Abstract
Human flavin-containing monooxygenase 3 (FMO3)-mediated microsomal oxygenation activities, levels of FMO3 protein and FMO3 mRNA, and its modification were investigated in Japanese livers genotyped for the FMO3 gene. Significant correlations were observed for benzydamine N-oxygenation or methyl p-tolyl sulfide S-oxygenation activities (in the range of ~20- to ~40-fold) and the FMO3 levels determined immunochemically in liver microsomes (r2 = 0.73 – 0.75, p < 0.0001, n = 16). Preincubation with the reducing agent ascorbate revealed that FMO3 activities in some liver samples were suppressed. Microsomal FMO3 protein contents (~40-fold) were correlated with FMO3 mRNA levels (r2 = 0.55, p = 0.0010, n = 16), but the FMO3 haplotypes did not affect FMO3 mRNA expression (~100-fold) under the present conditions. On the contrary, FMO3 mRNA levels were multivariately correlated with trans-acting factors, i.e. hepatic nuclear factor 4 (HNF-4) mRNA and nuclear factor Y box-binding protein (NF-Y) mRNA (r2 = 0.31, p = 0.0017, n = 37). These results suggest that considerable individual differences in FMO3 levels may exist in Japanese livers. The liver-enriched transcription factor HNF-4 appears to be a determinant of FMO3 expression in livers, as well as the ubiquitous factor NF-Y.
Keywords: FMO3, benzydamine, methyl p-tolyl sulfide, HNF-4, NF-Y
Introduction
The flavin-containing monooxygenase (FMO, EC 1.14.13.8) is an NADPH-dependent enzyme that catalyzes the oxygenation of a wide variety of nucleophilic compounds containing sulfur or nitrogen atoms. 1,2) FMO3 is considered to be a predominant functional form expressed in adult human liver.3,4) FMO3 plays a role in processing some drugs (e.g., the anticancer drug tamoxifen, the pain medication codeine, and the antifungal drug ketoconazole), the addictive chemical nicotine found in tobacco, and diet-derived trimethylamine.5,6) Ranges in FMO3 protein expression (>10-fold) have been reported in Caucasians.7–9) However, there are few reports on individual variance or causal factors regarding FMO3 for other ethnic groups, e.g. Japanese. Such interindividual variation due to environmental influences would be unlikely because FMO3 expression is unaffected by exogenous agents.5,10)
Interindividual differences in FMO3 levels or its catalytic function could be derived from genetic polymorphism of FMO3 11–13) and/or post-translational modification by environmental factors such as nitric oxide.14) We reported the human FMO3 haplotypes 15) in a Japanese cohort. Low expression of FMO3 could be associated with some variants in the 5′-upstream distal region comprising the FMO3 haplotypes, the putative hepatic nuclear factor 4 (HNF-4) binding site and CCAAT box could also be responsible cis-acting elements in common proximal region of the human FMO3 gene.16) Regarding trans-acting factors, HNF-4 and/or nuclear factor Y box-binding protein (NF-Y) may be candidates as causal factors in the FMO3 variation. Accordingly, hepatic levels of HNF-4 and/or NF-Y are of interest in considering individual differences of FMO3 contents or its function. On the other hand, it has been reported 14) that nitric oxide donors such as S-nitroso-N-acetyl-penicillamine (SNAP) suppress the FMO3 activity in vitro and that ascorbate, a sulfhydryl-reducing agent, can reverse FMO3 inhibition in human liver microsomes, as demonstrated by changes in drug oxygenation activities. The nitric oxide-mediated S-nitrosylation of FMO3 has not been fully examined in other liver samples.
Herein we report inter-individual variations of FMO3 catalytic activities and expression levels in liver microsomes from Japanese samples. FMO3 levels or activities are suggested to be less affected by nitrosation and generally independent of genetic variations of the FMO3 gene. In contrast, the correlations between the amounts of FMO3 mRNA and mRNAs encoding HNF-4, a liver-enriched transcription factor, and NF-Y, a ubiquitous factor binding to the CCAAT box, suggest that these transcription factors are involved in regulating expression of the FMO3 gene.
