Abstract
Background:
Cell surface sialylation is associated with tumor cell invasiveness in many cancers. Glioblastoma is the most malignant primary brain tumor and is highly infiltrative. ST3GAL1 sialyltransferase gene is amplified in a subclass of glioblastomas, and its role in tumor cell self-renewal remains unexplored.
Methods:
Self-renewal of patient glioma cells was evaluated using clonogenic, viability, and invasiveness assays. ST3GAL1 was identified from differentially expressed genes in Peanut Agglutinin–stained cells and validated in REMBRANDT (n = 390) and Gravendeel (n = 276) clinical databases. Gene set enrichment analysis revealed upstream processes. TGFβ signaling on ST3GAL1 transcription was assessed using chromatin immunoprecipitation. Transcriptome analysis of ST3GAL1 knockdown cells was done to identify downstream pathways. A constitutively active FoxM1 mutant lacking critical anaphase-promoting complex/cyclosome ([APC/C]-Cdh1) binding sites was used to evaluate ST3Gal1-mediated regulation of FoxM1 protein. Finally, the prognostic role of ST3Gal1 was determined using an orthotopic xenograft model (3 mice groups comprising nontargeting and 2 clones of ST3GAL1 knockdown in NNI-11 [8 per group] and NNI-21 [6 per group]), and the correlation with patient clinical information. All statistical tests on patients’ data were two-sided; other P values below are one-sided.
Results:
High ST3GAL1 expression defines an invasive subfraction with self-renewal capacity; its loss of function prolongs survival in a mouse model established from mesenchymal NNI-11 (P < .001; groups of 8 in 3 arms: nontargeting, C1, and C2 clones of ST3GAL1 knockdown). ST3GAL1 transcriptomic program stratifies patient survival (hazard ratio [HR] = 2.47, 95% confidence interval [CI] = 1.72 to 3.55, REMBRANDT P = 1.92x10-8; HR = 2.89, 95% CI = 1.94 to 4.30, Gravendeel P = 1.05x10-11), independent of age and histology, and associates with higher tumor grade and T2 volume (P = 1.46x10-4). TGFβ signaling, elevated in mesenchymal patients, correlates with high ST3GAL1 (REMBRANDT gliomacor = 0.31, P = 2.29x10-10; Gravendeel gliomacor = 0.50, P = 3.63x10-20). The transcriptomic program upon ST3GAL1 knockdown enriches for mitotic cell cycle processes. FoxM1 was identified as a statistically significantly modulated gene (P = 2.25x10-5) and mediates ST3Gal1 signaling via the (APC/C)-Cdh1 complex.
Conclusions:
The ST3GAL1-associated transcriptomic program portends poor prognosis in glioma patients and enriches for higher tumor grades of the mesenchymal molecular classification. We show that ST3Gal1-regulated self-renewal traits are crucial to the sustenance of glioblastoma multiforme growth.
Aberrant sialylation from dysregulated sialyltransferases mediates tumor cell motility, invasion, and metastasis in a variety of neoplasms. This has led to interest in targeting the sialylation pathway for therapy. We demonstrate its biological significance in glioblastoma multiforme, a disease where glioma-propagating cells (GPCs) have been established to contribute to tumor self-renewal and perpetuation (1). Although altered expression of various sialyltransferases and their modulation has been shown to mediate cellular processes in glioma (2), their mechanistic regulation of stem cell-like tumorigenic processes has yet to be explored. GPCs are clinically relevant as they relate to primary tumor phenotype, molecular profiles, and survival outcome (3–4). This is consistent with the large body of data establishing the molecular heterogeneity of gliomas where specific genomic abnormalities regulate disease progression and outcome (5). As such, the current practice of reliance on histopathology to diagnose and treat patients needs to be refined.
Several studies suggest that increased sialylation of glycoproteins is caused by the upregulation of sialyltransferases in cancer cells (6). The sialyltransferase gene, ST3GAL1, is amplified in a subset of glioblastomas and is enriched for major cell cycle processes (7). To date, ST3Gal1 sialylation has been implicated in cancers of the breast, colorectum, and bladder (8–10). In these studies, while α2,3-sialylation has been demonstrated to correspond to disease progression, no functional role has been ascribed. Furthermore, as brain tumors arise from a stem cell-like population that contributes to patient prognosis, we are keen to explore if the ST3GAL1 transcriptomic program can mediate pathways vital to self-renewal traits. This is timely as several anti-sialyltransferase inhibitors are in clinical trials, highlighting its potential as a therapeutic target (11–13).
We hypothesized that ST3Gal1 sialyltransferase contributes to glioma growth and invasiveness by promoting GPC survival. We further asked if stem cell regulatory modules are targets of ST3Gal1. We adopted a patient-centric approach by turning to major clinical databases for bioinformatical interrogation associated with elevated ST3GAL1 expression, followed by lab-driven validation. This approach provides greater statistical power of pathway prediction that would otherwise not be possible with our limited pool of GPCs, as with any such studies.
Methods
Tissue Collection and Primary GPC Culture
Graded brain tumor specimens were obtained with written informed consent, as part of a study protocol approved by the SingHealth Centralised Institutional Review Board A and the National Healthcare Group Domain-Specific Review Board A. GPC culture methods are described in Supplementary Methods (available online). All experiments were conducted with low-passage GPCs (within 10 passages) for which we previously demonstrated maintenance of phenotypic, transcriptomic, and karyotypic features similar to the primary tumor (14).
Intracranial Glioma Mouse Model
Mouse experimentation was performed according to protocols approved by the Institutional Animal Care and Use Committee. Implantation was carried out as previously described (14–15), using six- to eight-week-old male NOD/SCID gamma mice (NOD.Cg-Prkdc scid Il2rg tm1Wjl/SzJ, The Jackson Laboratory, Bar Harbor, ME). The following coordinates were used for stereotaxic implantation in the mouse brain: antero-posterior=+1.0mm; medio-lateral=+2.0mm; dorso-ventral=-2.5mm. Mice were killed by means of transcardiac perfusion with 4% paraformaldehyde upon presentation of neurological deficits with ataxia, cachexia, lethargy, or seizure. Hematoxylin and eosin staining was performed on 5-µm-thick paraffin sections.
Statistics
All grouped data were calculated as the mean ± standard deviation. All statistical tests were two-sided except where the assumption was made in advance that the results would only be in one direction (one-sided). This assumption was proven correct by the data. Mouse survival was assessed using the log-rank test in GraphPad Prism software (GraphPad Software, Inc., San Diego, CA). The statistical significance of correlation was evaluated using Spearman’s rank correlation test. Unless otherwise stated, all the experiments were performed at least three times. A P value of less than .05 was considered statistically significant. The Cox proportionality was verified using Schoenfeld residual test, and the assumption was not violated.
Microarray Data Processing and Statistical Analysis
The transcriptomic pattern of GPCs was quantified using microarray technologies established by Illumina Human Ref-8v2 bead chips or Affymetrix GeneChip Human Genome U133 Plus 2.0 Array. Detailed preprocessing of background corrected data from microarrays is presented in the Supplementary Methods (available online). Briefly, the standard processing steps were followed to summarize the expression values as described in R/lumi and R/Bioconductor packages (16–17). The summarized data were transformed on log2 scale to study the differential pattern across experimental conditions. A linear model was regressed to identify the differential transcripts using the recommended protocols in Linear Models for Microarray (limma) and RNA-Seq Data (18). The log2-fold change coefficient was estimated from the linear model and a positive or negative log2-FC represents an up- or downregulated gene, respectively, in the numerator condition. A false discovery rate (FDR)–adjusted P value of less than .05 was defined as statistically significant in microarray-based analysis of the present study.
