Abstract
Clubroot is a devastating disease caused by Plasmodiophora brassicae and results in severe losses of yield and quality in Brassica crops. Many clubroot resistance genes and markers are available in Brassica rapa but less is known in Brassica oleracea. Here, we applied the genotyping-by-sequencing (GBS) technique to construct a high-resolution genetic map and identify clubroot resistance (CR) genes. A total of 43,821 SNPs were identified using GBS data for two parental lines, one resistant and one susceptible lines to clubroot, and 18,187 of them showed >5× coverage in the GBS data. Among those, 4,103 were credibly genotyped for all 78 F2 individual plants. These markers were clustered into nine linkage groups spanning 879.9 cM with an average interval of 1.15 cM. Quantitative trait loci (QTLs) survey based on three rounds of clubroot resistance tests using F2 : 3 progenies revealed two and single major QTLs for Race 2 and Race 9 of P. brassicae, respectively. The QTLs show similar locations to the previously reported CR loci for Race 4 in B. oleracea but are in different positions from any of the CR loci found in B. rapa. We utilized two reference genome sequences in this study. The high-resolution genetic map developed herein allowed us to reposition 37 and 2 misanchored scaffolds in the 02–12 and TO1000DH genome sequences, respectively. Our data also support additional positioning of two unanchored 3.3 Mb scaffolds into the 02–12 genome sequence.
Keywords: cabbage, genotyping-by-sequencing, genetic linkage map, clubroot, QTL
1. Introduction
Since the completion of the first genome sequence of a plant, Arabidopsis thaliana,1 next-generation sequencing (NGS) technologies have facilitated genome sequencing and construction of reference genome sequences of many important crops.2 The reference genome sequences provide valuable information for understanding genomic variation and genome evolution. Based on the utility of reference genome sequences, detection of single-nucleotide polymorphisms (SNPs) has become a valuable tool for genomic selection in plant molecular breeding due to their genome-wide abundance. Accordingly, rapid and inexpensive SNP detection methods are required for genotyping large and diverse germplasm collections or breeding populations. Genotyping by sequencing (GBS) is one such highly efficient method for genome-wide SNP detection with relatively low cost and time demands, because genome-wide reduced representation sequencing is conducted using NGS technology.3
An important strength of GBS is that discovery of huge numbers of markers and genotyping of those markers occur simultaneously. GBS has been applied for construction of high-resolution genetic maps, association study for agricultural traits, and genomic prediction for development of breeding materials using populations of interest in several crop species, such as maize,3–5 soybean,6 barley,3,7 rice,8 and wheat.7 Finally, the SNP data deduced from GBS analysis can be used for genomics-assisted breeding. GBS approaches have largely been applied to crops with well-sequenced reference genomes. In further, GBS approach successfully adapted for genetic mapping without reference genome sequences.9
Brassica oleracea is one of the three basic diploid Brassica species belonging to the evolutionary U's triangle10 and is an economically important vegetable crop consumed worldwide. Brassica oleracea shows extremely diverse morphology and includes vegetables such as cabbages, broccoli, cauliflowers, kohlrabi, kalian, and kales. Recently, two reference genome sequences of B. oleracea were reported; two reference pseudochromosome sequences, of 385 and 447 Mb, were independently built by positioning and ordering of de novo-assembled scaffold sequences using 1,227 PCR-based markers11,12 and 15,909 GBS-based markers,13 respectively.
Clubroot is a devastating disease caused by Plasmodiophora brassicae that results in severe losses of yield and quality not only in cabbage but also in other Brassica crops.14 The pathogen induces gall formation on the root, leading to suppressed transport of nutrients and water, and finally abnormal growth and yield loss.15 Because the pathogen can survive in soil as resting spores without host plants for long periods, it is difficult to control by cultural practices or chemical treatments.16 Therefore, development of cultivars with clubroot resistance (CR) has been considered the most effective method to minimize yield loss caused by clubroot disease.
In B. rapa, at least eight race-specific CR loci have been introduced from various European fodder turnips,17 and the markers for Crr1, Crr2, Crr3, Crr4, CRa, CRb, CRc, and CRk18–21 have been identified in five different chromosomes. To increase efficiency in marker-assisted selection (MAS), fine mapping was carried out for each locus in several B. rapa studies.22–25 In contrast to B. rapa, completely resistant accessions have only rarely been identified in B. oleracea.17,26 In B. rapa, the CR traits are controlled in a qualitative27,28 and quantitative manner20,21,29 depending on genotypes studied, whereas genetic studies suggest that CR are controlled by many minor QTLs in B. oleracea.17,30 DNA markers linked to CR loci in B. oleracea have been developed by several research groups, but most of these studies did not disclose specific sequence information for the various marker types. To establish a CR breeding system in B. oleracea, it is important to identify the major CR genes active to various CR races.
In this study, we generated large-scale marker data sets using GBS analysis of 80 plant samples. The robust SNP marker sets developed in this study were valid for construction of a high-density genetic map and for QTL mapping of CR traits. In addition, the resulting high-resolution linkage map provided valuable information for identification of incorrectly anchored scaffold sequences and further improvement of reference genome assembly for B. oleracea. Finally, the QTLs identified herein, and the genetic map will be valuable for B. oleracea crop improvement.
2. Materials and methods
2.1. Plant materials
Two cabbage (B. oleracea L. var. capitata) inbred lines, C1176 and C1220, were selected as parents to develop a mapping population. The two lines show different susceptibility to clubroot disease caused by P. brassicae; C1176 is susceptible, whereas C1220 is resistant. The mapping population consisted of 78 F2 plants generated by crossing between C1220 and C1176. The 78 F2 plants were vernalized and self-pollinated to produce F3 progenies for CR tests. All plant materials examined in this study were obtained from Joeun Seeds Co. (Chungcheongbuk-Do, Korea).
Genomic DNAs from parental lines and F2 plants were extracted from 2 g samples of young leaves, following the modified cetyltrimethylammonium bromide (CTAB) protocol.31 The quality and quantity of the DNA were examined using a NanoDrop ND-1000 (Thermo Fisher Scientific Inc., USA).
2.2. Library construction and sequencing
Genomic DNAs of all 80 plants (two parents and 78 F2) were double digested with two different restriction endonucleases, ApeKI and MspI. The former recognizes a 5-bp sequence (GCWGC) and creates a 3-bp overhang, while the latter recognizes 4-bp sequence (CCGG) and creates a 2-bp overhang. A set of barcode adapters that recognize ApeKI-compatible sequences with variable barcode sequences (4–9 bp) for each different plant and a set of common adapters that recognize MspI-compatible sequences were ligated to the digested DNA of each plant. A total of 40 plant samples were pooled as a batch for further library construction steps, such as PCR amplification with adaptor-specific primers, and sequencing on two single lanes of Illumina Hi-seq 2000. DNA digestion, adapter ligation, library construction, and sequencing were carried out by Macrogen Co. (Korea).