Materials and methods
Chemicals
Benzydamine-HCl, methyl p-tolyl sulfide, methyl p-tolyl sulfoxide, and SNAP were purchased from Sigma-Aldrich (St. Louis, MO, USA). Olanzapine and sodium L-ascorbate were obtained from Wako Pure Chemicals (Osaka, Japan). Benzydamine N-oxide and olanzapine N-oxide (>99.0% purity) were generous gifts from Dr. A. E. Rettie (University of Washington, Seattle, WA, USA) and Eli Lilly and Company (Indianapolis, IN, USA). The other chemicals and reagents used were obtained in the highest grade commercially available.
DNA analysis for the FMO3 gene
The use of human livers for this study was approved by the Ethics Committees of Showa Pharmaceutical University, St. Marianna University, and Vanderbilt University School of Medicine. Genomic DNA samples were prepared from 37 Japanese livers after pathological examination of specimens isolated during hepatic surgery or after death (5 to 74-year-old males and females) as described previously.17,18) Polymerase chain reaction (PCR) for the all exons, exon-intron junctions, and upstream of the human FMO3 gene was conducted as described previously.15,19) The sequence of the complete human FMO3 gene described in GenBank (Accession Number AL021026) was used as a reference.
FMO3, HNF-4α, and NF-YA mRNA analysis
Hepatic total RNA was isolated from 37 samples using ~100 mg liver samples with an RNeasy Midi Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. cDNAs for mRNA measurement of FMO3, HNF-4α, and NF-YA were synthesized from total RNA (0.5 μg) using High-Capacity cDNA Reverse Transcription Kits for RT-PCR (Applied Biosystems) according to the manufacturer’s instructions. Quantitative real-time PCR reactions for FMO3 (assay identification number, Hs00199368_m1, Applied Biosystems), HNF-4α (Hs00230853_m1), and NF-YA (Hs00242929_m1) mRNA levels were performed using commercial TaqMan® chemistry (Applied Biosystems). Briefly, a reaction mixture contained cDNA from 20 ng total RNA, TaqMan® Gene Expression Master Mix, and 10 mM TaqMan® Gene Expression Assay (Applied Biosystems) in a final volume of 50 μl. The PCR conditions consisted of initial denaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 1 min. Standard curves were constructed with a plasmid containing the FMO3, HNF-4α, or NF-YA cDNA (each of which was constructed) ranging from 1.0 × 101 to 1.0 × 107 copies. The expression levels of FMO3, HNF-4α, and NF-YA mRNAs were estimated from the standard curves and normalized to the amount of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA (quantified using TaqMan® Gene Expression Assay (Hs99999905_m1)), referenced as the housekeeping gene.
FMO3 protein analysis
Human liver microsomes were prepared from 16 livers as described previously18) (restricted number due to limited amounts of the samples). In addition, pooled human liver microsomes were obtained from BD Gentest (Woburn, MA). Membrane fractions expressing recombinant FMO3 (wild-, V257M-, and R205C-type FMO3) were prepared from the Escherichia coli pellets by a series of fractionations and high-speed centrifugation steps described previously.19,20) The liver microsomes or bacterial membrane fractions were resuspended in one volume of 10 mM Tris-HCl buffer (pH 7.4) containing 0.1 mM EDTA and 20% glycerol (v/v). Expression levels of FMO3 were determined by immunoquantitation using a commercial monoclonal anti-FMO3 antibody (BD Gentest) by comparison with a standard of human FMO3 (BD Gentest).19,20)
Analysis of FMO3-mediated drug oxygenation activities
