||
| GG vs CC | Proximal | 3 | 4,841/4,823 | 1.137 | 0.820–1.575 | 0.441 | 0 | 0.408 | Fixed | None | 0.601 |
| Distal | 4 | 5,557/5,552 | 1.418 | 1.017–1.977 | 0.039** | 0 | 0.676 | Fixed | None | 0.770 | |
| CG + GG vs CC | Proximal | 3 | 4,841/4,823 | 1.095 | 0.882–1.360 | 0.411 | 0 | 0.481 | Fixed | None | 0.987 |
| Distal | 3 | 4,841/4,823 | 1.073 | 0.857–1.344 | 0.537 | 59.415 | 0.085 | Random | None | 0.414 | |
Notes:
Number of studies included in the meta-analysis.
Significant result.
Abbreviations: SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Figures 1–3 demonstrate the pooled associations between three nonsynonymous CASR polymorphisms and colorectal cancer risk in forest plot.
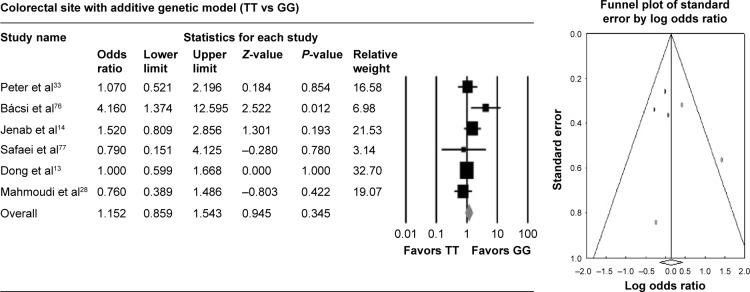
Figure 1.
Association of rs1801725 polymorphism with colorectal cancer risk by additive genetic model.
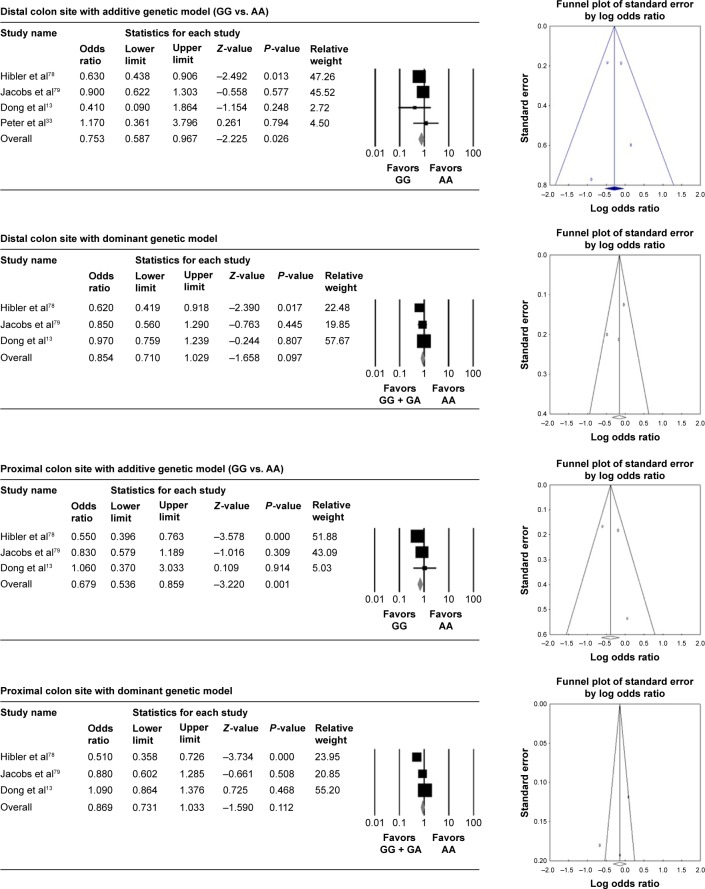
Figure 2.
Association of rs1042636 polymorphism with colorectal cancer risk stratified by cancer sites and three genetic models.
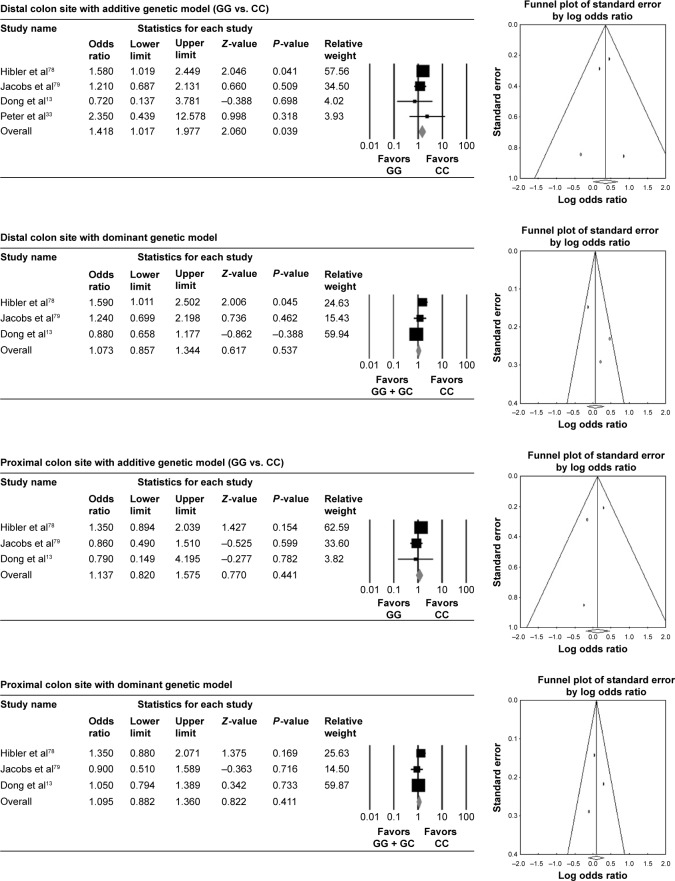
Figure 3.
Association of rs1801726 polymorphism with colorectal cancer risk stratified by cancer sites and three genetic models.
T allele polymorphisms of rs1801725 did not show any association with colorectal cancer risk compared with the wild-type homozygous GG genotype. With the additive genetic model (TT vs GG), the pooled OR was 1.152 (95% CI: 0.859–1.543, I2: 25.769) (Table 2, Figure 1).
The colorectal cancer risk was significantly reduced in GG genotype of rs1042636 compared with the wild type in both proximal and distal colon sites with additive genetic model (OR =0.679 [95% CI: 0.536–0.859], I2: 42.519) in proximal colon and (OR =0.753 [95% CI: 0.587–0.967], I2: 0) in distal colon. With the dominant genetic model, the association was not significant (Table 2, Figure 2). GG genotype of rs1801726 showed increased colorectal cancer risk in the distal colon site with additive genetic model (OR =1.418 [95% CI: 1.017–1.977], I2: 0) (Table 2, Figure 3).
