Abstract
The present study used a whole-genome, NGS resequencing-based mQTL-seq (multiple QTL-seq) strategy in two inter-specific mapping populations (Pusa 1103 × ILWC 46 and Pusa 256 × ILWC 46) to scan the major genomic region(s) underlying QTL(s) governing pod number trait in chickpea. Essentially, the whole-genome resequencing of low and high pod number-containing parental accessions and homozygous individuals (constituting bulks) from each of these two mapping populations discovered >8 million high-quality homozygous SNPs with respect to the reference kabuli chickpea. The functional significance of the physically mapped SNPs was apparent from the identified 2,264 non-synonymous and 23,550 regulatory SNPs, with 8–10% of these SNPs-carrying genes corresponding to transcription factors and disease resistance-related proteins. The utilization of these mined SNPs in Δ (SNP index)-led QTL-seq analysis and their correlation between two mapping populations based on mQTL-seq, narrowed down two (CaqaPN4.1: 867.8 kb and CaqaPN4.2: 1.8 Mb) major genomic regions harbouring robust pod number QTLs into the high-resolution short QTL intervals (CaqbPN4.1: 637.5 kb and CaqbPN4.2: 1.28 Mb) on chickpea chromosome 4. The integration of mQTL-seq-derived one novel robust QTL with QTL region-specific association analysis delineated the regulatory (C/T) and coding (C/A) SNPs-containing one pentatricopeptide repeat (PPR) gene at a major QTL region regulating pod number in chickpea. This target gene exhibited anther, mature pollen and pod-specific expression, including pronounced higher up-regulated (∼3.5-folds) transcript expression in high pod number-containing parental accessions and homozygous individuals of two mapping populations especially during pollen and pod development. The proposed mQTL-seq-driven combinatorial strategy has profound efficacy in rapid genome-wide scanning of potential candidate gene(s) underlying trait-associated high-resolution robust QTL(s), thereby expediting genomics-assisted breeding and genetic enhancement of crop plants, including chickpea.
Keywords: chickpea, mQTL-seq, pod number, SNP, wild accessions
1. Introduction
Chickpea (Cicer arietinum L.) is the third most vital food legume with a genome size of ∼720 Mb.1,2 The current climatic conditions are adversely affecting the world-wide chickpea productivity to a large extent by highly variable phenotypic plasticity in conjunction with multiple abiotic and biotic stresses, including drought, terminal heat stress, salinity, Fusarium wilt and Ascochyta blight.2–5 Therefore, the primary goal of present chickpea genomics is focused towards minimizing the effect of climate change and biotic/abiotic stresses upon chickpea yield and productivity by developing genetically tailored high-yielding stress-tolerant chickpea cultivars. However, most of the stress tolerance and yield component traits targeted for chickpea genetic enhancement usually have complex genetic architecture and are regulated by multiple major and/or minor genes/QTLs (quantitative trait loci). It is thus imperative to delineate the potential genes/alleles underlying QTLs associated with various traits of interest (yield contributing and stress-tolerant traits) by fine mapping/map-based cloning prior to their effective deployment in marker-assisted genetic enhancement of chickpea. So far, very limited attention has been paid towards fine mapping of trait-regulatory QTLs and their subsequent utilization in marker-assisted genetic improvement of chickpea.6–13 The narrow genetic base including low marker genetic polymorphism especially between parental accessions of diverse intra-specific mapping populations along with inadequate accessibility of high-density, intra-specific genetic linkage maps are the major bottlenecks in identification and fine mapping of QTLs in chickpea. In this perspective, an alternative genome-wide strategy is crucial for rapid molecular mapping of trait-associated high-resolution QTLs/genes, which in turn will benefit marker-assisted breeding in chickpea.
To accelerate molecular mapping of QTLs in chickpea, traditional QTL mapping strategy14–16 that primarily relies on genome-wide discovery and large-scale genotyping of simple sequence repeat (SSR) and single-nucleotide polymorphism (SNP) markers in individuals of diverse inter-/intra-specific mapping populations (recombinant inbred and near-isogenic lines) by using multiple high-throughput genotyping assays, including NGS (next-generation sequencing)-based GBS (genotyping-by-sequencing) assay, are found to be most fruitful in chickpea.9–13, 17–23 This approach successfully identified a number of low-resolution major QTLs governing various yield contributing (i.e. flowering and maturation time, seed size/100-seed weight, double podding and seed/pod number per plant) and abiotic/biotic stress tolerance traits (viz. salinity and drought tolerance root traits, Fusarium wilt, Ascochyta blight and Botrytis gray mold) in chickpea.1,2, 8–13, 22–41 In light of the added advantages and its broader applicability vis-à-vis the diverse aforesaid traditional QTL mapping approaches, the high-throughput NGS-based QTL-seq has recently been used fruitfully as a cost-effective strategy for rapid genome-wide scanning and mapping of major QTLs governing multiple qualitative and quantitative traits such as seedling vigour and blast resistance in rice, flowering time in cucumber, fruit weight and locule number in tomato, and 100-seed weight in chickpea.42–45 Nonetheless, most of the major QTLs identified through QTL-seq are being localized within sizeable long genetic marker intervals of chromosomes, which requires large-scale validation in multiple mapping populations and subsequent fine mapping through traditional QTL mapping. This will ensure the effective practical utilization of these well-validated robust QTLs/genes in marker-assisted genetic improvement of chickpea.
Simultaneously to expedite the above-mentioned, a multiple QTL-seq (mQTL-seq) approach that involves QTL-seq analysis in multiple mapping populations derived from the common parental accessions could be an attractive genome-wide strategy. Essentially, mQTL-seq deals with NGS-based whole-genome resequencing of DNA bulks (showing two extreme contrasting phenotypic trait values) of progenies that are derived from each of the multiple segregating mapping population with at least single common parental accession (detailed strategy illustrated in Fig. 1). Collectively, the utility of this strategy is evident from the large-scale validation of major QTLs derived from QTL-seq of each preliminary/advanced generation mapping population in diverse genetic backgrounds (multiple mapping populations) along with its efficiency to scale down each QTL-seq originated QTLs into potential candidate genes regulating diverse agronomic traits in chickpea. Henceforth, implementing this mQTL-seq approach can be beneficial for quick genome-wide scanning and fine mapping of trait-associated major genes harbouring robust QTLs (validated in diverse mapping populations) with optimal expense of resources in chickpea. This will provide much needed inputs for molecular dissection of complex quantitative traits culminating into genomics-assisted crop improvement in chickpea at a faster pace.
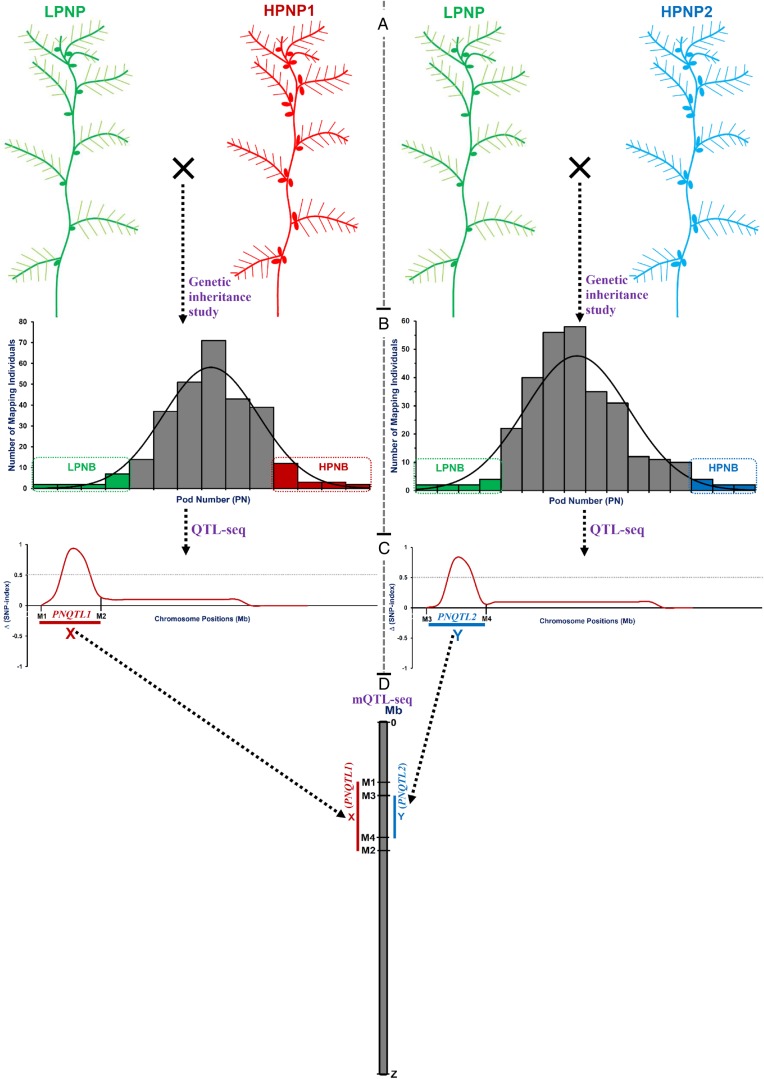
Figure 1.