Accession Number
The Gene Expression Omnibus accession number for the microarray data is GSE51413.
Please see the online Supplementary Methods for the methods used for all other assays and bioinformatical procedures.
Results
ST3GAL1 Expression in Self-Renewing Progenitors and Association With Tumor Grade
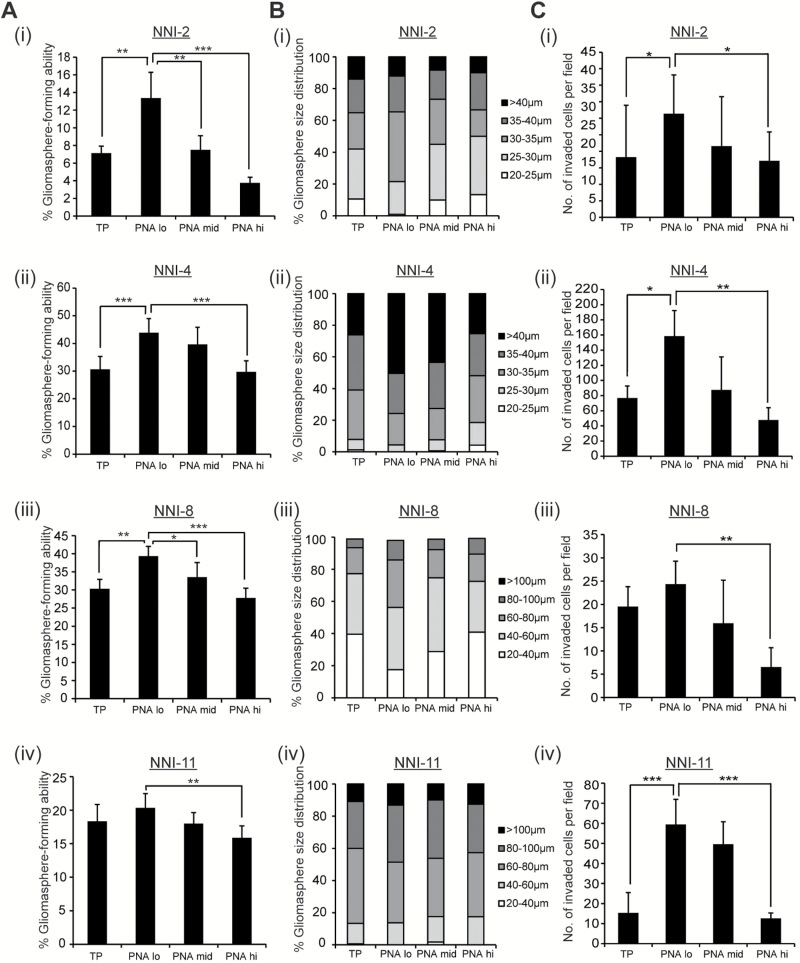
GPCs were stained with PNA and analyzed for self-renewal capacity. The Peanut Agglutinin (PNA)-lo fraction enriched for the highest gliomasphere-forming ability and stem cell frequency (Figure 1, A, i-iv; Supplementary Figure 1A, available online), largest spheres (Figure 1, B, i-iv) and greatest invasive potential (Figure 1, C, i-iv). Gene Set Enrichment Analysis (GSEA) revealed a statistically significant enrichment of the stemness module in the PNA-lo fraction (P = .04, FDR = .111) (Supplementary Figure 1B, available online). PNA staining had no impact on viability of the PNA-stained or PNA-hi fractions (data not shown). Thus, PNA-lo enriches for GPCs with in vitro self-renewal and invasive capacity.
Figure 1.
PNA expression in glioma-propagating cells (GPCs). A, i-iv) Gliomasphere-forming frequency was evaluated in total unsorted population (TP) and PNA-sorted, patient-derived GPCs (NNI-2, 4, 8, and 11). B, i-iv) Gliomasphere size distribution was measured in TP and PNA-sorted cells (NNI-2, 4, 8, and 11). C, i-iv) Invasive capacity was determined in TP and PNA-sorted GPCs (NNI-2, 4, 8, and 11). *P < .05; **P < .01; ***P < .001. For statistical analysis, two-sided Student’s t test was used. GPCs = glioma-propagating cells.
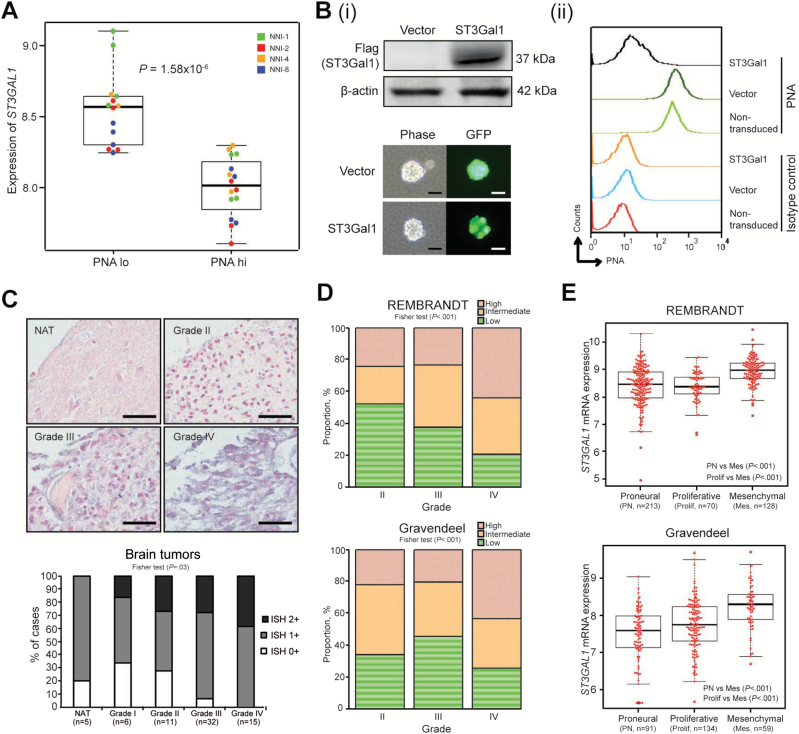
To determine the gene responsible for PNA binding, we derived a gene signature that could stratify the PNA-hi and -lo fractions (Supplementary Table 1, available online), as well as stratify patient survival in REMBRANDT and Gravendeel clinical databases (19–20). ST3GAL1, a sialyltransferase, was most statistically significant in the six-gene signature (P = 2.74x10-5). ST3GAL1 mRNA expression inversely associated with PNA staining in our GPCs (Figure 2A). ST3Gal1 overexpression in NNI-8 GPCs induced a PNA-lo phenotype (Figure 2B). Furthermore, overexpression of a ST3Gal1 loss-of-function mutant, where His299 at motif 3 of the enzyme was substituted with alanine (21), resulted in elevated PNA median fluorescence intensity compared with wild-type ST3Gal1, thereby implicating ST3Gal1 catalytic activity in mediating the PNA binding pattern in patient-derived GPCs (Supplementary Figure 1C, available online). Moreover, in patient tumors, we observed a similar inverse association between PNA and ST3Gal1 staining patterns (Supplementary Figure 1D, available online).
Figure 2.