Systematic reviews of the association of CASR polymorphisms with cancer risks
From 19 studies that reported CASR polymorphisms and cancer risks, we extracted significant SNPs associated with several cancer risks that could not be assessed by meta-analysis for future research targets stratified by cancer type and cancer site (Table 3).
Table 3.
Significant SNPs or haplo/diplotype of CASR found in selected studies stratified by cancer sites
| Cancer (specified by included studies) | SNP/haplotype/diplotype | Genotype | Case | Control | OR | 95% CI | P-value | Cofactor other than CASR | References |
|---|---|---|---|---|---|---|---|---|---|
| Colorectum | rs1801725 (G/T) | GG + GT vs TT | 278 | 260 | 4.01 | 1.33–12.07 | 0.026 | – | Bácsi et al76 |
| rs1801725 (G/T) | GG vs TT | 105 | 105 | 0.56 | 0.31–0.99 | 0.04 | Safaei et al77 | ||
| rs2270916 (T/C) | TT vs CC | 420 | 815 | 2.11 | 1.27–3.51 | NA | With low Ca intake | Kim et al34 | |
| rs10934578 (T/G) | TT vs GG | 420 | 815 | 1.84 | 1.12–3.00 | NA | With low Ca intake | Kim et al34 | |
| rs12485716 (G/A) | GG vs AA | 420 | 815 | 1.89 | 1.14–3.11 | NA | With low Ca intake | Kim et al34 | |
| rs4678174 (T/C) | TT vs CC | 420 | 815 | 1.73 | 1.06–2.83 | NA | With low Ca intake | Kim et al34 | |
| rs1042636 (A/G) | AA vs GG | 1,439 | 0 | 0.63 | 0.47–0.85 | 0.002 (0.104)* | – | Hibler et al78 | |
| rs1042636 (A/G) | AA vs AG + GG | 1,439 | 0 | 0.61 | 0.45–0.83 | 0.002 (0.091)* | – | Hibler et al78 | |
| Proximal colon | rs12485716 (G/A) | GG vs GA + AA | 1,600 | 1,949 | 0.84 | 0.71–1.00 | NA | – | Dong et al13 |
| rs4678174 (T/C) | TT vs TC + CC | 1,600 | 1,949 | 0.83 | 0.70–0.98 | NA | – | Dong et al13 | |
| rs4678174 (T/C) | TT vs CC | 1,600 | 1,949 | 0.83 | 0.69–0.99 | NA | – | Dong et al13 | |
| rs10934578 (T/G) | TT vs GG | 1,600 | 1,949 | 1.35 | 1.01–1.81 | NA | – | Dong et al13 | |
| rs2270916 (T/C) | TT vs CC | 1,600 | 1,949 | 0.43 | 0.19–0.97 | NA | – | Dong et al13 | |
| rs4678174 (T/C), rs2270916 (T/C) | Haplotype CC/TT | 1,600 | 1,949 | 0.80 | 0.67–0.97 | NA | – | Dong et al13 | |
| rs17203502 (A/G) | AA + AG vs GG | 1,802 | 2,874 | 0.55 | 0.40–0.78 | 0.001 (0.036)* | – | Jacobs et al79 | |
| rs1501900 (A/T) | AA vs TT | 1,802 | 2,874 | 0.71 | 0.54–0.94 | 0.017 (0.514)* | – | Jacobs et al79 | |
| AA vs AT + TT | 1,802 | 2,874 | 0.71 | 0.52–0.98 | 0.035 (0.744)* | – | |||
| rs17282022 (A/G) | AA + AG vs GG | 1,802 | 2,874 | 0.62 | 0.45–0.85 | 0.003 (0.136)* | – | Jacobs et al79 | |
| rs3845918 (A/G) | AA vs GG | 1,802 | 2,874 | 1.30 | 1.01–1.66 | 0.041 (0.789)* | – | Jacobs et al79 | |
| AA vs AG + GG | 1,802 | 2,874 | 1.51 | 1.12–2.02 | 0.006 (0.257)* | – | |||
| rs4678013 (G/T) | GG vs TT | 1,802 | 2,874 | 0.69 | 0.52–0.90 | 0.007 (0.285)* | – | Jacobs et al79 | |
| GG vs GT + TT | 1,802 | 2,874 | 0.69 | 0.51–0.94 | 0.020 (0.566)* | – | |||
| rs6764205 (C/T) | CC vs CT + TT | 1,802 | 2,874 | 1.42 | 1.06–1.91 | 0.020 (0.565) | – | Jacobs et al79 | |
| rs1042636 (A/G) | AA vs GG | 1,439 | 0 | 0.55 | 0.40–0.77 | <0.001 (0.022)* | – | Hibler et al78 | |
| AA vs AG + GG | 1,439 | 0 | 0.51 | 0.36–0.73 | <0.001 (0.011)* | – | |||
| rs12635478 (A/C) | AA vs CC | 1,439 | 0 | 0.82 | 0.69–0.97 | 0.017 (0.523) | – | Hibler et al78 | |
| AA vs AC + CC | 1,439 | 0 | 0.74 | 0.59–0.92 | 0.008 (0.299)* | – | |||
| rs3749208 (C/T) | CC vs TT | 1,439 | 0 | 0.82 | 0.69–0.97 | 0.020 (0.563)* | – | Hibler et al78 | |
| CC vs CT + TT | 1,439 | 0 | 0.74 | 0.59–0.92 | 0.008 (0.30)* | – | |||
| Distal colon | rs1801725 (G/T)–rs1042636 | Diplotype GAC- | 410 | 369 | 0.56 | 0.36–0.88 | NA | – | Peters et al33 |
| (A/G)–rs1801726 (C/G) | GAG/GAC-GAC | ||||||||
| rs10222633 (A/G) | AA vs AG + GG | 1,802 | 2,874 | 0.69 | 0.48–0.98 | 0.036 (0.757)* | – | Jacobs et al79 | |
| rs1802757 (C/T) | CC vs CT + TT | 1,802 | 2,874 | 0.68 | 0.47–1.00 | 0.050 (0.850)* | – | Jacobs et al79 | |
| rs1042636 (A/G) | AA vs GG | 1,439 | 0 | 0.63 | 0.44–0.91 | 0.015 (0.478)* | – | Hibler et al78 | |
| AA vs AG + GG | 1,439 | 0 | 0.62 | 0.42–0.92 | 0.017 (0.511)* | – | |||
| rs1801726 (C/G) | CC vs GG | 1,439 | 0 | 1.58 | 1.02–2.45 | 0.042 (0.802)* | – | Hibler et al78 | |
| CC vs CG + GG | 1,439 | 0 | 1.59 | 1.01–2.50 | 0.048 (0.841)* | – | |||
| Rectum | rs1801725 (A/T) | AA vs TT | 32 | 0 | 0.107 | 0.018–0.635 | 0.012 | ERBB2, EGFR, p53, ras coexpressed | Speer et al26 |
| rs1801726 (C/G) | CC vs GG | 1,802 | 2,874 | 0.53 | 0.29–0.96 | 0.036 (0.755)* | – | Jacobs et al79 | |
| rs17282008 (C/G) | CC vs GG | 1,802 | 2,874 | 1.31 | 1.01–1.72 | 0.045 (0.820)* | – | Jacobs et al79 | |
| rs4678174 (T/C) | TT + TC vs CC | 1,802 | 2,874 | 0.60 | 0.37–0.98 | 0.041 (0.794)* | – | Jacobs et al79 | |
| rs7644390 (C/T) | CC vs CT + TT | 1,802 | 2,874 | 1.38 | 1.00–1.91 | 0.050 (0.847)* | – | Jacobs et al79 | |
| Prostate | rs1801726 (C/G) | CC vs GG | 458 | 248 | 0.16 | 0.03–0.74 | 0.01 | – | Schwartz et al30 |
| rs17251221 (G/A) | GG vs AA | 370 | 1,647 | 2.32 | 1.24–4.36 | <0.01 | – | Jorde et al31 | |
| rs6438705 (G/A) | GG vs AA | 113 | 1,244 | 0.65 | 0.42–0.99 | 0.04 | – | Shui et al35 | |
| rs13083990 (T/C) | TT vs CC | 113 | 1,244 | 0.65 | 0.47–0.89 | 0.008 | Low plasma 25(OH)D, low Ca intake | Shui et al35 | |
| rs2270916 (T/C) | TT vs CC | 113 | 1,244 | 1.55 | 1.09–2.20 | 0.01 | Low plasma 25(OH)D, | Shui et al35 | |
| rs1801725 (G/T) | GG vs TT | 73 | 614 | 0.54 | 0.31–0.95 | 0.03 | Low plasma 25(OH)D | Shui et al35 | |
| rs1979869 (C/T) | CC vs TT | 73, 74 | 614, 829 | 0.59 | 0.38–0.94 | 0.03 | Low plasma 25(OH)D, low Ca intake | Shui et al35 | |