Rates of N-oxygenation of benzydamine21) and S-oxygenation of methyl p-tolyl sulfide22) were determined using HPLC methods described previously.20) Rates of N-oxygenation of olanzapine were also determined23) by UV detection (256 nm) with minor modification. A typical incubation mixture consisted of 100 mM potassium phosphate buffer (pH 8.4), an NADPH-generating system (0.25 mM NADP+, 2.5 mM glucose 6-phosphate, and 0.25 unit/ml glucose 6-phosphate dehydrogenase), substrate, and human liver microsomes (0.10 mg protein) or E. coli membranes expressing FMO3 (1.0 pmol FMO3 equivalent) in a final volume of 0.20 ml, unless otherwise stated. In some experiments, recombinant FMO3 (5.0 μM) was pretreated with a nitric oxide donor, SNAP (5.0 mM), in 100 mM potassium phosphate buffer (pH 8.4) at 30 °C for 10 min. In some cases, human liver microsomes or recombinant FMO3 pretreated with SNAP were preincubated with ascorbate (10 mM) at 15 °C for 30 min. To confirm the contribution of FMO3 in the drug oxygenation activities, some human liver microsomes were preheated at 45 °C for 5 min24) in the absence of the NADPH-generating system. For FMO3 activity, incubations were carried out at 37 °C for 10 min in the presence of the NADPH-generating system. Incubations were terminated by adding 0.20 ml of ice-cold acetonitrile. The aqueous supernatant was centrifuged at 2,000 g for 10 min and subjected to HPLC equipped with an octadecylsilane (C18) column (4.6 mm × 150 mm, 5 μm, Capcellpak, Shiseido, Tokyo, Japan).
Statistical analysis
Linear regression analysis was performed to compare the rates of FMO3-mediated drug oxygenation and amounts of protein or mRNA for FMO3 with the program InStat (GraphPad Software, San Diego, CA, USA). Univariate or multivariate linear regression analysis was also performed to compare FMO3 mRNA levels with contents of HNF-4α and/or NF-YA. Analysis of variance was used to determine statistically significance.
Results
Individual variations of FMO3 catalytic activities
The buffer strength and pH conditions for FMO3-mediated activities were determined using recombinant FMO3 and pooled liver microsomes as enzymes sources (Figure 1). A 100 mM phosphate buffer of pH 8.4 gave apparently optimal activities for recombinant FMO3 for benzydamine N-oxygenation, olanzapine N-oxygenation, or methyl p-tolyl sulfide S-oxygenation (Figures 1A,1B). The 100 mM phosphate buffer (pH 8.4) was also suitable for benzydamine N-oxygenation in human liver microsomes (Figures 1C, 1D)
Figure 1. Effects of pH (A,C) and ion strength (B,D) of phosphate buffer on the drug oxygenation activities of recombinant FMO3 (A,B) and pooled human liver microsomes (C,D).
Rates of N-oxygenation of benzydamine (1 mM) (○) and olanzapine (300 μM) (□) and S-oxygenation of methyl p-tolyl sulfide (1 mM) (△) were determined in 100 mM potassium phosphate buffer (pH 5.4 – 9.0, in A,C) and 10–200 mM potassium phosphate buffer (pH 8.4, in B,D). Means and range are shown for duplicate determinations
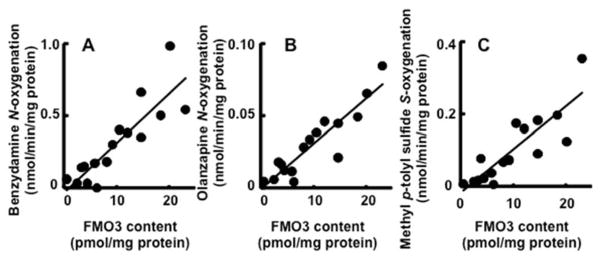
To investigate interindividual differences in the FMO3 activities, the substrates benzydamine, olanzapine, and methyl p-tolyl sulfide were incubated with individual human liver microsomal samples (Figure 2). Significant correlation coefficients were obtained between benzydamine N-oxygenation activities (in the range of ~30-fold) and FMO3 contents (~40-fold) (r2 = 0.75, p < 0.0001, n = 16, Figure 2A) and olanzapine N-oxygenation activities (~20-fold) and the FMO3 contents (r2 = 0.84, p < 0.0001, n = 16, Figure 2B). A similar significant correlation coefficient was obtained between methyl p-tolyl sulfide S-oxygenation activities (~40-fold) and FMO3 contents (r2 = 0.73, p < 0.0001, n = 16) (Figure 2C). Under these conditions, correlation coefficients (r2) among these oxygenation activities ranged from 0.47 to 0.83 (p < 0.01).