ST3Gal1 expression in high-grade gliomas. A) ST3GAL1 mRNA expression was determined in flow-sorted PNA-lo and -hi expression groups of four glioma-propagating cells (GPCs): NNI-1, 2, 4, and 8; P = 1.58x10-6. B, i) Upper panel, flag-tagged ST3Gal1 protein was overexpressed in NNI-8 GPCs; lower panel, underwent puromycin selection. Efficient lentiviral transduction efficiency was visualized through the green fluorescent protein (GFP) tag incorporated in the vectors. Scale bar denotes 50 μm; (ii) flow histograms representing PNA fluorescence with controls defined in figure. C) Upper panel, ST3GAL1 mRNA expression was evaluated by in situ hybridization in five normal-adjacent cerebral tissue (NAT) specimens and 64 glioma specimens from grades I-IV. Representative images are shown. Lower panel, staining intensity was quantified as ISH0+, ISH1+, and ISH2+, in order of increasing positive signal. Fisher test P = .04. Scale bar denotes 50 μm. D) ST3GAL1 mRNA expression was evaluated in two independent clinical glioma databases, REMBRANDT and Gravendeel (Fisher test P values, REMBRANDT P < .001; Gravendeel P < .001). E) ST3GAL1 mRNA expression was determined in molecular subclasses of REMBRANDT and Gravendeel databases. P values are indicated in the figures. For statistical analysis, two-sided Student’s t test was used. GPCs = glioma-propagating cells; PNA = Peanut Agglutinin.
As sialyltransferase enzymes are accurately reflected by their transcription profiles—furthermore, no reliable commercial antibody against ST3Gal1 exists for immunoblot detection (22–23)—we examined ST3GAL1 mRNA expression by in situ hybridization in a collection of 69 clinical specimens, of which 64 were gliomas of varying grades I-IV, while the remaining five were normal adjacent cerebral tissue. We observed a higher intensity of ST3GAL1 staining with increasing tumor grades (Figure 2C). Furthermore, ST3Gal1 staining of invasive tumors (NNI-19 and -25) revealed extensive staining at the infiltrative edge, in contrast to less invasive tumors (NNI-8 and -21) (Supplementary Figure 2A, available online). As independent verification, we evaluated ST3GAL1 mRNA expression in REMBRANDT and Gravendeel clinical databases (19–20). Similarly, we observed higher ST3GAL1 expression in tumors of higher grades (Figure 2D) and preferential enrichment in the mesenchymal patient cohort of five clinical databases tested (Figure 2E; Supplementary Figure 2B, available online). Our findings indicate that ST3GAL1 expression is clinically important and may serve as a prognostic marker in high-grade gliomas.
Effect of TGFβ Signaling on ST3GAL1 Expression
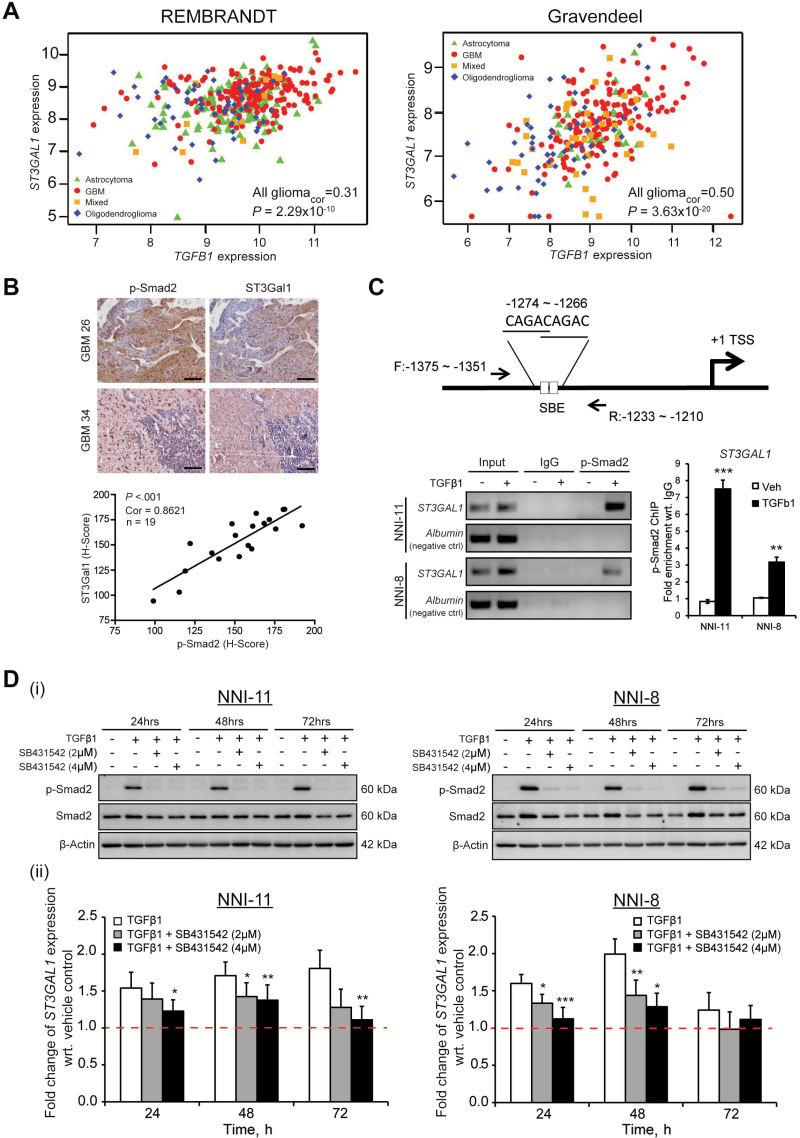
To determine the upstream mechanism triggering ST3GAL1 transcription and activation, we relied on a patient-centric bioinformatics approach. ST3GAL1 mRNA expression was ranked and categorized into three groups as high, intermediate, and low according to their terciles. GSEA was performed on the genome-wide transcriptome profiles of high and low ST3GAL1 patient groups. The core markers of top-ranking enriched genesets were extracted and Reactome pathway analysis was performed on the core markers to detect pathway clusters. ST3GAL1 clustered along with TGFB1 that provided us with a lead (enrichment P < .05) (Supplementary Figure 3A; Supplementary Tables 2 and 3, available online). To test our hypothesis that TGFβ1 may act as a trigger for ST3Gal1 activation, we examined the correlation between TGFB1 and ST3GAL1 mRNA expression. We found a statistically significant linear relationship, where ST3GAL1 expression increases with increasing TGFB1 (REMBRANDT all gliomacor = .31, P = 2.29x10-10; Gravendeel all gliomacor = .50, P = 3.63x10-20) (Figure 3A). We further verified this observation immunohistochemically in an independent collection of 19 primary glioblastoma tumors. Similarly, we found that increasing p-Smad2 staining (TGFβ pathway) correlated with increasing ST3Gal1 staining (cor = .86, P = 1x10-4) (Figure 3B).
Figure 3.
Effect of TGFβ signaling on ST3GAL1 expression. A) TGFB1 and ST3GAL1 mRNA expression were evaluated in REMBRANDT, left panel, and Gravendeel, right panel, clinical databases. The statistical significance of correlation was evaluated using Spearman’s rank correlation test (REMBRANDT all gliomacor = .31, P = 2.29x10-10; Gravendeel all gliomacor = .50, P = 3.63x10-20). B) TGFβ (p-Smad2 staining) and ST3Gal1 staining were verified immunohistochemically in 19 patient glioblastoma multiforme (GBM) tumors. Upper panel, representative images of two patient tumors; GBM 26 and GBM 34 are shown. Lower panel, images were quantified using the H-score method. The statistical significance of correlation was evaluated using Spearman’s rank correlation test (cor = .86, P < .001). Scale bar denotes 100 μm. C) Upstream promoter regions of the ST3GAL1 gene and location of primers used for the chromatin immunoprecipitation (ChIP)–quantiative polymerase chain reaction (qPCR) assay are illustrated. NNI-11 and NNI-8 GPCs were treated with 200 pM TGFβ1 for 24 hours. ChIP assays were performed with anti-p-Smad2 antibody, and ChIP-enriched DNA was amplified using the indicated primers. The bar graphs represent relative enrichment levels normalized to the respective IgG control. Albumin was used as the negative control. **P < .01; ***P < .001; vs vehicle control. D, i) TGFβ1 treatment with/without SB431542 at 2 and 4 μM in two GPCs, NNI-11 (left panel) and NNI-8 (right panel) was evaluated by probing for p-Smad2 and Smad2 protein levels. D, ii) ST3GAL1 transcript expression was determined in the presence of TGFβ1 with/without SB431542 in NNI-11 (left panel) and NNI-8 (right panel) GPCs. *P < .05; **P < .01; ***P < .001, vs vehicle control. For statistical analysis, the one-sided Student’s t test was used, where the assumption was made in advance that the results would only be in one direction (one-sided). This assumption was proven correct by the data. GBM = glioblastoma multiforme; GPCs = glioma-propagating cells; SBE = Smad binding element; TSS = transcription start site.