2.3. Raw sequence data processing and SNP genotyping
Raw paired-end (PE) sequences were filtered based on criteria for sequence quality and length using the program clc_quality_trim V4.3.0.114910 (http://www.clcbio.com). Sequence reads that had <20 in sequence quality score and 50 bp of sequence length were removed, and then barcode sequences were eliminated. Filtered reads, which were ∼93 bp on average, were aligned to the reference sequences of B. oleracea var. capitata homozygous line 02–12 (02–12 reference genome) reported by Liu et al.12 using the Burrows–Wheeler alignment tool (BWA)32 with default parameters, and properly paired reads were sorted by the parameter of maximum insert size (800 bp). Read grouping and removal of PCR duplicates were done using Picard (http://picard.sourceforge.net). Genome analysis toolkit (GATK) was used to perform local realignment of reads to correct misalignments due to the presence of INDELs. Finally, UnifiedGenotyper,33 which is one of the utilities in the GATK package, was used for detection of SNPs and calling of SNP genotypes. Additional filtering and genotyping processes were carried out with user-friendly option changes in the web-interfaced GBS Genotyping Pipeline, and the final genotype data were graphically displayed using GBS Browser (http://www.phyzen-lab.com/oleracea) developed by Phyzen Genomics Institute (Korea).
2.4. Molecular marker analysis
We analysed 32 markers for genotyping as position markers for comparison with the GBS genotyping results: 2 EST-based dCAPS, 9 whole-genome resequencing-based dCAPS,34 3 EST-SSRs,35 16 genomic SSRs,20,36–40 and 2 polymorphic markers based on miniature inverted transposable element (MITE) insertion polymorphism (MIP)41,42 (Supplementary Table S1). The amplicons of the markers were analysed by visualization on a UV trans-illuminator after electrophoresis and stained with ethidium bromide using 9% non-denaturing polyacrylamide gels or 1% agarose gels depending on amplified fragment size.
2.5. Genetic map construction
Detected GBS-SNP markers were grouped and named based on the position on the reference chromosome sequences. Genotype results of all markers were listed for each F2 individual in order of position on pseudochromosomes. The genotyping scores of GBS markers and PCR-based markers were integrated, and linkage analysis and map construction were performed using JoinMap version 4.1 with the same parameters as in previous studies.34,35 Genetic and physical map positions showed the same order for most GBS-SNPs, but some SNPs were assigned different positions between the genetic map and the physical map. When the genetic mapping position of the GBS-SNP did not coincide with the physical mapping at the pseudomolecule reference genome sequence, we considered these differential positions as misallocated. These areas were named as sequence variance blocks (v-blocks) if the positions of genetic map and physical map were differential for more than three contiguous SNPs, and as small variance blocks (s-blocks) if only one or two contiguous SNPs showed different positions between the genetic and physical maps (Supplementary Fig. S1A and B). Additionally, we divided the pseudomolecule into different blocks if it was interrupted by v-blocks or showed 10–15% recombination between adjacent markers (Supplementary Fig. S1C and B). We labelled each block in alphabetical order, and the information for each block on the reference genome is described in Supplementary Table S2.
2.6. Clubroot resistance test and QTL analysis
Inoculum preparation and resistance tests were performed by the Korea Research Institute of Chemical Technology (Daejeon, Korea). Two isolates of P. brassicae, YC and GN, which were collected in Yeoncheon and Gangneung, Korea, respectively, were determined as Race 2 and 9, respectively,43 based on Williams classification.44 The pathogen was inoculated onto roots of Chinese cabbage, and the induced clubs were collected from infected roots and stored at −20°C until needed. For preparation of inocula, the clubs were ground in distilled water using a blender, and homogenized tissue was sifted through four-layered gauze. The resting spore suspensions of YC and GN isolates were diluted with sterile distilled water for inoculation. The final resting spore concentration of YC and GN was adjusted to 1.4 × 108 and 0.4 × 108 spores/ml, respectively.
A total of three inoculation tests were performed, two with GN isolate in 2012 and 2014, and one with YC isolate in 2014. All analyses were carried out under the same conditions with 10–15 F2 : 3 plants of each individual F2 line randomly selected for genotyping analysis. The F3 seeds were sown and grown in 5 × 8 plastic pots for 2 weeks in a greenhouse, then the F3 plants were inoculated by applying 5 ml spore suspension at the bottom of the stem base using pipettes. Each plastic pot (40 plants) received 80 ml spore suspension, and the inoculated plants were moved into a dew chamber with the temperature set at 20°C. After incubation for 1 week, the plants were supplied with water from the bottom of the pots for 3 days and then water was withhold for another 4 days. The inoculated plants were transferred to a greenhouse at 20–25°C. The plants were evaluated for clubroot infection 5 weeks after the transfer. After the roots were completely washed, the severity of the clubroot symptoms was scored as follows; 0 = no visible clubs, 1 = a few slight clubs usually confined to lateral roots, 2 = moderate clubbing on lateral roots, 3 = larger clubs in main roots and slight clubs on lateral roots, and 4 = severe clubbing on main roots and lateral roots.
A disease index for each F2 individual was calculated as the mean score of 10–15 F3 seedlings. QTLs for resistance against P. brassicae were evaluated using composite interval mapping (CIM) analysis with 0.5 cM scan intervals using the QGene program. CIM was performed with LOD (logarithm of odds) threshold values that were estimated using 1,000 permutation tests at 5% significance.
2.7. Comparative analysis of CR loci
The genomes of B. oleracea and B. rapa share a set of 24 conserved chromosomal blocks.45 The complete B. oleracea draft genome also demonstrates generally strong conservation with B. rapa in large segments at the pseudomolecule level.12,13 Based on comparison with the chromosomal block reported in Liu et al.,12 syntenic blocks were compared between B. rapa and B. oleracea. We compared the loci of reported CR genes and QTLs in B. rapa20–23,46,47 and B. oleracea30 to our CR-QTLs, using basic local alignment search tool (BLAST).
3. Results
3.1. Sequence production and alignment
A total of ∼824 million PE reads (83.2 Gb) were generated using the Illumina sequencing platform for 80 plants, with an average of 10.3 million reads per plant sample (Table 1). After removing low-quality reads and barcode sequences, 45.9 Gb of sequence data remained, with an average read length of 93.5 bp. Among 80 samples, the read amounts ranged between 256 and 897 Mb (Fig. 1A). Based on mapping on the 02–12 genome sequence,12 we classified the raw data as discarded reads (because of lack of paired read alignment on the reference genome sequence), unmapped reads, or properly aligned reads. Although the amount of data produced from each plant sample was different, the proportion of discarded reads, unmapped reads, and properly aligned reads was similar for each individual plant (Fig. 1B). In total, 66.4% (30.5 Gb) of the filtered sequence data was mapped on the reference genome, which accounted for an average of 380.9 Mb per each plant. From the mapped sequence, we did not use the reads on multi-copy regions and instead used only unique mapped reads for further analysis, representing 40.5% (a total of 18.6 Gb, which accounted for an average of 232.4 Mb per plant) of raw reads.