Figure 2. Correlations between drug oxygenation activities and FMO3 contents in liver microsomes.
Rates of N-oxygenation of benzydamine (1 mM) (A) and olanzapine (300 μM) (B) and S-oxygenation of methyl p-tolyl sulfide (1 mM) (C) were determined in individual human liver microsomes (n=16).
Recovery of nitrosylation-modulated FMO3 protein with ascorbate
The effects of the nitric oxide donor SNAP on the drug oxygenation activities of recombinant FMO3 were examined. Because of reported S-nitrosylation14), recombinant FMO3 systems harboring Val257Met and Arg205Cys mutations (found in Japanese) were also used as enzyme sources (Figure 3A). Pretreatment of recombinant FMO3 proteins with SNAP resulted in a significant decrease of benzydamine N-oxygenation activities. There was no apparent difference among the variant FMO3 proteins in terms of suppression by nitric oxide. The suppression was significantly reversed following preincubation of the proteins with ascorbate. When pooled human liver microsomes were pretreated with ascorbate, no effects were seen. Among the Japanese livers tested, four liver samples showed increased benzydamine N-oxygenation activities after pretreatment with ascorbate: HL4, HL5, HL16, and HL28 significantly had 9.5-, 2.3-, 1.6-, and 1.9-fold higher activities, respectively (Figure 3B). The recovery of the FMO3 activities of samples HL4, HL5, and HL28 following ascorbate treatment was confirmed by the significant 2.7-, 1.5-, and 1.3-fold increases in methyl p-tolyl sulfide S-oxygenation activities, respectively (results not shown).
Figure 3. Effects of SNAP and ascrobate on FMO3-mediated benzydamine N-oxygenation activities with the recombinant FMO3 and human liver microsomes.
FMO3 activities were determined with recombinant FMO3 (wild-, V257M-, and R205C-type) pretreated with or without SNAP and/or ascorbate (A). Benzydamine N-oxygenation activities in liver microsomes were determined, pretreated with or without ascorbate (B). Some of the liver microsomes were pretreated at 45 °C for 5 min to define the contribution of FMO3 in the enhanced activities. Means and SD are shown for triplicate determinations.
FMO3 levels are independent of genetic variation of the FMO3 gene
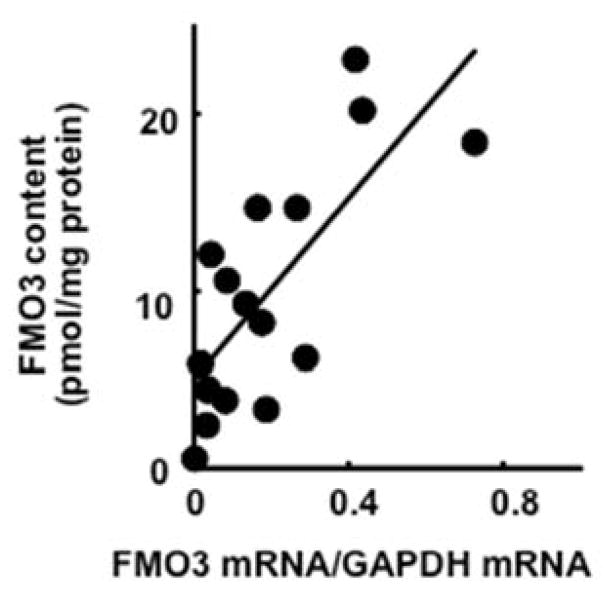
Because FMO3 protein expression is the key factor in drug oxygenation activities (Figure 2), variation of FMO3 protein levels was further investigated. A significant correlation coefficient was obtained between the levels of FMO3 protein and FMO3 mRNA (relative to GAPDH) (in the range of ~100-fold) (r2 = 0.55, p = 0.0010, n = 16, Figure 4).