(A) Development of multiple mapping populations by inter-crossing of parental accessions (with at least one common parent) contrasting for a particular agronomic trait such as pod number. LPNP: low pod number mapping parent (green colour) and HPNP1 and 2: high pod number mapping parents 1 (red colour) and 2 (blue colour). LPNP is considered the common parent. (B) Genetic inheritance study on normal frequency distribution of target trait (such as pod number) in mapping populations for selection of homozygous individuals (constituting the bulks) from each mapping population exhibiting two extreme contrasting phenotypic trait values (like low and high pod number). LPNB and HPNB: low and high pod number bulks. (C) The Δ (SNP index)-based QTL-seq analysis (following detailed strategies of Takagi et al.42 and Das et al.44) using NGS-genome resequences of parents and bulks from each of the multiple mapping populations to identify major genomic regions that underlie QTLs governing particular agronomic trait in chickpea. The major QTLs identified from each of the multiple mapping populations are localized at varying physical sequence intervals (Mb) spanned by flanking SNP markers on the chromosomes. For instance, QTL-seq analysis in two contrasting pod number mapping populations identified two genomic regions harbouring two major pod number QTLs (PNQTL1 and PNQTL2) that spanned X and Y-Mb sequence intervals between M1 and M2 as well as M3 and M4 flanking SNP markers, respectively, on the same chromosomes. (D) Integration of QTL-seq-derived individual QTL outcomes from multiple mapping populations (with at least one common parent) called ‘mQTL-seq (multiple QTL-seq)’ for large-scale validation of these identified major QTLs in multiple genetic backgrounds to scan the robust QTLs, and further to narrow down the longer robust QTL intervals into shorter genomic intervals for delineation of candidate gene(s) regulating the trait under study. The integration of QTL-seq-derived two pod number QTLs (PNQTL1 and PNQTL2) identified from two mapping populations enabled to validate these QTLs in two diverse genetic backgrounds to ascertain their robustness for pod number regulation in chickpea. Further, the mQTL-seq analysis scaled-down a longer QTL interval (spanned X Mb) into a shorter pod number robust QTL interval (spanned Y Mb) flanking within M3 and M4 SNP markers on the chromosome; this can serve as a potential target genomic region for rapid identification and delineation of candidate gene(s) regulating pod number in chickpea. This figure is available in black and white in print and in colour at DNA Research online.
As a proof of concept, we made an effort to implement an NGS-based genome-wide high-throughput mQTL-seq approach in two F5 mapping populations of C. arietinum and C. reticulatum inter-specific crosses (Pusa 1103 × ILWC 46 and Pusa 256 × ILWC 46) for identifying the major genomic regions harbouring the robust QTLs associated with pod number per plant in chickpea. The integration of mQTL-seq with QTL region-specific association analysis and differential expression profiling delineated a potential novel candidate gene and natural allelic variants at the major QTL interval governing pod number in chickpea.
2. Materials and methods
2.1. Development of inter-specific mapping populations and their phenotyping for pod number
Two inter-specific F5 mapping populations of Pusa 1103× ILWC 46 (population size, 102) and Pusa 256× ILWC 46 (population size, 98) derived from high (cultivated C. arietinum desi accessions Pusa 1103 and Pusa 256 with 129 and 120 pods/plant, respectively) and low (wild C. reticulatum accession ILWC 46 with 10 pods/plant) pod number-containing parental accessions were developed using single seed descent method. The wild accession ILWC 46 serves as a common parent for both the populations generated. The mapping individuals and parental accessions of both populations were grown (following random complete block design with two replications) and phenotyped in the real field conditions at two diverse geographical locations of India (CSKHPKV, Palampur: latitude 32.1°N and longitude 76.5°E and NBPGR, New Delhi: 28.6°N and 77.2°E) for two consecutive years. A set of 15–20 representative plants were selected from each mapping individual and parental accession of both populations, and pod number (PN) was measured by counting the average number of fully developed pods per plant at maturity stage. The mean, standard deviation, coefficient of variation (CV%), broad-sense heritability (H2) and frequency distribution of PN (based on ANOVA) were measured in both populations individually as per Kujur et al.10,11 and Bajaj et al.23 To determine the trait inheritance pattern, the effects/interactions among accessions/genotypes (G) and environments (E) (i.e. experimental years and geographical locations) were measured. The broad-sense heritability among accessions [H2 = σ2g/(σ2g + σ2ge/n + σ2e/nr)] was measured based on σ2g (genetic), σ2ge (G x E) and σ2e (error) variance with n (number of experimental years/environments) = 2 and r (number of replicates) = 2. Additionally, 92 desi and kabuli chickpea accessions representing a PN-specific association panel were also grown in the field and phenotyped for pod number following aforesaid methods.
2.2. Whole-genome resequencing and mQTL-seq analysis
According to the preliminary clues obtained from our traditional QTL mapping analysis, 10 of each high and low pod number-containing homozygous mapping individuals (a total of 20 individuals) representing two utmost ends of PN normal frequency distribution curve were selected individually from the mapping populations of Pusa 1103 × ILWC 46 and Pusa 256 × ILWC 46 for QTL-seq analysis. The homozygous genetic constitution of these selected 20 individuals from each of the two mapping populations for either of the high and low pod number trait was ascertained by utilizing their PN field phenotyping information and genotyping data of 96 genome-wide SSR markers in QTL mapping. The isolation and quantification of genomic DNA, pooling of equal concentration DNA to constitute low pod number bulk (LPNB) and high pod number bulk (HPNB), construction and sequencing of pair-end sequencing libraries (100-bp read length) using HiSeq2000 (Illumina Inc., San Diego, CA, USA) NGS platform and generation of high-quality genomic sequences were performed according to Das et al.44 The BWA with default parameters was used to align and map the high-quality sequence reads onto the reference kabuli genome.46 Subsequently, normalization of the mappable sequence reads based on depth of read coverage among mapping parents and individuals constituting LPNB and HPNB was performed. The high-quality homozygous SNPs (minimum sequence read depth: 10 with SNP base quality ≥20) between two parental accessions as well as among mapping individuals constituting the LPNB and HPNB were discovered and further structurally and functionally annotated with respect to reference genome as per Kujur et al.12,13
A SNP index and Δ (SNP index)-based QTL-seq approach was deployed individually in two mapping populations to scan major PN QTLs in chickpea following the earlier defined recommended parameters.42–45, 47 The Δ (SNP index) was estimated according to subtraction of SNP index (proportion of sequence reads supported the SNPs, which are entirely different from the reference kabuli genome sequences) between LPNB and HPNB. The SNP index was estimated as ‘0’ and ‘1’ based on the representation of genomic fragments derived from Pusa 1103/Pusa 256 and ILWC 46, respectively, in entire high-quality sequence reads generated. A sliding window approach with 1-Mb window size and 10-kb increment was utilized to measure the average distribution of Δ (SNP index) of SNPs physically mapped across eight kabuli chromosomes in a given genomic interval. The SNP index plots were generated for two mapping populations individually by plotting the Δ (SNP index) of LPNB and HPNB and their corresponding SNP index within the specified window size in the graphs. The statistical confidence intervals of Δ (SNP index) with a given sequence read depth under the null hypothesis of no QTLs were calculated to assure the accuracy of QTLs derived from QTL-seq as per Takagi et al.,42 Lu et al.43 and Das et al.44
2.3. QTL region-specific association analysis
The major genomic regions underlying PN QTLs identified from two mapping populations individually using QTL-seq were compared/correlated to identify the consensus robust QTLs that are well-validated across these diverse populations. One of the selected novel genomic region harbouring robust PN QTL was sequenced in low and high pod number-containing homozygous mapping individuals (constituting LPNB and HPNB in QTL-seq analysis) and parental accessions from each of the two mapping populations using the multiplexed amplicons sequencing protocol (as per manufacturer's instructions) of TruSeq Custom Amplicon v1.5 in Illumina MiSeq next-generation sequencer. The mapping of high-quality amplicon sequence reads onto the reference kabuli genome and detection of high-quality SNPs among parental accessions and mapping individuals was performed as per Saxena et al.9 and Kujur et al.12,48 The SNPs discovered at the sequenced genomic region harbouring robust PN QTL was structurally and functionally annotated following Kujur et al.12,48 The physically mapped SNPs revealing differentiation between high and low pod number-containing parental accessions and individuals of two mapping populations at QTL region of interest were genotyped in the genomic DNA of 92 desi and kabuli chickpea accessions belonging to a PN-specific association panel using Illumina GoldenGate assay as per Saxena et al.9 and Bajaj et al.23 The replicated multi-location/years PN field phenotyping data, population structure (K = 2) statistics, principal component analysis (PCA) and kinship matrix of 92 accessions were obtained from Kujur et al.13 The association analysis was performed by correlating the genotyping information of SNPs at robust PN QTL interval with aforesaid phenotyping and diversity statistics using general linear model (GLM), mixed linear model (MLM at optimum level of compression with P3D method) and mixed model approach of EMMA49 following the methods of Kujur et al.13 and Kumar et al.50 The Bonferroni correction of P-value was used for each trait-associated SNPs at 5% significance level to eliminate the confounding effect of population structure and correct the false discovery rate (FDR) based on multiple comparisons. The informative SNPs-carrying candidate gene associated (R2 = correlation potential of significant SNPs with traits) with PN at significant cut-off P ≤ 10−5 was screened by combining the outcomes of GLM and MLM with EMMA and FDR correction.