To test for interaction between TGFβ pathway and ST3GAL, we carried out a chromatin immunoprecipitation (ChIP) experiment using anti-p-Smad2 antibody and designed primer pairs flanking all novel, putative Smad binding element (SBE) motifs in the ST3GAL1 promoter (24). p-Smad2 binding enriched the site (-1274 to -1266) by approximately eight- and three-fold, in NNI-11 and NNI-8 cell lines, respectively (Figure 3C). Exogenous addition of TGFβ1 protein resulted in an induction of ST3GAL1 transcription in a time-dependent manner in both NNI-11 and NNI-8 GPCs, which was mitigated upon addition of TGFβ pathway small molecule inhibitor SB431542 (Figure 3Dii). We verified an active TGFβ signaling status by immunoblot analysis (Figure 3Di). To substantiate our findings, we assessed ST3Gal1 expression and its effect on PNA intensity as a functional readout (Supplementary Figure 3, B, C, and D, available online). TGFβ1 addition activated ST3GAL1 transcription and resulted in lower PNA intensity (Supplementary Figure 3, C and D, available online). In contrast, SB431542 treatment reduced ST3GAL1 transcription, accompanied by elevated PNA intensity, to levels comparable with the vehicle control, thus ruling out autocrine mode of regulation by TGFβ.
Role of ST3Gal1 in Glioma Cell Survival
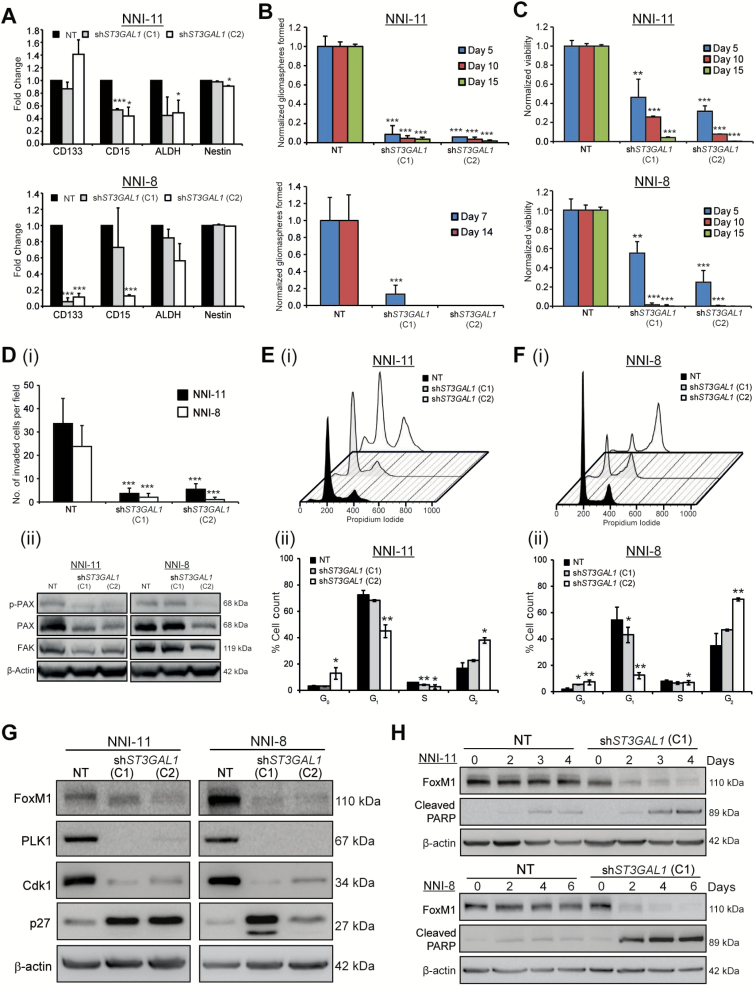
Lentiviral-mediated ST3GAL1 knockdown in NNI-11 and NNI-8 GPCs (Supplementary Figure 4A, available online) resulted in a statistically significant decrease in CD15-expressing cells (P < .05), while CD133-expressing cells were also reduced in NNI-8 (P < .001) (Figure 4A). Consistently, we observed downregulation of the stemness marker Olig2 and upregulation of differentiation markers glial fibrillary acidic protein (GFAP) and neuron-specific class III β-tubulin (TuJ1) upon ST3GAL1 knockdown, suggesting that ST3Gal1 plays a role in promoting GPC self-renewal (Supplementary Figure 4B, available online). Furthermore, we detected statistically significant downregulation of three of four genes representing a core set of neurodevelopmental transcription factors (OLIG2, POU3F2, SALL2, SOX2) recently identified to be essential for GBM propagation (P < .05) (Supplementary Figure 4C, available online) (25). A similar reduction in GPC frequency (Figure 4B), proliferation (Figure 4C), and invasive capacity (Figure 4Di) was observed. As tumor cells invade, they often lead to the breakdown of extracellular matrix proteins (6). We demonstrate that Paxillin (PAX) and focal adhesion kinase (FAK) cell adhesion proteins were reduced upon gene knockdown (Figure 4Dii). Our data implicate a GPC prosurvival role for ST3Gal1.
Figure 4.
Role of ST3Gal1 in glioma cell survival. A) Effect of ST3GAL1 knockdown on common glioma-propagating cell (GPC) markers CD133, CD15, and ALDH; nestin was determined by flow cytometry in NNI-11 (upper panel) and NNI-8 (lower panel) GPCs. NT represents nontargeting vector, while C1 and C2 represent two different shST3GAL1 clones. *P < .05; ***P < .001; vs NT control. For statistical analysis, two-sided Student’s t test was used. (B) Effect of ST3GAL1 knockdown on gliomasphere formation was determined in NNI-11 (upper panel) and NNI-8 (lower panel) GPCs. ***, p<.001; versus NT control at each corresponding day. (C) Effect of ST3GAL1 knockdown on viability was determined in NNI-11 (upper panel) and NNI-8 (lower panel) GPCs. **, P<.01; ***, P<.001; vs NT control at each corresponding day. D, i) The effect of ST3GAL1 knockdown on invasive potential was determined in NNI-11 and NNI-8 GPCs. ***P < .001; vs NT control of corresponding GPCs. D, ii) Cell adhesion proteins p-PAX, PAX, and FAK were evaluated after ST3GAL1 knockdown in NNI-11 (left panel) and NNI-8 (right panel) GPCs. E) Cell cycle profile was (i) evaluated by flow cytometry and (ii) quantified upon ST3GAL1 knockdown in NNI-11 GPCs. *P < .05; **P < .01; vs NT control at each corresponding cell cycle phase. F) Cell cycle profile was (i) evaluated by flow cytometry and (ii) quantified upon ST3GAL1 knockdown in NNI-8 GPCs. *P < .05; **P < .01; vs NT control at each corresponding cell cycle phase. G) Cell cycle proteins were assessed in NNI-11 (left panel) and NNI-8 (right panel) GPCs after ST3GAL1 knockdown and compared with the loading control β-actin. H) Extent of apoptosis, defined by cleaved poly ADP ribose polymerase (PARP) level, and FoxM1 protein levels were determined upon ST3GAL1 knockdown over four to six days in NNI-11 (upper panel) and NNI-8 (lower panel) GPCs. For statistical analysis, unless otherwise specified, one-sided Student’s t test was used, where the assumption was made in advance that the results would only be in one direction (one-sided). This assumption was proven correct by the data. GPCs = glioma-propagating cells.