Figure 4.
Correlations between expression levels of FMO3 protein and of FMO3 mRNA in human livers (n=16).
FMO3 mRNA was accordingly used as a surrogate marker for the FMO3 protein expression. Effects of haplotypes of the FMO3 gene on FMO3 mRNA expression levels in livers (n=37) were investigated. We previously reported seven FMO3 haplotypes, which were determined in the 5′-upstream and coding regions of the FMO3 gene.15) The expression levels of FMO3 mRNA were classified into wild-, heterozygous- or homozygous-genotype groups based on haplotypes 1 and 3 because of possible altered mRNA levels of haplotype 3 15) (Table 1). No relationship was observed between the FMO3 expression levels and the FMO3 genotypes in the present samples. When genotypes were analyzed based on only wild-haplotype 1, no relationship was similarly obtained (results not shown).
Table 1.
FMO3 mRNA expression in livers genotyped for the FMO3 gene
| Genotype | FMO3 mRNA /GAPDH mRNA
|
|
|---|---|---|
| Mean ± SD | n | |
| haplotype 1/1 | 0.30 ± 0.27 | 4 |
| haplotype 1/3 | 0.67 ± 0.44 | 4 |
| haplotype 1/others * | 0.55 ± 0.30 | 15 |
| haplotype 3/3 | 0.26 | 1 |
| haplotype 3/others * | 0.12 ± 0.10 | 3 |
| others */others * | 0.48 ± 0.39 | 10 |
Others: FMO3 haplotypes 2 and 4–7 as reported.15)
Correlation with trans-acting factors HNF-4α and NF-YA
Correlation between FMO3 mRNA levels and HNF-4α or NF-YA mRNA levels in individual liver samples (n=37) was investigated (Figure 5). The multivariate (HNF-4α + NF-YA) correlation coefficient (r2 = 0.31, p = 0.0017, n = 37) was higher than the univariate (HNF-4α or NF-YA) correlation coefficients. In comparing univariate coefficients, the liver-enriched transcription factor HNF-4α (r2 = 0.28, p = 0.0008, n = 37) appears to be the determinant factor regulating individual differences in FMO3 expression, as well as the ubiquitous factor NF-YA (r2 = 0.24, p = 0.0019, n = 37).
Figure 5. Correlations between expression levels of FMO3 mRNA and HNF-4.
α (A) or NF-YA mRNA (B) in human livers.
Relationships between FMO3 and HNF-4α or NF-YA mRNA levels were analyzed with livers (n=37). The univariate and multivariate correlation coefficients are shown in text.
Discussion
We first investigated the relationship between liver microsomal FMO3 catalytic activities (under apparently optimal conditions) and the FMO3 protein levels. Typical liver microsomal N- and S-oxygenation activities were generally correlated with FMO3 protein concentrations in liver samples (Figure 2). The FMO3 protein levels were also significantly correlated with FMO3 mRNA levels (Figure 4). A greater than ten-fold range in interindividual FMO3 protein expression or its catalytic function was seen in Japanese, as in the case of Caucasians.7,8)
We also investigated the modulation of FMO3 function by nitric oxide, because cytochrome P450s and flavin monooxygenases have been reported as targets of nitric oxide in a study with liver microsomes from Koreans 14,25) (another sulfhydryl-reducing agent, dithiothreitol, 14) could not be used in our experimental conditions because of its inhibitory effects on the marker drug oxygenation activities). 3-Morpholinosydnonimine was also tested as a nitric oxide donor, but little decrease in the FMO3 activities was observed in our preliminary experiments. Consequently, SNAP and/or ascorbate were used with samples. At least three livers samples had FMO3 activities elevated by ascrobate treatment (Figure 3); however, these findings should be interpreted as minor contributions to the variation of FMO3 in human livers, with apparently no changes in correlation coefficients between modified FMO3 activities and proteins (results not shown). The most ascorbate-enhanced activities were seen with the liver sample HL-4, harboring the homozygous wild FMO3 genotype. These results in human liver microsomes may imply that the position(s) of nitrosation would not be at the genetic variant Arg205Cys or Val257Met positions, consistent with similarly decreased activities of these recombinant FMO3 proteins after nitrosation (Figure 3).
