Abstract
Genetic studies suggest that the immune system is the greatest genetic contributor to multiple sclerosis (MS) susceptibility. Yet, these immune-related genes do not explain why inflammation is limited to the CNS in MS. We hypothesize that there is an underlying dysregulation in the CNS of MS patients that makes them more vulnerable to CNS inflammation. The sparsity of CNS-related genes associated with MS suggests that epigenetic changes in the CNS may play a role. Thus, a miRNA profiling study was performed in NAWM of MS patients and control subjects to determine if specific CNS pathways can be identified that may be altered due to miRNA-mediated post-transcriptional dysregulation. There were 15 differentially expressed miRNAs found in the MS patients’ NAWM. Pathway analysis indicated that the MAPK pathway and pathways associated with the blood-brain barrier were predicted to be significantly affected by these miRNAs. Using target predication and mRNA analysis, an inverse relationship was found between miR-191 and BDNF, SOX4, FZD5 and WSB1. The pathway and target analysis of the MS-associated miRNAs suggests that MS patients’ CNS is more prone to inflammation and less capable of repair, yet enriched in neuroprotective mechanisms.
Keywords: Multiple Sclerosis, miRNA, Brain-derived neurotrophic factor
Introduction
The cause of multiple sclerosis (MS) is still unknown, and the epidemiological data indicates that events in childhood contribute significantly to this typically adult-onset disease, making it even more challenging to define elements that directly influence disease susceptibility [1]. Most of the genetic studies indicate that the immune system is the greatest genetic contributor to disease susceptibility [2]. However, these immune-related genes do not explain why the central nervous system (CNS) is the specific target of inflammation in MS. We hypothesize that there is an underlying dysregulation in the CNS of MS patients that makes them more vulnerable to CNS inflammation and/or damage.
Historically, viruses have been postulated to be the environmental factor that dictates susceptibility to MS. This remains a valid theory, although no specific virus has been identified and it appears that it is more likely that an altered immune response to a virus, not a specific virus, may predispose individuals to the development of MS. Given that the CNS harbors numerous latent viruses, it is reasonable to speculate that the episodic interaction between the CNS and immune system to minimize virus reactivation continually shapes our immune response and may inadvertently prime adaptive immune cells to myelin antigens.
An alternative hypothesis is that the CNS in MS patients is inherently vulnerable to inflammation due to altered gene expression or pathway. There have been several microarray studies that have analyzed gene expression in MS lesions and normal appearing white matter (NAWM) in brain [3-6]. Although differential gene expression has been observed, we still do not know if altered gene expression in the CNS specifically contributes to the onset of MS. To date, no miRNA studies have been performed on NAWM in the CNS of MS patients. However, miRNA studies on immune cells of MS patients have been successful in defining alter pathways in T cells that favor CNS inflammation [7-9]. While numerous studies have focused on the pathway analysis of CNS plaques in MS [10], there are very limited studies of the NAWM [5,6]. These studies found molecular changes in pathways associated with cellular homeostasis, neuroprotection for oxidative stress, and neuroglial differentiation. In this study, we have performed a comprehensive analysis of miRNA expression in NAWM of MS patients and control subjects to determine if specific CNS pathways can be identified that may be altered due to miRNA-mediated post-transcriptional dysregulation. miRNAs are small non-coding RNA that bind to the 3’ UTR of partially complementary mRNA, inhibiting specific mRNAs from being efficiently translated into protein. Pathway analysis of the differentially expressed miRNA can identify potential targets for investigation, making it possible to identify susceptibility factors for MS.
Experimental Procedure
Human Subjects
Brain specimens from MS patients (n= 16) and healthy donors (HD, n=5) were obtained through the UCLA Human Brain and Spinal Fluid Resource Center and Dr. David Pitt.
Brain samples were kept at −70oC until RNA isolation. Table I lists information on the donors.
Table 1.