To identify possible downstream targets of ST3Gal1 activation, we established microarray gene expression data from GPCs transduced with nontargeting vector control, or shST3GAL1. The differential gene list is highly enriched for cell cycle gene modules, specifically involving mitotic progression (Supplementary Figure 5A; Supplementary Table 4, available online). FOXM1 emerged as the statistically significantly modulated upstream gene (P = 2.25x10-5); furthermore, its expression positively associated with patient ST3GAL1 status (REMBRANDT P = 1.41x10-52, Gravendeel P = 6.56x10-22) (Supplementary Figure 5, B and C, available online). We thus evaluated the cell cycle profile of ST3GAL1-knockdown GPCs. We observed that the gene knockdown resulted in a G2/M cell cycle arrest, with concomitant increase in the level of sub-G0 apoptotic cells (Figure 4, E, F, and H). Importantly, FoxM1 was reduced, accompanied by a similar reduction in levels of its well-established transcriptional targets polo-like kinase 1 (PLK1) and cyclin-dependent kinase 1 (CDK1) (Figure 4G) (26). In contrast, the cyclin-dependent kinase inhibitor (CDKI), p27Kip1, which is normally reduced as cells exit G1, was elevated upon ST3GAL1 knockdown. To verify that FoxM1 (and its transcriptional targets PLK1 and CDK1) degradation was specifically because of ST3Gal1 activity and not a consequence of cell cycle arrest with subsequent apoptosis, we analyzed nocodazole-treated GPCs (Supplementary Figure 5D, available online). The GPCs demonstrated G2/M arrest; however, we did not detect any degradation of FoxM1 or its transcriptional targets. These data implicate ST3Gal1 in mitotic progression, likely mediated by the FoxM1 transcription factor.
Role of FoxM1 in ST3Gal1 Signaling
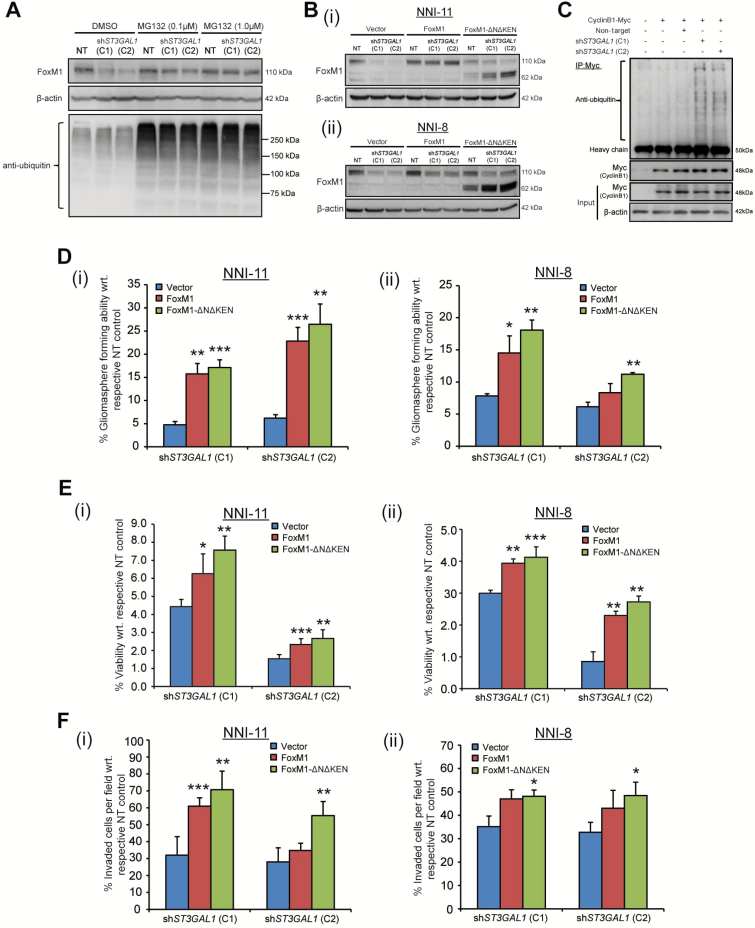
We focused our efforts on FoxM1 because its overexpression has been shown to induce hyperplasia of human epithelial cells through a mechanism involving expansion of the stem cell/progenitor pool (27). Furthermore, FoxM1 was recently implicated in a GPC prosurvival role in glioblastoma (28). We observed the degradation of FoxM1 upon ST3GAL1 knockdown, which was abrogated by increasing doses of MG132, a proteasome inhibitor that blocks the proteolytic activity of the 26S proteasome complex (Figure 5A). We confirmed that this regulation was mediated at the protein and not mRNA level. The depletion of FoxM1 protein was observed at 48 hours post-transduction, while statistically significant reduction of FOXM1 mRNA only occurred at 60 hours (P < .01) (Supplementary Figure 6A, available online), thus ruling out transcription-initiated FoxM1 degradation. This is an important distinction as FoxM1 also auto-regulates itself via a positive feedback loop mediated by its own transcript level (29). Physical interaction between ST3Gal1 and FoxM1 was absent, suggesting intermediary players (data not shown). As FoxM1 degradation can be mediated by the APC/C-Cdh1 adaptor proteins binding to the N-terminal region, we created a stable mutant of FoxM1 lacking its destruction (D)-box and KEN sequences (FoxM1-ΔNΔKEN) (30). Upon ST3GAL1 knockdown, endogenous FoxM1 degraded (110kDa); in contrast, the FoxM1ΔNΔKEN mutant protein was stable (62kDa) (Figure 5B). Moreover, increased APC/C-Cdh1 activity was observed that corresponded with increased ubiquitination of its well-known substrate cyclin B1 (Figure 5C). We also detected an increase in the nonphosphorylated form of Cdh1 upon ST3GAL1 knockdown, indicative of APC activation, in contrast to nocodazole-arrested cells that accumulate the inactive form of phosphorylated Cdh1 (Supplementary Figure 6B, available online) (31). Consistent with our hypothesis, FoxM1 or FoxM1ΔNΔKEN overexpression rescued the gliomasphere-forming ability, viability, and invasive potential of ST3GAL1-knockdown GPCs (Figure 5, D-F) but not of vector control-transduced cells (data not shown). These data indicate that ST3Gal1 regulates GPC survival through indirect control of FoxM1 protein degradation by the APC/C-Cdh1 complex.
Figure 5.