| rs7637874 (C/T) | CC vs TT | 74 | 829 | 1.62 | 1.11–2.35 | 0.01 | Low Ca intake | Shui et al35 | |
| Breast | rs17251221 (G/A) | GG vs AA | 403 | 2,256 | 1.948 | 1.216–3.120 | 0.007 | – | Jorde et al31 |
| GG vs GA + AA | 217 | 231 | 10.957 | 1.374–87.393 | 0.007 | – | Li et al32 | ||
| Pancreas | rs3804592 (G/A) | GG vs AA | 628 | 1,193 | 0.81 | 0.043 | – | Anderson et al53 | |
| Neuroblastoma | rs1801725 (G/T), rs1042636 (A/G), rs1801726 (C/G) | Haplotype TAC | 65 | 0 | 2.74 (HR) | 1.20–6.25 | 0.016 | – | Masvidal et al29 |
Notes:
P-values were adjusted for multiple comparisons using a modification of P for correlated tests developed by Conneely and Boehnke.80 ACT
Abbreviations: SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; NA, not applicable; HR, hazard ratio; PACT, P-value adjusted for correlated tests.
CASR SNPs
Having a T allele of rs1801725 is associated with clinical stage 4 (P=0.002) and the histological subgroup of undifferentiated neuroblastomas (P=0.046).29 Patients with this polymorphism had significantly lower overall survival rates (P=0.022) and event-free survival rates (P=0.01) than those who had GG homozygotes.
African–American prostate cancer patients having advanced disease were approximately six times less carrying the homozygote minor allele of rs1801726 than were controls (P=0.01).30
The polymorphism of rs17251221 demonstrated a noticeable association with prostate and breast cancer risk; carrying a homozygote variant increases the risk of breast and prostate cancer considerably.31,32
Haplotype and diplotypes
Colorectal adenoma risk was associated with diplotype (GAC/GAG) of rs1801725, rs1042636, and rs1801726 (OR =0.56 [95% CI: 0.36–0.88]).33 The polymorphism of rs1801726 on this diplotype reduced distal colon adenoma risk by half compared with the diplotype only composed of wild types (GAC/GAC). The haplotype (CC) of rs4678174 and rs2270916 was associated with cancer risk compared with the wild-type haplotype (TT) in the proximal colon (OR =0.80 [95% CI: 0.67–0.97]).13 TAC haplotype of CASR rs1801725, rs1042636, and rs1801726 was compared with the wild-type GAC haplotype, and the increased incidence of stage 4 neuroblastoma (OR =5.52 [95% CI: 1.78–17.18]) and inferior overall survival (hazard ratio =2.74 [95% CI: 1.20–6.25]) was reported with TAC haplotype.29
Diet effects and CASR polymorphisms
The polymorphisms of rs2270916, rs10934578, rs12485716, and rs4678174 were not associated with colorectal cancer risk;34 however, with low calcium intake, the genetic association was significant. This correlation was also valid in a study for prostate cancer;35 several SNPs were significant only under low calcium levels or low plasma vitamin D levels.
Quality score assessment
The quality score of each study was graded: 13 studies were graded 8 and over and six studies were under 8 (Table S2), and overall included studies are well designed: 13 studies have over 500 research subjects and 12 studies have population-based recruiting methods.
Publication bias
As a widely accepted tool for publication bias, Egger’s linear regression methods and funnel plot were used. Overall, Egger’s linear regression methods and funnel plots in rs1801725, rs1042636, and rs1801726 polymorphisms did not detect publication bias (Table 2, Figures 1–3).
Discussion
In this review, we presented the novel findings of significant association between CASR rs1042636, rs1801726, and rs17251221 polymorphisms; rs1042636 decreased the colorectal cancer risk in proximal and distal sites, but rs1801726 increased the risk in distal colon site. The rs17251221 considerably increased the cancer risk in prostate and breast. The CASR encodes a polypeptide of 1,078 amino acids with seven membrane spanning helixes characteristic of G protein-coupled receptors (GPCRs).36,37 GPCRs have been known to have a direct link with cellular transformation with the discovery of MAS oncogene.38 Wild-type GPCRs could become oncogenic by the excessive exposure to local or circulating agonists.39–41 The G protein-coupled CaSR, through which calcium mediates its carcinogenesis, has been implicated in parathyroid gland cancer.42 CaSR is also distributed through the entire gastrointestinal tract43–46 and reacts to the calcium concentrations in the lumen of the colon as well as circulating concentrations.47,48 Evidence from several studies49–51 suggests that risk factors differ by site within the colorectum, and molecular and functional differences result in different susceptibility to exposures and environment, such as diet. Thus, colorectal cancer risk was analyzed by proximal and distal colon sites in our research.
The CASR gene carries three common nonsynonymous SNPs, each expressed at a much different allele frequency in three ethnic populations: rs1801725 (A986S) in Europeans (minor allele frequency: 13.3%), rs1042636 (R990G) in Asians (minor allele frequency: 50.4%), and rs1801726 (Q1011E) in Africans (minor allele frequency: 23.3%).52
The most frequent SNP in the Caucasian ethnicity, rs1801725, did not show any association with colorectal cancer risk. This finding is consistent with studies included in this systematic review on pancreatic53 and prostate cancers35,54 in Caucasians. The functional significance of this variant is small by amino acid substitution,55,56 such that the outcome of cancer risk could be negligible.13 The study of Masvidal et al29 is the only one to demonstrate that having a T allele of rs1801725 is associated with later stage with significantly low overall and event-free survival in patients with neuroblastoma.