Normal Appearing White Matter Sample Information.
| CASE | AGEa | Genderb | Postmortem intervalc | Cause of death | MS |
|---|---|---|---|---|---|
| 1 | 62 | M | NAd | Surgical complication | No |
| 2 | 82 | M | 6 | Heart Attack | No |
| 3 | 54 | F | 21 | NA | No |
| 4 | 80 | M | 9 | NA | No |
| 5 | 48 | M | 20 | Drug Overdose | No |
| 6 | 36 | F | 5 | NA | Yes |
| 7 | 55 | F | 4 | Sepsis/pneumona | Yes |
| 8 | 38 | F | 8 | Pneumonia | Yes |
| 9 | 59 | M | NA | Myocardial infarct | Yes |
| 10 | 50 | F | 15 | NA | Yes |
| 11 | 68 | M | 18 | NA | Yes |
| 12 | 53 | F | 23 | NA | Yes |
| 13 | 54 | F | 24 | NA | Yes |
| 14 | 32 | F | 8 | NA | Yes |
| 15 | 74 | M | 24 | NA | Yes |
| 16 | 59 | F | 12 | Pneumonia | Yes |
| 17 | 46 | M | 16 | NA | Yes |
| 18 | 69 | M | 4 | Sepsis | Yes |
| 19 | 72 | F | 12 | NA | Yes |
| 20 | 58 | F | NA | Pneumonia | Yes |
| 21 | 52 | F | 3 | NA | Yes |
No significant difference in mean age between control and MS patients
There was a higher percentage of males in the control compared to the MS patients.
No significant difference in mean postmortem interval between control and MS patients.
NA – Information is not available
RNA Isolation
Normal appearing white matter from the periventricular region was homogenized under a dry ice bed, lysed in mirVana Lysis Buffer (Life Technologies) and further processed according to manufacturer's specifications. RNA concentration was quantified using a Nanodrop spectrophotometer (ThermoScientific, Wilmington, DE) and RNA integrity and quality was assessed via the RNA 6000 Nano Chip Agilent bioanalyzer (Agilent Technologies, Santa Clara, CA) only samples with an RNA Integrity Number (RIN) >7 were used for further processing. Samples were stored at −70°C until analysis.
Nanostring miRNA Array Profiling and Analysis
100 mg of RNA with a 260/280 ratio higher than 1.9 and a 260/230 ratio higher than 1.8 were processed with the nCounter Human miRNA Assay Vs 1 and the expression of approximately 800 miRNA was evaluated with the nCounter System. Data was analyzed with the support of the OSUCCC Bioinformatics Core. Molecule counts reported by NanoString were first robust multiple-array average (RMA) normalized to the spike-in positive control and then housekeeping genes.
DNA Microarray Profiling and Analysis
RNA (1 μg) was reverse transcribed, in vitro transcribed, labeled and hybridized to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array at the Ohio State University Comprehensive Cancer Center (OSUCC) Microarray facility. Raw data were normalize with the RMA algorithm implemented in the “Expression File Creator” module and significant differences were identified using the Comparative Marker Selection Analysis Module from the GenePattern software package. Statistically significant changes in miRNA expression were identified using t-test (p value) and corrected for multiple comparisons using Bonferroni correction.
miRNA Target Prediction and Pathway Analysis
To identify potential mRNA targets for the differentially expressed miRNA in MS patients NAWM, TargetScan 6.2 was utilized. To identify cellular pathways which were potentially altered due to the differentially expressed miRNA, DNA Intelligence Analysis (DIANA Tools) miRPath 2.0 was utilized. In Table 2, pathways that included >30 predicted mRNA targets were included.
Table 2.
