Abstract
Full-length (FL) cDNA sequences provide the most reliable evidence for the presence of genes in genomes. In this report, detailed gene structures of barley, whole genome shotgun (WGS) and additional transcript data of the cultivar Haruna Nijo were quality controlled and compared with the published Morex genome information. Haruna Nijo scaffolds have longer total sequence length with much higher N50 and fewer sequences than those in Morex WGS contigs. The longer Haruna Nijo scaffolds provided efficient FLcDNA mapping, resulting in high coverage and detection of the transcription start sites. In combination with FLcDNAs and RNA-Seq data from four different tissue samples of Haruna Nijo, we identified 51,249 gene models on 30,606 loci. Overall sequence similarity between Haruna Nijo and Morex genome was 95.99%, while that of exon regions was higher (99.71%). These sequence and annotation data of Haruna Nijo are combined with Morex genome data and released from a genome browser. The genome sequence of Haruna Nijo may provide detailed gene structures in addition to the current Morex barley genome information.
Keywords: genome sequencing, full-length cDNA, Hordeum, RNA-Seq
1. Introduction
Barley (Hordeum vulgare L.) is used for many purposes including human food, malting and animal feed. Coupled with its wide adaptability in environments ranging from the highlands of Africa at the equator to the Arctic Regions in Scandinavia, barley is the fourth most important cereal crop (http://faostat.fao.org, 7 November 2015, date accessed). In addition, barley is an ancient crop that was domesticated ca. 10,000 yrs ago in the Fertile Crescent.1 Decades of collection and curation of barley germplasm have resulted in substantial germplasm collections including: IPK (Germany), USDA-ARS (USA) and Okayama University (Japan).2 To fully exploit these in situ and ex situ collections for barley breeding and gene discovery, barley genome sequences of multiple genotypes are needed.
To generate the first genome sequence of barley, the International Barley Sequencing Consortium (IBSC) conducted BAC fingerprinting, BAC end sequencing, whole genome shotgun (WGS) sequencing, RNA-Seq analysis and genetic mapping and integrated these data to develop a gene-based genome sequence of the North American six-row spring malting barley cultivar Morex.3 Morex traces to barley germplasm of Manchurian origin with six-row inflorescence, which is not common for malting barleys other than the USA. Currently, Morex is the reference genome for barley genetics studies, and additional efforts are in progress to improve the Morex genome sequence.4,5 Three other barley genomes including Bowman, Barke and a Tibetan hulless genotype have been WGS sequenced3,6 and provide additional sequence information for comparison to Morex.
Haruna Nijo is a Japanese malting barley cultivar exhibiting excellent malting quality for brewing beer. Historically, Japanese malting barleys have been developed by crossing European malting barley cultivars and Japanese landraces and selecting for malting quality and adaptation to Japanese environments. Haruna Nijo was also developed by this approach in 1979 and has been extensively used as a foundation genotype of current Japanese breeding. Haruna Nijo has a high thermostability of β-amylase,7 and it also shows different Bmy1 allele for β-amylase activity from Morex.8 It is also shown that Haruna Nijo has a high malt extract (related to beer production) by QTL analysis.9 To enhance the utility of Haruna Nijo for breeding, a suite of genomics resources were developed including: an EST collection, transcript map construction,10,11 a BAC library,12 454-based WGS sequencing3 and full-length (FL) cDNA sequencing and analysis13,14 (available at http://barleyflc.dna.affrc.go.jp/bexdb/, 7 November 2015, date accessed, with functions of expression profiling and genome browsing).
In the case of barley genome resources, the Morex reference genome and the Haruna Nijo FLcDNA sequences provide unique resources in different genotypes. Thus, to enhance the utility of both resources they need to be compared and integrated. Sequence comparisons of the two different malting barley haplotypes will be much more useful than single haplotype information, as they provide the basis of structural and functional allelic diversity. Previous comparisons of WGS data and FLcDNA sequences from Haruna Nijo and the genome sequence from Morex demonstrated a large amount of sequence polymorphisms between the two genotypes.3 For example, the genetic distance (dissimilarity) between Morex and Haruna Nijo by 1,536 SNPs of Illumina GoldenGate Assay is 0.50, which is larger than the difference (0.48) between Morex and Akashinriki (Japanese food barley).15 Thus, the Haruna Nijo FLcDNA and WGS sequences may provide useful gene annotations for the reference genotype Morex and a resource for future breeding and gene discovery activities.
Our overall goal is to provide genome information of Haruna Nijo to annotate gene structures on the genome sequence of Morex. The four specific objectives of this study were to (i) generate high-quality genomic sequence of Haruna Nijo, (ii) map the precise position of genes identified in the Haruna Nijo genome, (iii) compare gene information between Haruna Nijo and Morex, and (iv) develop gene models based on the Haruna Nijo sequence data.