The rs1042636 (R990G) variant, which is frequently found in the Asian population, seems functionally relevant, as evidenced by cross-species evolutionary conservation.57 Based on physical properties, the change from positively charged arginine (R) to hydrophilic glycine (G) at codon 990 results in different functionality.58 This property is consistent with the results of this meta-analysis that GG genotype showed a decreased cancer risk by 25% compared to the wild-type AA genotype in the distal colon and by 32% in the proximal colon.
According to a report by the Center for Disease Control in 2011, Africans had the highest rate of colorectal cancer, followed by Caucasian, Hispanic, Asian/Pacific Islander, and American Indian/Alaska Native.59,60 The results of our study that represent decreasing cancer risk by variant rs1042636 (high frequency in Asian) and increasing cancer risk by variant rs1801726 (high frequency in African) might explain part of the colorectal cancer risk by genetic causality.
One of the major risk factors of colorectal cancer is diet.61 Specifically, calcium and dairy product intake have been studied, and high calcium intake is associated with decreased colorectal cancer risk.62–67 According to the study by Kim et al34 on colorectal cancer and Shui et al35 on prostate cancer, several SNPs are significant only under low calcium intake or low plasma vitamin D level and that SNPs of CASR are under strong influence of epigenetic factors and regulation of calcium and vitamin D intake is a vital factor in tumorigenesis. In fact, methylation of CASR was shown in 69% of colorectal cancer tissues and 90% of lymph node metastatic tissues and was strongly associated with reduced CaSR expression.68 Both prostate and breast cancers of high mortality are strongly related to bone metastasis.69 Approximately 75% of patients who develop advanced breast cancer will have secondary tumors in the bone, while in the case of prostate cancer, ~90% of patients who die of advanced prostate cancer develop bone metastases.70,71 Overexpression of CaSR can serve as a major target of calcium in facilitating the formation and growth of skeletal metastasis of prostate and breast cancers.
One of the important aspects of CaSR research is that CaSR is highly correlated with the response of chemotherapeutics. CaSR signaling regulates the expression of thymidylate synthase and survivin and facilitates 5-fluorouracil treatment, which is one of the drugs of choice in colon cancer chemotherapy.72,73 The treatment of paclitaxel, a mitotic inhibitor used in chemotherapy is also related with CaSR. Knocking down the tumor suppressor gene BRAC1 leads to a downregulation of CaSR expression and results in upregulation of survivin which reduced the cancer cell’s sensitivity.74
Therefore, CASR gene polymorphisms can be the research target for the cancer causality and improvement of chemotherapeutics.
The limitations of this study should be acknowledged. First, most of the studies were mainly on colorectal cancers in Caucasians, ethnic factors could not be evaluated in the meta-analysis. Second, the total number of cases and controls is ~10,000, which is not enough for a meta-analysis of genetic association study under Venice guidelines75 to elucidate robust evidence. Third, several studies were performed under hospital-based control population, which could modulate population characteristics by selection bias.
Conclusion
In summary, CASR polymorphisms are highly associated with cancer risks in various sites. The evaluation of CASR in clinical aspect as a cancer biomarker and in therapeutics should consider the ethnicity, environment and diet effects concomitantly. Further research stratified by cancer site, environmental impact, and ethnicity should be undertaken.
Supplementary materials
The literature search and selection process by PRISMA flow diagram: 19 studies were included for meta-analysis and systematic review.
Abbreviation: SNP, single-nucleotide polymorphism.
Table S1.
Methodological tool of quality assessment of individual studies included for CASR polymorphisms and cancer risk
| Criteria | Quality score |
|---|---|
| Representativeness of cases | |
| Consecutive/randomly selected from case population with clearly defined sampling frame | 2 |
| Consecutive/randomly selected from case population without clearly defined sampling frame or with extensive inclusion/exclusion criteria | 1 |
| No method of selection described | 0 |
| Representativeness of controls | |
| Controls were consecutive/randomly drawn from the same sampling frame (ward/community) as cases | 2 |
| Controls were consecutive/randomly drawn from a different sampling frame as cases | 1 |
| Not described | 0 |
| Ascertainment of cancer diagnosis | |
| Clearly described objective criteria for diagnosis of asthma | 2 |
| Diagnosis of asthma by patient self-report or by patient history | 1 |
| Not described | 0 |
| Ascertainment of controls | |
| Controls were tested to screen out cancer | 2 |
| Controls were subjects who did not report cancer; no objective testing | 1 |
| Not described | 0 |
| Genotyping examination | |
| Genotyping done under “blinded” condition | 1 |
| Unblinded or not mentioned | 0 |
| Hardy–Weinberg equilibrium | |
| Hardy–Weinberg equilibrium in control group | 2 |
| Hardy–Weinberg disequilibrium in control group | 1 |
| No checking for Hardy–Weinberg equilibrium | 0 |
| Association assessment | |
| Assess association between genotypes and cancers with appropriate statistics and adjustment for confounders | 2 |
| Assess association between genotypes and cancers with appropriate statistics without adjustment for confounders | 1 |
| Inappropriate statistics used | 0 |
Table S2.
Results of comprehensive quality assessment of included studies for the meta-analysis and systematic review
| References | Representativeness of cases | Representativeness of controls | Ascertainment of cancer diagnosis | Ascertainment of controls | Genotyping examination | HWE | Association assessment | Total score |
|---|---|---|---|---|---|---|---|---|
| Speer et al1 | 1 | 2 | 0 | 1 | 0 | 2 | 1 | 7 |
| Peters et al2 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 12 |
| Fuszek et al3 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 5 |
| Bácsi et al4 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 9 |
| Dong et al5 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 11 |
| Jenab et al6 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 11 |
| Jacobs et al7 | 2 | 2 | 1 | 1 | 0 | 0 | 2 | 8 |
| Schwartz et al8 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 12 |
| Szendroi et al9 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 10 |
| Safaei et al10 | 2 | 2 | 1 | 2 | 0 | 1 | 1 | 9 |
| Fedirko et al11 | 2 | N/A | 1 | N/A | 0 | 0 | 2 | 5 |
| Shui et al12 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 12 |
| Hibler et al13 | 2 | N/A | 1 | N/A | 0 | 2 | 2 | 7 |
| Anderson et al14 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 10 |
| Kim et al15 | 2 | 1 | 0 | 1 | 0 | 2 | 1 | 7 |
| Jorde et al16 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 11 |
| Masvidal et al17 | 2 | N/A | 1 | N/A | 0 | 2 | 1 | 6 |
| Mahmoudi et al18 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 10 |
| Li et al19 | 2 | 1 | 1 | 2 | 0 | 2 | 2 | 10 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; N/A, not applicable.