Pathways predicted to be targeted by MS-associated CNS miRNA.
| Pathway | # of Genes (Union) | p-value (Union) |
|---|---|---|
| MAPK signaling | 82 | 1.18 × 10−7 |
| Focal adhesion | 66 | 2.56 × 10−7 |
| Regulation of actin cytoskeleton | 68 | 1.14 × 10−6 |
| Adherens junction | 30 | 8.98 × 10−6 |
| ECM-receptor interaction | 30 | 1.75 × 10−4 |
| Wnt signaling | 46 | 4.10 × 10−4 |
| ErbB signaling | 31 | 4.95 × 10−4 |
| TGFβ signaling | 31 | 8.59 × 10−4 |
| Ubiquitin-mediated proteolysis | 39 | 4.38 × 10−3 |
| Neuroactive ligand-receptor | 34 | 0.011 |
| Insulin signaling | 39 | 0.016 |
| Jak-STAT signaling | 41 | 0.03 |
| Glycan structure-biosynthesis | 33 | 0.041 |
| Tight junctions | 33 | 0.049 |
Real-time PCR
Real-time PCR was performed using the Taqman Gene Expression assay (Applied Biosystems). Results were analyzed with the comparative threshold cycle (Ct) method, by which data were normalized with internal control gene, Hprt1.
Results
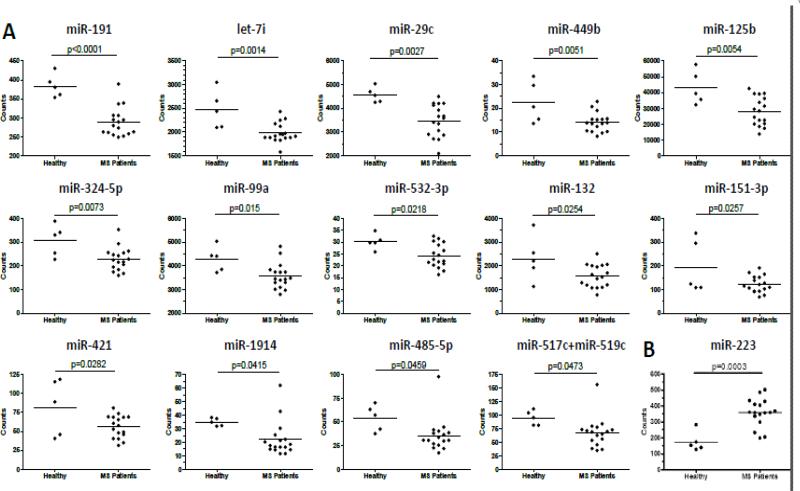
Nanostring Technology miRNA analysis was performed on NAWM of MS patients and control brain tissues (Table 1) to determine if there was differential expression of miRNA that may shed light on why the CNS is vulnerable to inflammation in MS patients. The NAWM samples were taken primarily from brain regions in which MS lesions are common, such as the periventricular area, but there was no evidence of a lesion in the MS samples used in this study. This strategy provided RNA samples that were from an area known to be susceptible to inflammation, making it a highly relevant site for investigation. Lesion sites were avoided because changes in the CNS due to inflammation would compromise identification of inherent differences in the CNS between MS patients and control that may be relevant to disease susceptibility. Total RNA was extracted from 16 MS patient samples and 5 control samples acquired from individuals who died of a non-CNS disease and showed no gross CNS abnormalities (Table I). Over 1000 human miRNA were analyzed by Nanostring nCounter technology which is a system to directly detect and enumerate RNA transcripts using miRNA-specific capture and color-coded reporter probes [11]. This system is highly sensitive and accurate since it is not dependent on enzymatic amplification to detect the RNA. Based on analysis of RNA counts and t-test, 14 miRNAs were significantly decreased in the MS patients and one miRNA was increased (Figure 1).
Figure 1. miRNA profiling of NAWM from MS patients and control.
Total RNA was extracted from NAWM and Nanostring Technology was used to quantitate miRNA in the samples. (A) Significantly down-regulated miRNA in MS patients. (B) miR-223 was the only significantly up-regulated miRNA in MS patients.