2. Materials and methods
2.1. Plant materials and nucleic acid isolation
For DNA and RNA isolation from seedling tissues, seeds of Haruna Nijo were germinated on moist filter paper in Petri dishes at 20°C in the dark. For DNA isolation, shoots of ca. 5 cm were harvested and high-quality genomic DNA was isolated by DNeasy Plant Mini Kit (QIAGEN K.K., Japan). For RNA isolation, seedling shoot and seedling root tissues were sampled from plants exhibiting 5 cm shoots. For the RNA samples from immature spike and immature seed samples, barley seeds were planted in pot with soil mixture (N: 120 mg l−1; P: 100 mg l−1; K: 160 mg l−1, pH 5.5) and grown in the greenhouse at 20/15°C day/night temperature under natural light conditions. Immature spike samples were harvested from the leaf sheath 5 days before heading. Immature seeds were sampled 35 days after flowering (soft dough stage). Total RNA from immature seeds was isolated by TRIzol® Reagent (Life Technologies, Japan) following the Plant RNA Isolation protocol. All RNA samples were purified by RNeasy Plant Mini Kit (QIAGEN K.K., Japan), and DNA was removed by RNase-Free DNase Set (QIAGEN K.K., Japan).
2.2. Library development
2.2.1. 454 long paired-end library
The high-quality genomic DNA was fragmented in 8 k and 20 k by the Hydroshear (Digilab Inc., Holliston, MA, USA). The library was developed by a library preparation kit (Roche diagnostics, Japan) for long paired-end libraries (8 k and 20 k). Circularization was performed according to the Roche Paired End Library Protocol, using Roche circularization adapters (Paired End Library Preparation Method Manual 20 kb and 8 kb Span; Roche Diagnostics, October 2009, Steps 3.1–3.7.3). Subsequently, the circularized fragments were fragmented again by nebulization to develop a sequencing library.
2.2.2. Illumina HiSeq sequencing library
DNA (2 µg) was fragmented by nebulization. Libraries were prepared according to the manufacturer's instruction ‘Preparing Samples for Paired-End Sequencing, Part # 1005063 Rev. A June 2008’ of TruSeq DNA Sample Prep Kit (Illumina Japan). DNA fragments were size selected (500 bp) following the Low-Throughput Protocol of TruSeq DNA Sample Prep Kit (Illumina Japan). The quality of the library (fragment length distribution) was checked by Agilent Bioanalyzer High Sensitivity DNA Assay (Agilent Technologies, Japan) and KAPA Library Quantification Kit (KK4835, Kapa Biosystems, MA, USA).
2.2.3. MiSeq RNA sequencing library
Libraries for RNA-Seq analysis were developed from each RNA sample using TruSeq RNA Sample Prep Kit V2 (Illumina Japan). The protocol of TruSeq RNA Sample Preparation V2 Guide (Illumina Japan) was used with modification of fragment isolation from agarose gel electrophoresis and elution in EB (elution buffer) following the method in the TruSeq Sample Preparation Guide (Illumina Japan). The library was quantified with Agilent High Sensitivity DNA Kit (Agilent Technologies Japan) and Qubit 2.0 Fluorometer (Life Technologies Japan).
2.3. Sequencing
2.3.1. 454 FLX Titanium platform
Each library was emulsion PCR amplified. The PCR-amplified fragments on beads were washed, and the bead number was counted using a Coulter Counter Z1 single threshold instrument (Beckman Coulter Japan). The appropriate number of beads was applied on a pico titre plate according to the manufacturer's protocol. The FLX Titanium platform was used for sequencing (average read length 500 bp). The pyrosequencing reaction data were base-called to generate sff format files using the software installed on the analysis server of the 454 sequencer (Roche diagnostics, Japan).
2.3.2. Illumina HiSeq platform
The shotgun library was sequenced on an Illumina HiSeq 2000 to produce 2 × 101 paired-end reads. Raw data processing, base calling and quality control were performed with manufacture's standard pipeline. The quality of the output sequences was inspected using the FastQC program (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/, 7 November 2015, date accessed).
2.3.3. Illumina MiSeq RNA sequencing
The RNA-Seq library was sequenced with MiSeq Reagent Kit V3 (2 × 300 bp cycles) on MiSeq NGS system according to the MiSeq System User Guide (Illumina Japan) and fastq files were generated from both ends of the fragments.
2.4. Assembly, sequence trimming and repeat masking
2.4.1. WGS assembly
Illimina PE reads, 454 single-end reads and 454 long paired-end reads were hybrid assembled by the de novo assemble algorithm of CLC Assembly Cell ver. 3.2.2 installed on a linux server with a main memory of 256 Gb.
2.4.2. Sequence trimming and repeat masking
Human, fungi (33 species) and microbial (2,777 species) nucleotide sequence data were obtained from NCBI. Each assembled contig sequence was queried on sequences of these species, and the contigs showing >80% identity and >50% of cumulative coverage by blastn16 were assumed as alien sequences against the barley genome. For repeat masking, fasta files of Triticeae repeat sequence database (TREP Release 10: n = 1,717) were downloaded from GrainGenes (http://wheat.pw.usda.gov/ITMI/Repeats/, 7 November 2015, date accessed). In addition, new repetitive elements were generated from the Haruna Nijo genome assembly by RepeatModeler (http://www.repeatmasker.org/RepeatModeler.html, 7 November 2015, date accessed). These two libraries were used for repeat masking the Haruna Nijo assembly by censor17 with -mode norm mode. For the analysis of repeat distribution, only results of repeat masking by the TREP library were used. For comparison, genomes of Triticum aestivum,18 Triticum urartu19 and Aegilops tauschii20 were repeat masked in a similar manner as the Haruna Nijo genome.