References
- 1.Speer G, Cseh K, Mucsi K, et al. Calcium-sensing receptor A986S polymorphism in human rectal cancer. Int J Colorectal Dis. 2002;17(1):20–24. doi: 10.1007/s003840100359. [DOI] [PubMed] [Google Scholar]
- 2.Peters U, Chatterjee N, Yeager M, et al. Association of genetic variants in the calcium-sensing receptor with risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2004;13(12):2181–2186. [PubMed] [Google Scholar]
- 3.Fuszek P, Speer G, Nagy Z, Papp J, Lakatos PL, Lakatos P. Lack of association between the calcium-sensing receptor (CaSR) A986S polymorphism and colorectal cancer. Gastroenterology. 2003;124(4):A551–A552. [Google Scholar]
- 4.Bácsi K, Hitre E, Kósa JP, et al. Effects of the lactase 13910 C/T and calcium-sensor receptor A986S G/T gene polymorphisms on the incidence and recurrence of colorectal cancer in Hungarian population. BMC Cancer. 2008;8:317. doi: 10.1186/1471-2407-8-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong LM, Ulrich CM, Hsu L, et al. Genetic variation in calcium-sensing receptor and risk for colon cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2755–2765. doi: 10.1158/1055-9965.EPI-08-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenab M, McKay J, Bueno-de-Mesquita HB, et al. Vitamin D receptor and calcium sensing receptor polymorphisms and the risk of colorectal cancer in European populations. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2485–2491. doi: 10.1158/1055-9965.EPI-09-0319. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs ET, Martínez ME, Campbell PT, et al. Genetic variation in the retinoid X receptor and calcium-sensing receptor, and risk of colorectal cancer in the Colon Cancer Family Registry. Carcinogenesis. 2010;31(8):1412–1416. doi: 10.1093/carcin/bgq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz GG, John EM, Rowland G, et al. Prostate cancer in African–American men and polymorphism in the calcium-sensing receptor. Cancer Biol Ther. 2010;9(12):994–999. doi: 10.4161/cbt.9.12.11689. [DOI] [PubMed] [Google Scholar]
- 9.Szendroi A, Speer G, Tabak A, et al. The role of vitamin D, estrogen, calcium sensing receptor genotypes and serum calcium in the pathogenesis of prostate cancer. Can J Urol. 2011;18(3):5710–5716. [PubMed] [Google Scholar]
- 10.Safaei A, Arbabi-Aval E, Arkani M, et al. Association of Calcium-sensing Receptor(CASR rs1801725) with Colorectal Cancer. Zahedan Journal of Research in Medical Sciences. 2012;14(7):45–48. [Google Scholar]
- 11.Fedirko V, Riboli E, Tjønneland A, et al. Prediagnostic 25-hydroxyvitamin D, VDR and CASR polymorphisms, and survival in patients with colorectal cancer in western European ppulations. Cancer Epidemiol Biomarkers Prev. 2012;21(4):582–593. doi: 10.1158/1055-9965.EPI-11-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shui IM, Mucci LA, Wilson KM, et al. Common genetic variation of the calcium-sensing receptor and lethal prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22(1):118–126. doi: 10.1158/1055-9965.EPI-12-0670-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibler EA, Hu C, Jurutka PW, Martinez ME, Jacobs ET. Cancer Epidemiol Biomarkers Prev. 2012;21(2):368–375. doi: 10.1158/1055-9965.EPI-11-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson LN, Cotterchio M, Knight JA, et al. Genetic variants in vitamin d pathway genes and risk of pancreas cancer; results from a population-based case-control study in ontario, Canada. PLoS One. 2013;8(6):e66768. doi: 10.1371/journal.pone.0066768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KZ, Shin A, Kim J, et al. Association between CASR polymorphisms, calcium intake, and colorectal cancer risk. PLoS One. 2013;8(3):e59628. doi: 10.1371/journal.pone.0059628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorde R, Schirmer H, Njølstad I, et al. Serum calcium and the calcium-sensing receptor polymorphism rs17251221 in relation to coronary heart disease, type 2 diabetes, cancer and mortality: the Tromsø Study. Eur J Epidemiol. 2013;28(7):569–578. doi: 10.1007/s10654-013-9822-y. [DOI] [PubMed] [Google Scholar]
- 17.Masvidal L, Iniesta R, Casala C, et al. Polymorphisms in the calcium-sensing receptor gene are associated with clinical outcome of neuroblastoma. PLoS One. 2013;8(3):e59762. doi: 10.1371/journal.pone.0059762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudi T, Karimi K, Arkani M, et al. Parathyroid hormone gene rs6256 and calcium sensing receptor gene rs1801725 variants are not associated with susceptibility to colorectal cancer in Iran. Asian Pac J Cancer Prev. 2014;15(15):6035–6039. doi: 10.7314/apjcp.2014.15.15.6035. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Kong X, Jiang L, et al. A Genetic Polymorphism (rs17251221) in the Calcium-Sensing Receptor is Associated with Breast Cancer Susceptibility and Prognosis. Cell Physiol Biochem. 2014;33:165–172. doi: 10.1159/000356659. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This research was supported by Basic Science Research Program (2014R1A1A2055734) and ICT & Future Planning (2014M3C1B3064644) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education and Brain Korea 21 Plus Program in 2014.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Brennan S, Conigrave A. Regulation of cellular signal transduction pathways by the extracellular calcium-sensing receptor. Curr Pharm Biotechnol. 2009;10:270–281. doi: 10.2174/138920109787847484. [DOI] [PubMed] [Google Scholar]
- 2.Lamprecht S, Lipkin M. Cellular mechanisms of calcium and vitamin D in the inhibition of colorectal carcinogenesis. Ann N Y Acad Sci. 2001;952:73–87. doi: 10.1111/j.1749-6632.2001.tb02729.x. [DOI] [PubMed] [Google Scholar]
- 3.Brembeck F, Rosario M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Hiani YE, Lehen’kyi V, Ouadid-Ahidouch H, et al. Activation of the calcium-sensing receptor by high calcium induced breast cancer cell proliferation and TRPC1 cation channel over-expression potentially through EGFR pathways. Arch Biochem Biophys. 2009;486:58–63. doi: 10.1016/j.abb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Kapur K, Johnson T, Beckmann ND, et al. Genome-Wide Meta-Analysis for Serum Calcium Identifies Significantly Associated SNPs near the Calcium-Sensing Receptor (CASR) Gene. Plos genetics. 2010;6(7):1–12. doi: 10.1371/journal.pgen.1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamauchi M, Sugimoto T, Yamaguchi T, et al. Association of polymorphic alleles of the calcium-sensing receptor gene with the clinical severity of primary hyperparathyroidism. Clin Endocrinol (Oxf) 2001;55(3):373–379. doi: 10.1046/j.1365-2265.2001.01318.x. [DOI] [PubMed] [Google Scholar]
- 7.Corbetta S, Eller-Vainicher C, Filopanti M, et al. R990G polymorphism of the calcium-sensing receptor and renal calcium excretion in patients with primary hyperparathyroidism. Eur J Endocrinol. 2006;155(5):687–692. doi: 10.1530/eje.1.02286. [DOI] [PubMed] [Google Scholar]