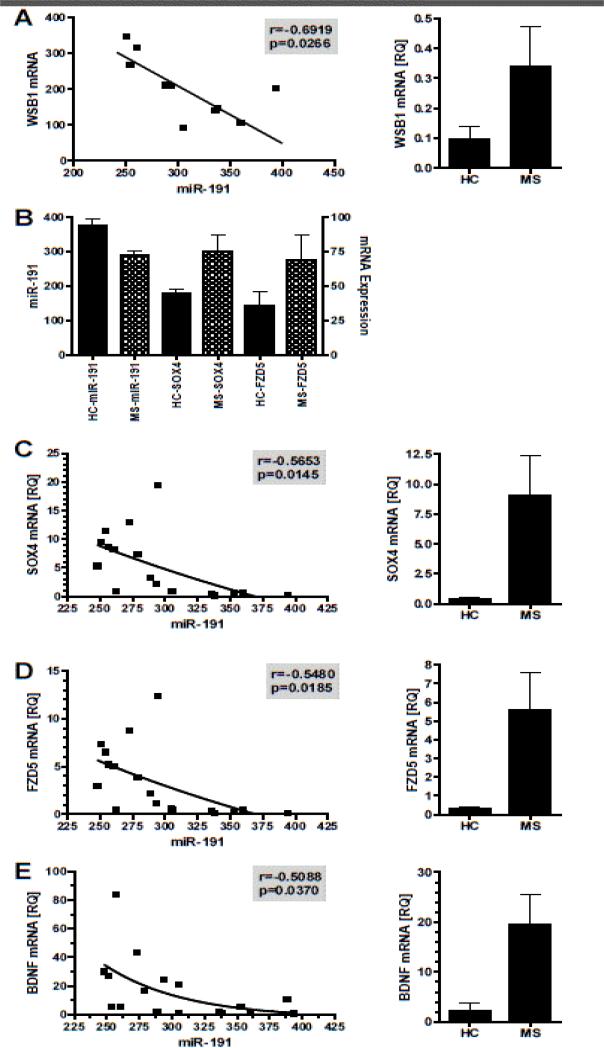
Using DNA Intelligent Analysis [12], an online pathway analysis program, we determined which molecular pathways in the NAWM were most likely altered due to the differentially expressed miRNA in the MS patients. Table II lists the pathways predicted to be significantly affected by the miRNAs in Figure 1. The mitogen-activated protein kinase (MAPK) pathway had the most significant p-value with 86 genes predicted to be targeted by one or more of MS-associated CNS miRNA. It is also important to note that several pathways that would be associated with an altered blood-brain barrier were included, such as focal adhesion, adherens junction, ECM-receptor interaction, and tight junctions. To determine if these miRNA may be regulating the level of specific genes expressed in these pathways, a subset of the RNA samples (n=10) were analyzed by mRNA microarray. TargetScan 6.2 was used to predict specific mRNA targets of miR-191, the most significantly down-regulated miRNA. Since miRNA negatively regulate gene expression of specific target mRNA, we would predict that decreased miR-191 would result in increased mRNA levels of predicted targets and thus have a negative correlation. There were 54 predicted targets for miR-191 and we found one, WSB1, with a statistically significant negative correlation (Figure 2A). WSB1 is involved in ubiquitin-mediated proteolysis [13], one of the pathways identified in the DNA Intelligent Analysis (Table 2). Since we only had mRNA data on 10 of the original 21 RNA samples, there was insufficient power to determine significant correlations for most mRNA. Therefore, we analyzed if there was an inverse relationship between miR-191 and the mean expression level of the 54 predicted mRNA targets identified by TargetScan 6.2. The mean mRNA levels of SOX4 and FZD5 were both increased in the MS mRNA samples compared to the control samples (Figure 2B), suggesting that decreased miR-191 in the MS patients’ NAWM samples may be responsible for the increase in SOX4 and FZD5. To determine if there was a relationship between miR-191 and SOX4 levels, real-time PCR was used to quantitate SOX4 transcripts in the brain samples. SOX4 mRNA was elevated in the NAWM of MS patients compared to control subjects, and there was an inverse correlation between miR-191 and SOX4 mRNA levels (Figure 2C). A previous study confirmed that miR-191 directly regulated SOX4 [14]. Similarly, FZD5 transcript levels were determined by real-time PCR from the same brain samples, illustrating the FZD5 mRNA levels were lower in MS patients, and FZD5 mRNA levels negatively correlated with miR-191 levels (Figure 2D).