2.5. FLcDNA/RNA-Seq mapping
2.5.1. FLcDNA mapping
Of the two independent projects,13,14 5,006 FLcDNAs were retrieved from NCBI13 and 23,614 FLcDNAs14 were downloaded from the bex-db (http://barleyflc.dna.affrc.go.jp/bexdb/).21 Morex genome assembly (contigs) was retrieved from the PGSB PlantsDB database (http://pgsb.helmholtz-muenchen.de/plant/plantsdb.jsp, 7 November 2015, date accessed).3 FLcDNAs were mapped both on the Haruna Nijo scaffolds and on the Morex contigs using blast+ with the parameters ‘-task blastn -evalue 10e−10 -lcase_masking’, and with est2genome22 in the EMBOSS package with the parameters ‘-align –mode both -gappenalty 8 -mismatch 6 -minscore 10’. FLcDNAs mapped to scaffolds with >95% identity and >50% coverage were accepted. Mapped regions with the highest identity were adopted for each FLcDNA. Overlapped FLcDNA sequences were clustered onto a locus on Haruna Nijo genome.
2.5.2. RNA-Seq analysis
After trimming of low-quality nucleotide and adapter sequences by Trimmomatic,23 reads derived from rRNA sequences were discarded by Bowtie2.24 A set of programs, Bowtie2,24 TopHat225 and Cufflinks,26 were used to map reads on the Haruna Nijo scaffolds. Gene structures derived from FLcDNA mapping were utilized to develop consensus gene models of the four library reads. To predict ORFs on the gene structures, blastx searches were conducted to RefSeq and UniProtKB data sets. From the best hit of blastx results, the most reliable ORFs were determined with at least 70 amino acids in length. To assign gene function, InterProScan 527 was conducted on the predicted ORFs. Based on the InterPro domain, GO terms were assigned to each ORF, and GOslim28 (http://agbase.msstate.edu/cgi-bin/tools/goslimviewer_select.pl, 7 November 2015, date accessed) was conducted.
2.5.3. RNA gene prediction
From the silva database29 (http://www.arb-silva.de/, 7 November 2015, date accessed), 17 barley rRNA sequences were downloaded. They were mapped on the Haruna Nijo scaffolds using blast+ with the parameters ‘-task blastn -evalue 0.01 -lcase_masking -num_descriptions 100’ and blast hits with >98% identity and >50% coverage were assumed as rRNAs. tRNA genes were predicted using the tRNAscan-SE ver. 1.3.1 program.30 Any tRNAs that were annotated as ‘possible pseudogenes’ were not counted.
2.5.4. Genome sequence comparison between Haruna Nijo and Morex
The Haruna Nijo scaffolds were mapped on the Morex contigs using megablast with the parameters ‘-evalue 10e−10 -num_descriptions 5’. Since several query sequences were hit on the same target sequence, the best hit regions were identified. If more than one Morex contig was positioned on a Haruna Nijo scaffold, these contigs were aligned on the scaffold allowing 10% overlap of the contig sequences.
2.5.5. Morex gene mapping
High confidence (HC) and low confidence (LC) genes of Morex were downloaded from the PGSB barley genome database and mapped to the Haruna Nijo scaffolds by GMAP31 with >95% identity and >90% coverage.
3. Results and discussion
3.1. Assembly of the Haruna Nijo genome sequence
We analysed different platforms of WGS sequences of Haruna Nijo by hybrid assembly. A total of 305 Gbp of sequence data were generated by Illumina HiSeq paired-end reads and 454 Titanium long paired-end reads (8 K and 20 K) (Table 1). The hybrid assembly of Illumina and 454 reads, which also include the published 454 Titanium single-end reads (25Gbp),3 generated a total of 2,055,601,874 bp in 1,753,384 scaffolds (contigs and gaps). After sequence trimming, the Haruna Nijo genome size was 2,005,970,762 bp in 1,712,236 scaffolds (Table 2). The N50 was ∼3.5 kb, which is 2.5 times larger than the published Morex WGS contigs.3 The average sequence length in the Haruna Nijo scaffolds was longer than the Morex WGS contigs (Supplementary Fig. S1). Compared with Morex WGS contigs, which were assembled with only Illumina paired-end reads, the longer Haruna Nijo WGS assembly is likely due to assembling the Illumina paired-end reads and 454 longer single-end reads.
Table 1.
Sources of WGS sequencing data for Haruna Nijo
| Method | Read | No. of reads | No. of bases (bp) |
|---|---|---|---|
| Illumina | Paired end (PE) | 3,005,632,276 | 298,945,920,500 |
| 454 | PE 20 kb | 14,242,510 | 4,569,072,624 |
| 454 | PE 8 kb | 2,955,788 | 1,016,746,348 |
| 454 | Single end 500 bp3 | 70,936,197 | 25,410,875,001 |
Table 2.
