Abstract
Background
Anti-tumor necrosis factor (anti-TNF) biologic associated psoriasis has been reported in inflammatory bowel disease (IBD) patients. However, little is known regarding its pathogenesis.
Aims
To identify potential genetic predispositions to anti-TNF associated psoriasis in IBD patients
Methods
This retrospective chart review included IBD patients enrolled in a prospective registry. Cases of anti-TNF associated psoriasis and idiopathic psoriasis unrelated to anti-TNF exposure were confirmed by an expert dermatologist. All patients were genotyped on the Illumina Immunochip. A weighted genetic risk score ascertaining genetic predisposition towards psoriasis was calculated and overall genetic predisposition as well as differential distribution of individual polymorphisms was compared across the three groups.
Results
Our study included 724 IBD patients who initiated anti-TNF therapy and did not develop psoriasis, 35 patients with anti-TNF associated psoriasis, and 38 patients with idiopathic psoriasis. Anti-TNF users who developed psoriasis had a modest but statistically significantly greater psoriasis genetic risk score than anti-TNF controls (mean 0.64 vs. 0.61, p=0.04), and had a similar genetic risk score as those with idiopathic psoriasis (0.64 vs. 0.62, p=0.22). Two loci associated with NOS2 and ETS1 genes achieved p-value < 0.05 when comparing anti-TNF associated psoriasis to anti-TNF controls. Three loci were significantly different between anti-TNF associated psoriasis and idiopathic psoriasis including a polymorphism near NOS2 encoding for inducible nitric oxide synthase that is produced by dendritic cells in skin lesions in psoriasis.
Conclusions
Patients with anti-TNF associated psoriasis had a modestly greater genetic predisposition towards psoriasis but no single causative polymorphism was identified.
Keywords: Crohn’s disease, ulcerative colitis, psoriasis, anti-TNF
INTRODUCTION
Inflammatory bowel diseases (IBD) comprising Crohn’s disease (CD) and ulcerative colitis (UC) are chronic immunologically mediated diseases that often have an onset during young adulthood and are characterized by a protracted life-long course of remissions and relapses1. Monoclonal antibodies to tumor necrosis factor α (anti-TNF; infliximab (IFX), adalimumab (ADA), certolizumab pegol (CZP), golimumab (GLM)) have revolutionized our management of IBD. They have substantially improved our ability to achieve disease remission, reduced the need for hospitalizations and surgery and improved patients’ health-related quality of life2-4. However, emergence of treatment-related adverse effects is an important concern regarding the long-term safety of these drugs to both physicians and patients.
Safety of anti-TNF agents has thus far primarily focused on treatment-related infections and malignancies5, 6. However, a group of adverse events that are increasingly being recognized to occur with use of anti-TNF therapies are paradoxical immunologic phenomena7, 8 and emergence of immune-mediated diseases following initiation of anti-TNF therapy. They often require additional immunosuppression or treatment with corticosteroids and sometimes necessitate cessation of the offending anti-TNF drug for resolution. They also have a penchant to recur with a second anti-TNF agent in a sizeable proportion of patients8. One such complication that is peculiarly paradoxical owing to the established efficacy of anti-TNF agents to treat it is the development of psoriasis. Several cases series report the occurrence of this complication associated with anti-TNF therapy9-14. While occurring in fewer than 10% of patients initiating anti-TNF therapy, the consequences of such an adverse effect can be substantial. In a large series of cases, nearly half the patients required withdrawal of the offending anti-TNF agent due to the rash and over half recurred with the second anti-TNF agent11 though in other studies this proportion was lower15. The exact mechanism of this adverse event, which represents an immunologic paradox as anti-TNF therapy is highly effective for the treatment of psoriasis, remains unknown with several hypotheses postulated10, 12, 13. Some have suggested that this phenomenon may represent a genetic predisposition towards psoriasis given the known overlap in risk loci between various immune mediated diseases16-18 while others have suggested an alteration in the TNF-α inhibition of plasmacytoid dendritic cells and a subsequent increase in interferon-α production8, 12, 14.
With increasing availability of agents targeting diverse mechanisms of inflammation in CD and UC, identification of the biological basis of treatment emergent-adverse effects will allow for selection of the appropriate patient population for specific therapies as well as better understand the disease pathogenesis and shared biologic mechanisms behind the different immune mediated diseases. Consequently, we performed this study with the following aims: (1) To use a large cohort of well-characterized patients with CD and UC initiating anti-TNF therapy to identify those who developed psoriasis associated with anti-TNF therapy; (2) Compare genetic predisposition towards psoriasis among anti-TNF users who develop anti-TNF associated psoriasis with those who did not develop this, and identify specific genetic polymorphisms predisposing towards this complication; and (3) Compare genetic risk score and distribution of psoriasis risk loci between those who developed psoriasis after anti-TNF exposure to those with idiopathic psoriasis.