Figure 2. Analysis of potential targets of miR-191.
Using mRNA expression levels of the NAWM and miRNA predication analysis, the potential targets of miR-191 were identified. (A) A correlation analysis was performed between the mRNA expression of WSB1 in the mRNA microarray and miR-191. (B) Two additional mRNA, SOX4 and FZD4, had an inverse relationship with miR-191 expression. Independent real-time PCR analysis of SOX4 (C), FZD5 (D) and BDNF (E) mRNA illustrated an inverse correlation with miR-191, as well as increased levels of these transcripts in NAWM of MS patient brains compared to control.
SOX4 plays a role in both Wnt and TGFβ signaling pathways, while FZD5 is involved in the Wnt signaling pathway, all pathways identified by the DNA Intelligent Analysis (Table 2).
It had previously been shown that BDNF was a direct target of miR-191 [15]. The mRNA microarray analysis had no data on BDNF expression and therefore, we performed real-time PCR on the NAWM samples to analyze BDNF mRNA levels to determine if there was a correlation between miR-191 and BDNF expression.
As shown in Figure 2E, BDNF mRNA levels were reduced in the NAWM of MS patients and there was a statistically significant inverse correlation between miR-191 and BDNF, suggesting that reduced miR-191 resulted in increased levels of BDNF. BDNF has been postulated to be a potential therapy for reducing neurodegeneration in MS [16]. However, it has also been demonstrated that trkB signaling via BDNF on astrocytes promotes nitric oxide production and neurodegeneration [17]. Thus, enhanced BDNF levels may have both beneficial and detrimental effects in MS.
Discussion
The cause of MS is unknown and there are few known susceptibility factors. In the current study, a miRNA profiling analysis of NAWM was performed to determine if there was evidence of inherent dysregulation of cellular pathways in the CNS of MS patients. To our knowledge, this is the first study to use miRNA to study changes in normal appearing white matter in MS. Two previous studies used mRNA microarray analysis to address this question. Lindberg et al. [6] found an increase in genes associated with cellular immune responses and neural homeostasis, concluding that MS pathogenesis is a generalized process involving the entire CNS. A similar study by Graumann et al. [5] found upregulation of HIF-1α and PI3K/Akt pathways, consistent with our observation that the MAPK signaling pathway is the most highly altered pathway as predicted by the miRNA profiling. Graumann et al. [5] postulated that the cellular pathway changes in NAWM suggest activation of neuroprotective mechanisms to preserve the cellular functions of the CNS from hypoxic insult. Together, these three studies find that the NAWM in MS patients has distinct changes in cellular pathways.
What is not known from these three studies is whether these changes preceded the onset of MS or were initiated in response to inflammation and CNS damage. It has been postulated that epigenetic regulation of gene expression, including miRNAs, may have long-term consequences on the central nervous system [18]. Environmental factors, such as stress or infection, induce epigenetic changes in the CNS that may change how the CNS responds to insults later in life. This is consistent with the observation that environmental factors in early life influences ones susceptibility to MS. Although our goal was to determine if miRNA profiling of NAWM could identify dysregulated pathways in the CNS of MS patients as a mechanism to understand why the CNS of MS patients is vulnerable to inflammation, the use of post-mortem samples from patients who have had MS for many years limits our ability to make such conclusions. However, this study provides a foundation for understanding differences inthe NAWM of MS patients that may be useful for the development of neuroprotective strategies to minimize disease progression.