- 8.Scillitani A, Guarnieri V, Battista C, et al. Primary hyperparathyroidism and the presence of kidney stones are associated with different haplotypes of the calcium-sensing receptor. J Clin Endocrinol Metab. 2007;92(1):277–283. doi: 10.1210/jc.2006-0857. [DOI] [PubMed] [Google Scholar]
- 9.Hu J, Spiegel AM. Structure and function of the human calcium-sensing receptor: insights from natural and engineered mutations and allosteric modulators. J Cell Mol Med. 2007;11(5):908–922. doi: 10.1111/j.1582-4934.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yun FH, Wong BY, Chase M, et al. Genetic variation at the calcium-sensing receptor (CASR) locus: implications for clinical molecular diagnostics. Clin Biochem. 2007;40(8):551–561. doi: 10.1016/j.clinbiochem.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 11.O’Seaghdha CM, Wu H, Yang Q, et al. Meta-analysis of genome-wide association studies identifies six new Loci for serum calcium concentrations. Plos genetics. 2013;9(9):1–13. doi: 10.1371/journal.pgen.1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou YH, Woon PY, Chen WC, et al. A genetic polymorphism (rs17251221) in the calcium-sensing receptor gene (CASR) is associated with stone multiplicity in calcium nephrolithiasis. PLoS One. 2011;6(9):e25227. doi: 10.1371/journal.pone.0025227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong LM, Ulrich CM, Hsu L, et al. Genetic variation in calcium-sensing receptor and risk for colon cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2755–2765. doi: 10.1158/1055-9965.EPI-08-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenab M, McKay J, Bueno-de-Mesquita HB, et al. Vitamin D receptor and calcium sensing receptor polymorphisms and the risk of colorectal cancer in European populations. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2485–2491. doi: 10.1158/1055-9965.EPI-09-0319. [DOI] [PubMed] [Google Scholar]
- 15.Thakkinstian AMM, Minelli C, Gibson P, et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–211. doi: 10.1093/aje/kwi184. [DOI] [PubMed] [Google Scholar]
- 16.van Tulder MW, Assendelft WJ, Koes BW, et al. Spinal radiographic findings and nonspecific low back pain. A systematic review of observational studies. Spine. 1997;22(4):427–434. doi: 10.1097/00007632-199702150-00015. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, McElduff P, D’Este C, et al. Does dietary calcium have a protective effect on bone fractures in women? A meta-analysis of observational studies. Br J Nutr. 2004;91(04):625–634. doi: 10.1079/BJN20031085. [DOI] [PubMed] [Google Scholar]
- 18.LeBlanc ES, Janowsky J, Chan BK, et al. Hormone replacement therapy and cognition: systematic review and meta-analysis. JAMA. 2001;285(11):1489–1499. doi: 10.1001/jama.285.11.1489. [DOI] [PubMed] [Google Scholar]
- 19.Harris RP, Helfand M, Woolf SH, et al. Current methods of the US preventive services task force: a review of the process. Am J Prev Med. 2001;20(3 suppl):21–35. doi: 10.1016/s0749-3797(01)00261-6. [DOI] [PubMed] [Google Scholar]
- 20.Davey SG, Egger M. Meta-analyses of randomised controlled trials. Lancet. 1997;350(9085):1182. doi: 10.1016/s0140-6736(05)63833-0. [DOI] [PubMed] [Google Scholar]
- 21.Attia J, Thakkinstian A, D’Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56(4):297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 22.Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25(20):3443–3457. doi: 10.1002/sim.2380. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315(7109):629. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sterne JAC, Sutton AJ, Ioannidis JPA, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:1–8. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 25.Speer G, Cseh K, Fuszek P, et al. The role of estrogen receptor, vitamin D receptor and calcium receptor genotypes in the pathogenesis of colorectal cancer. Orv Hetil. 2001;142(18):947–951. [PubMed] [Google Scholar]
- 26.Speer G, Cseh K, Mucsi K, et al. Calcium-sensing receptor A986S polymorphism in human rectal cancer. Int J Colorectal Dis. 2002;17(1):20–24. doi: 10.1007/s003840100359. [DOI] [PubMed] [Google Scholar]
- 27.Rao S, Welsh L, Cunningham D, et al. Correlation of overall survival with gene expression profiles in a prospective study of resectable esophageal cancer. Clin Colorectal Cancer. 2011;10(1):48–56. doi: 10.3816/CCC.2011.n.007. [DOI] [PubMed] [Google Scholar]
- 28.Mahmoudi T, Karimi K, Arkani M, et al. Parathyroid hormone gene rs6256 and calcium sensing receptor gene rs1801725 variants are not associated with susceptibility to colorectal cancer in Iran. Asian Pac J Cancer Prev. 2014;15(15):6035–6039. doi: 10.7314/apjcp.2014.15.15.6035. [DOI] [PubMed] [Google Scholar]
- 29.Masvidal L, Iniesta R, Casala C, et al. Polymorphisms in the calcium-sensing receptor gene are associated with clinical outcome of neuroblastoma. PLoS One. 2013;8(3):e59762. doi: 10.1371/journal.pone.0059762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz GG, John EM, Rowland G, et al. Prostate cancer in African–American men and polymorphism in the calcium-sensing receptor. Cancer Biol Ther. 2010;9(12):994–999. doi: 10.4161/cbt.9.12.11689. [DOI] [PubMed] [Google Scholar]
- 31.Jorde R, Schirmer H, Njølstad I, et al. Serum calcium and the calcium-sensing receptor polymorphism rs17251221 in relation to coronary heart disease, type 2 diabetes, cancer and mortality: the Tromsø Study. Eur J Epidemiol. 2013;28(7):569–578. doi: 10.1007/s10654-013-9822-y. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Kong X, Jiang L, et al. A Genetic Polymorphism (rs17251221) in the Calcium-Sensing Receptor is Associated with Breast Cancer Susceptibility and Prognosis. Cell Physiol Biochem. 2014;33(1):165–172. doi: 10.1159/000356659. [DOI] [PubMed] [Google Scholar]
- 33.Peters U, Chatterjee N, Yeager M, et al. Association of genetic variants in the calcium-sensing receptor with risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2004;13(12):2181–2186. [PubMed] [Google Scholar]
- 34.Kim KZ, Shin A, Kim J, et al. Association between CASR polymorphisms, calcium intake, and colorectal cancer risk. PLoS One. 2013;8(3):e59628. doi: 10.1371/journal.pone.0059628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shui IM, Mucci LA, Wilson KM, et al. Common genetic variation of the calcium-sensing receptor and lethal prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22(1):118–126. doi: 10.1158/1055-9965.EPI-12-0670-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aida K, Koishi S, Inoue M, et al. Familial hypocalciuric hypercalcemia associated with mutation in the human Ca(2+)-sensing receptor gene. J Clin Endocrinol Metab. 1995;80(9):2594–2598. doi: 10.1210/jcem.80.9.7673400. [DOI] [PubMed] [Google Scholar]