Table 1.
Overview of GBS sequence data and alignment to the reference sequence
| Total | Average/plant | |
|---|---|---|
| Raw data | ||
| Reads | 823,976,286 | 10,299,704 |
| Bases (bp) | 83,221,604,886 | 1,040,270,061 |
| After filtering and removing barcodes | ||
| Reads | 491,664,470 | 6,145,806 |
| Bases (bp) | 45,888,404,252 | 573,605,053 |
| Mapped reads on reference genome | ||
| All mapped | ||
| Reads | 326,477,878 | 4,080,973 |
| Bases (bp) | 30,471,082,942 | 380,888,537 |
| (66.4%) | ||
| Unique mapped | ||
| Reads | 199,164,830 | 2,489,560 |
| Bases (bp) | 18,588,604,200 | 232,357,552 |
| (40.5%) | ||
Figure 1.
Distribution of sequencing data for each sample after filtering and mapping ratio of generated data. This figure is available in black and white in print and in colour at DNA Research online.
3.2. SNP discovery and genotyping
Brassica oleracea genome underwent whole-genome triplication ∼15.9 million yrs ago.12,48 To avoid incorrect genotyping by mismapping of reads in paralogous regions instead of the original position, we used the correctly mapped PE reads for which both reads were uniquely mapped with correct direction and interval on the reference genome. A total of 43,821 SNPs were detected between the two parental lines, and 18,187 of those remained after removing SNPs whose the minimum mapping depth was lower than five (Table 2). The SNPs were distributed throughout the nine chromosomes, with the most SNPs (3,261) on chromosome C03 and the fewest (1,458) on chromosome C02.
Table 2.
Summary of SNPs detected between C1220 and C1176
| Chr | SNPs between C1220 and C1176 |
Genotyped SNPs in 78 F2 population (Genetic bins)a | |
|---|---|---|---|
| Detected | Filtered | ||
| C01 | 4,381 | 1,805 | 410 (79) |
| C02 | 4,053 | 1,458 | 306 (64) |
| C03 | 7,151 | 3,261 | 865 (132) |
| C04 | 4,149 | 1,834 | 432 (87) |
| C05 | 4,478 | 1,924 | 441 (68) |
| C06 | 5,360 | 2,103 | 434 (84) |
| C07 | 4,787 | 1,953 | 455 (78) |
| C08 | 5,532 | 2,094 | 388 (71) |
| C09 | 3,930 | 1,755 | 372 (72) |
| Total | 43,821 | 18,187 | 4,103 (735) |
aGrouping same genotyping results with adjacent markers.
Among 18,187 SNP sites, we selected 4,103 SNPs that were genotyped for all 78 F2 plants as the final data set for genetic mapping, to reduce uncertainty of placement potentially conferred by loci derived from missing data. We grouped GBS markers showing the same genotype as genetic bins, or loci, to simplify the genetic map (e.g. the first locus of linkage group C1, C1a-1(7), represents seven GBS-SNPs C1a-1 to C1a-7 in tandem) (Table 2). We noted the number of SNPs and bins per each 3 Mb along the nine chromosomes, revealing biased distribution for SNP density and genetic recombination bins. Generally, SNP density and genetic recombination bins were richer in euchromatic regions near the telomere but rare in heterochromatic regions near the centromere of each chromosome (Fig. 2).
Figure 2.
Distribution of GBS-SNPs and bins in 3 Mb intervals along nine pseudochromosomes of 02–12 genome sequences. This figure is available in black and white in print and in colour at DNA Research online.
3.3. High-density genetic mapping and refinement of two reference genomes sequence
A genetic map was built based on F2 genotyping results of 735 GBS loci represented by 4,103 markers and 32 PCR-based reported markers used for identifying linkage groups (Table 3, Fig. 3). All genotyped markers were allocated into nine linkage groups. Nine linkage groups spanned 879.9 cM with an average distance of 1.15 cM between neighbouring loci. The biggest gap in the genetic map was 15.8 cM in linkage group C02.
Table 3.
Distribution of GBS loci and PCR-based markers on the cabbage genetic map
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| GBS bin (GBS-SNPs) | ||||||||||
| Derived from target chr. | 74 (382) | 55 (262) | 124 (833) | 79 (392) | 66 (434) | 77 (405) | 69 (427) | 65 (371) | 70 (359) | 679 (3,876) |
| Derived from other chr. | 2 (7) | 8 (24) | 11 (49) | 3 (8) | 8 (37) | 4 (16) | 7 (43) | 2 (11) | 11 (43) | 56 (238) |
| Total | 76 (389) | 63 (286) | 135 (882) | 82 (400) | 74 (471) | 81 (421) | 76 (470) | 67 (382) | 81 (402) | 735 (4,103) |
| PCR-based markers | 4 | 9 | 4 | 14 | 0 | 0 | 1 | 0 | 0 | 32 |
| Total | 80 | 72 | 139 | 96 | 74 | 81 | 77 | 67 | 81 | 767 |
| Length (cM) | 97.5 | 106.1 | 139.5 | 120.3 | 85.1 | 77.9 | 68 | 67.4 | 118.1 | 879.9 |
| Average interval (cM) | 1.22 | 1.47 | 1.00 | 1.25 | 1.15 | 0.96 | 0.88 | 1.01 | 1.46 | 1.15 |
Figure 3.
Genetic linkage map of cabbage constructed using 735 GBS loci, representing 4,103 SNP markers, and 32 PCR-based markers. Markers in blue (gray) are PCR-based markers and in red (light gray) are loci derived from other pseudochromosomes. QTLs identified in inoculation tests with GN in 2012, 2014, and with YC in 2014 are shown as red (dark gray on C02 and left dark gray on C03), green (light gray), and pink (right dark gray on C03) bars, respectively. The numbers in parentheses after locus names or genetic bins refer to the number of co-segregating SNP markers. The position of the peak LOD score in each QTL is indicated by an arrowhead. This figure is available in black and white in print and in colour at DNA Research online.
The genetic map displayed overall, but with some exceptions, contiguously collinear marker order, because markers were named based on the backbone of reference genome sequence. All linkage groups included several variance blocks (v- or s-blocks), which showed disordered markers with different order or derived from different chromosomes. Chromosome C05 and C09 had only one v-block, whereas C07 included five v-blocks (Supplementary Table S2). In addition, chromosomes C02, C03, C06, C08, and C09 included at least one s-block. According to our linkage map data, we suggest that all s- and v-blocks likely should be re-allocated into the proper positions. Linkage groups C3 and C9 contained the most complex structure. Both included 11 genetic loci derived from other pseudochromosomes (Table 3 and see the marker names in red in Fig. 3). C3 included 49 GBS markers derived from seven v-blocks, C1-v4, C7-v5, C6-v1, C6-v3, C7-v1, C1-v3, and C2-v2 derived from other chromosomes (Supplementary Table S3). Most s- and v-blocks were assigned to other chromosomes, except C3-v1 and C6-s1 block, which were misallocated into different positions in the original chromosome (Supplementary Table S3 and Fig. 3).