METHODS
Study Population
The population for this study consisted of patients enrolled in the Prospective Registry for Inflammatory Bowel Diseases Study at Massachusetts General Hospital (PRISM). This registry, described in previous publications from our center19-21, is an ongoing prospective registry of patients initiated in 2005 recruiting patients aged 18 years or older with a diagnosis of CD, UC, or indeterminate colitis. After providing informed consent, all patients are interviewed by a trained research co-coordinator who gathered information on disease including phenotype, location, and prior medical and surgical treatments. Disease location and behavior in CD was classified by the Montreal classification while disease extent was similarly described in UC. Current and past use of immunomodulators (azathioprine, 6-mercaptopurine, methotrexate) and anti-TNF agents (IFX, ADA, CZP, GLM) was assessed and updated until December 2014. All patients were of European ancestry.
Assessment of anti-TNF associated psoriasis
All medical records of enrolled patients were reviewed by two authors (PV, JSJ) to identify cases of anti-TNF associated psoriasis. Patients were divided into three groups: individuals with anti-TNF exposure with subsequent psoriasis development, individuals with anti-TNF use but no psoriasis, and individuals with psoriasis without a history of anti-TNF exposure (idiopathic psoriasis). In a small subset of patients (n=6), development of psoriasis coincided with anti-TNF exposure but it was not possible to determine a clear correlation with medication exposure for this group, therefore, they were excluded from analysis. For patients with suspected anti-TNF associated psoriasis who did not undergo dermatology evaluation at our institution during the course of their clinical care, we confirmed the assessment of anti-TNF associated psoriasis with review by a Board-certified dermatologist (DK). A diagnosis of anti-TNF associated psoriasis was made based on morphology and distribution of lesions, lack of history of psoriasis prior to anti-TNF exposure, skin biopsy findings when performed, and response to topical therapy and/or cessation of anti-TNF agent. For those with anti-TNF associated psoriasis, psoriasis type (guttate, plaque, inverse, palmoplantar), and anti-TNF agent associated with psoriasis induction were recorded.
Genotyping
Buffy coat was extracted from approximately 10mL of blood drawn from all consented patients. Genotyping was performed on the Illumina Immunochip platform at the Broad Institute (Cambridge, MA). The Immunochip is a custom designed Immunochip with nearly 200,000 fine mapping loci that have been putatively associated with inflammatory or immune mediated diseases22. We extracted information on polymorphisms at the most recently described list of 36 psoriasis risk loci23. A psoriasis genetic risk score (GRS) ascertaining predisposition towards psoriasis was calculated by multiplying the odds ratio for association with psoriasis based on data from Tsoi et al. multiplied by the number of copies of the risk allele23 [GRS = ∑ (Odds ratio SNP × number of risk alleles (0,1, or 2))]. This incorporates both the strength of the association of each SNP with the disease of interest as well as the number of such polymorphisms in each patient. On sensitivity analysis, we calculated this risk score by using the log of the odds ratio as well as by merely summing the number of risk alleles assigning equal weight to all loci. While it is acknowledged that the distribution of risk loci may be different in our cohort, due to the larger size of the Tsoi et al. study provides more robust estimates of effect sizes in calculating the genetic risk score.
Statistical Analysis
Continuous variables were summarized using means and standard deviations while categorical variables were described as proportions and compared using the chi-square test. Statistical analysis was performed using Stata 13.1 (StataCorp, College Station, TX). Extraction of genotypic data and calculation of the genetic risk score was performed using Plink V1.0724. All SNPs met the Hardy-Weinberg equilibrium threshold of p > 0.001, genotyping call rate > 95% and genotyping success rate > 90%. First, we compared the genetic risk score of anti-TNF associated psoriasis to the no psoriasis group using the t-test where a p-value < 0.05 indicated statistical significance. Next, we examined differential distribution of the 36 psoriasis risk loci between the anti-TNF associated psoriasis and no psoriasis group where a nominal p-value < 0.05 indicated plausibility of association while a p-value adjusted for multiple comparison testing (i.e. < 0.001) indicated independent statistical significance. Similar analyses were performed comparing anti-TNF associated psoriasis to idiopathic psoriasis. All genetic analysis utilized the allelic model. The study was approved by the Institutional Review Board of Partners Healthcare and all patients provided full informed consent.