The analysis of miRNA in MS has largely been used to identify a biomarker or understand peripheral immune responses in MS [8,19,9,20,21]. However, very few miRNA studies have been done to understand their role in the CNS in MS. A miRNA profiling study analyzing active and inactive MS lesions found that miRNAs may be regulating macrophage function in lesions, promoting the phagocytosis to clear myelin and debris [22]. miRNAs have been found to influence the integrity of the blood brain barrier and MS patients have an altered miRNA signature on their endothelial cells that may compromise the blood brain barrier in MS patients [23]. In the present study, NAMW was analyzed and most differentially expressed miRNAs were down-regulated (Figure 1). miR-191 was the most significantly down-regulated miRNA and mRNA correlation analysis indicated that WSB1 mRNA may be a target of miRNA-191. Very little is known about the function of WSB1, also known as WD repeat and SOCS box containing 1, an ubiquitin ligase that plays a role in hypoxia and the proliferation of neuroblastomas [24,25]. Two additional potential genes identified as targets of miR-191 were SOX4 and FZD5. FZD5 is a receptor for Wnt7 that has been shown to play a role in neuron development, survival and synaptogenesis [26-28].
SOX4 is a transcription factor expressed in many cell types and a previous study found that SOX4 is regulated by miR-191 [14]. SOX4 is expressed in oligodendrocyte precursors and over-expression of SOX4 in oligodendrocytes prevents myelination [29]. A similar study found that over-expression of SOX4 in radial glia and astrocytes results in cerebellar malformation [30], suggesting that SOX4 is a negative regulator of CNS maturation. Thus, increased expression of SOX4, due to reduced miR-191, in the NAWM of MS patients may result in phenotypic changes in oligodendrocytes and astrocytes which in turn make the NAWM of MS patients more vulnerable to inflammation. Alternatively, the changes in miR-191 and SOX4 in the NAWM in MS patients may be in response to regional inflammation and damage, and MS patients’ oligodendrocytes and astrocytes may be less effective at responding appropriately to insults. A known target of miR-191, BDNF [15], was found to have a negative correlation in the NAWM. Initially, one would anticipate that increased BDNF would be beneficial for MS patients since it promotes the growth and survival of neurons, and thus up regulation of BDNF via miR-191 may be protecting the NAWM from inflammation. Many studies in models of MS support this idea [31]. Yet, it has also been found that BDNF signalling via TrkB on astrocytes may enhance nitric oxide production and neuro degeneration in experimental autoimmune encephalomyelitis [17]; therefore, the role of BDNF in inflammatory demyelinating disease in not fully understood.
The only miRNA that was significantly increased in NAWM of MS patients was miR-223. Several potential targets of miR-223 were identified, including FBXW7, APC, SLC8A1, SLC4A4 and IL6ST (data not shown), but we have not been able to confirm these targets. Decreased expression of these molecules due to increased miR-223 would affect the differentiation and function of neurons, oligodendrocytes and microglia [32-36]. Decreased FBXW7 and APC would suggest that neuron and oligodendrocyte differentiation would be impaired [32,33]. miR-223 has been shown to be enriched in microglia relative to other CNS cell types [37]. Decreased SLC8A1 via miR-223 would theoretically result in decreased respiratory burst by microglia [34]. In addition, miR-223 has been shown to reduce glutamate-mediated excitotoxicity in a stroke model [38]. Thus, increased expression of miR-223 in NAWM of MS patients may prevent optimal differentiation of neurons and microglia, yet provide neuroprotection from insults.
This pilot study demonstrates that miRNA are differentially expressed in the NAWM of MS patients and a larger with well-defined MS patient samples is warranted to conclusively define miRNA dysregulation in the CNS of MS patients. Pathway and target analysis indicates that there may be defects in the differentiation and function of various neural cell types that may make the NAWM of MS patients more prone to inflammation and less capable of repair. However, the data also suggests that neuroprotective mechanisms may be enriched in the NAWM of MS patients. It is unclear whether these changes are inherent in the CNS of MS patients or induced due to disease. However, further study of NAWM in MS patients may provide insights into pathways that may be therapeutically manipulated to protect the CNS from inflammation and minimize disease progression.
Acknowledgment
NIH grants R21 NS078390 and RO1 NS067441-01A1 (AELR) and National Multiple Sclerosis Society grant RG 4742-A-14 (MKR).