Comparative analysis between the genetic map and the reference genome revealed the new positions for all 27 v-blocks and 10 s-blocks and several inversions of some alphabetically named blocks (Fig. 4A). The positions of GBS markers between chromosomes C04, C05, and C06 and linkage group C4, C5, and C6 suggested that there was less collinearity in those chromosomes than in the other chromosomes.
Figure 4.
Comparison between the loci in the genetic map and physical position of SNPs in nine pseudochromosomes of (A) 02–12 and (B) TO1000DH genome sequences. Misallocated scaffolds denoted with stars were compared with their corresponding sequences in the other reference genomes (Fig. 7). This figure is available in black and white in print and in colour at DNA Research online.
Based on the relative ratio between physical and genetic distance, possible heterochromatin and euchromatin regions could be inferred (represented by black and red dotted lines between chromosomes and linkage groups, respectively in Fig. 4). Presumed heterochromatin regions from chromosomes C01, C02, C03, C07, C08, and C09 occupied large portions of pseudochromosomes, but showed low frequency of recombination in genetic maps and also low density of SNPs and bins on each pseudochromosome (Fig. 2). Many v-blocks and s-genotyped markers in pseudochromosomes tended to be localized near heterochromatin regions in linkage groups (Fig. 4). Conversely, the regions showing high frequency of recombination (represented by red dotted lines in Fig. 4) were considered as euchromatin.
Another cabbage reference sequence was reported by Parkin et al.13 using the kale-like TO1000DH genotype (TO1000DH genome sequence). When we performed GBS read mapping on the TO1000DH genome sequence backbone (Fig. 4B and Supplementary Table S4), the genetic map consisted of nine linkage groups with 4,338 GBS markers (Supplementary Table S4). The genetic map identified two misanchored sequence scaffolds (v-blocks), which were mapped on the other chromosomes instead of the original position of sequences in the pseudomolecule of TO1000DH genome (Fig. 4B). We compared the v-block sequences in one unique genome with the corresponding sequence in the other reference genome sequence (denoted as star in Fig. 4). Approximately 1.5 Mb sequence of a region (39.5–41.0 Mbp) including the C7-v5 block in 02–12 genome which mapped into linkage group C3 in our map did not show corresponding sequence in chromosome C07 of TO1000DH. However, ∼673 Kb of the sequence in that region corresponded to the C3a block in chromosome C03 of TO1000DH (Fig. 5A), which is coincident with our map position (stars in Fig. 4). The remaining ∼860 Kb sequence could not be placed due to lack of GBS markers. The two mispositioned scaffolds of TO1000DH were identified in two v-blocks are derived from the heterochromatin regions with rare SNPs and a low recombination rate. Approximately 300 Kb of the C2-v block in chromosome C02 of TO1000DH was not present in the corresponding sequence of 02–12, and the mapping data suggested that the sequence should be repositioned in linkage group C8 (Figs 4B and 5B).
Figure 5.
Comparison of reference sequences containing V-blocks between 02–12 and TO1000DH genomes. (A) Only flanking sequences of the C7-v5 block in the 02–12 genome, estimated to be in euchromatin regions with abundant GBS markers, were found in the corresponding sequences of the TO1000DH genome. (B) Corresponding sequence of the C2-v block region in TO1000DH, which seemed to be in heterochromatin regions with sparse GBS markers, was not observed in chromosome C02 of the 02–12 genome. This figure is available in black and white in print and in colour at DNA Research online.
3.4. Clubroot resistance assays and QTL analysis
Three independent inoculation trials were performed for clubroot resistance, with GN isolate in 2012 and with GN and YC isolates in 2014. The disease index for F2 plants was determined by calculating the average value of the clubroot disease indices obtained from 10–15 F2 : 3 progeny plants in each inoculation test. Disease symptoms caused by YC isolate (Race 2) were much severer than those caused by GN isolate (Race 9) (Fig. 6). The disease symptoms induced by GN isolate were slightly different between the two independent tests in different years, possibly due to different environmental conditions.
Figure 6.
Disease index distribution of F2 population, evaluated by average scores resulted from inoculated F2 : 3 plants. This figure is available in black and white in print and in colour at DNA Research online.
QTL analyses were performed independently for each of the three tests. We detected significant QTLs, based on LOD scores higher than the thresholds calculated in the permutation tests; LOD threshold values for the tests with GN isolate in 2012 and 2014 were 2.982 and 2.862, respectively, and the value with YC in 2014 was 2.914. From the two tests with GN isolate, two significant QTL regions were repeatedly detected; CRQTL-GN_1 on chromosome C02 and CRQTL-GN_2 on chromosome C03 (Fig. 3). In the first test, CRQTL-GN_1 had higher values for LOD score, additive effect and variance explained compared with CRQTL-GN_2 (Table 4). The contrary results were detected in the second test. The test with YC isolate identified only a signle QTL region, named CRQTL-YC, which was in chromosome C03 with a location similar to CRQTL-GN_2. CRQTL-YC showed the largest QTL effect, 8.723 LOD score, for clubroot resistance and explained 47.1% of the phenotypic variation (Table 4).
Table 4.
QTLs for resistance to Plasmodiophora brassicae, position of the QTL on the map, LOD scores, additive and dominant effects, and percentage of variance explained
| Inoculation tests | QTL name | Linkage group | Marker interval | Nearest marker of peak LOD score | LODa | Additive effectb | Variance explained (%)c |
|---|---|---|---|---|---|---|---|
| GN (Race 9) in 2012 | CRQTL-GN_1 | C2 | C2d-1(2)–C2g-1(1) | C2h-1(4) | 4.907 | 0.813 | 29.7 |
| CRQTL-GN_2 | C3 | C3a-37(6)–C3b-14(6) | C3b-3(8) | 3.717 | −0.736 | 23.5 | |
| GN (Race 9) in 2014 | CRQTL-GN_1 | C2 | C2f-36(1)–C3-s1 | C2h-5(4) | 3.402 | 0.58 | 22 |
| CRQTL-GN_2 | C3 | C3a-1(11)–C3b-11(1) | C3a-34(2) | 4.698 | −0.686 | 29.1 | |
| YC (Race 2) in 2014 | CRQTL-YC | C3 | C3a-1(11)–C3b-153(3) | C3a-65(8) | 8.723 | −0.827 | 47.1 |
aPeak LOD score of the QTL.
bAdditive or dominant effect of C1176 allele.
cPercentage of variance explained at the peak of QTL.