- 37.Brown EM, Gamba G, Riccardi D, et al. Cloning and characterization of an extracellular Ca(2+)-sensing receptor from bovine parathyroid. Nature. 1993;366(6455):575–580. doi: 10.1038/366575a0. [DOI] [PubMed] [Google Scholar]
- 38.Young D, Waitches G, Birchmeier C, et al. Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell. 1986;45:711–719. doi: 10.1016/0092-8674(86)90785-3. [DOI] [PubMed] [Google Scholar]
- 39.Dai Q, Shrubsole MJ, Ness RM, et al. The relation of magnesium and calcium intakes and a genetic polymorphism in the magnesium transporter to colorectal neoplasia risk. Am J Clin Nutr. 2007;86(3):743–751. doi: 10.1093/ajcn/86.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutkind JS, Novotny EA, Brann MR, et al. Muscarinic acetylcholine receptor subtypes as agonist-dependent oncogenes. Proc Natl Acad Sci U S A. 1991;88(11):4703–4707. doi: 10.1073/pnas.88.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Julius D, Livelli TJ, Jessell TM, et al. Ectopic expression of the serotonin 1c receptor and the triggering of malignant transformation. Science. 1989;244(4908):1057–1062. doi: 10.1126/science.2727693. [DOI] [PubMed] [Google Scholar]
- 42.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 43.Buchan AM, Squires PE, Ring M, et al. Mechanism of action of the calcium-sensing receptor in human antral gastrin cells. Gastroenterology. 2001;120(5):1128–1139. doi: 10.1053/gast.2001.23246. [DOI] [PubMed] [Google Scholar]
- 44.Cheng SX, Okuda M, Hall AE, et al. Expression of calcium-sensing receptor in rat colonic epithelium: evidence for modulation of fluid secretion. Am J Physiol Gastrointest Liver Physiol. 2002;283(1):G240–G250. doi: 10.1152/ajpgi.00500.2001. [DOI] [PubMed] [Google Scholar]
- 45.Gama L, Baxendale-Cox LM, Breitwieser GE. Ca2+-sensing receptors in intestinal epithelium. Am J Physiol. 1997;273(4 pt 1):C1168–C1175. doi: 10.1152/ajpcell.1997.273.4.C1168. [DOI] [PubMed] [Google Scholar]
- 46.Mitsuma T, Rhue N, Kayama M, et al. Distribution of calcium sensing receptor in rats: an immunohistochemical study. Endocr Regul. 1999;33(2):55–59. [PubMed] [Google Scholar]
- 47.Chattopadhyay N, Cheng I, Rogers K, et al. Identification and localization of extracellular Ca(2+)-sensing receptor in rat intestine. Am J Physiol. 1998;274(1 pt 1):G122–G130. doi: 10.1152/ajpgi.1998.274.1.G122. [DOI] [PubMed] [Google Scholar]
- 48.Lamprecht SA, Lipkin M. Chemoprevention of colon cancer by calcium, vitamin D and folate: molecular mechanisms. Nat Rev Cancer. 2003;3(8):601–614. doi: 10.1038/nrc1144. [DOI] [PubMed] [Google Scholar]
- 49.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113(10):779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 50.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101(5):403–408. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 51.McMichael AJ, Potter JD. Host factors in carcinogenesis: certain bile-acid metabolic profiles that selectively increase the risk of proximal colon cancer. J Natl Cancer Inst. 1985;75(2):185–191. [PubMed] [Google Scholar]
- 52.NCBI dbSNP: Short Genetic Variation. 2015. [Accessed September 12, 2015]. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=1801725.
- 53.Anderson LN, Cotterchio M, Knight JA, et al. Genetic variants in vitamin d pathway genes and risk of pancreas cancer; results from a population-based case-control study in ontario, Canada. PLoS One. 2013;8(6):e66768. doi: 10.1371/journal.pone.0066768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szendroi A, Speer G, Tabak A, et al. The role of vitamin D, estrogen, calcium sensing receptor genotypes and serum calcium in the pathogenesis of prostate cancer. Can J Urol. 2011;18(3):5710–5716. [PubMed] [Google Scholar]
- 55.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunyaev S, Ramensky V, Koch I, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10(6):591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 57.Garrett JE, Capuano IV, Hammerland LG, et al. Molecular cloning and functional expression of human parathyroid calcium receptor cDNAs. J Biol Chem. 1995;270(21):12919–12925. doi: 10.1074/jbc.270.21.12919. [DOI] [PubMed] [Google Scholar]
- 58.Miyata T, Miyazawa S, Yasunaga T. Two types of amino acid substitutions in protein evolution. J Mol Evol. 1979;12(3):219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- 59.The Surveillance, Epidemiology and End Results Program as Submitted to the National Cancer Institute. 2013. [Accessed October 30, 2014]. Available from: http://www.cdc.gov/cancer/colorectal/statistics/race.htm.
- 60.National Program of Cancer Registries as Submitted to the National Cancer Institute. 2013. [Accessed October 30, 2014]. Available from: http://www.cdc.gov/cancer/colorectal/statistics/race.htm.
- 61.Garland CF, Garland FC. Do sunlight and vitamin D reduce the likelihood of colon cancer? Int J Epidemiol. 2006;35(2):217–220. doi: 10.1093/ije/dyi229. [DOI] [PubMed] [Google Scholar]
- 62.Huncharek M, Muscat J, Kupelnick B. Colorectal cancer risk and dietary intake of calcium, vitamin D, and dairy products: a meta-analysis of 26,335 cases from 60 observational studies. Nutr Cancer. 2009;61(1):47–69. doi: 10.1080/01635580802395733. [DOI] [PubMed] [Google Scholar]
- 63.Larsson SC, Bergkvist L, Rutegard J, et al. Calcium and dairy food intakes are inversely associated with colorectal cancer risk in the cohort of Swedish men. Am J Clin Nutr. 2006;83(3):667–673. doi: 10.1093/ajcn.83.3.667. [DOI] [PubMed] [Google Scholar]
- 64.Park SY, Murphy SP, Wilkens LR, et al. Calcium and vitamin D intake and risk of colorectal cancer: the multiethnic cohort study. Am J Epidemiol. 2007;165(7):784–793. doi: 10.1093/aje/kwk069. [DOI] [PubMed] [Google Scholar]
- 65.Shin A, Li H, Shu XO, et al. Dietary intake of calcium, fiber and other micronutrients in relation to colorectal cancer risk: results from the Shanghai women’s health study. Int J Cancer. 2006;119(12):2938–2942. doi: 10.1002/ijc.22196. [DOI] [PubMed] [Google Scholar]
- 66.Wu K, Willett WC, Fuchs CS, et al. Calcium intake and risk of colon cancer in women and men. J Natl Cancer Inst. 2002;94(6):437–446. doi: 10.1093/jnci/94.6.437. [DOI] [PubMed] [Google Scholar]
- 67.Cho E, Smith-Warner SA, Spiegelman D, et al. Dairy foods, calcium, and colorectal cancer: a pooled analysis of 10 cohort studies. J Natl Cancer Inst. 2004;96(13):1015–1022. doi: 10.1093/jnci/djh185. [DOI] [PubMed] [Google Scholar]
- 68.Hizaki K, Yamamoto H, Taniguchi H, et al. Epigenetic inactivation of calcium-sensing receptor in colorectal carcinogenesis. Mod Pathol. 2011;24(6):876–884. doi: 10.1038/modpathol.2011.10. [DOI] [PubMed] [Google Scholar]
- 69.Coleman RE. Skeletal complications of malignancy. Cancer. 1997;80(8 suppl):1588–1594. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1588::aid-cncr9>3.3.co;2-z. [DOI] [PubMed] [Google Scholar]