We investigated the effects of genetic polymorphism of the FMO3 gene on FMO3 expression levels. In Caucasians, the upstream mutations (−2650C>G, −2543T>A, and −2177G>C) might give a higher FMO3 expression than for wild-type FMO3 in a reported reporter assay.12) Accordingly, we compared FMO3 mRNA levels and genotypes, especially wild-type haplotype 1 and the corresponded haplotype 3 15) in Japanese (Table 1). Consequently, FMO3 levels were generally independent of genetic variation of the FMO3 gene in the present analysis.
We also investigated transcriptional regulation of the FMO3 in a Japanese population. Alternative processing of the FMO3 gene has been considered as one of the sources of variation of FMO3 in Caucasians.26) Since alternative processing events at exons 3, 4, 7, and 8 of the FMO3 gene have been reported,26) we confirmed consistent FMO3 mRNA levels in our samples (determined with the primer sets for exons 5–6 of the FMO3 gene) in comparison with those for exons 3–4, 7–8, or 8–9 sets of the FMO3 gene. In our preliminary studies, similar levels of FMO3 mRNA in human livers were observed when differently designed primers were used, with correlation coefficients (r2) of > 0.85 between the results with exons 5–6 primers and other sets of primers in FMO3 mRNA determinations (results not shown), suggesting apparently no alternative processing events.
Our previous findings with HepG2 cell-based reporter assays suggested putative HNF-4 binding sites and the CCAAT box of the FMO3 gene as possible cis-acting elements.16) Klick and Hines27) have suggested octamer-binding protein 1 (Oct-1) or albumin promoter D-site binding protein (DBP) as factors for the putative HNF-4 binding site and also NF-Y as a transcriptional factor for the FMO3 gene. NF-Y has been suggested to be the protein that binds the CCAAT box of FMO3, based on the supershift seen with a NF-Y antibody and a competition assay.27) In contrast, relatively high correlation of HNF-4α mRNA with FMO3 levels was observed in examination of 37 individual Japanese livers (Figure 5). HNF-4 could be an important trans-acting element for (the putative HNF-4 binding site of) the FMO3 gene. The contribution of HNF-4α has been supported by the recent finding28) that endogenous FMO3 mRNA expression can be enhanced by expression of an HNF-4α vector in HepG2 cells. NF-YA mRNA expression was significantly (although not strongly) correlated with the FMO3 levels in individual human liver mRNA (Figure 5). Because multivariate (HNF-4α plus NF-YA) correlation coefficients were higher than the univariate (HNF-4α or NF-YA) individual correlation coefficients, the present results collectively suggest that both HNF-4 and NF-Y may be the responsible trans-acting elements involved with the proximal region of the human FMO3 gene. Consequently, these results are of interest in the context that HNF-4 has been known to be specifically expressed in human adult liver.
In conclusion, the present results suggest that there is considerable individual variation in FMO3 levels and activities in Japanese livers, in an apparently similar manner as Caucasians. FMO3 levels are suggested to be generally independent of genetic variation in the FMO3 gene or of modification by nitric oxide but co-regulated by trans-acting factors such as HNF-4, one of the liver-enriched transcription factors, and NF-Y, a ubiquitous factor binding to the CCAAT box of the FMO3 gene.
Acknowledgments
This work was supported in part by the Ministry of Education, Science, Sports and Culture of Japan, and United States Public Health Service grants R37 CA090426 and P30 ES000267.