3.5. Comparative genetic analysis of CR loci
We next compared the QTL regions identified in this study with reported CR loci in B. rapa and B. oleracea (Table 5). Chromosome C02 harbouring CRQTL-GN_1 exhibited strong collinearity with A02 in B. rapa,12,13 where the CRc gene, the linked marker for which is m6R,21 was mapped (Fig. 7A). In addition, two QTLs in B. oleracea, PbBo(Anju)1 and PbBo(Anju)2 as a major and a minor QTL, respectively, were identified in linkage group O02, which also represents chromosome C02, in Nagaoka et al.30 PbBo(Anju)2 is also linked to the m6R marker, and KBrH059L13 and BRMS-228 are the closely linked markers to PbBo(Anju)1. BLAST searches with the m6R marker revealed that that CRc and the PbBo(Anju)2 locus were between the C2b-9(6) and C2b-15(9) GBS markers, which were not included in CRQTL-GN_1. While the position of KBrH059L13 in the reference genome was found near the C2h-5(4) locus, BRMS-228 could not be found in the pseudochromosomes, but rather in unanchored scaffold000122_p2. Therefore, unused reads for alignment on pseudochromosomes were mapped to the scaffold with the same criteria, and two different blocks that showed different genotyping results were detected and named scaffold000122_p2a and scaffold 000122_p2b. A single SNP marker found in scaffold000122_p2b had same genotyping results as the C3-s1 locus (Supplementary Fig. S2A). The region between C2h-5(4) and C3-s1 overlapped with CRQTL-GN_1 (Fig. 7A).
Table 5.
Clubroot-resistant genes and QTLs reported in Brassica rapa and Brassica oleracea, and positional comparison with the genetic map in this study
| CR genes or QTLs | Linked markers or sequence | Reference | Position in B. rapa |
Chr. in B. oleracea | Closest markers in the genetic map | ||
|---|---|---|---|---|---|---|---|
| Chr. | Start | End | |||||
| Pb(Anju)2a, CRcb | m6R | Sakamoto et al.21, Nagaoka et al.30 |
A02 | 2,112,653 | 2,113,153 | C02 | C2b-9(6), C2b-15(9) |
| Pb(Anju)1a | KBrH059L13 BRMS-228 |
Suwabe et al.20, Nagaoka et al.30 |
A02 A02 |
24,842,317 25,208,934 |
24,842,620 25,209,181 |
C02 Scaffold000122_p2 |
C2h-5(4), C3-s1c |
| Pb(Anju)3a | BRMS-330 | Nagaoka et al.30 | A03 | 1,699,749 | 1,699,978 | C03 | C3a-73(6), C3a-79(4) |
| CRa, CRbb | HC352b B4732 B1005 |
Matsumoto et al.46, Kato et al.23 |
A03 A03 A03 |
23,614,238 24,348,058 24,376,820 |
23,614,816 24,348,275 24,377,054 |
C03 | C7-v5-1(6)c, C3b-183(20) |
| Crr3, CRkb | BrSTS61 | Saito et al.22 | A03 | 15,161,430 | 15,161,655 | Scaffold000040 | |
| Crr1b | BSA7 | Hatakeyama et al.49 | A08 | 10,766,540 | 10,766,858 | C03 | C3e-19(14) |
aQTLs identified in B. oleracea.
bCR loci identified in B. rapa.
cMarkers represented v-block or s-genotyped marker.
Figure 7.
Comparison of syntenic regions containing identified CRQTLs in this study; (A) linkage group C02 harbouring CRQTL-GN_1, and (B) linkage group C03 including CRQTL-GN_2 and CRQTL-YC with pseudochromosomes of B. oleracea (02–12 genotype) and B. rapa. QTL names in red (Pb-Anju1, 2, and 3) were identified in B. oleracea, and CR genes in black (CRa, CRb, CRc, CRk, Crr1, and Crr3) were identified in B. rapa. This figure is available in black and white in print and in colour at DNA Research online.
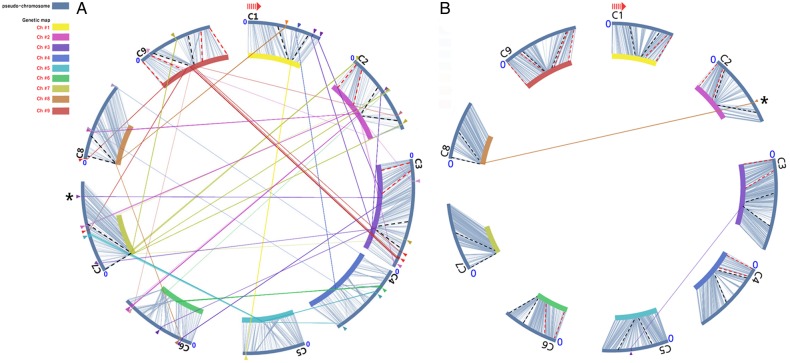
Chromosome C03 containing two similar QTLs, CRQTL-GN_2 and CRQTL-YC, show syntenic counterpart regions present in chromosome A03, A06, and A08 in B. rapa12,13 (Fig. 7B). The syntenic region of A03 harbours the loci for Crr3, CRk, CRa, and CRb. The locus of CRa is linked to HC352b sequence,46 and CRb is linked to B4732 and B1005 markers.23 All of those marker sequences could be found near C7-v5 block represented by C7-v5–1(6) in this study. C7-v5-1(6) was also linked to the same locus as C3b-183(20) (Fig. 3). To find the Crr1 locus present in A08, linked BSA7 marker49 sequences were searched in our genetic map, and C3e-19(14) in chromosome C03 was found to be the closest locus. In addition, BrSTS61 marker linked to Crr3 and CRk22 was found in unanchored scaffold000040. Genotyping of SNPs in scaffold00040 revealed that its position could be between C3c and C3d blocks (Supplementary Fig. S2B). The position of BRMS-330 marker linked to PbBo(Anju)330 was found in our QTLs CRQTL-GN_2 and CRQTL-YC, in chromosome C03. Although we could not find the sequence information for BRMS-125, which is linked to Crr4 and contained in A06,20 the syntenic counterpart of A06 to C03 was not related to QTL regions (Fig. 7B).
4. Discussion
4.1. GBS browser and GBS-based genetic mapping
In a previous study, we identified ∼670,000 SNPs between two elite cabbage breeding lines using 20× genome coverage whole-genome resequencing.34 Here, a total of 43,821 GBS-based SNPs were detected between parental lines, 15-fold fewer than detected by resequencing. Although the detection power was reduced, GBS-based genotyping provided a high-density genetic map at low cost in a highly efficient manner.