2.4. Differential gene expression profiling
To infer the gene regulation, differential expression profiling of SNPs-containing gene annotated at the major genomic interval harbouring a novel robust PN QTL (validated by both QTL-seq and QTL region-specific association analysis) was performed. The vegetative (leaf and root) and reproductive (flower bud, ovary, anther and mature pollen) tissues and two-pod (∼5 mm beginning pod size at 5–10 days after anthesis/DAA and ∼2 cm full pod at 15–20 DAA) developmental stages were collected (at least with three biological replicates) from 40–60 days old healthy plants of low and high pod number-containing homozygous mapping individuals (constituting LPNB and HPNB in QTL-seq analysis) and parental accessions from each of the two mapping populations under study. The RNA isolated from all the tissues/stages of mapping individuals and parents were amplified with desirable gene-specific primers and internal control elongation factor 1-alpha (EF1α) using semi-quantitative and quantitative RT–PCR assays. The differential expression level of gene observed in diverse tissues/developmental stages of high and low pod number-containing mapping individuals and parents was compared/correlated following Bajaj et al.22
3. Results and discussion
3.1. Genetic inheritance pattern of pod number in two inter-specific mapping populations
We observed a significant difference of PN (5-237) with 13% CV and 81% H2 in 102 individuals and parental accessions of an inter-specific F5 mapping population of Pusa 1103 × ILWC 46 (Table 1). A wider phenotypic variation for PN (5-229) with 14.8% CV and 80% H2 was detected in another 98 individuals and parents of an inter-specific F5 mapping population of Pusa 256 × ILWC 46 (Table 1). The continuous variation as well as normal frequency distribution of PN trait was observed in these two mapping populations, which indicates the quantitative genetic inheritance pattern of target trait under study. The pod number is a complex yield component quantitative trait in chickpea, which is known to be governed by multiple abiotic (drought, heat, cold and salinity) and biotic (pod bores and other diseases) stress factors and often influenced by climate/growing conditions. To overcome these intricacies, present study selectively used three contrasting parental accessions-derived inter-specific mapping populations exhibiting wider phenotypic variability and higher heritability (consistent phenotypic expression) for pod number across geographical locations/years for molecular mapping of major QTLs regulating pod number in chickpea through mQTL-seq.
Table 1.
Statistical measures of pod number estimated in parental accessions and individuals of two inter-specific F5 mapping populations
| Mapping populations | Parental accessions |
F5 mapping individuals |
Broad-sense heritability (H2%) | |||
|---|---|---|---|---|---|---|
| Pusa 1103 (Mean ± S.D.) | ILWC 46 (Mean ± S.D.) | Mean ± S.D. | Range | Coefficient of variation (CV%) | ||
| Pusa 1103 × ILWC 46 | 129 ± 2.2 | 29 ± 1.9 | 145.5 ± 18.9 | 5.0–237.0 | 13.0 | 81 |
| Pusa 256 (Mean ± S.D.) | ILWC 46 (Mean ± S.D.) | Mean ± S.D. | Range | Coefficient of variation (CV%) | ||
| Pusa 256 × ILWC 46 | 125 ± 2.4 | 29 ± 1.5 | 129.1 ± 19.1 | 5.0–229.0 | 14.8 | 80 |
3.2. NGS-based whole-genome resequencing for QTL-seq
The NGS-based high-throughput whole-genome resequencing of two high and low pod number-containing parental accessions as well as bulks (LPNB and HPNB) of two inter-specific mapping populations [(Pusa 1103 × ILWC 46) and (Pusa 256 × ILWC 46)] were generated on an average of 81.5 million high-quality sequence reads (ranging from 80.7 to 83.5 million reads) with a ∼11.6-fold sequencing depth coverage. Notably, 82.1% (varying from 82.3 to 83.6%) sequence reads of these were mapped (minimum mapping quality: 30) to unique physical locations of kabuli reference genome with a 69% mean coverage. The high-quality uniquely mapped sequence reads generated from parental accessions and bulks (LPNB and HPNB) of two mapping populations were normalized based on depth of read coverage to reduce the potential biasness of read depth in the studied samples. Subsequently, the sequencing depth coverage (fold) and genome coverage (%) of uniquely mapped non-redundant sequence reads in mapping parents and bulks were estimated individually, with the average coverage of ∼11.6-fold and 64.1% (474.2 Mb) of kabuli chickpea genome (with an estimated genome size of ∼740 Mb), respectively (Supplementary Table S1). The normalized individual sequence reads of mapping parents and bulks were compared with reference kabuli genomic sequences (pseudomolecules and scaffolds) to mine the valid homozygous SNPs for QTL-seq analysis. The sequencing data generated in the present investigation was submitted to NCBI-sequence read archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra) with accession number SRR2228974 under BioProject ID: PRJNA294404 (submission ID: SUB1081829). The comparative assessment on desi and kabuli draft chickpea genome assemblies provided clues regarding high-quality and uniform whole-genome sequence assembly of kabuli, including its large size (Mb) chromosome pseudomolecule and scaffolds than desi chickpea genome.51 Henceforth, kabuli rather than the desi genome sequence was preferably utilized as a reference for whole-genome resequencing-based SNPs mining and QTL-seq analyses in mapping parents and bulks of chickpea.
3.3. Discovery and annotation of genome-wide SNPs in a mapping population of Pusa 1103 × ILWC 46
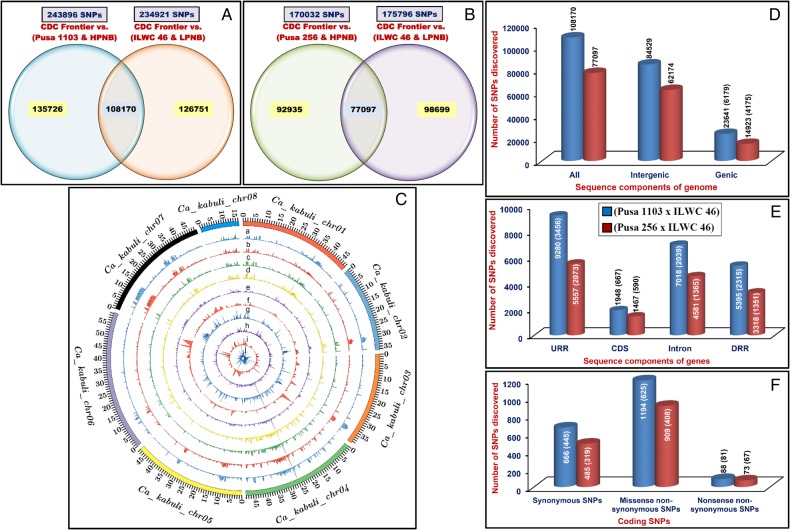
The comparative genome sequence data analysis of high (Pusa 1103 and HPNB) and low (ILWC 46 and LPNB) pod number-containing mapping parents and bulks with reference genomic sequence (pseudomolecule) of kabuli accession (CDC Frontier) identified 243,896 and 234,921 high-quality homozygous SNPs (with read depth ≥10 and SNP base quality ≥20) (Supplementary Tables S2 and S3, Fig. 2A and B). Of these, 108,170 SNPs irrespective of monomorphic/polymorphic allele types were found to be common between high (Pusa 1103 and HPNB) and low (ILWC 46 and LPNB) pod number-containing mapping parents and bulks based on their congruent physical positions (bp) on the reference kabuli genome (with a mean map density of 4.8 kb) (Table 2, Supplementary Table S4, Fig. 2A). These selected SNPs were further used for QTL-seq analysis. The 54,731 (50.6% of identified 108 170 SNPs) SNPs were physically mapped on eight chromosomes of kabuli genome with a mean map density of 6.3 kb that varied from 4.3 (chromosome 7) to 9.1 (chromosome 6) kb (Table 2, Fig. 2B). At a genome-wide level, the density of physically mapped SNPs was maximum on chromosome 7 (235.1 SNPs/Mb) followed by chromosome 4 (213.2 SNPs/Mb) and minimum on chromosome 6 (109.9 SNPs/Mb), with a mean of 157.6 SNPs/Mb (Supplementary Fig. S1A). The remaining 53,439 (49.4%) SNPs were mapped on the unanchored scaffolds of kabuli genome with an average map density of 3.2 kb (Table 2). All the 243,896 and 234,921 high-quality homozygous SNPs were submitted to NCBI dbSNP (http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=NIPGR) with SNP submission (SS) accession numbers (1940124817-1945872952; Supplementary Tables S2 and S3) for unrestricted public access.