References
- 1.Ziegler DM. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab Rev. 2002;34:503–511. doi: 10.1081/dmr-120005650. [DOI] [PubMed] [Google Scholar]
- 2.Phillips IR, Shephard EA. Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol Sci. 2008;29:294–301. doi: 10.1016/j.tips.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Cashman JR. Quantitive analysis of FMO gene mRNA levels in human tissues. Drug Metab Dispos. 2006;34:19–26. doi: 10.1124/dmd.105.006171. [DOI] [PubMed] [Google Scholar]
- 4.Yeung CK, Adman ET, Rettie AE. Functional characterization of genetic variants of human FMO3 associated with trimethylaminuria. Arch Biochem Biophys. 2007;464:251–259. doi: 10.1016/j.abb.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cashman JR, Zhang J. Human flavin-containing monooxygenases. Annu Rev Pharmacol Toxicol. 2006;46:65–100. doi: 10.1146/annurev.pharmtox.46.120604.141043. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki H, Shimizu M. Genetic polymorphism of the flavin-containing monooxygenase 3 (FMO3) associated with trimethylaminuria (fish odor syndrome): Observations from Japanese patients. Curr Drug Metab. 2007;8:487–491. doi: 10.2174/138920007780866825. [DOI] [PubMed] [Google Scholar]
- 7.Overby LH, Carver GC, Philpot RM. Quantitation and kinetic properties of hepatic microsomal and recombinant flavin-containing monooxygenases 3 and 5 from humans. Chem Biol Interact. 1997;106:29–45. doi: 10.1016/s0009-2797(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 8.Koukouritaki SB, Simpson P, Yeung CK, Rettie AE, Hines RN. Human hepatic flavin-containing monooxygenases 1 (FMO1) and 3 (FMO3) developmental expression. Pediatr Res. 2002;51:236–243. doi: 10.1203/00006450-200202000-00018. [DOI] [PubMed] [Google Scholar]
- 9.Hines RN. Developmental and tissue-specific expression of human flavin-containing monooxygenases 1 and 3. Expert Opin Drug Metab Toxicol. 2006;2:41–49. doi: 10.1517/17425255.2.1.41. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell SC. Flavin mono-oxygenase (FMO)--the ‘other’ oxidase. Curr Drug Metab. 2008;9:280–284. doi: 10.2174/138920008784220682. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Shephard EA. Mutation, polymorphism and perspectives for the future of human flavin-containing monooxygenase 3. Mutat Res. 2006;612:165–171. doi: 10.1016/j.mrrev.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Koukouritaki SB, Poch MT, Cabacungan ET, McCarver DG, Hines RN. Discovery of novel flavin-containing monooxygenase 3 (FMO3) single nucleotide polymorphisms and functional analysis of upstream haplotype variants. Mol Pharmacol. 2005;68:383–392. doi: 10.1124/mol.105.012062. [DOI] [PubMed] [Google Scholar]
- 13.Hisamuddin IM, Yang VW. Genetic polymorphisms of human flavin-containing monooxygenase 3: implications for drug metabolism and clinical perspectives. Pharmacogenomics. 2007;8:635–643. doi: 10.2217/14622416.8.6.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryu S-D, Yi H-G, Cha Y-N, Kang J-H, Kang J-S, Jeon Y-C, Park H-K, Yu T-M, Lee J-N, Park C-S. Flavin-containing monooxygenase activity can be inhibited by nitric oxide-mediated S-nitrosylation. Life Sciences. 2004;75:2559–2572. doi: 10.1016/j.lfs.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Allerston CK, Shimizu M, Fujieda M, Shephard EA, Yamazaki H, Phillips IR. Molecular evolution and balancing selection in the flavin-containing monooxygenase 3 gene (FMO3) Pharmacogenet Genomics. 2007;17:827–839. doi: 10.1097/FPC.0b013e328256b198. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu M, Murayama N, Nagashima S, Fujieda M, Yamazaki H. Complex mechanism underlying transcriptional control of the haplotyped flavin-containing monooxygenase 3 (FMO3) gene in Japanese: Different regulation between mutations in 5′-upstream distal region and common element in proximal region. Drug Metab Pharmacokinet. 2008;23:54–58. doi: 10.2133/dmpk.23.54. [DOI] [PubMed] [Google Scholar]