References
- 1.Simpson S, Jr, Taylor BV, van der Mei I. The role of epidemiology in MS research: Past successes, current challenges and future potential. Mult Scler. 2015;21:969–977. doi: 10.1177/1352458515574896. [DOI] [PubMed] [Google Scholar]
- 2.Oksenberg JR. Decoding multiple sclerosis: an update on genomics and future directions. Expert Rev Neurother. 2013;13:11–19. doi: 10.1586/14737175.2013.865867. [DOI] [PubMed] [Google Scholar]
- 3.Whitney LW, Becker KG, Tresser NJ, Caballero-Ramos CI, Munson PJ, et al. Analysis of gene expression in mutiple sclerosis lesions using cDNA microarrays. Ann Neurol. 1999;46:425–428. doi: 10.1002/1531-8249(199909)46:3<425::aid-ana22>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 4.Tajouri L, Mellick AS, Ashton KJ, Tannenberg AE, Nagra RM, et al. Quantitative and qualitative changes in gene expression patterns characterize the activity of plaques in multiple sclerosis. Brain Res Mol Brain Res. 2003;119:170–183. doi: 10.1016/j.molbrainres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Graumann U, Reynolds R, Steck AJ, Schaeren-Wiemers N. Molecular changes in normal appearing white matter in multiple sclerosis are characteristic of neuroprotective mechanisms against hypoxic insult. Brain Pathol. 2003;13:554–573. doi: 10.1111/j.1750-3639.2003.tb00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindberg RL, De Groot CJ, Certa U, Ravid R, Hoffmann F, et al. Multiple sclerosis as a generalized CNS disease – comparative microarray analysis of normal appearing white matter and lesions in secondary progressive MS. J Neuroimmunol. 2004;152:154–167. doi: 10.1016/j.jneuroim.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Junker A, Hohlfeld R, Meinl E. The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol. 2011;7:56–59. doi: 10.1038/nrneurol.2010.179. [DOI] [PubMed] [Google Scholar]
- 8.Guerau-de-Arellano M, Smith KM, Godlewski J, Liu Y, Winger R, et al. Micro-RNA dysregulation in multiple sclerosis favours proinflammatory T-cell-mediated autoimmunity. Brain. 2011;134:3578–3589. doi: 10.1093/brain/awr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerau-de-Arellano M, Alder H, Ozer HG, Lovett-Racke A, Racke MK. miRNA profiling for biomarker discovery in multiple sclerosis: from microarray to deep sequencing. J Neuroimmunol. 2012;248:32–39. doi: 10.1016/j.jneuroim.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinman L, Zamvil S. Transcriptional analysis of targets in multiple sclerosis. Nat Rev Immunol. 2003;3:483–492. doi: 10.1038/nri1108. [DOI] [PubMed] [Google Scholar]
- 11.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 12.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, et al. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem. 2008;283:4682–4689. doi: 10.1074/jbc.M708873200. [DOI] [PubMed] [Google Scholar]
- 14.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, et al. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010;70:8077–8087. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 15.Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 2013;34:1889–1899. doi: 10.1093/carcin/bgt107. [DOI] [PubMed] [Google Scholar]
- 16.Kerschensteiner M, Stadelmann C, Dechant G, Wekerle H, Hohlfeld R. Neurotrophic cross-talk between the nervous and immune systems: implications for neurological diseases. Ann Neurol. 2003;53:292–304. doi: 10.1002/ana.10446. [DOI] [PubMed] [Google Scholar]
- 17.Colombo E, Cordiglieri C, Melli G, Newcombe J, Krumbholz M, et al. Stimulation of the neurotrophin receptor TrkB on astrocytes drives nitric oxide production and neurodegeneration. J Exp Med. 2012;209:521–535. doi: 10.1084/jem.20110698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babenko O, Kovalchuk I, Metz GA. Epigenetic programming of neurodegenerative diseases by an adverse environment. Brain Res. 2012;1444:96–111. doi: 10.1016/j.brainres.2012.01.038. [DOI] [PubMed] [Google Scholar]