Figure 2.
Genome-wide distribution pattern of SNPs identified by comparing the whole-genome resequencing data of high and low pod number-containing parental accessions and homozygous individuals (constituting bulks) from each of two mapping populations [(Pusa 1103 × ILWC 46) and (Pusa 256 × ILWC 46)] with respect to kabuli chickpea (CDC Frontier). SNPs differentiating high and low-pod number-containing parental accessions and homozygous bulks of two mapping populations [A: (Pusa 1103 × ILWC 46) and B: (Pusa 256 × ILWC 46)] from CDC Frontier are illustrated by venn diagrams. HPNB: high pod number bulk and LPNB: low pod number bulk. (C) The relative genomic distribution of SNPs physically mapped on eight chromosomes of kabuli chickpea genome are depicted by a Circos circular ideogram. The outermost circles denote the different physical size (Mb) of eight chromosomes coded with multiple colours as per the pseudomolecule size reported in kabuli chickpea genome.46 SNPs (Pusa 1103 and HPNB) vs. CDC Frontier (a), (ILWC 46 and LPNB) vs. CDC Frontier (b), (Pusa 256 and HPNB) vs. CDC Frontier (c), and (ILWC 46 and LPNB) vs. CDC Frontier (d) are indicated. (e) Total SNPs, including non-synonymous (f) and regulatory SNPs (g) common between high (Pusa 1103 and HPNB) and low (ILWC 46 and LPNB) pod number-containing parental accessions and homozygous mapping individuals. (h) Total SNPs, including non-synonymous (i) and regulatory SNPs (j) common between high (Pusa 256 and HPNB) and low (ILWC 46 and LPNB) pod number-containing parental accessions and homozygous mapping individuals. Frequency of SNPs discovered from the intergenic and various coding (synonymous and non-synonymous) and non-coding (introns and regulatory regions) sequence components of protein-coding genes (D, E and F) annotated on kabuli chickpea genome. Parenthesis designates the number of SNPs-containing genes. The CDS (coding sequences), URR (upstream regulatory region) and DRR (downstream regulatory region) of genes were defined as per the gene annotation of kabuli genome. This figure is available in black and white in print and in colour at DNA Research online.
Table 2.
Genomic distribution of SNPs physically mapped on eight chromosomes and unanchored scaffolds of kabuli chickpea genome
| Chromosomes | Size (Mb) of kabuli chromosomes (pseudomolecules) | Number (%) of common SNPs mapped |
Average map density (kb) |
||
|---|---|---|---|---|---|
| (Pusa 1103 and HPNB) vs. (ILWC 46 and LPNB) | (Pusa 256 and HPNB) vs. (ILWC 46 and LPNB) | (Pusa 1103 and HPNB) vs. (ILWC 46 and LPNB) | (Pusa 256 and HPNB) vs. (ILWC 46 and LPNB) | ||
| Ca_Kabuli_Chr01 | 48.4 | 6,191 (11.3) | 5,643 (16.2) | 7.8 | 8.6 |
| Ca_Kabuli_Chr02 | 36.6 | 5,232 (9.5) | 3,475 (10.0) | 7.0 | 10.5 |
| Ca_Kabuli_Chr03 | 40.0 | 5,712 (10.4) | 3,505 (10.1) | 7.0 | 11.4 |
| Ca_Kabuli_Chr04 | 49.2 | 10,491 (19.2) | 7,331 (21.1) | 4.7 | 6.7 |
| Ca_Kabuli_Chr05 | 48.2 | 6,770 (12.4) | 3,659 (10.5) | 7.1 | 13.2 |
| Ca_Kabuli_Chr06 | 59.5 | 6,540 (11.9) | 4,761 (13.7) | 9.1 | 12.5 |
| Ca_Kabuli_Chr07 | 49.0 | 11,507 (21.0) | 4,564 (13.1) | 4.3 | 10.7 |
| Ca_Kabuli_Chr08 | 16.5 | 2,288 (4.2) | 1,775 (5.1) | 7.2 | 9.3 |
| Total | 347.2 | 54,731 (50.6) | 34,713 (45.0) | 6.3 | 10.0 |
| Unanchored scaffolds | 171.5 | 53,439 (49.4) | 42,384 (55.0) | 3.2 | 4.0 |
| Total | 518.7 | 108,170 | 77,097 | 4.8 | 6.7 |
The structural annotation of SNPs exhibited the occurrence of 84,529 (78.1% of total mined 108,170 SNPs) and 23,641 (21.9%) SNPs in the intergenic and different sequence components of 6,179 genes, respectively (Fig. 2D). A maximum (9,280 SNPs, 39.2%) and minimum (1,948 SNPs, 8.2%) frequency of SNPs were observed in the upstream regulatory regions (URRs) and CDS of genes, respectively (Fig. 2E). Notably, 445 and 706 coding SNPs-containing genes exhibited synonymous (666 SNPs) and non-synonymous (1,282 SNPs) (missense and nonsense) substitutions, respectively (Fig. 2F). The non-synonymous SNPs comprise 1,194 missense and 88 nonsense SNPs in the 625 and 81 genes, respectively. The average frequency of SNPs within genes was estimated as 3.8 SNPs/gene. The functional annotation of 6,179 SNPs-carrying genes revealed correspondence of ∼9.6 and 8.4% of SNPs to transcription factors and disease resistance-related proteins, respectively. The numerous informative SNPs differentiating desi (Pusa 1103), kabuli (CDC Frontier) and wild (ILWC 46) accessions, structurally and functionally annotated at a genome-wide scale, can be deployed for multi-dimensional high-throughput genotyping applications in chickpea.
3.4. Discovery and annotation of genome-wide SNPs in a mapping population of Pusa 256 × ILWC 46
We identified 170,032 and 175,796 high-quality genome-wide homozygous SNPs differentiating the high (Pusa 256 and HPNB) and low (ILWC 46 and LPNB) pod number-containing mapping parents and bulks from reference kabuli accession (CDC Frontier) (Supplementary Table S5 and S6, Fig. 2B and C). Of these, 77,097 SNPs identified to be common between high (Pusa 256 and HPNB) and low (ILWC 46 and LPNB) pod number-containing mapping parents and bulks following the aforesaid criteria were further used in QTL-seq analysis (Supplementary Table S7, Fig. 2C). All these 77,097 SNPs were mapped on the kabuli genome with an average map density of 6.7 kb (Table 2). The 34,713 (45% of identified 770,987 SNPs) SNPs mined were physically mapped on eight chromosomes of kabuli genome with a mean map density of 10 kb that ranged from 6.7 (chromosome 4) to 13.2 (chromosome 5) kb (Table 2, Fig. 2B). At a whole-genome level, the density of physically mapped SNPs was highest on chromosome 4 (149.1 SNPs/Mb) followed by chromosome 1 (116.7 SNPs/Mb) and minimum on chromosome 5 (75.9 SNPs/Mb), with a mean of 99.9 SNPs/Mb (Supplementary Fig. S1B). The rest 42,384 (55%) SNPs were mapped on the unanchored scaffolds of kabuli genome with an average map density of 4.0 kb (Table 2). All these 170,032 and 175,796 high-quality homozygous SNPs were submitted to NCBI dbSNP (http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=NIPGR) with SNP submission (SS) accession numbers (1945872954–1947070701, Supplementary Tables S5 and S6) for unrestricted use.