RESULTS
Study Population
Our study population consisted of 765 patients who had initiated anti-TNF therapy among whom 35 developed anti-TNF associated psoriasis (4.5%). Our cohort additionally included 38 patients who had idiopathic psoriasis. Table 1 compares the characteristics of anti-TNF users who developed psoriasis to those who did not. There was no difference in the gender, or racial distribution between the two groups. However, those with anti-TNF associated psoriasis were diagnosed with IBD at a younger age (21 years) compared to anti-TNF users without psoriasis (27 years, p=0.002). Approximately between one-quarter and one-third of patients in each group had UC (p=0.34). There was also no significant difference in the type of anti-TNF used, and psoriasis was observed with all three anti-TNF agents though most commonly with infliximab, likely reflecting larger population at risk with the earlier availability of this drug. Table 2 compares the characteristics of the patients with anti-TNF associated psoriasis to those with idiopathic psoriasis. As noted, there was no difference in gender, or racial distribution of both groups. However, anti-TNF associated psoriasis patients were more likely to have been diagnosed at a younger age compared to idiopathic psoriasis. As well, there was a trend towards more perianal involvement or complicated phenotype in CD and pancolitis in UC. There was no difference in the type of psoriasis with plaque-type being the most common in both groups. However, plaque psoriasis was more frequent in the anti-TNF associated psoriasis group compared to idiopathic psoriasis (p=0.01).
Table 1. Comparison of characteristics of patients with anti-tumor necrosis factor antibodies (anti-TNF) with or without development of psoriasis.
| Characteristics | Anti-TNF associated psoriasis (n = 35) (%) |
Anti-TNF controls (n = 724) (%) |
p value |
|---|---|---|---|
|
Age at diagnosis (in
years) (Mean (SD) |
21 (10) | 27 (12) | 0.002 |
| Gender | 0.38 | ||
| Male | 39 | 46 | |
| Female | 61 | 54 | |
| Race | 0.98 | ||
| White | 95 | 95 | |
| Black | 0 | 2 | |
| Hispanic | 5 | 1 | |
| Asian | 0 | 2 | |
| Native American | 0 | 1 | |
| IBD Type | 0.34 | ||
| UC | 24 | 31 | |
| CD | 76 | 65 | |
| IC | 0 | 4 | |
|
Disease Location
(CD only) |
0.14 | ||
| Ileal | 16 | 24 | |
| Colonic | 76 | 56 | |
| Ileocolonic | 8 | 20 | |
|
Disease behavior
(CD only) |
0.80 | ||
| Inflammatory | 39 | 43 | |
| Stricturing | 29 | 23 | |
| Penetrating | 32 | 33 | |
|
Perianal disease
(CD only) |
24 | 22 | 0.79 |
|
Disease extent –
Pancolitis (UC only) |
87 | 59 | 0.27 |
| TNF Exposure** | |||
| Certolizumab | 22 | 14 | 0.15 |
| Infliximab | 49 | 54 | 0.53 |
| Adalimumab | 29 | 32 | 0.69 |
Race was not available in 106 patients in the no psoriasis group;
For TNF associated psoriasis group, this reflects the drug they were on at time of psoriasis. For anti-TNF controls, these numbers reflect the number of patients exposed to certolizumab, infliximab or adalimumab and consequently can add up to more than the total number of patients in cases of users of multiple anti-TNF agents.
Table 2. Comparison of characteristics of patients with anti-tumor necrosis factor biologic associated psoriasis and idiopathic psoriasis.