Results of whole-genome assembly of Haruna Nijo and Morex3
| Data | Haruna Nijo | Morex |
|---|---|---|
| Total length (bp) | 2,005,970,672 | 1,868,648,155 |
| N50 bp | 3,539 | 1,425 |
| No. of contigs/scaffolds | 1,712,236 | 2,670,738 |
| No. of ‘N’ | 225,899,677 | 50,995,654 |
| No. of non-‘N’ | 1,780,070,995 | 1,817,652,501 |
| GC (%) | 45.0 | 44.4 |
3.2. Repeat content of the Haruna Nijo genome
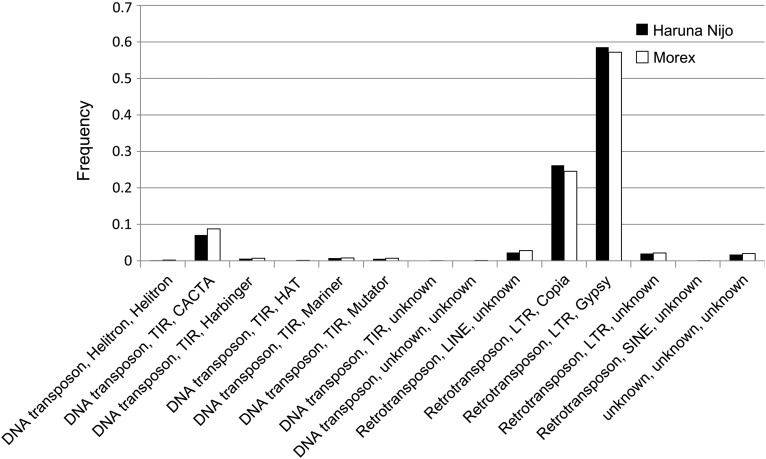
Previous studies have shown that the barley genome contains ∼80% repeated elements.3 In the published WGS assemblies,3 the relative frequency was reduced to ∼60%, due to the degradation of redundant repeated sequences. On the assembly of Haruna Nijo WGS data, repeat analysis by the Triticeae repeat library (TREP) detected 55.9% as repeated sequences. After deleting unknown genomic regions (‘N’ sites), 60.8% of the assembled sequences were classified as repeated sequences. The composition and distribution of repeat sequence categories were similar between Haruna Nijo and Morex (Fig. 1). Noteworthy, Gypsy, Copia retrotransposons and the CACTA DNA transposon showed higher frequencies than those normally observed in other Triticeae species. When we compared repeat categories from Haruna Nijo, T. aestivum (common wheat) cv. Chinese Spring, T. urartu and A. tauschii, the frequency of Copia in Haruna Nijo was higher than those in the wheat genomes, while Haruna Nijo contained more CACTA and less Gypsy than those in wheat genomes (Supplementary Fig. S2).
Figure 1.
Distribution of TREP repeat categories on the Haruna Nijo and Morex barley genome assemblies. Frequency of each repeat content was calculated by the result of repeat masking using the TREP library. Total bases of each repeat region on the genomes were divided by those of all repeat regions.
To identify unique repeat sequences in barley, de novo repeat sequences in the Haruna Nijo scaffolds were detected by RepeatModeler, which identified a total of 979 consensus sequences. The de novo repeat library and the TREP library were used for repeat masking. As a result, 74.8% of the assembled sequences were masked (‘N’s were not masked and thus deleted from the calculation). The repeat masking by the de novo library identified 68.1% repeat sequences, which were much more than the masked repeats (60.8%) detected by the TREP library.
3.3. FLcDNA mapping
FLcDNA mapping on genome sequence provides reliable exon vs. intron structures on gene models. A non-redundant set of 28,620 FLcDNA sequences13,14 was mapped to the Haruna Nijo scaffolds. Blastn mapping revealed that 27,784 FLcDNAs (97%) showed sequence similarities with the Haruna Nijo scaffolds. According to the results of the FLcDNA mapping to scaffolds, we analysed mapping coverage of 5′-end of each FLcDNA on Haruna Nijo scaffolds. The ratio of completely mapped FLcDNAs, which were mapped from the first nucleotide sequence, was <10%. This shows that most of the FLcDNAs do not have the complete sequences on the 5′-end side exon. To analyse additional mapped FLcDNAs, we relaxed the mapping position to the first 10 nucleotide sequences. Then, the ratio of mapped FLcDNA was raised to 80% (Supplementary Fig. S3). GC contents and CG-skew were analysed on the FLcDNAs having mapped 5′-ends within 10 bp. The typical high GC contents and CG-skewed peaks at the transcription start sites were identified as previously observed in rice and Arabidopsis32 (Supplementary Fig. S4).