The structural annotation of SNPs showed the presence of 62,174 (80.6% of total mined 77,097 SNPs) and 14,923 (19.4%) SNPs in the intergenic and various sequence components of 4,175 genes, respectively (Fig. 2D). The URRs and CDS of genes contained maximum (5,557 SNPs, 37.2%) and minimum (1,467 SNPs, 9.8%) frequency of SNPs, respectively (Fig. 2E). Notably, 319 and 475 coding SNPs-carrying genes revealed synonymous (485 SNPs) and non-synonymous (982 SNPs) substitutions, respectively (Fig. 2F). The non-synonymous SNPs contained 909 missense and 73 nonsense SNPs in the 408 and 67 genes, respectively. The average frequency of SNPs within genes was estimated to be 3.6 SNPs/gene. The functional annotation of 4,175 SNPs-carrying genes exhibited that ∼7.3 and 5.3% of SNPs were belonging to transcription factors and disease resistance-related proteins, respectively. These functionally relevant SNPs, discriminating desi (Pusa 256), kabuli (CDC Frontier) and wild (ILWC 46) accessions annotated in diverse sequence components of genome/genes, have tremendous practical utility towards establishing efficient marker-trait association and rapid detection of potential genes/QTLs governing important agronomic traits in chickpea.
3.5. Molecular mapping of QTL-seq-derived major PN QTLs in a mapping population of Pusa 1103 × ILWC 46
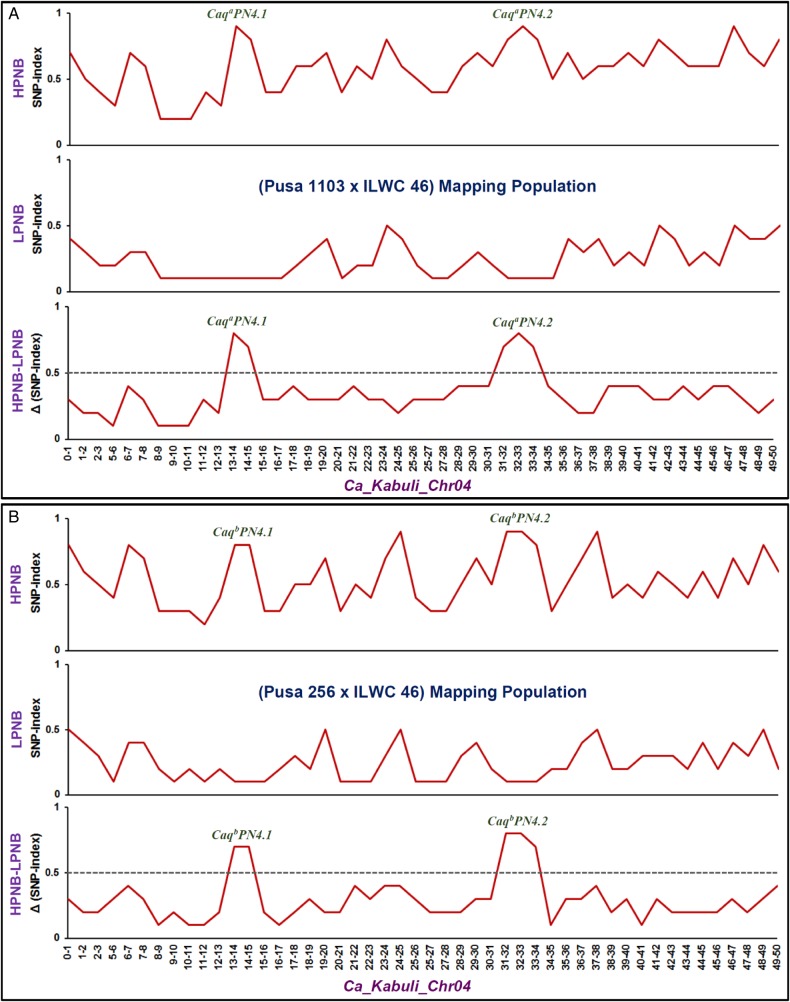
The SNP index of individual SNPs showing differentiation between high (Pusa 1103 and HPNB) and low (ILWC 46 and LPNB) pod number-containing mapping parents and bulks was assessed. We estimated an average SNP index across a 1-Mb genomic interval individually in Pusa 1103 and HPNB as well as ILWC 46 and LPNB using a 10-kb sliding window approach. Further, these SNP indexes were plotted against eight chromosomes of kabuli reference genome. The Δ (SNP index) was measured through combining the SNP index information of HPNB and LPNB, and plotted against the genomic positions (Mb) of kabuli reference genome (Fig. 3A). Two major genomic regions (CaqaPN4.1: 13690423 to 14558233 bp and CaqaPN4.2: 31916540 to 33716608 bp) on chromosome 4 demonstrating the average SNP index of higher than 0.8 in HPNB and lower than 0.2 in LPNB were detected (Fig. 3A) following the SNP index estimation criteria as defined in QTL-seq analysis.42–44 The major genomic regions harbouring PN QTLs identified by QTL-seq were assured by Δ (SNP index) value that is significantly different from 0 at 99% significance level. Our comprehensive analysis of these target genomic regions inferred that high and low pod number-containing mapping individuals constituting the HPNB and LPNB comprised majority of the SNP alleles from high (Pusa 1103) and low (ILWC 46) pod number-containing mapping parental accessions, respectively. Taken together, our findings by QTL-seq in an inter-specific mapping population (Pusa 1103 × ILWC 46) ascertained the presence of two major QTLs (CaqaPN4.1 and CaqaPN4.2) governing PN at the 867.8 Kb [13690423 (SNP_20990A) to 14558233 (SNP_21161A) bp] and 1.80 Mb [31916540 (SNP_27349A) to 33716608 (SNP_28364A) bp] genomic intervals on chromosome 4 of chickpea (Fig. 3A). The SNPs flanking the QTL-seq-derived major PN QTL (CaqaPN4.1 and CaqaPN4.2) were validated in accordance with their expected allelic discrimination by resequencing of PCR amplicons amplified from the parental accessions (Pusa 1103 and ILWC 46) and mapping individuals constituting the HPNB and LPNB.
Figure 3.
Graphs illustrating the SNP index and Δ (SNP index) of HPNB (high pod number bulk), LPNB (low pod number bulk) generated from mQTL-seq analysis in two inter-specific chickpea mapping populations [A: (Pusa 1103 × ILWC 46) and B: (Pusa 256 × ILWC 46)]. The X-axis designates the physical positions (Mb) of kabuli chickpea chromosome 4. The Y-axis denotes the SNP indexes that are measured according to 1-Mb physical interval with a 10-kb sliding window. The Δ (SNP index) was plotted using the statistical confidence intervals under null hypothesis of no QTL (P < 0.01) (indicated by a black dotted line) as per Takagi et al.42 and Das et al.44 Two candidate major genomic intervals underlying the robust PN QTLs (CaqaPN4.1, CaqaPN4.2, CaqbPN4.1 and CaqbPN4.2) identified from each of the two mapping populations using mQTL-seq were defined following the criteria of SNP index near to 1 and 0 in HPNB and LPNB, respectively, and the confidence value of significant Δ (SNP index) >0.5 (significance level at P < 0.01). This figure is available in black and white in print and in colour at DNA Research online.
3.6. Molecular mapping of QTL-seq-derived major PN QTLs in a mapping population of Pusa 256 × ILWC 46
The SNP index of individual SNPs revealing differentiation between high (Pusa 256 and HPNB) and low (ILWC 46 and LPNB) pod number-containing mapping parents and bulks was measured. The estimation of average SNP index (across a 5-Mb genomic interval of a 10-kb sliding window) and Δ (SNP index) of HPNB and LPNB, and their plotting across eight kabuli chromosomes (following aforementioned strategies) identified two major genomic regions (CaqbPN4.1: 13770030 to 14407570 bp and CaqbPN4.2: 31916540 to 33195624 bp) on chromosome 4 exhibiting the average SNP index of higher than 0.9 in HPNB and lower than 0.1 in LPNB (Fig. 3B). The authenticity of these identified major genomic regions harbouring PN QTLs was confirmed by valid significant Δ (SNP index) value (at 99% significance level). The detail analysis of these target genomic regions indicated the presence of most of the SNP alleles from Pusa 256 and ILWC 46 in high and low pod number-containing mapping individuals constituting the HPNB and LPNB, respectively. Collectively, the QTL-seq in an inter-specific mapping population (Pusa 256 × ILWC 46) identified two major QTLs (CaqbPN4.1 and CaqbPN4.2) regulating PN at the 637.5 kb [13770030 (SNP_15561B) to 14407570 (SNP_15696B) bp] and 1.28 Mb [31916540 (SNP_18917B) to 33195624 (SNP_19254B) bp] genomic intervals on chromosome 4 of chickpea (Fig. 3B). The SNPs flanking the QTL-seq-derived major PN QTL (CaqbPN4.1 and CaqbPN4.2) were validated according to their expected allelic discrimination by resequencing of PCR amplicons amplified from the parental accessions (Pusa 256 and ILWC 46) and mapping individuals constituting the HPNB and LPNB.