Role of FoxM1 in ST3Gal1 signaling. A) FoxM1 protein levels were assessed upon ST3GAL1 knockdown in NNI-11 glioma-propagating cells (GPCs) or in the presence of increasing concentrations (0.1 and 1 μM) of MG132, a proteasome inhibitor. Extent of ubiquitination was also assessed. B, i) NNI-11 and (ii) NNI-8 GPCs were lentivirally transduced with vector control, FOXM1 or FOXM1-ΔNΔKEN overexpression constructs, followed by ST3GAL1 knockdown (C1 and C2) or NT control. FoxM1 protein levels were evaluated and compared with β-actin loading control. C) APC/C-Cdh1 activity was determined in cells transduced with NT or shST3GAL1 (C1 and C2) vectors. Cyclin B1 (Myc-tagged) was immunoprecipitated, and the extent of ubiquitination assessed. D) Ability of FOXM1 and FOXM1-ΔNΔKEN overexpression constructs to rescue gliomasphere formation after ST3GAL1 knockdown in (i) NNI-11 and (ii) NNI-8 GPCs was evaluated. *P < .05; **P < .01; ***P < .001; vs NT control. E) Ability of FOXM1 and FOXM1-ΔNΔKEN overexpression constructs to rescue viability after ST3GAL1 knockdown in (i) NNI-11 and (ii) NNI-8 GPCs was evaluated. *P < .05; **P < .01; ***P < .001; vs NT control. F) Ability of FOXM1 and FOXM1-ΔNΔKEN overexpression constructs to rescue invasive potential after ST3GAL1 knockdown in (i) NNI-11 and (ii) NNI-8 GPCs was evaluated. *P < .05; **P < .01; ***P < .001; vs NT control. For statistical analysis, the one-sided Student’s t test was used, where the assumption was made in advance that the results would only be in one direction (one-sided). This assumption was proven correct by the data. C1 and C2 represent 2 independent ST3GAL1 knockdown clones. APC/C-Cdh1 = anaphase-promoting complex/cyclosome-Cdh1; GPCs = glioma-propagating cells; NT = nontargeting.
Role of ST3Gal1 in Orthotopic Mouse Xenograft Model and Patient Prognosis
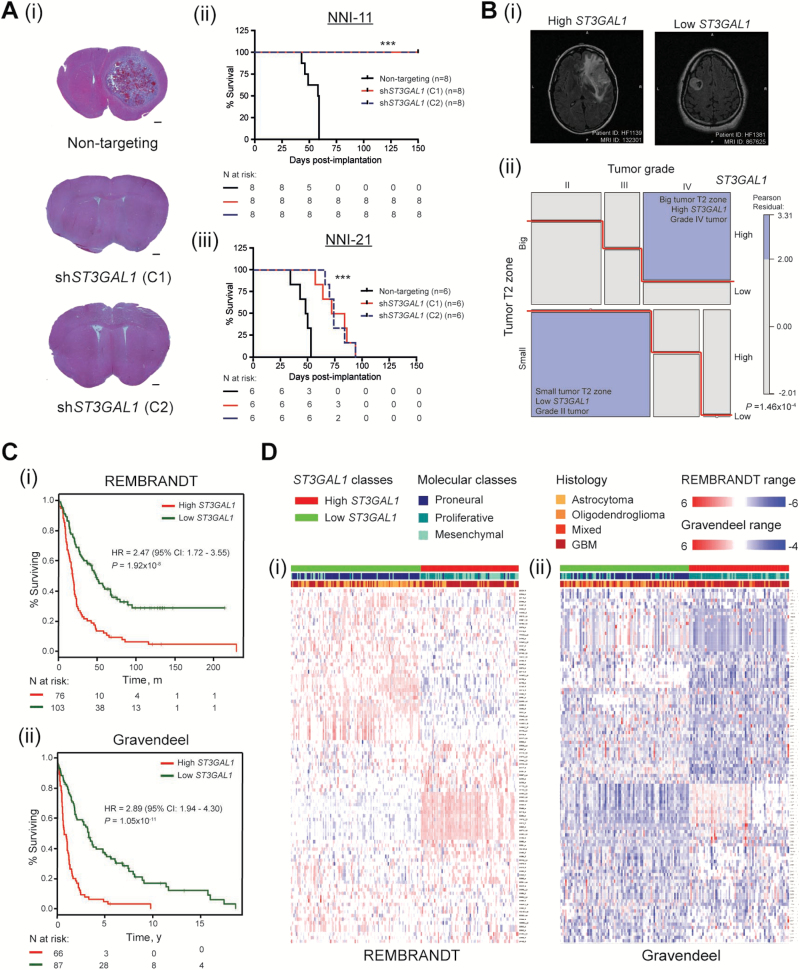
To determine the role of ST3Gal1 in brain tumor formation, we stereotaxically implanted mesenchymal (NNI-11) and proneural (NNI-21) GPCs transduced with either nontargeting control vector (NT) or two different clones of shST3GAL1 vectors (C1, C2). The molecular classification of GPCs was determined according to Lottaz et al. (32). We demonstrate in Figure 6A that the median survival differences of mice implanted with NNI-21 upon ST3GAL1 knockdown were 29 (C1, clone 1) and 25 (C2, clone 2) days, whereas for NNI-11 the mice remained alive for up to 150 days (P < .001 for both NNI-11 and NNI-21) (Figure 6A), suggesting that mesenchymal tumors are more sensitive to ST3Gal1 inhibition.
Figure 6.
Role of ST3Gal1 in orthotopic mouse xenograft model and patient prognosis. A) NOD-SCID gamma (NSG) mice were intracranially implanted with NNI-11 (mesenchymal) and NNI-21 (proneural) glioma-propagating cells (GPCs) transduced with nontargeting or two independent shST3GAL1 clones, C1 and C2. A, i) Representative coronal sections of NNI-11 mice brains are shown (n = 3 shown in figure). Scale bar denotes 500 μm. Kaplan-Meier survival plots of animal groups implanted with (ii) NNI-11 (groups of 8 in 3 arms: nontargeting, C1, and C2) and (iii) NNI-21 (groups of 6 in 3 arms: nontargeting, C1, and C2) are shown. ***P < .001. B) Interaction of ST3GAL1 status with tumor grade and T2 volume was determined. A three-way contingency analysis was performed using the log-linear model. A graphical representation was constructed using a mosaic plot to illustrate the interactions between these categorical variables. Mosaic plots were generated using R statistical packages called “vcd” and “vcdExtra”. High ST3GAL1 represents the patient cohort with upregulated ST3GAL1-associated gene signature, and vice versa. B, i) Representative magnetic resonance scans and (ii) dark purple boxes represent enriched interaction criteria; P = 1.46x10-4. C) The ST3GAL1-associated gene signature stratifies survival in (i) REMBRANDT (hazard ratio [HR] = 2.47, 95% confidence interval [CI] = 1.72 to 3.55, P = 1.92x10-8) and (ii) Gravendeel (HR = 2.89, 95% CI = 1.94 to 4.30, P = 1.05x10-11) clinical glioma databases. D) High and low ST3GAL1 patient cohorts were analyzed for the primary tumor molecular classification scheme specified by Phillips et al. (proneural, proliferative, mesenchymal) and histologies (astrocytoma, oligodendroglioma, mixed, glioblastsoma multiforme) in (i) REMBRANDT and (ii) Gravendeel databases. The corresponding heatmaps of the ST3GAL1-associated gene signature candidates are shown. Numbers of patients at risk in each group at various time points are indicated in the figure. Kaplan-Meier survival curves were analyzed using the log-rank test with R survival package. GPCs = glioma-propagating cells; MR = magnetic resonance; NOD-SCID = nonobese diabetic/severe combined immunodeficiency; NSG = NOD-SCID gamma.