The longer Haruna Nijo scaffolds may contribute to the efficiency in FLcDNA mapping. The number of mapped FLcDNAs under the threshold of 50% mapping coverage was 26,240 in the Haruna Nijo scaffolds and 24,261 in the Morex contigs (Table 3). These numbers included FLcDNAs that mapped to either the Haruna Nijo scaffolds (1,985) or the Morex contigs (379). However, if the threshold of mapping coverage was raised to 95%, the number of mapped FLcDNAs decreased to 14,044 on the Morex contigs, while 19,485 were still mapped on the Haruna Nijo scaffolds.
Table 3.
FLcDNA mapping on genome assemblies
| Coverage (%) | No. of hit FLcDNAs (ratio/total) |
|
|---|---|---|
| Haruna Nijo | Morex3 | |
| 50 | 26,241 (0.917) | 24,621 (0.860) |
| 70 | 22,835 (0.798) | 19,926 (0.696) |
| 80 | 21,429 (0.749) | 17,706 (0.619) |
| 90 | 20,153 (0.704) | 15,361 (0.537) |
| 95 | 19,485 (0.681) | 14,044 (0.491) |
We also analysed the coverage of FLcDNA mapped reads on the Haruna Nijo scaffolds and the Morex contigs by comparing the mapped exon numbers of FLcDNAs between Haruna Nijo and Morex. Of the 24,255 FLcDNA gene models, 19,207 (79.2%) were mapped with the same number of exons both on the Haruna Nijo scaffolds and on the Morex contigs. Of these, 3,066 gene models had more exons in Haruna Nijo than that in Morex, while 1,982 gene models had more exons in Morex than Haruna Nijo. The start position analysis also showed that the Haruna Nijo scaffolds (6,842) had more mapped 5′-end sequences than the Morex contigs (1,721).
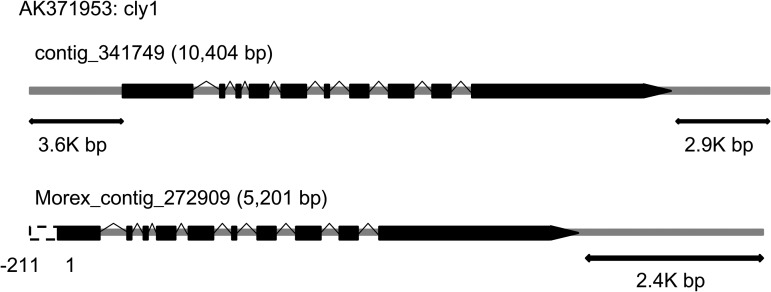
These results indicated that the available genome sequences of Morex and Haruna Nijo are different, and there may be more cases where FLcDNAs mapped with higher coverage on Haruna Nijo genomes. For example, the entire FLcDNA (AK371953) for the cleistogamy1 (cly1 for closed flowering) gene,33 which was mapped on a Haruna Nijo scaffold, while the 5′ region was not mapped on a Morex contig (Fig. 2). Moreover, flanking regions for the FLcDNA (3.6 Kb for upstream and 2.9 Kb for downstream regions) were present on the Haruna Nijo scaffold to provide the opportunity to analyse the cis-element(s) of the gene.
Figure 2.
Gene structure of closed flowering locus cly1 (AK371953). In the Haruna Nijo genome, the complete gene structures of cly1 including upstream and downstream regions were identified. In the Morex genome,3 the first exon and upstream regions were truncated because of insufficient length of the contig.
3.4. Gene annotation
The IBSC presented 26,159 HC (high confidence) and 53,220 LC (low confidence) gene models by using RNA-Seq-based transcript and FLcDNAs.3 This number of genes was larger than that of the Haruna Nijo FLcDNAs. Actually, of the 24,243 HC Morex gene models (in GFF file), only 7,645 had corresponding Haruna Nijo FLcDNAs, indicating that the FLcDNAs sequence data were not deep enough to construct a comprehensive set of barley gene models. To obtain more comprehensive transcript sequences of Haruna Nijo, RNA-Seq data from four libraries of seedling shoot, seedling root, immature spike and immature seed were collected (Table 4). As reference gene structures, 26,240 gene models on 18,226 loci determined by FLcDNA mapping with 50% coverage (Table 3) were used for mapping RNA-Seq reads. Sequences of each RNA-Seq library mapped 73.2–81.0% to each other, indicating that the concordant pair alignment rates were not much different among the libraries (Table 4). Sequences from all four libraries were combined by cuffmerge26 and 30,606 loci (51,249 gene models) were estimated. Of these, 12,390 loci and 5,635 alternative variants were added on the Haruna Nijo gene models.
Table 4.
Sequencing and mapping results of Haruna Nijo RNA-Seq data
| Sample | No. of read pairs | Aligned pairs | Concordant pair alignment rate (%) |
|---|---|---|---|
| Leaf | 4,977,702 | 3,738,352 | 75.0 |
| Root | 3,981,366 | 2,921,944 | 73.2 |
| Seed | 7,024,371 | 5,691,954 | 81.0 |
| Spike | 4,292,946 | 3,399,764 | 79.1 |
We compared the Haruna Nijo gene models with the Morex HC and LC gene models.3 We mapped 78.0% of Morex HC gene models and 72.2% of Morex LC gene models on Haruna Nijo gene models with 95% coverage and 90% identity. Of these, 1,446 Morex gene models were mapped on multiple Haruna Nijo gene models. Clustering analysis showed that 21,381 Haruna Nijo gene models overlapped with Morex gene models on the Haruna Nijo scaffolds, while 9,225 gene models did not overlap each other.