GBS analysis encounters bioinformatic, statistical, and computational challenges, because the large data set is frequently accompanied by a large number of erroneous SNP calling if the data are not properly filtered.8 To find suitable conditions for precise genotyping, the data set generated in this study was analysed under various conditions with various filtering options using a built-in GBS browser. In the browser, we could change the options to optimize the output (Supplementary Fig. S3). The average depth of read mapping was 11×, and we utilized SNPs derived from the regions with 5–100× read mapping depth. We filtered out SNPs derived from reads with <5× coverage to reduce uncertain genotyping due to low-coverage sequences. We also removed SNPs derived from high-depth reads, over than 100× coverage, to alleviate malalignment of reads derived from duplicated genome or repeat sequences. We further changed the criteria for genotyping of heterozygous loci and selected threshold value <0.9. We applied a stringent condition to select high-confidence GBS markers only when a genotype was present in all 80 samples with proper read mapping depth. Afterwards, we validated the genotyping results of the GBS markers by comparison to the genotyping results of 32 PCR-based markers. Consequently, we were able to obtain a high-density genetic map composed of 4,103 SNP markers even though both parental lines are cabbage breeding lines with good agriculture traits and narrow genetic diversity.
GBS analysis performed by Parkin et al.13 identified 826 bins in B. oleracea, which was more than identified in this study (679 bins). Our fewer bin numbers are probably due to the difference in genetic diversity of parental lines as well as the number of segregating progeny we used for GBS analysis. We used two elite breeding lines with narrow genetic diversity as parental lines, whereas Parkin et al.13 analysed the population between doubled haploid (DH) kale-like and DH broccoli lines. We also analysed 78 F2 plants for mapping. If we increase the number of F2 individuals for fine mapping, we may identify more recombination bins.
4.2. Our GBS-based high-resolution map improves the reference genome sequence
High-quality reference genome sequences provide information for identification of genes and regulatory elements, and thus, it is helpful for researchers to understand genomic variation.2 The genomes of A. thaliana8 and Oryza sativa50 were assembled by physical map-based approaches, which involve sequencing of minimally overlapping bacterial artificial chromosome (BAC) clones and filling gaps by various attempts.51 Thus, the reference sequence covers almost the complete whole genome. However, most of the recent plant genome sequences were primarily assembled by whole-genome shotgun sequencing strategies using NGS technologies.2
We performed GBS analysis based on the backbone of two reference genome sequences, 02–12 genome sequence reported by Liu et al.12 and TO1000DH genome sequence reported by Parkin et al.13 to construct a high-density genetic map. The pseudomolecules of 02–12 were constructed based on the genetic mapping of 1,227 PCR-based markers such as SSRs and SNPs.11 The TO1000DH genome sequence was developed by genotyping based on 15,509 GBS-based SNPs for anchoring of scaffold sequences. The TO1000DH genome sequence comprises much larger pseudomolecules (447 Mb) than the pseudochromosomes of 02–12 genome sequence (385 Mb). Here, we utilized 40.5% of the total filtered GBS reads for genotyping by excluding multi-mapped (25.9%) and un-mapped (33.6%) reads. Unmapped reads might be derived from outside of the reference pseudomolecules, probably from unanchored scaffolds or unassembled repeat sequences. In addition, 11.9 Gb of reads (25.9% of data produced in this study) showed multiple hits to the reference sequence, which is probably due to mapping of reads derived from paralogous regions associated with whole-genome triplication events in the Brassiceae tribe52,53 or repetitive sequence regions.
We also expected improvement of the 02–12 genome sequence12 by discovery and ordering of unanchored scaffold sequences.54 We were able to position two unanchored scaffolds, scaffold000040 and scaffold000122_p2, which are 2,345,746 bp and 983,585 bp, respectively, into the pseudomolecule (Supplementary Fig. S2). The present 02–12 genome sequence includes ∼150 Mb of unanchored scaffolds. In our analysis, >33% of GBS reads were remained as unmapped (Table 1) that could be utilized as guidance for mapping of the remaining unanchored scaffolds, similar to efforts made in barley and wheat genomes.7,54 Likewise, the genome of Drosophila simulans was developed using GBS approach, in which 30% of the unassembled scaffolds were newly assigned to linkage groups.55
Furthermore, the 37 and 2 scaffold sequences in v-blocks of the 02–12 and TO1000DH genome sequences, respectively, could be updated via proper repositioning (Fig. 5). The v-blocks are usually derived from repeat-rich and recombination-deficient heterochromatin regions. Comparison of two v-blocks in both reference genome sequences identified the proper position of 673 Kb (C7-v5 block, Fig. 5A) among the 1,533 Kb mispositioned 02–12 genome sequence. The remaining 860 Kb sequence could not be placed due to lack of GBS markers (Fig. 5A). The C2-v blocks in TO1000DH identified ∼300 Kb additional sequence that mapped in other C8 chromosome. However, we could not find the corresponding 300 Kb sequence in the 02–12 genome sequence (Fig. 5B). We assume that the lack of corresponding sequence in the 02–12 genome sequence is because this sequence is derived from a repeat-rich heterochromatin region. Moreover, we identified some misordered scaffolds in same chromosome. For example, the C2e block seemed to be within the C2c block, and the C6f, C6e, and C6 g blocks were likely to be inverted (Supplementary Table S3). The benefits of having both genome sequences are innumerable for genomics studies such as this one. Refining and completing the genome sequences is an ongoing task, and our analyses suggest potential errors to address. However, it remains possible that such differences are derived from utilization of different mapping populations in different B. oleracea genotypes with chromosomal-level reorganization.
4.3. Clubroot resistance loci differ in the B. rapa and B. oleracea genomes
We identified two QTLs (CRQTL-GN_1 and CRQTL-GN_2) for resistance to GN isolate (Race 9) and one QTL (CRQTL-YC) for resistance to YC isolate (Race 2). Although the LOD values of the two QTLs for GN isolate were different between two tests, both QTLs were detected repeatedly in independent inoculation tests, suggesting that both QTLs are strong candidates for major QTLs against Race 9. CRQTL-YC showed the highest LOD score among all trials and also overlapped with CRQTL-GN_2. This finding implies that the CRQTL-YC locus is probably responsible for resistance against both Race 2 and Race 9. Although resistance to the YC isolate was detected only as a single resistance QTL, we think that the resistance against race 2 is likely controlled by this major QTL, CRQTL-YC, and also affected by many minor quantitative loci, because the disease index distribution of the F2 population did not follow the Mendelian single-gene segregation ratio (Fig. 6).
The comparison of genetic loci between our QTLs in B. oleracea and eight CR loci in B. rapa revealed no coincidence in chromosomal positions, even though CRa, CRb, CRk, Crr1, Crr3, and Crr4 in B. rapa are known to be involved in resistance to Race 2,17,21,29,46 the same isolate as YC. Although CRa and CRb in B. oleracea were also reported as resistance genes against Race 2 using swede cv Wilhelmsburger as a resistance source,56 these results raise the possibility that none of the reported CR genes in B. rapa and B. oleracea are related to resistance in our cabbage materials, and that they have a different genetic origin of resistance genes. Because genetic loci for Race 9 resistance have not been identified in either species, our results regarding CRQTL-GN_1 and 2 can provide valuable resources for further research.