3.7. QTL region-specific trait association mapping
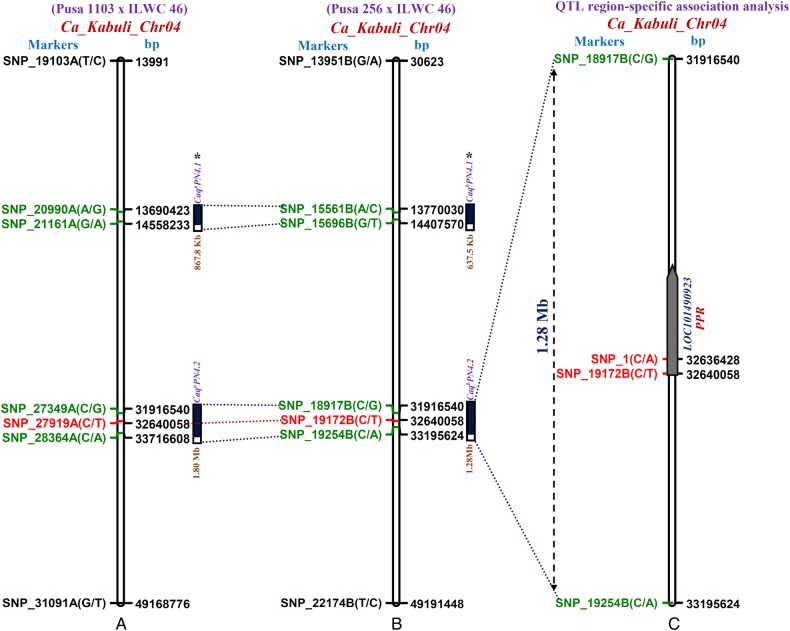
The four major genomic regions underlying PN QTLs (CaqaPN4.1, CaqaPN4.2, CaqbPN4.1 and CaqbPN4.2) detected in two mapping populations of Pusa 1103 × ILWC 46 and Pusa 256 × ILWC 46 using QTL-seq were compared and correlated (Fig. 4A and B). This led to identification of two consensus major genomic regions with short physical intervals of 637.5 kb [CaqbPN4.1: 13770030 (SNP_15561B) to 14407570 (SNP_15696B) bp] and 1.28 Mb [CaqbPN4.2: 31916540 (SNP_18917B) to 33195624 (SNP_19254B) bp] on chromosome 4 harbouring PN QTLs (Fig. 4A and B). These QTLs, being well-validated across two diverse chickpea mapping populations under study, were considered as robust QTLs. The scale-down of longer major PN QTL intervals identified by QTL-seq in individual mapping population into shorter QTL regions by mQTL-seq implicates the significance of mQTL-seq over QTL-seq approach for high-resolution molecular mapping along with fine mapping of major potential genomic regions underlying robust trait-regulatory QTLs in chickpea. Based on congruent physical position, one of the robust QTL (CaqbPN4.1) of these exhibited correspondence with a priorly identified known major QTL regulating pod number/plant was mapped on an intra-specific genetic linkage map of chickpea (Fig. 4A and B).41 The remaining one robust PN QTL (CaqbPN4.2) was considered to be novel (not previously identified) and thus selected by us to delineate potential candidate gene-regulating pod number through QTL region-specific high-resolution trait association mapping. A wider practical applicability of mQTL-seq strategy for rapid detection of major as well as robust high-resolution PN QTLs using two F5 inter-specific chickpea mapping populations (with at least one common parental accession) was evident. More recently, such QTL-seq approach has been deployed in multiple mapping populations for scanning the major QTLs controlling fruit weight and locule number in tomato.45 Collectively, this suggests the accuracy and robustness of mQTL-seq as an approach for rapid delineation of high-resolution major QTLs governing traits of agronomic importance at a genome-wide scale in chickpea.
Figure 4.
The integration of Δ (SNP index)-based QTL-seq-derived four pod number major QTLs in two mapping populations [A: (Pusa 1103 × ILWC 46) and B: (Pusa 256 × ILWC 46)] narrowed down one novel major genomic region harbouring a robust pod number QTL (CaqbPN4.2) into 1.28 Mb sequence interval [between flanking SNP (SNP_18917B and SNP_19254B) markers] indicated by green font on kabuli chickpea chromosome 4. This QTL region was subsequently scaled down into the regulatory (C/T) and coding (C/A) SNPs-containing one candidate PPR (pentatricopeptide repeat) gene associated strongly with pod number by combining mQTL-seq with QTL region-specific association analysis (C). *Known pod number QTLs (CaqaPN4.1 and CaqbPN4.1) documented previously by Varshney et al.41 The details regarding SNPs flanking/localized at the major PN QTLs and SNPs in a PPR gene associated with PN are mentioned in the Supplementary Tables S4 and S7. This figure is available in black and white in print and in colour at DNA Research online.
The targeted resequencing of this 1.28 Mb delineated novel CaqbPN4.2 robust QTL region in high and low pod number-containing mapping parents (Pusa 1103 × ILWC 46) and bulks (HPNB and LPNB) detected 2,913 high-quality SNPs with an average SNP density of 1/439.4 bp. This contained 1,543 intergenic SNPs and 1,370 SNPs derived from the coding and non-coding sequence components of 448 genes. For QTL region-specific trait association analysis, the genotyping information of 2,913 SNPs mined and mapped on a 1.28 Mb (CaqbPN4.2) novel QTL region was correlated with PN field phenotyping data (PN: 21.9–204.5 with 33% CV and 70% H2) and diversity statistics (population structure, PCA, kinship) of 92 desi and kabuli chickpea accessions (PN-specific association panel). The comprehensive association analysis by effective integration of GLM and MLM outcomes with EMMA and FDR based on multiple-comparisons detected two SNPs (SNP_1 at 32636428 bp: C/A and SNP_19172B at 32640058 bp: C/T) in the CDS and URR of a pentatricopeptide repeat (PPR) protein-coding gene revealing strong association (P: 1.5–2.0 × 10−7 and R2 = 25–28%) with PN in chickpea (Fig. 4B and C). This is further ascertained by identification of similar PPR gene-derived regulatory SNP (SNP_19172B at 32640058 bp: C/T) with high Δ (SNP index) (0.8) at CaqbPN4.2 QTL region governing pod number based on our mQTL-seq analysis in two inter-specific mapping populations of chickpea (Fig. 3A and B). Therefore, this strong PN-associated PPR gene localized at a major PN novel QTL interval (CaqbPN4.2) was selected as one of the potential candidate for understanding its efficacy in pod number regulation through differential expression profiling.
3.8. Validation of a PN-associated PPR gene through differential expression profiling
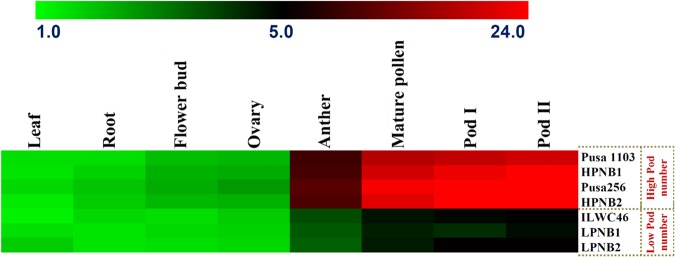
One strong PN-associated regulatory and coding SNPs-containing PPR gene delineated at 1.28 Mb major genomic region harbouring a novel CaqbPN4.2 robust QTL (validated by both mQTL-seq- and QTL region-specific association analysis) was selected for differential expression profiling. The primer-pair designed targeting this gene was used for amplification with the RNA isolated from the vegetative (leaf and root) and reproductive (flower bud, ovary, anther and mature pollen) tissues and two pod (∼5 mm beginning pod size at 5–10 days after anthesis/DAA and ∼2 cm full pod at 15–20 DAA) developmental stages of low and high pod number-containing homozygous mapping individuals (used in QTL-seq analysis) and parental accessions of two mapping populations using semi-quantitative and quantitative RT–PCR assays. A strong PN-associated PPR gene revealed tissue-specific expression in the anthers, mature pollens and pods of parental accessions and mapping individuals compared with their respective vegetative (leaf and root) and reproductive (flower bud and ovary) tissues (Fig. 5). This gene also exhibited pronounced differential up-regulation (∼6.7-folds, P ≤ 10−3) in anther, mature pollen and two pod developmental stages (compared with vegetative tissues) of all high and low pod number-containing parental accessions and individuals of two mapping populations under study (Fig. 5). Interestingly, a significant higher (∼3.5-fold, P ≤ 10−3) differential up-regulation of PPR gene in anther, mature pollen and pod developmental stages of high pod number-containing parental accessions and mapping individuals compared with low pod number-containing parental accessions and mapping individuals was evident (Fig. 5).