Blastx search on RefSeq/UniProt showed that 48,619 gene models of Haruna Nijo were similar to known protein sequences. In addition, 48,105 predicted ORFs on 28,422 gene models of 24,467 scaffolds were >70 aa in length. Of these, 44,133 ORFs had Met at the start positions. Noteworthy, 23,882 ORFs derived from 25,897 mapped FLcDNA contained Met at the start position.
InterProScan analysis showed that 26,956 ORFs on 19,438 loci contained 67,293 InterPro domains. While 2,760 InterPro domains were in the ‘REPEAT’ category, which may not have biological function, other 44,769 domains were ‘DOMAIN’ category (Supplementary Table S1). Moreover, GOslim analysis revealed that the gene content of Haruna Nijo (barley) and Nipponbare (rice) was quite similar (Supplementary Fig. S5).
We also predicted rRNA genes on Haruna Nijo scaffolds. Based on the silva database, nine scaffolds were identified as rRNA regions. In the case of rice, rDNA genes compose nucleolar-organizing region (NOR) in which rDNA are aligned in tandem. In wheat, the chromosome-based WGS analysis showed that the chromosome 6B contains 60% of the total rRNA gene (5,500 genes).34 However, these regions could not be assembled by NGS data35 due to the degeneration in the process of assembly. This was also true for the Haruna Nijo scaffolds which showed only nine scaffolds carrying rRNA sequences. The difficulty of assembling NOR was also reported in rice,36 but the NOR assembly in WGS might be more difficult in general.
tRNAs were also annotated by tRNAscan-SE. The spectrum of tRNA distribution across the genome was uneven but similar to the rice genome37 (Supplementary Fig. S6). It is reported that tRNALys sequences are extraordinarily abundant in wheat chromosome 6B due to the abundance of the retrotransposon Gypsy.35 The Haruna Nijo scaffolds also have higher rate of Gypsy; however, the number of tRNAs was smaller than that in wheat.
3.5. WGS assembly comparison between Haruna Nijo and Morex
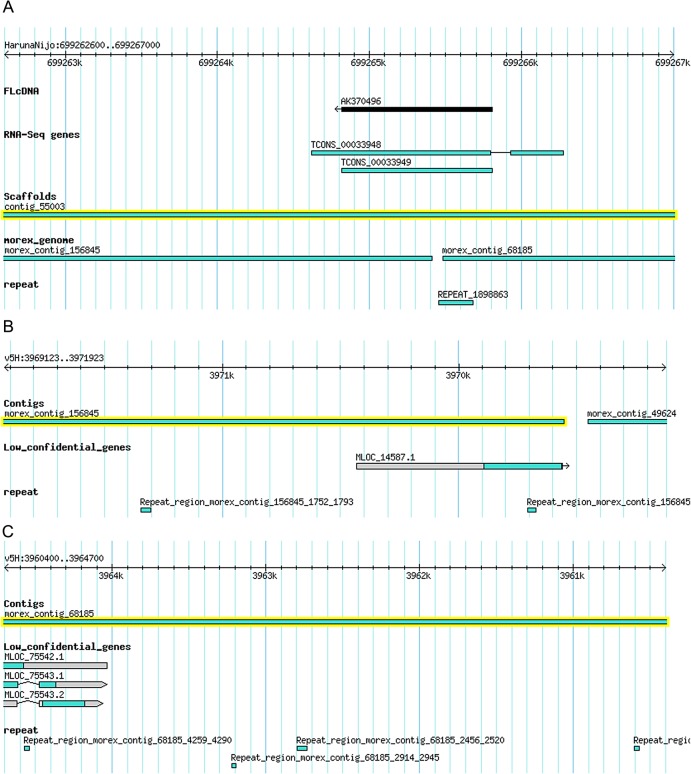
There are several published3 and ongoing efforts of sequencing BACs derived from Morex. BAC-based assemblies may provide longer sequences than WGS assemblies to estimate gene models on the genome. The final goal for developing Haruna Nijo gene models is to map them on the BAC-based Morex genome sequences. To understand the quality of current Haruna Nijo genome resources, sequences of the Haruna Nijo scaffolds and the Morex contigs were aligned by megablast. An average sequence identity between Haruna Nijo and Morex was estimated as 95.99% (Supplementary Table S2). We found that mapped regions of query were different when query and database sequences were exchanged between Haruna Nijo and Morex. This was caused by the difference of sequence length in two assemblies. In this analysis, we used the best hit of megablast for the calculation, and the other hits with lower coverage on Haruna Nijo scaffolds were discarded. Therefore, the total length of aligned regions was decreased. Genome comparison of Haruna Nijo and Morex identified the occasions that the genomic regions of Haruna Nijo and Morex do not match. An example of mismatch between two assemblies on chromosome 5H was that a region of the Haruna Nijo gene model of TCONS_00033948 or TCONS_00033949 (Fig. 3A), which is supported by a FLcDNA, AK370496 that encodes bZIP transcription factor family protein on a contig_55003 (scaffold), was divided into two loci of MLOC_14578.1 on morex_contig_156845 (Fig. 3B) and MLOC_75543 on morex_contig_68185 (Fig. 3C). Thus, we may find a better gene model if we compare multiple gene models derived from different assemblies.