To determine the role of ST3GAL1 transcriptomic program in patient prognosis, we interrogated the ST3GAL1-associated gene signature (Supplementary Table 5, available online) for patterns of association with individual patient gene expression data in REMBRANDT (n = 186) and Gravendeel (n = 156), using the Connectivity Map (Supplementary Tables 6 and 7, available online) (33). Activated ST3GAL1 refers to the molecular pattern of those patients with an inverse relation to the ST3GAL1 knockdown transcriptome program. We demonstrate that: 1) The ST3GAL1 gene signature stratifies survival in glioma patients, with activated ST3GAL1 signaling associating with poorer prognosis (HR = 2.47, 95% CI = 1.72 to 3.55, REMBRANDT P = 1.92x10-8; HR = 2.89, 95% CI = 1.94 to 4.30, Gravendeel P = 1.05x10-11) (Figure 6C). 2) This survival stratification is not confounded by current clinical indicators such as age and histology (Table 1), suggesting that the ST3GAL1 transcriptomic program contributes to the molecular heterogeneity of gliomas. This highlights the limitation of relying solely on histology to diagnose and subsequently guide patient treatment regimens. 3) ST3GAL1 activation statistically significantly associates with higher grade mesenchymal gliomas, while downregulation of the pathway is enriched in tumors of lower grades and the proneural molecular subtype (P < 2.20x10-16) (Figure 6D; Supplementary Table 8, available online). In magnetic resonance imaging (MRI), the T2-bright tissue volume represents a combination of edema and infiltrating tumor cells (34). As such, the volume of T2-bright tissue surrounding the primary tumor mass has been utilized as a surrogate for the extent of tumor cell invasion (35–36). Interestingly, the high ST3GAL1 patient cohort is statistically significantly enriched in tumors demonstrating a greater volume of peritumoral hyperintensity on T2-weighted MRI scans of patients (Pearson residuals P = 1.46x10-4), clearly indicating a role for ST3Gal1 in promoting tumor cell invasiveness (Figure 6B; Supplementary Table 9, available online).
Table 1.
Results from Cox regression analysis performed on three independent predictive glioma databases*
| Covariates | Univariate analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|
| HR (95% CI) | se(coef) | Pr(>|z|)* | HR (95% CI) | se(coef) | Pr(>|z|) | |
| REMBRANDTHigh sialylation | 2.59 (1.84 to 3.64) | 0.17 | <.001 | 1.57 (1.07 to 2.29) | 0.19 | .02 |
| Histology GBM | 4.95 (3.15 to 7.79) | 0.23 | <.001 | 3.55 (2.11 to 5.98) | 0.27 | <.001 |
| Histology mixed | 1.46 (0.51 to 4.15) | 0.53 | .48 | 1.19 (0.41 to 3.44) | 0.54 | .75 |
| Histology OD | 1.74 (0.97 to 3.2) | 0.30 | .06 | 1.32 (0.69 to 2.52) | 0.33 | .4 |
| Age | 1.02 (1.01 to 1.03) | 0.01 | .005 | 1.01 (0.10 to 1.02) | 0.01 | .18 |
| Gravendeel | ||||||
| High sialylation | 3.39 (2.35 to 4.91) | 0.19 | <.001 | 2.26 (1.48 to 3.46) | 0.22 | <.001 |
| Histology GBM | 2.26 (1.27 to 4.02) | 0.29 | .006 | 1.14 (0.60 to 2.18) | 0.33 | .68 |
| Histology mixed | 0.66 (0.32 to 1.36) | 0.37 | .26 | 0.54 (0.26 to 1.14) | 0.38 | .11 |
| Histology OD | 0.62 (0.33 to 1.19) | 0.33 | .15 | 0.44 (0.23 to 0.85) | 0.34 | .01 |
| Age | 1.03 (1.02 to 1.05) | 0.01 | <.001 | 1.03 (1.02 to 1.05) | 0.01 | <.001 |
| Freije | ||||||
| High sialylation | 4.59 (2.35 to 8.98) | 0.34 | <.001 | 2.73 (1.32 to 5.64) | 0.37 | .007 |
| Histology GBM | 2.80 (1.03 to 7.64) | 0.51 | .04 | 1.87 (0.64 to 5.44) | 0.54 | .25 |
| Histology mixed | 0.36 (0.09 to 1.52) | 0.73 | .17 | 0.39 (0.092 to 1.65) | 0.74 | .2 |
* Low ST3GAL1 patient cohort was considered a reference category to estimate the coefficient in Cox regression model.
Astrocytoma patients were treated as reference to estimate the coefficient in Cox regression model. CI = confidence interval; GBM = glioblastoma multiforme; HR = hazard ratio; OD = oligodendroglioma; Pr(<|z|) = two-sided Wald test P value; se(coef) = standard error of the coefficient.
Thus, ST3GAL1 activation portends poor prognosis in glioma patients, and ST3Gal1 represents a novel target for therapeutic intervention of malignant glioma progression.
Discussion
Malignant gliomas are devastating, in part because of the highly infiltrative and recurrent nature of the disease. Sialylation, the addition of 9-carbon sugars at the nonreducing ends of cell surface glycoproteins, has been shown to play major roles in tumor cell invasiveness. Our findings here are unique because we show for the first time how ST3Gal1 sialyltransferase is triggered by the TGFβ signaling pathway in the mesenchymal patient cohort and regulates brain tumor formation through APC/C-Cdh1–targeted control of FoxM1 protein degradation. Our work further highlights the role that GPCs play in the sustenance of brain tumor progression, and we provide strong evidence that the ST3GAL1 gene expression–based activation program prognosticates survival in patient databases, suggesting that targeting ST3Gal1-linked processes may provide viable therapeutic strategies.
We unraveled the role of ST3Gal1 by evaluating its co-expressed gene module for the ability to stratify PNA-sorted cellular fractions and, more importantly, for its ability to stratify patient survival groups. This patient-centric approach is advantageous as it offers greater statistical power in a clinical context to guide our lab decisions. Consistent with this, gene signatures derived from cancer stem–like cells have been demonstrated to contribute to patient prognostic outcome in several cancer types (4). Furthermore, our study has identified the functional cause behind the frequently observed stem cell–related PNA staining (37–38). In support, the ST3GAL1 copy number has been shown to be amplified in an analysis of oligoneural glioblastoma tumors from The Cancer Genome Atlas (TCGA) effort, in particular, linked with major cell cycle processes that have been found relevant in our study (7). Incidentally, we also observed that low ST3GAL1 in low-grade gliomas is statistically significantly associated with the IDH1 mutation status in an independent dataset (TCGA, P = .01, data not shown). Our findings emphasize the important contribution of stem-like GPCs to the primary tumor phenotype and underscore the value of targeting ST3Gal1-linked processes in self-renewing, cellular fractions.
We further identified the mesenchymal molecular subtype of glioma patients with association to TGFβ and ST3Gal1 signaling. Consistent with our finding, the mesenchymal molecular profile has previously been shown to enrich for the TGFβ pathway in both patient-derived GPCs and primary tumors (32). The finding that TGFβ1 triggers ST3GAL1 transcription and activity is important because a major glioma regulatory module is now implicated in ST3Gal1-mediated tumor cell invasiveness and tumorigenicity. Our ability to further stratify patient cohorts with ranked patterns of similarity between the ST3GAL1 gene module and individual patient gene expression profiles demonstrates that the ST3GAL1 transcriptomic program contributes to the molecular heterogeneity of the disease that cannot be accounted for by current clinical prognostic factors. This is important as histology remains the current guide to diagnose and subsequently treat patients. Our bioinformatics analyses strongly suggest that a closer examination of the ST3GAL1 gene signature may reveal prognostic and therapeutic candidates for further validation. This presents a novel paradigm to clinical management of the disease as now histologically identical tumors can be molecularly distinct and their response to specific pathway inhibitors may be predicted by the signaling modules implicated by their molecular profiles. We thus provide a means to identify patient subgroups most likely to benefit from ST3Gal1 inhibition therapeutic approaches.