Figure 5.
Differential expression profiles of one PPR gene delineated at a novel 1.28 Mb major genomic region underlying the robust pod number QTL (CaqbPN4.2) (detected by integrating mQTL-seq with QTL region-specific association analysis) in diverse vegetative (leaf and root) and reproductive (flower bud, ovary, anther and mature pollen) tissues and two pod developmental stages of low and high pod number-containing parental accessions and homozygous individuals of two mapping populations [(Pusa 1103 × ILWC 46) and (Pusa 256 × ILWC 46)]. The average log signal expression value of PPR gene in various tissues and developmental stages was specified at the top with a colour scale; in which green, black and red colours represent low, medium and high level of expression, respectively. The tissues/stages as well as homozygous mapping individuals and parental accessions utilized for expression profiling are indicated on the top and right sides of expression map, respectively. An endogenous control elongation factor-1 alpha was used in quantitative RT–PCR assay for normalization of the expression value across different tissues/developmental stages of parents and mapping individuals. HPNB: high pod number bulk and LPNB: low pod number bulk. Pod I: Pod development stage 1 (∼5 mm beginning pod size at 5–10 days after anthesis/DAA) and Pod II: Pod development stage 2 (∼2 cm full pod at 15–20 DAA). This figure is available in black and white in print and in colour at DNA Research online.
The integration of mQTL-seq (two inter-specific mapping populations, Pusa 1103 × ILWC 46 and Pusa 256 × ILWC 46) with QTL region-specific association analysis (92 association panel) and differential expression profiling (anther, mature pollen and pod developmental stages) has got functional relevance to identify potential novel regulatory and coding SNP allelic variants in one of the major PPR gene delineated at a novel robust QTL region (CaqbPN4.2) associated strongly with pod number in chickpea. The efficacy of the combinatorial approach integrating genetic and association mapping with expression profiling to scale down potential candidate gene and alleles at a major QTL region governing diverse agronomic traits, including seed weight and pod/seed number is well established in chickpea.9–13, 22,23 A pod number-regulating PPR gene identified in our study, showing >80% sequence conservation with Arabidopsis gene orthologue (At1g52620), is reportedly involved in controlling growth and development-related traits (pollination and embryogenesis) in multiple crop plants, including Arabidopsis and Phaseolus.52,53 Specifically, PPR gene family proteins have definite functional role in regulating proper pollen development by higher accumulation of transcripts in the mature pollen and efficient nuclear-organelle interactions to produce higher fertile seeds (embryogenesis) and pods in diverse plant species.54–62 Accordingly, we observed a higher expression of PPR gene transcripts in the mature pollen and two pod developmental stages of high pod number-containing parental accessions and homozygous mapping individuals vis-a-vis low pod number accessions and mapping individuals in chickpea. However, a detail molecular characterization and functional validation of this gene is essential to decipher its regulation specifically during pollen and pod development causing high pod number in chickpea. The PPR gene once validated in a large scale could be deployed for genomics-assisted crop improvement to develop genetically tailored varieties with higher pod/seed number and yield in chickpea.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
Funding to pay the Open Access publication charges for this article was provided by the National Institute of Plant Genome Research (NIPGR).
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the financial support for this study provided by a research grant from the Department of Biotechnology (DBT), Government of India (102/IFD/SAN/2161/2013-14). S.D. acknowledges the DBT for Junior Research Fellowship award. The authors also acknowledge the financial support provided by the Department of Agriculture and Co-operation (DAC), Government of India for developing wide cross populations and their precise phenotyping under the National Food Security Mission. We are thankful to the Editor and reviewers for critically evaluating the manuscript and providing constructive comments.
References
- 1.Kumar A., Choudhary A.K., Solanki R.K., Pratap A.. 2011, Towards marker-assisted selection in pulses: a review, Plant Breed., 130, 297–313. [Google Scholar]
- 2.Varshney R.K., Murali Mohan S., Gaur P.M. et al. . 2013, Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics, Biotechnol. Adv., 31, 1120–34. [DOI] [PubMed] [Google Scholar]
- 3.Gholipoor M. 2007, Potential effects of individual versus simultaneous climate change factors on growth and water use in chickpea, Int. J. Plant Prod., 1, 189–204. [Google Scholar]
- 4.Merila J., Hendry A.P.. 2014, Climate change, adaptation, and phenotypic plasticity: the problem and the evidence, Evol. Appl., 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jha U.C., Chaturvedi S.K., Bohra A., Basu P.S., Khan M.S., Barh D.. 2014, Abiotic stresses, constraints and improvement strategies in chickpea, Plant Breed., 133, 163–78. [Google Scholar]
- 6.Varshney R.K., Gaur P.M., Chamarthi S.K. et al. . 2013, Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance trait in JG 11, an elite and leading variety of chickpea (Cicer arietinum L.), Plant Genome, 6, doi:10.3835/plantgenome2013.07.0022. [Google Scholar]
- 7.Varshney R.K., Mohan S.M., Gaur P.M. et al. . 2014, Marker-assisted backcrossing to introgress resistance to Fusarium wilt race 1 and Ascochyta blight in C214, an elite cultivar of chickpea, Plant Genome, 7, doi:10.3835/plantgenome2013.10.0035. [Google Scholar]
- 8.Ali L., Madrid E., Varshney R.K. et al. . 2014, Mapping and identification of a Cicer arietinum NSP2 gene involved in nodulation pathway, Theor. Appl. Genet., 127, 481–8. [DOI] [PubMed] [Google Scholar]
- 9.Saxena M.S., Bajaj D., Das S. et al. . 2014, An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea, DNA Res., 21, 695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kujur A., Bajaj D., Saxena M.S. et al. . 2013, Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping, DNA Res., 20, 355–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kujur A., Bajaj D., Saxena M.S. et al. . 2014, An efficient and cost-effective approach for genic microsatellite marker-based large-scale trait association mapping: identification of candidate genes for seed weight in chickpea, Mol. Breed., 34, 241–65. [Google Scholar]
- 12.Kujur A., Upadhyaya H.D., Shree T. et al. . 2015, Ultra-high density intra-specific genetic linkage maps accelerate identification of functionally relevant molecular tags governing important agronomic traits in chickpea, Sci. Rep., 5, 9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kujur A., Bajaj D., Upadhyaya H.D. et al. . 2015, A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea, Sci. Rep., 5, 11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuinstra M.R., Ejeta G., Goldsbrough P.B.. 1997, Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci, Theor. Appl. Genet., 95, 1005–11. [Google Scholar]
- 15.Monforte A.J., Tanksley S.D.. 2000, Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery, Genome, 43, 803–13. [PubMed] [Google Scholar]
- 16.Loudet O., Gaudon V., Trubuil A., Daniel-Vedele F.. 2005, Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family, Theor. Appl. Genet., 110, 742–53. [DOI] [PubMed] [Google Scholar]
- 17.Gaur R., Azam S., Jeena G. et al. . 2012, High-throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.), DNA Res., 19, 357–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiremath P.J., Kumar A., Penmetsa R.V. et al. . 2012, Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes, Plant Biotechnol. J., 10, 716–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roorkiwal M., Sawargaonkar S.L., Chitikineni A. et al. . 2013, Single nucleotide polymorphism genotyping for breeding and genetics applications in chickpea and pigeonpea using the BeadXpress platform, Plant Genome, 6, doi:10.3835/plantgenome2013.05.0017. [Google Scholar]