The Brassicaceae family evolved from rearrangement of shared 24 chromosomal blocks (A–X blocks), which are denoted as the ancestral translocation Proto-Calepineae Karyotype (tPCK). Hundreds of species in the Brassiceae tribe are derived from whole genome triplication of the tPCK, followed by genome rearrangement.52,53,57 The fate of paralogous gene function can be differentiated in other Brassica species. We evaluated paralogous genomic blocks harbouring our QTL regions and compared the blocks to the clubroot resistance loci in B. rapa. Crr3 and CRk were found in the F block, CRa and CRb were found in the J block of chromosome A03, and Crr1 and Crr2 found in the U blocks of chromosome A08 and A01, respectively, of B. rapa. These F, J, and U blocks were not related to our CRQTL regions. The CRc gene was identified in the R block of chromosome A02 of B. rapa, and our CRQTL-YC and CRQTL-GN_2 as well as Pb-Anju3 are identified from the R block of chromosome C03 of B. oleracea (Fig. 6B). It is likely that the paralogous gene of CRc in the R block of chromosome C03 can be the candidate clubroot resistance gene in B. oleracea.
Among five QTLs identified in B. oleracea by Nagaoka et al.,30 who used Race 4 for inoculation tests, PbBo(Anju)1, which had a maximum LOD score, and PbBo(Anju)3, regarded as a minor effect QTL, seemed to overlap with CRQTL-GN_1 and CRQTL-GN_2 (or CRQTL-YC), respectively. This, despite the difference in pathogen tested, two overlapping QTLs were identified, suggesting that CRQTL-GN_1 and CRQTL-GN_2 could confer resistance to Races 2, 4, and Races 2, 4, 9, respectively. In the future, cloning of CR genes and QTL analyses using more diverse races are required to characterize the relationship between clubroot resistance and the breeding materials in both B. oleracea and B. rapa.
4.4. Conclusions
We performed GBS for high-resolution mapping using an F2 population between two elite cabbage breeding lines with narrow genetic diversity. We constructed a high-resolution genetic map using 4,103 stringently filtered GBS-SNPs and 32 reference markers. Based on comparison of the high-resolution map and the position of each marker on the reference genome, we identified 37 putatively mislocated scaffolds and their suggested revised positions and also could anchor two unassigned scaffolds, thereby improving the reference genome sequence of cabbage. Moreover, two QTL regions were identified for clubroot resistance against two different P. brassicae isolates. We thus demonstrated that GBS can successfully be applied for rapid genotyping of many individuals for molecular breeding and for genetics and genomics research in cabbage.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This research was supported by the Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-3-SB430) of the Ministry of Agriculture, Food and Rural Affairs (MAFRA) in the Republic of Korea and the Next Biogreen 21 program (PJ01101101) of the Rural Development Administration (RDA) and the Korea Forest Service (KFS). Funding to pay the Open Access publication charges for this article was provided by Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-3-SB430) of the Ministry of Agriculture, Food and Rural Affairs (MAFRA) in the Republic of Korea.
Supplementary Material
Acknowledgements
We thank all lab-members in Laboratory of Functional Crop Genomics and Biotechnology, Seoul National University for their technical assistance.
References
- 1.Initiative A.G. 2000, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana, Nature, 408, 796. [DOI] [PubMed] [Google Scholar]
- 2.Feuillet C., Leach J.E., Rogers J., Schnable P.S., Eversole K.. 2011, Crop genome sequencing: lessons and rationales, Trends Plant Sci., 16, 77–88. [DOI] [PubMed] [Google Scholar]
- 3.Elshire R.J., Glaubitz J.C., Sun Q. et al. 2011, A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species, PLoS ONE, 6, e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crossa J., Beyene Y., Kassa S. et al. 2013, Genomic prediction in maize breeding populations with genotyping-by-sequencing, Genes Genomes Genetics, 3, 1903–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romay M.C., Millard M.J., Glaubitz J.C. et al. 2013, Comprehensive genotyping of the USA national maize inbred seed bank, Genome Biol., 14, R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonah H., Bastien M., Iquira E. et al. 2013, An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping, PLoS ONE, 8, e54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poland J.A., Brown P.J., Sorrells M.E., Jannink J.-L.. 2012, Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach, PLoS ONE, 7, e32253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spindel J., Wright M., Chen C. et al. 2013, Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations, Theor. Appl. Genet., 126, 2699–716. [DOI] [PubMed] [Google Scholar]
- 9.Mascher M., Wu S., Amand P.S., Stein N., Poland J.. 2013, Application of genotyping-by-sequencing on semiconductor sequencing platforms: a comparison of genetic and reference-based marker ordering in barley, PLoS ONE, 8, e76925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.U, N. 1935, Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization, J. Jpn Bot., 7, 389–452. [Google Scholar]
- 11.Wang W., Huang S., Liu Y. et al. 2012, Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata), BMC Genomics, 13, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S., Liu Y., Yang X. et al. 2014, The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes, Nat. Commun., 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkin I.A., Koh C., Tang H. et al. 2014, Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea, Genome Biol., 15, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crute I., Gray A., Crisp P., Buczacki S.. 1980, Variation in Plasmodiophora brassicae and resistance to clubroot disease in brassicas and allied crops-a critical review, Plant Breed. Abst., 50, 91–104. [Google Scholar]
- 15.Dixon G., Robinson D.. 1986, The susceptibility of Brassica oleracea cultivars to Plasmodiophora brassicae (clubroot), Plant Pathol., 35, 101–7. [Google Scholar]
- 16.Voorrips R.E. 1995, Plasmodiophora brassicae: aspects of pathogenesis and resistance in Brassica oleracea, Euphytica, 83, 139–46. [Google Scholar]
- 17.Piao Z., Ramchiary N., Lim Y.P.. 2009, Genetics of clubroot resistance in Brassica species, J. Plant Growth Regul., 28, 252–64. [Google Scholar]
- 18.Hirai M., Harada T., Kubo N., Tsukada M., Suwabe K., Matsumoto S.. 2004, A novel locus for clubroot resistance in Brassica rapa and its linkage markers, Theor. Appl. Genet., 108, 639–43. [DOI] [PubMed] [Google Scholar]
- 19.Piao Z., Deng Y., Choi S., Park Y., Lim Y.. 2004, SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis), Theor. Appl. Genet., 108, 1458–1465. [DOI] [PubMed] [Google Scholar]
- 20.Suwabe K., Tsukazaki H., Iketani H. et al. 2006, Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance, Genetics, 173, 309–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto K., Saito A., Hayashida N., Taguchi G., Matsumoto E.. 2008, Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis), Theor. Appl. Genet., 117, 759–67. [DOI] [PubMed] [Google Scholar]