FoxM1 has many roles in cancer biology, including regulation of tumorigenicity through expansion of the stem/progenitor cell pool (27). Our study has revealed the role of ST3Gal1-mediated FoxM1 degradation through the APC/C-Cdh1 complex. Conceivably, ST3Gal1 depletion elevates the activity of the APC/C-Cdh1 complex as cells enter mitosis, which then targets the N-terminal of FoxM1 protein for subsequent degradation, thus arresting GPCs at the G2/M phase. Consequently, this arrest becomes a signal for cell fates such as apoptosis and cellular differentiation, as our findings have shown. Zhang et al. have implicated a role for FoxM1 in the maintenance of the GPC state, where its depletion disrupts β-catenin nuclear localization, inducing cellular differentiation and ultimately tumor involution (28). Our work provides another key mechanism through which ST3Gal1, a major invasiveness factor, impacts tumorigenic growth by regulating the stem cell/progenitor pool.
Our study is not without limitations. Although we implicated FoxM1 downstream of ST3Gal1 activity, our current data indicates partial rescue of ST3GAL1 knockdown cells in the presence of FoxM1 or FoxM1ΔNΔKEN overexpression (Figure 5, D-F). Further work will be needed to identify proteins directly sialylated by ST3Gal1 to effect tumorigenic growth. Collectively, our study provides biologically significant insight into the role of TGFβ pathway–mediated ST3Gal1 regulation in promoting GPC self-renewal and tumorigenicity.
Funding
This work was supported by the National Medical Research Council (Singapore) grants awarded to B. T. Ang and C. Tang (CIRGnov074, CIRG12nov024, and NMRC/CSA/0058/2013).
Supplementary Material
The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; nor the decision to submit the manuscript for publication. We thank Professor Yin Bun Cheung, Centre for Quantitative Medicine, Duke-NUS Graduate Medical School, and Singapore Clinical Research Institute, for his advice on statistical interpretation and Professor David Virshup of Duke-NUS Graduate Medical School for critical reading of the manuscript. This work was supported by the A*STAR Computational Resource Centre through the use of its high-performance computing facilities. The image data used in this research were obtained from The Cancer Imaging Archive sponsored by The Cancer Imaging Program (Division of Cancer Treatment and Diagnosis/National Cancer Institute/National Institutes of Health).
References
- 1. Rich JN, Eyler CE. Cancer stem cells in brain tumor biology. Cold Spring Harb Symp Quant Biol. 2008;73:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moskal JR, Kroes RA, Dawson G. The glycobiology of brain tumors: disease relevance and therapeutic potential. Expert Rev Neurother. 2009;9(10):1529–1545. [DOI] [PubMed] [Google Scholar]
- 3. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 4. Shats I, Gatza ML, Chang JT, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2011;71(5):1772–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dall’Olio F, Chiricolo M. Sialyltransferases in cancer. Glycoconj J. 2001;18(11–12): 841–850. [DOI] [PubMed] [Google Scholar]
- 7. Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71(9):3387–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burchell J, Poulsom R, Hanby A, et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9(12):1307–1311. [DOI] [PubMed] [Google Scholar]
- 9. Schneider F, Kemmner W, Haensch W, et al. Overexpression of sialyltransferase CMP-sialic acid:Galbeta1,3GalNAc-R alpha6-Sialyltransferase is related to poor patient survival in human colorectal carcinomas. Cancer Res. 2001;61(11):4605–4611. [PubMed] [Google Scholar]
- 10. Videira PA, Correia M, Malagolini N, et al. ST3Gal.I sialyltransferase relevance in bladder cancer tissues and cell lines. BMC Cancer. 2009;9:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Becker R, Eichler MK, Jennemann R, Bertalanffy H. Phase I clinical trial on adjuvant active immunotherapy of human gliomas with GD2-conjugate. Br J Neurosurg. 2002;16(3):269–275. [DOI] [PubMed] [Google Scholar]
- 12. Chiang CH, Wang CH, Chang HC, et al. A novel sialyltransferase inhibitor AL10 suppresses invasion and metastasis of lung cancer cells by inhibiting integrin-mediated signaling. J Cell Physiol. 2010;223(2):492–499. [DOI] [PubMed] [Google Scholar]
- 13. Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. [DOI] [PubMed] [Google Scholar]
- 14. Chong YK, Toh TB, Zaiden N, et al. Cryopreservation of neurospheres derived from human glioblastoma multiforme. Stem Cells. 2009;27(1):29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng FS, Toh TB, Ting EH, et al. Progenitor-like Traits Contribute to Patient Survival and Prognosis in Oligodendroglial Tumors. Clin Cancer Res. 2012;18(15):4122–4135. [DOI] [PubMed] [Google Scholar]
- 16. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. [DOI] [PubMed] [Google Scholar]
- 17. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. [DOI] [PubMed] [Google Scholar]
- 20. Madhavan S, Zenklusen JC, Kotliarov Y, et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeanneau C, Chazalet V, Auge C, et al. Structure-function analysis of the human sialyltransferase ST3Gal I: role of n-glycosylation and a novel conserved sialylmotif. J Biol Chem. 2004;279(14):13461–13468. [DOI] [PubMed] [Google Scholar]
- 22. Datta AK, Paulson JC. The sialyltransferase “sialylmotif” participates in binding the donor substrate CMP-NeuAc. J Biol Chem. 1995;270(4):1497–1500. [DOI] [PubMed] [Google Scholar]
- 23. Taniguchi A, Hioki M, Matsumoto K. Transcriptional regulation of human Galbeta1,3GalNAc/Galbeta1, 4GlcNAc alpha2,3-sialyltransferase (hST3Gal IV) gene in testis and ovary cell lines. Biochem Biophys Res Commun. 2003;301(3):764–768. [DOI] [PubMed] [Google Scholar]
- 24. Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. [DOI] [PubMed] [Google Scholar]
- 25. Suva ML, Rheinbay E, Gillespie SM, et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laoukili J, Kooistra MR, Bras A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–136. [DOI] [PubMed] [Google Scholar]
- 27. Gemenetzidis E, Elena-Costea D, Parkinson EK, et al. Induction of human epithelial stem/progenitor expansion by FOXM1. Cancer Res. 2010;70(22):9515–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang N, Wei P, Gong A, et al. FoxM1 Promotes beta-Catenin Nuclear Localization and Controls Wnt Target-Gene Expression and Glioma Tumorigenesis. Cancer Cell. 2011;20(4):427–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochem Pharmacol. 2013;85(5):644–652. [DOI] [PubMed] [Google Scholar]
- 30. Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008;7(17):2720–2726. [DOI] [PubMed] [Google Scholar]
- 31. Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11(5):1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lottaz C, Beier D, Meyer K, et al. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70(5):2030–2040. [DOI] [PubMed] [Google Scholar]
- 33. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. [DOI] [PubMed] [Google Scholar]
- 34. Kelly PJ, Daumas-Duport C, Kispert DB, et al. Imaging-based stereotaxic serial biopsies in untreated intracranial glial neoplasms. J Neurosurg. 1987;66(6):865–874. [DOI] [PubMed] [Google Scholar]
- 35. Aghi M, Gaviani P, Henson JW, et al. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin Cancer Res. 2005;11(24 Pt 1):8600–8605. [DOI] [PubMed] [Google Scholar]
- 36. Szeto MD, Chakraborty G, Hadley J, et al. Quantitative metrics of net proliferation and invasion link biological aggressiveness assessed by MRI with hypoxia assessed by FMISO-PET in newly diagnosed glioblastomas. Cancer Res. 2009;69(10):4502–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rietze RL, Valcanis H, Brooker GF, et al. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature. 2001;412(6848):736–739. [DOI] [PubMed] [Google Scholar]
- 38. Salner AL, Obbagy JE, Hellman S. Differing stem cell self-renewal of lectin-separated murine bone marrow fractions. J Natl Cancer Inst. 1982;68(4):639–641. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.