- 19.Martinelli-Boneschi F, Fenoglio C, Brambilla P, Sorosina M, Giacalone G, et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci Lett. 2012;508:4–8. doi: 10.1016/j.neulet.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73:729–740. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 21.Søndergaard HB, Hesse D, Krakauer M, Sørensen PS, Sellebjerg F. Differential microRNA expression in blood in multiple sclerosis. Mult Scler. 2013;19:1849–1857. doi: 10.1177/1352458513490542. [DOI] [PubMed] [Google Scholar]
- 22.Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 23.Reijerkerk A, Lopez-Ramirez MA, van het Hof B, Drexhage JAR, Kamphuis WW, et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: Implications for multiple sclerosis. J Neurosci. 2013;13:6857–6863. doi: 10.1523/JNEUROSCI.3965-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen QR, Bilke S, Wei JS, Greer BT, Steinberg SM, et al. Increased WSB1 copy number correlates with its over-expression which associates with increased survival in neuroblastoma. Genes Chromosomes Cancer. 2006;45:856–862. doi: 10.1002/gcc.20349. [DOI] [PubMed] [Google Scholar]
- 25.Moehlenbrink J, Bitomsky N, Hofmann TG. Hypoxia suppresses chemotherapeutic drug-induced p53 Serine 46 phosphorylation by triggering HIPK2 degradation. Cancer Lett. 2010;292:119–124. doi: 10.1016/j.canlet.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Burns CJ, Zhang J, Brown EC, Van Bibber AM, Van Es J, et al. Investigation of Frizzled-5 during embryonic neural development in mouse. Dev Dyn. 2008;237:1614–1626. doi: 10.1002/dvdy.21565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Wang Y, Smallwood PM, Nathans J. An essential role for Frizzled5 in neuronal survival in the parafascicular nucleus of the thalamus. J Neurosci. 2008;28:5641–5653. doi: 10.1523/JNEUROSCI.1056-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahores M, Gibb A, Salinas PC. Frizzled-5, a receptor for the synaptic organizer Wnt7a, regulates activity-mediated synaptogenesis. Development. 2010;137:2215–2225. doi: 10.1242/dev.046722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potzner MR, Griffel C, Lütjen-Drecoll E, Bösl MR, Wegner M, et al. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27:5316–5326. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoser M, Baader SL, Bösl MR, Ihmer A, Wegner M, et al. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci. 2007;27:5495–5505. doi: 10.1523/JNEUROSCI.1384-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lühder F, Gold R, Flügel A, et al. Brain-derived neurotrophic factor in neuroimmunology: lessons learned from multiple sclerosis patients and experimental autoimmune encephalomyelitis models. Arch Immunol Ther Exp. 2013;61:95–105. doi: 10.1007/s00005-012-0211-0. [DOI] [PubMed] [Google Scholar]
- 32.Hoeck JD, Jandke A, Blake SM, Nye E, Spencer-Dene B, et al. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci. 2010;13:1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 33.Lang J, Maeda Y, Bannerman P, Xu J, Horiuchi M, et al. Adenomatous polyposis coli regulates oligodendroglial development. J Neurosci. 2013;33:3113–3130. doi: 10.1523/JNEUROSCI.3467-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newell EW, Stanley EF, Schlichter LC. Reversed Na+/Ca2+ exchange contributes to Ca2+ influx and respiratory burst in microglia. Channels (Austin) 2007;1:366–376. doi: 10.4161/chan.5391. [DOI] [PubMed] [Google Scholar]
- 35.Rickmann M, Orlowski B, Heupel K, Roussa E. Distinct expression and subcellular localization patterns of Na+/HCO3-cotransporter (SLC 4A4) variants NBCe1-A and NBCe1-B in mouse brain. Neuroscience. 2007;146:1220–1231. doi: 10.1016/j.neuroscience.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 36.Spooren A, Kolmus K, Laureys G, Clinckers R, De Keyser J, et al. Interleukin-6, a mental cytokine. Brain Res Rev. 2011;67:157–183. doi: 10.1016/j.brainresrev.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33:5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc Natl Acad Sci U S A. 2012;109:18962–18967. doi: 10.1073/pnas.1121288109. [DOI] [PMC free article] [PubMed] [Google Scholar]