- 17.Inoue K, Yamazaki H, Imiya K, Akasaka S, Guengerich FP, Shimada T. Relationship between CYP2C9 and 2C19 genotypes and tolbutamide methyl hydroxylation and S-mephenytoin 4′-hydroxylation activities in livers of Japanese and Caucasian populations. Pharmacogenetics. 1997;7:103–113. doi: 10.1097/00008571-199704000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki H, Okayama A, Imai N, Guengerich FP, Shimizu M. Interindividual variation of cytochrome P450 2J2 expression and catalytic activities in liver microsomes from Japanese and Caucasian populations. Xenobiotica. 2006;36:1201–1209. doi: 10.1080/00498250600944318. [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki H, Fujita H, Gunji T, Zhang J, Kamataki T, Cashman JR, Shimizu M. Stop codon mutations in the flavin-containing monooxygenase 3 (FMO3) gene responsible for trimethylaminuria in a Japanese population. Mol Genet Metab. 2007;90:58–63. doi: 10.1016/j.ymgme.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu M, Yano H, Nagashima S, Murayama N, Zhang J, Cashman JR, Yamazaki H. Effect of genetic variants of the human flavin-containing monooxygenase 3 (FMO3) on N- and S-oxygenation activities. Drug Metab Dispos. 2007;35:328–330. doi: 10.1124/dmd.106.013094. [DOI] [PubMed] [Google Scholar]
- 21.Yeung CK, Rettie AE. Benzydamine N-oxygenation as a measure of flavin-containing monooxygenase activity. Methods Mol Biol. 2006;320:157–162. doi: 10.1385/1-59259-998-2:157. [DOI] [PubMed] [Google Scholar]
- 22.Stevens JC, Melton RJ, Zaya MJ, Engel LC. Expression and characterization of functional dog flavin-containing monooxygenase 1. Mol Pharmacol. 2003;63:271–275. doi: 10.1124/mol.63.2.271. [DOI] [PubMed] [Google Scholar]
- 23.Ring BJ, Catlow J, Lindsay TJ, Gillespie T, Roskos LK, Cerimele BJ, Swanson SP, Hamman MA, Wrighton SA. Identification of the human cytochromes P450 responsible for the in vitro formation of the major oxidative metabolites of the antipsychotic agent olanzapine. J Pharmacol Exp Ther. 1996;276:658–666. [PubMed] [Google Scholar]
- 24.Yanni SB, Annaert PP, Augustijns P, Bridges A, Gao Y, Benjamin DK, Jr, Thakker DR. Role of flavin-containing monooxygenase in oxidative metabolism of voriconazole by human liver microsomes. Drug Metab Dispos. 2008;36:1119–1125. doi: 10.1124/dmd.107.019646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan ET, Ullrich V, Daiber A, Schmidt P, Takaya N, Shoun H, McGiff JC, Oyekan A, Hanke CJ, Campbell WB, Park CS, Kang JS, Yi HG, Cha YN, Mansuy D, Boucher JL. Cytochromes P450 and flavin monooxygenases--targets and sources of nitric oxide. Drug Metab Dispos. 2001;29:1366–1376. [PubMed] [Google Scholar]
- 26.Lattard V, Zhang J, Cashman JR. Alternative processing events in human FMO genes. Mol Pharmacol. 2004;65:1517–1525. doi: 10.1124/mol.65.6.1517. [DOI] [PubMed] [Google Scholar]
- 27.Klick DE, Hines RN. Mechanisms regulating human FMO3 transcription. Drug Metab Rev. 2007;39:419–442. doi: 10.1080/03602530701498612. [DOI] [PubMed] [Google Scholar]
- 28.Klick DE, Shadley JD, Hines RN. Differential regulation of human hepatic flavin containing monooxygenase 3 (FMO3) by CCAAT/enhancer-binding protein β (C/EBPβ) liver inhibitory and liver activating proteins. Biochem Pharmacol. 2008;76:268–278. doi: 10.1016/j.bcp.2008.05.002. [DOI] [PubMed] [Google Scholar]