- 20.Deokar A.A., Ramsay L., Sharpe A.G. et al. . 2014, Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly, BMC Genomics, 15, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaganathan D., Thudi M., Kale S. et al. . 2015, Genotyping-by-sequencing based intra-specific genetic map refines a “QTL-hotspot” region for drought tolerance in chickpea, Mol. Genet. Genomics, 290, 559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj D., Saxena M.S., Kujur A. et al. . 2015, Genome-wide conserved non-coding microsatellite (CNMS) marker-based integrative genetical genomics for quantitative dissection of seed weight in chickpea, J. Exp. Bot., 66, 1271–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajaj D., Upadhyaya H.D., Khan Y. et al. . 2015, A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea, Sci. Rep., 5, 9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho S., Kumar J., Shultz J.F., Anupama K., Tefera F., Muehlbauer F.J.. 2002, Mapping genes for double podding and other morphological traits in chickpea, Euphytica, 125, 285–92. [Google Scholar]
- 25.Rakshit S., Winter P., Tekeoglu M. et al. . 2003, DAF marker tightly linked to a major locus for Ascochyta blight resistance in chickpea (Cicer arietinum L.), Euphytica, 132, 23–30. [Google Scholar]
- 26.Cobos M.J., Fernandez M., Rubio J. et al. . 2005, Linkage map of chickpea (Cicer arietinum L.) based on populations from Kabuli × Desi crosses: location of genes for resistance to Fusarium wilt race 0, Theor. Appl. Genet., 110, 1347–53. [DOI] [PubMed] [Google Scholar]
- 27.Cobos M., Rubio J., Strange R.N., Moreno M.T., Gil J., Millán T.. 2006, A new QTL for Ascochyta blight resistance in an RIL population derived from an interspecific cross in chickpea, Euphytica, 149, 105–11. [Google Scholar]
- 28.Cobos M.J., Winter P., Kharrat M. et al. . 2009, Genetic analysis of agronomic traits in a wide cross of chickpea, Field Crops Res., 111, 130–6. [Google Scholar]
- 29.Lichtenzveig J., Bonfil D.J., Zhang H.B., Shtienberg D., Abbo S.. 2006, Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight, Theor. Appl. Genet., 113, 1357–69. [DOI] [PubMed] [Google Scholar]
- 30.Tar'an B., Warkentin T.D., Tullu A., Vanderberg A.. 2007, Genetic mapping of Ascochyta blight resistance in chickpea (Cicer arietinum) using a simple sequence repeat linkage map, Genome, 50, 26–34. [DOI] [PubMed] [Google Scholar]
- 31.Radhika P., Gowda S.J.M., Kadoo N.Y. et al. . 2007, Development of an integrated intraspecific map of chickpea (Cicer arietinum L.) using two recombinant inbred line populations, Theor. Appl. Genet., 115, 209–16. [DOI] [PubMed] [Google Scholar]
- 32.Anbessa Y., Tar'an B., Warkentin T.D., Tullu A., Vandenberg A.. 2009, Genetic analyses and conservation of QTL for Ascochyta blight resistance in chickpea (Cicer arietinum L.), Theor. Appl. Genet., 119, 757–65. [DOI] [PubMed] [Google Scholar]
- 33.Gowda S.J.M., Radhika P., Kadoo N.Y., Mhase L.B., Gupta V.S.. 2009, Molecular mapping of wilt resistance genes in chickpea, Mol. Breed., 24, 177–83. [Google Scholar]
- 34.Anuradha C., Gaur P.M., Pande S. et al. . 2011, Mapping QTL for resistance to Botrytis grey mould in chickpea, Euphytica, 182, 1–9. [Google Scholar]
- 35.Aryamanesh N., Nelson M.N., Yan G., Clarke H.J., Siddique K.H.M.. 2010, Mapping a major gene for growth habit and QTLs for Ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum, Euphytica, 173, 307–19. [Google Scholar]
- 36.Gowda C.L.L., Upadhyaya H.D., Dronavalli N., Singh S.. 2011, Identification of large-seeded high-yielding stable kabuli chickpea germplasm lines for use in crop improvement, Crop Sci., 5, 198–209. [Google Scholar]
- 37.Rehman A.U., Malhotra R.S., Bett K., Tar'an B., Bueckert R., Warkentin T.D.. 2011, Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress, Crop Sci., 51, 450–63. [Google Scholar]
- 38.Vadez V., Krishnamurthy L., Thudi M. et al. . 2012, Assessment of ICCV2 × JG62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTLs for seed yield and yield components, Mol. Breed., 30, 9–21. [Google Scholar]
- 39.Sabbavarapu M.M., Sharma M., Chamarthi S.K. et al. . 2013, Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.), Euphytica, 193, 121–33. [Google Scholar]
- 40.Stephens A., Lombardi M., Cogan N.O.I. et al. . 2014, Genetic marker discovery, intraspecific linkage map construction and quantitative trait locus analysis of Ascochyta blight resistance in chickpea (Cicer arietinum L.), Mol. Breed., 33, 297–313. [Google Scholar]
- 41.Varshney R.K., Thudi M., Nayak S.N. et al. . 2014, Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.), Theor. Appl. Genet., 127, 445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi H., Abe A., Yoshida K. et al. . 2013, QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations, Plant J., 74, 174–83. [DOI] [PubMed] [Google Scholar]
- 43.Lu H., Lin T., Klein J. et al. . 2014, QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber, Theor. Appl. Genet., 127, 1491–9. [DOI] [PubMed] [Google Scholar]
- 44.Das S., Upadhyaya H.D., Bajaj D. et al. . 2015, Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea, DNA Res., 22, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Illa-Berenguer E., Van Houten J., Huang Z., van der Knaap E.. 2015, Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq, Theor. App. Genet., 128, 1329–42. [DOI] [PubMed] [Google Scholar]
- 46.Varshney R.K., Song C., Saxena R.K. et al. . 2013, Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement, Nat. Biotechnol., 31, 240–6. [DOI] [PubMed] [Google Scholar]
- 47.Abe A., Kosugi S., Yoshida K. et al. . 2012, Genome sequencing reveals agronomically important loci in rice using MutMap, Nat. Biotechnol., 30, 174–8. [DOI] [PubMed] [Google Scholar]
- 48.Kujur A., Bajaj D., Upadhyaya H.D. et al. . 2015, Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea, Front. Plant. Sci., 6, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang H.M., Zaitlen N.A., Wade C.M. et al. . 2008, Efficient control of population structure in model organism association mapping, Genetics, 178, 1709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar V., Singh A., Amitha Mithra S.V. et al. . 2015, Genome-wide association mapping of salinity tolerance in rice (Oryza sativa), DNA Res., 22, 133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruperao P., Chan C.K., Azam S. et al. . 2014, A chromosomal genomics approach to assess and validate the desi and kabuli draft chickpea genome assemblies, Plant Biotechnol. J., 12, 778–86. [DOI] [PubMed] [Google Scholar]
- 52.Cushing D.A., Forsthoefel N.R., Gestaut D.R., Vernon D.M.. 2005, Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis, Planta, 221, 424–36. [DOI] [PubMed] [Google Scholar]
- 53.Abid G., Sassi K., Muhovski Y. et al. . 2012, Identification and analysis of differentially expressed genes during seed development using suppression subtractive hybridization (SSH) in Phaseolus vulgaris, Plant Mol. Biol. Rep., 30, 719–30. [Google Scholar]
- 54.Bentolila S., Alfonso A.A., Hanson M.R.. 2002, A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants, Proc. Natl. Acad. Sci. USA, 99, 10887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kazama T., Toriyama K.. 2003, A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice, FEBS Lett., 544, 99–102. [DOI] [PubMed] [Google Scholar]
- 56.Koizuka N., Imai R., Fujimoto H. et al. . 2003, Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish, Plant J., 34, 407–15. [DOI] [PubMed] [Google Scholar]
- 57.Saha D., Prasad A.M., Srinivasan R.. 2007, Pentatricopeptide repeat proteins and their emerging roles in plants, Plant Physiol. Biochem., 45, 521–34. [DOI] [PubMed] [Google Scholar]
- 58.Schmitz-Linneweber C., Small I.. 2008, Pentatricopeptide repeat proteins: a socket set for organelle gene expression, Trends Plant Sci., 13, 663–70. [DOI] [PubMed] [Google Scholar]
- 59.Ngangkham U., Parida S.K., Dey S.K. et al. . 2010, Genic markers for WA cytoplasm based male sterility and its fertility restoration in rice, Mol. Breed., 26, 275–92. [Google Scholar]
- 60.Su N., Hu M.L., Wu D.X. et al. . 2012, Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production, Plant Physiol., 159, 227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barkan A., Small I.. 2014, Pentatricopeptide repeat proteins in plants, Annu. Rev. Plant Biol., 65, 415–42. [DOI] [PubMed] [Google Scholar]
- 62.Manna S. 2015, An overview of pentatricopeptide repeat proteins and their applications, Biochimie, 113, 93–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.