- 22.Saito M., Kubo N., Matsumoto S., Suwabe K., Tsukada M., Hirai M.. 2006, Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa, Theor. Appl. Genet., 114, 81–91. [DOI] [PubMed] [Google Scholar]
- 23.Kato T., Hatakeyama K., Fukino N., Matsumoto S.. 2013, Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa, Breed. Sci., 63, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao F., Hirani A.H., Liu J. et al. 2014, Fine mapping a clubroot resistance locus in Chinese cabbage, J. Am. Soc. Hort. Sci., 139, 247–52. [Google Scholar]
- 25.Zhang T., Zhao Z., Zhang C. et al. 2014, Fine genetic and physical mapping of the CRb gene conferring resistance to clubroot disease in Brassica rapa, Mol. Breed., 34, 1173–83. [Google Scholar]
- 26.Crisp P., Crute I., Sutherland R. et al. 1989, The exploitation of genetic resources of Brassica oleracea in breeding for resistance to clubroot (Plasmodiophora brassicae), Euphytica, 42, 215–26. [Google Scholar]
- 27.Kuginuki Y., Ajisaka H., Yui M., Yoshikawa H., Hida K.-i., Hirai M.. 1997, RAPD markers linked to a clubroot-resistance locus in Brassica rapa L, Euphytica, 98, 149–54. [Google Scholar]
- 28.Zhong Yun P., Yong Joo P., Su Ryun C. et al. 2002, Conversion of an AFLP marker linked to clubroot resistance gene in Chinese cabbage into a SCAR marker, Hort. Environ. Biotechnol., 43, 653–9. [Google Scholar]
- 29.Chen J., Jing J., Zhan Z., Zhang T., Zhang C., Piao Z.. 2013, Identification of Novel QTLs for Isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa, PLoS ONE, 8, e85307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaoka T., Doullah M., Matsumoto S. et al. 2010, Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea, Theor. Appl. Genet., 120, 1335–46. [DOI] [PubMed] [Google Scholar]
- 31.Allen G., Flores-Vergara M., Krasynanski S., Kumar S., Thompson W.. 2006, A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide, Nat. Protocols, 1, 2320–5. [DOI] [PubMed] [Google Scholar]
- 32.Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows–Wheeler transform, Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A., Hanna M., Banks E. et al. 2010, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data, Genome Res., 20, 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J., Izzah N.K., Jayakodi M. et al. 2015, Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage, BMC Plant Biol., 15, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izzah N.K., Lee J., Jayakodi M. et al. 2014, Transcriptome sequencing of two parental lines of cabbage (Brassica oleracea L. var. capitata L.) and construction of an EST-based genetic map, BMC Genomics, 15, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe A., Moule C., Trick M., Edwards K.. 2004, Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species, Theor. Appl. Genet., 108, 1103–12. [DOI] [PubMed] [Google Scholar]
- 37.Piquemal J., Cinquin E., Couton F. et al. 2005, Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers, Theor. Appl. Genet., 111, 1514–23. [DOI] [PubMed] [Google Scholar]
- 38.Choi S.R., Teakle G.R., Plaha P. et al. 2007, The reference genetic linkage map for the multinational Brassica rapa genome sequencing project, Theor. Appl. Genet., 115, 777–92. [DOI] [PubMed] [Google Scholar]
- 39.Louarn S., Torp A.M., Holme I., Andersen S.B., Jensen B.D.. 2007, Database derived microsatellite markers (SSRs) for cultivar differentiation in Brassica oleracea, Genet. Resourc. Crop Evol., 54, 1717–25. [Google Scholar]
- 40.Cheng X., Xu J., Xia S. et al. 2009, Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus, Theor. Appl. Genet., 118, 1121–31. [DOI] [PubMed] [Google Scholar]
- 41.Sampath P., Lee S.-C., Lee J. et al. 2013, Characterization of a new high copy Stowaway family MITE, BRAMI-1 in Brassica genome, BMC Plant Biol., 13, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sampath P., Murukarthick J., Izzah N.K. et al. 2014, Genome-wide comparative analysis of 20 miniature inverted-repeat transposable element families in Brassica rapa and B. oleracea, PLoS ONE, 9, e94499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SuJung J., SunAh S., KyoungSoo J., YongHo C., JinCheol K., GyungJa C.. 2011, Resistance of cultivars of Chinese cabbage to Plasmodiophora brassicae isolates of several races collected in Korea, Kor. J. Hort. Sci. Technol., 29, 610–6. [Google Scholar]
- 44.Williams P.H. 1966, A system for the determination of races of Plasmodiophora brassicae that infect Cabbage and Rutabaga, Phytopathology, 56, 624–6. [Google Scholar]
- 45.Schranz M.E., Lysak M.A., Mitchell-Olds T.. 2006, The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes, Trends Plant Sci., 11, 535–42. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto E., Yasui C., Ohi M., Tsukada M.. 1998, Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis), Euphytica, 104, 79–86. [Google Scholar]
- 47.Suwabe K., Tsukazaki H., Iketani H. et al. 2003, Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L, Theor. Appl. Genet., 107, 997–1002. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Wang H., Wang J. et al. 2011, The genome of the mesopolyploid crop species Brassica rapa, Nat. Genet., 43, 1035–9. [DOI] [PubMed] [Google Scholar]
- 49.Hatakeyama K., Suwabe K., Tomita R.N. et al. 2013, Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L, PLoS ONE, 8, e54745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.International Rice Genome Sequencing Project. 2005, The map-based sequence of the rice genome, Nature, 436, 793–800. [DOI] [PubMed] [Google Scholar]
- 51.Yang T.-J., Yu Y., Nah G. et al. 2003, Construction and utility of 10-kb libraries for efficient clone-gap closure for rice genome sequencing, Theor. Appl. Genet., 107, 652–60. [DOI] [PubMed] [Google Scholar]
- 52.Town C.D., Cheung F., Maiti R. et al. 2006, Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy, Plant Cell, 18, 1348–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang T.-J., Kim J.S., Kwon S.-J. et al. 2006, Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa, Plant Cell, 18, 1339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poland J.A., Rife T.W.. 2012, Genotyping-by-sequencing for plant breeding and genetics, Plant Genome, 5, 92–102. [Google Scholar]
- 55.Andolfatto P., Davison D., Erezyilmaz D. et al. 2011, Multiplexed shotgun genotyping for rapid and efficient genetic mapping, Genome Res., 21, 610–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landry B.S., Hubert N., Crete R., Chang M.S., Lincoln S.E., Etoh T.. 1992, A genetic map for Brassica oleracea based on RFLP markers detected with expressed DNA sequences and mapping of resistance genes to race 2 of Plasmodiophora brassicae (Woronin), Genome, 35, 409–20. [Google Scholar]
- 57.Cheng F., Mandáková T., Wu J., Xie Q., Lysak M.A., Wang X.. 2013, Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa, Plant Cell, 25, 1541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.