Figure 3.
Example screenshots of (A) Haruna Nijo contig_55003, (B) Morex contig_156845 and (C) Morex contig_68185 under the platform of Gbrowse. Separated genomic regions of Morex which correspond to the single FLcDNA region in Haruna Nijo. This figure is available in black and white in print and in colour at DNA Research online.
We also analysed conserved genic regions between Haruna Nijo and Morex by using mapped regions of FLcDNAs. The difference between Haruna Nijo scaffolds and FLcDNA sequences was 0.05% (1 bp of 2000bp in exon), which is still five times higher than the value of 0.01% in rice genome.37 However, the sequence difference of exon regions between Haruna Nijo and Morex (identified by Haruna Nijo FLcDNAs) was 0.29%, indicating that the sequence quality of scaffolds in Haruna Nijo was high compared with the haplotype difference between Haruna Nijo and Morex.
3.6. Conclusion
Based on this study, there are several advantages of using the Haruna Nijo sequence resources. The Haruna Nijo scaffolds provide a high-quality genome sequence for the barley research community. In particular, the FLcDNA sequences, which are available only for Haruna Nijo, provide a key resource. In addition, the gene structures of Haruna Nijo provide supplementary data sets to the Morex genome sequence. Finally, the haplotype of Haruna Nijo is popular among the malting barley cultivars in the world and can be used as an alternative source of alleles for important traits in barley.
4. Availability
The short genomic reads used in the study are deposited at DDBJ-SRA under accession ID PRJDB4103. The scaffolds for the Hordeum vulgare cv. Haruna Nijo are available at http://barleyflc.dna.affrc.go.jp/bexdb/pages/harunanijo_index.jsp for download. Genome browsing of the Haruna Nijo gene models are available on GBrowse [(http://gmod.org/wiki/GBrowse)38 at http://barleyflc.dna.affrc.go.jp/gb2/gbrowse/HarunaNijo_genome/.21]. All scaffolds were concatenated by 100 ‘Ns’ from the longer scaffolds. Haruna Nijo scaffolds, FLcDNAs, RNA-Seq genes, repeat information, Morex contigs and Morex HC/LC genes are displayed (Fig. 3). From the Haruna Nijo genome browser, FLcDNA annotation data and Morex genome browser of bex-db could be accessed (Fig. 4). Moreover, users can access the Haruna Nijo genome from FLcDNA information of bex-db.
Figure 4.
Example screenshots of bex-db (http://barleyflc.dna.affrc.go.jp/bexdb/) for a FLcDNA (AK370496). This figure is available in black and white in print and in colour at DNA Research online.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
Funding to pay the Open Access publication charges for this article was provided by Okayama University to Kazuhiro Sato, corresponding author.
Supplementary Material
Acknowledgements
We would like to express our thanks to Prof. Gary Muehlbauer, University of Minnesota for his critical reading and linguistic editing of the manuscript. Barley seeds were provided through the National Bioresource Project of Barley, MEXT of Japan. Analysis of Illumina HiSeq 2000 sequence reads was performed by the Japan Advanced Plant Science Research Network (http://www.psr-net.riken.jp/index.html). This work was partly supported by the scientific technique research promotion program for agriculture, forestry fisheries and food industry to K.S.