- 70.Roodman GD. Mechanisms of bone metastasis. Discov Med. 2004;4(22):144–148. [PubMed] [Google Scholar]
- 71.Liao J, Schneider A, Datta NS, et al. Extracellular calcium as a candidate mediator of prostate cancer skeletal metastasis. Cancer Res. 2006;66(18):9065–9073. doi: 10.1158/0008-5472.CAN-06-0317. [DOI] [PubMed] [Google Scholar]
- 72.Liu G, Hu X, Chakrabarty S. Vitamin D mediates its action in human colon carcinoma cells in a calcium-sensing receptor-dependent manner: downregulates malignant cell behavior and the expression of thymidylate synthase and survivin and promotes cellular sensitivity to 5-FU. Int J Cancer. 2010;126:631–639. doi: 10.1002/ijc.24762. [DOI] [PubMed] [Google Scholar]
- 73.Liu G, Hu X, Varani J, et al. Calcium and calcium sensing receptor modulates the expression of thymidylate synthase, NAD(P)H:quinone oxidoreductase 1 and survivin in human colon carcinoma cells: promotion of cytotoxic response to mitomycin C and fluorouracil. Mol Carcinog. 2009;48(3):202–211. doi: 10.1002/mc.20470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Promkan M, Liu G, Patmasiriwat P, et al. BRCA1 suppresses the expression of survivin and promotes sensitivity to paclitaxel through the calcium sensing receptor (CaSR) in human breast cancer cells. Cell Calcium. 2011;49:79–88. doi: 10.1016/j.ceca.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 75.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37(1):120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 76.Bácsi K, Hitre E, Kósa JP, et al. Effects of the lactase 13910 C/T and calcium-sensor receptor A986S G/T gene polymorphisms on the incidence and recurrence of colorectal cancer in Hungarian population. BMC Cancer. 2008;8:317. doi: 10.1186/1471-2407-8-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Safaei A, Arbabi-Aval E, Arkani M, et al. Association of Calcium-sensing Receptor(CASR rs1801725) with Colorectal Cancer. Zahedan Journal of Research in Medical Sciences. 2012;14(7):45–48. [Google Scholar]
- 78.Hibler EA, Hu C, Jurutka PW, Martinez ME, Jacobs ET. Cancer Epidemiol Biomarkers Prev. 2012;21(2):368–375. doi: 10.1158/1055-9965.EPI-11-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacobs ET, Martínez ME, Campbell PT, et al. Genetic variation in the retinoid X receptor and calcium-sensing receptor, and risk of colorectal cancer in the Colon Cancer Family Registry. Carcinogenesis. 2010;31(8):1412–1416. doi: 10.1093/carcin/bgq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conneely KN, Boehnke M. Meta-analysis of genetic association studies and adjustment for multiple testing of correlated SNPs and traits. Genet Epidemiol. 2010;34(7):739–746. doi: 10.1002/gepi.20538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuszek P, Speer G, Nagy Z, Papp J, Lakatos PL, Lakatos P. Lack of association between the calcium-sensing receptor (CaSR) A986S polymorphism and colorectal cancer. Gastroenterology. 2003;124(4):A551–A552. [Google Scholar]
- 82.Fedirko V, Riboli E, Tjønneland A, et al. Cancer Epidemiol Biomarkers Prev. 2012;21(4):582–593. doi: 10.1158/1055-9965.EPI-11-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The literature search and selection process by PRISMA flow diagram: 19 studies were included for meta-analysis and systematic review.
Abbreviation: SNP, single-nucleotide polymorphism.
Table S1.
Methodological tool of quality assessment of individual studies included for CASR polymorphisms and cancer risk
| Criteria | Quality score |
|---|---|
| Representativeness of cases | |
| Consecutive/randomly selected from case population with clearly defined sampling frame | 2 |
| Consecutive/randomly selected from case population without clearly defined sampling frame or with extensive inclusion/exclusion criteria | 1 |
| No method of selection described | 0 |
| Representativeness of controls | |
| Controls were consecutive/randomly drawn from the same sampling frame (ward/community) as cases | 2 |
| Controls were consecutive/randomly drawn from a different sampling frame as cases | 1 |
| Not described | 0 |
| Ascertainment of cancer diagnosis | |
| Clearly described objective criteria for diagnosis of asthma | 2 |
| Diagnosis of asthma by patient self-report or by patient history | 1 |
| Not described | 0 |
| Ascertainment of controls | |
| Controls were tested to screen out cancer | 2 |
| Controls were subjects who did not report cancer; no objective testing | 1 |
| Not described | 0 |
| Genotyping examination | |
| Genotyping done under “blinded” condition | 1 |
| Unblinded or not mentioned | 0 |
| Hardy–Weinberg equilibrium | |
| Hardy–Weinberg equilibrium in control group | 2 |
| Hardy–Weinberg disequilibrium in control group | 1 |
| No checking for Hardy–Weinberg equilibrium | 0 |
| Association assessment | |
| Assess association between genotypes and cancers with appropriate statistics and adjustment for confounders | 2 |
| Assess association between genotypes and cancers with appropriate statistics without adjustment for confounders | 1 |
| Inappropriate statistics used | 0 |
Table S2.
Results of comprehensive quality assessment of included studies for the meta-analysis and systematic review
| References | Representativeness of cases | Representativeness of controls | Ascertainment of cancer diagnosis | Ascertainment of controls | Genotyping examination | HWE | Association assessment | Total score |
|---|---|---|---|---|---|---|---|---|
| Speer et al1 | 1 | 2 | 0 | 1 | 0 | 2 | 1 | 7 |
| Peters et al2 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 12 |
| Fuszek et al3 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 5 |
| Bácsi et al4 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 9 |
| Dong et al5 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 11 |
| Jenab et al6 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 11 |
| Jacobs et al7 | 2 | 2 | 1 | 1 | 0 | 0 | 2 | 8 |
| Schwartz et al8 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 12 |
| Szendroi et al9 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 10 |
| Safaei et al10 | 2 | 2 | 1 | 2 | 0 | 1 | 1 | 9 |
| Fedirko et al11 | 2 | N/A | 1 | N/A | 0 | 0 | 2 | 5 |
| Shui et al12 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 12 |
| Hibler et al13 | 2 | N/A | 1 | N/A | 0 | 2 | 2 | 7 |
| Anderson et al14 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 10 |
| Kim et al15 | 2 | 1 | 0 | 1 | 0 | 2 | 1 | 7 |
| Jorde et al16 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 11 |
| Masvidal et al17 | 2 | N/A | 1 | N/A | 0 | 2 | 1 | 6 |
| Mahmoudi et al18 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 10 |
| Li et al19 | 2 | 1 | 1 | 2 | 0 | 2 | 2 | 10 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; N/A, not applicable.