References
- 1.Pourkheirandish M., Hensel G., Kilian B. et al. 2015, Evolution of the grain dispersal system in barley, Cell, 162, 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Sato K., Flavell A., Russell J., Börner A., Valkoun J.. 2014, In: Kumlehn J., Stein N. (eds), Biotechnological approaches to barley improvement, biotechnology in agriculture and forestry. Springer: Heidelberg, pp.21–36. [Google Scholar]
- 3.The International Barley Genome Sequencing Consortium. 2012, A physical, genetic and functional sequence assembly of the barley genome, Nature, 491, 711–6. [DOI] [PubMed] [Google Scholar]
- 4.Ariyadasa R., Mascher M., Nussbaumer T. et al. 2014, A sequence-ready physical map of barley anchored genetically by two million SNPs, Plant Physiol., 164, 412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascher M., Muehlbauer G.J., Rokhsar D.S. et al. 2013, Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ), Plant J., 76, 718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng X., Long H., Wang Z. et al. 2015, The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau, Proc. Natl Acad. Sci. USA, 112, 1095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaneko T., Zhang W.S., Ito K. et al. 2001, Worldwide distribution of β-amylase thermostability in barley, Euhytica, 121, 225–8. [Google Scholar]
- 8.Gong X., Westcott S., Zhang X. et al. 2013, Discovery of novel Bmy1 alleles increasing β-amylase activity in Chinese landraces and Tibetan wild barley for improvement of malting quality via MAS, PLoS ONE, 8, e72875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins H., Panozzo J., Logue S. et al. 2003, Mapping and validation of chromosome regions associated with high malt extract in barley (Hordeum vulgare L.), Aust. J. Agr. Res., 54, 1223–40. [Google Scholar]
- 10.Sato K., Nankaku N., Takeda K.. 2009, A high-density transcript linkage map of barley derived from a single population, Heredity (Edinb), 103, 110–7. [DOI] [PubMed] [Google Scholar]
- 11.Close T.J., Bhat P.R., Lonardi S. et al. 2009, Development and implementation of high-throughput SNP genotyping in barley, BMC Genomics, 10, 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saisho D., Myoraku E., Kawasaki S., Sato K., Takeda K.. 2007, Construction and characterization of a bacterial artificial chromosome (BAC) library for Japanese malting barley ‘Haruna Nijo’, Breed. Sci., 57, 29–38. [Google Scholar]
- 13.Sato K., Shin I.T., Seki M. et al. 2009, Development of 5006 full-length CDNAs in barley: a tool for accessing cereal genomics resources, DNA Res., 16, 81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto T., Tanaka T., Sakai H. et al. 2011, Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries, Plant Physiol., 156, 20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato K., Close T.J., Bhat P. et al. 2011, Development of genetic map and alignment of recombinant chromo-some substitution lines from a cross of EST donors by high accuracy SNP typing in barley, Plant Cell Physiol., 52, 728–37. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990, Basic local alignment search tool, J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 17.Jurka J., Klonowski P., Dagman V., Pelton P.. 1996, CENSOR-a program for identification and elimination of repetitive elements from DNA sequences, Comput. Chem., 20, 119–21. [DOI] [PubMed] [Google Scholar]
- 18.The International Wheat Genome Sequencing Consortium. 2014, A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome, Science, 345, 1251788. [DOI] [PubMed] [Google Scholar]
- 19.Ling H.Q., Zhao S., Liu D. et al. 2013, Draft genome of the wheat A-genome progenitor Triticum urartu, Nature, 496, 87–90. [DOI] [PubMed] [Google Scholar]
- 20.Jia J., Zhao S., Kong X. et al. 2013, Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation, Nature, 496, 91–5. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T., Sakai H., Fujii N. et al. 2013, bex-db:Bioinformatics workbench for comprehensive analysis of barley-expressed genes, Breeding Sci., 63, 430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mott R. 1997, EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA, Comput. Appl. Biosci., 13, 477–8. [DOI] [PubMed] [Google Scholar]
- 23.Bolger A.M., Lohse M., Usadel B.. 2014, Trimmomatic: a flexible trimmer for Illumina Sequence Data, Bioinformatics, 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Salzberg S.L.. 2012, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. 2013, ToHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C., Williams B.A., Pertea B. et al. 2010, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotech., 28, 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P., Binns D., Chang H.Y. et al. 2014, InterProScan 5: genome-scale protein function classification, Bioinformatics, 30, 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy F.M., Wang N., Magee G.B. et al. 2006, AgBase: a functional genomics resource for agriculture, BMC Genomics, 7, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C., Pruesse E., Yilmaz P. et al. 2013, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 41, D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe T.M., Eddy S.R.. 1997, tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence, Nucleic Acids Res., 25, 955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu T.D., Watanabe C.K.. 2005, GMAP: a genomic mapping and alignment program for mRNA and EST sequences, Bioinformatics, 21, 1859–75. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka T., Koyanagi K.O., Itoh T.. 2009, Highly diversified molecular evolution of downstream transcription start sites in Oryza sativa and Arabidopsis thaliana, Plant Physiol., 149, 1316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair S.K., Wang N., Turuspekov Y. et al. 2010, Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage, Proc. Natl Acad. Sci. USA, 107, 490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flavell R.B., O'Dell M.. 1976, Ribosomal RNA genes on homoeologous chromosomes of groups 5 and 6 in hexaploid wheat, Heredity, 37, 377–85. [Google Scholar]
- 35.Tanaka T., Kobayashi F., Joshi G.P. et al. 2014, Next-generation survey sequencing and the molecular organization of wheat chromosome 6B, DNA Res., 21, 103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuno H., Sasaki T., Matsumoto T.. 2008, Characterization of internal structure of the nucleolar organizing region in rice (Oryza sativa L.), Cytonegetic Genome Res., 121, 282–5. [DOI] [PubMed] [Google Scholar]
- 37.International Rice Genome Sequencing Project. 2005, The map-based sequence of the rice genome, Nature, 436, 793–800. [DOI] [PubMed] [Google Scholar]
- 38.Stein L.D., Mungall C., Shu S.Q. et al. 2002, The Generic genome browser: a building block for a model organism system database, Genome Res., 12, 1599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.