| Characteristic | Anti-TNF associated psoriasis (n = 35) (%) |
Idiopathic psoriasis (n = 38) (%) |
p value |
|---|---|---|---|
|
Age at diagnosis (in
years) (Mean (SD) |
21 (10) | 37 (17) | < 0.001 |
| Gender | 0.59 | ||
| Male | 39 | 45 | |
| Female | 61 | 55 | |
| Race | 0.32 | ||
| White | 95 | 89 | |
| Black | 0 | 8 | |
| Hispanic | 5 | 0 | |
| Asian | 0 | 3 | |
| Native American | 0 | 0 | |
| IBD Type | 0.43 | ||
| UC | 24 | 32 | |
| CD | 76 | 63 | |
| IC | 0 | 5 | |
|
Disease Location
(CD only) |
0.05 | ||
| Ileal | 16 | 36 | |
| Colonic | 76 | 36 | |
| Ileocolonic | 8 | 28 | |
|
Disease behavior
(CD only) |
0.06 | ||
| Inflammatory | 39 | 72 | |
| Stricturing | 29 | 14 | |
| Penetrating | 32 | 14 | |
|
Perianal disease
(CD only) |
24 | 0 | 0.002 |
|
Disease extent –
Pancolitis (UC only) |
87 | 37 | 0.06 |
| TNF Exposure | |||
| Certolizumab pegol | 9 (22) | N/A | |
| Infliximab | 20 (49) | N/A | |
| Adalimumab | 12 (29) | N/A | |
| Psoriasis Type | |||
| Inverse | 6 (17) | 6 (16) | 0.90 |
| Guttate | 3 (9) | 0 (0) | N/A |
| Plaque | 18 (51) | 9 (24) | 0.01 |
| Palmoplantar | 5 (14) | 3 (8) | 0.41 |
| Nails | 0 (0) | 1 (3) | N/A |
| Scalp | 10 (29) | 17 (45) | 0.16 |
Genetic Analysis
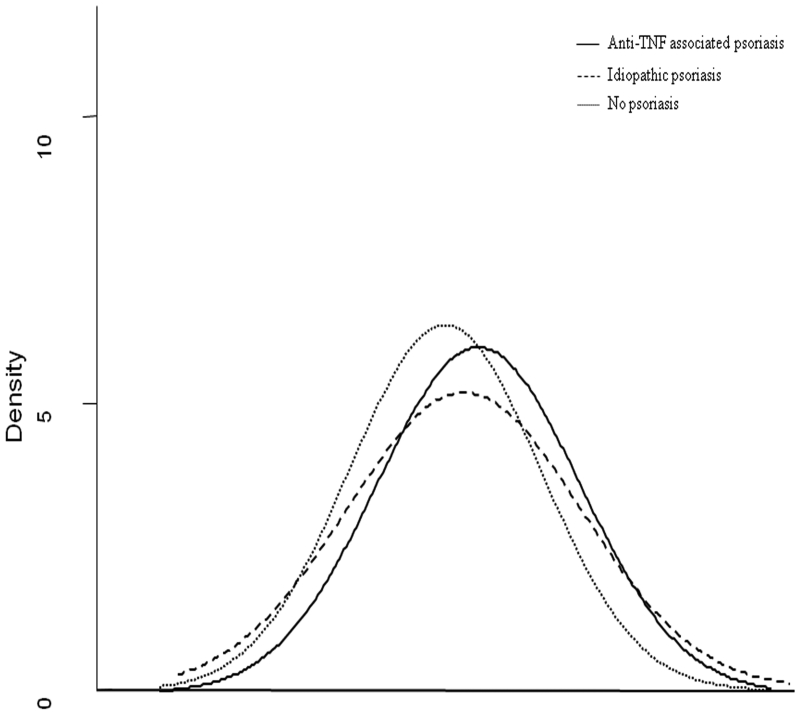
Figure 1 demonstrates the distribution in genetic risk score between anti-TNF users who developed psoriasis and those who did not. The mean psoriasis genetic burden score using the cumulative sum of odds ratios was higher in those who developed psoriasis compared to those who did not (p=0.04). This difference persisted as marginally significant in a count of the number of risk alleles (p=0.06) or using log odds ratio (p=0.07). In contrast, there was no difference in genetic burden between those who developed anti-TNF associated psoriasis and patients with idiopathic psoriasis (p=0.22) (Figure 1).
Figure 1. Comparison of genetic predisposition towards psoriasis among anti-TNF users, anti-TNF associated psoriasis, and idiopathic psoriasis.
A genetic risk score was calculated by using a cumulative sum of polymorphisms in the 36 risk alleles for psoriasis. The figure demonstrates a greater genetic predisposition towards psoriasis among anti-TNF associated psoriasis (n=35, black line) compared to anti-TNF users without this complication (n=730, dotted line) (p=0.04) and comparable to idiopathic psoriasis (n=38, dashed line) (p=0.22)
We then examined differential distribution of individual genetic polymorphisms predisposing towards psoriasis between anti-TNF associated psoriasis and anti-TNF user controls. Overall, two SNPs met our nominal significance threshold at p < 0.05 (Supplemental Table 1). The risk allele (A) for rs28998802 (NOS2) had a minor allele frequency of 0.22 among those with anti-TNF associated psoriasis compared to 12% among anti-TNF users who did not develop psoriasis (Odds ratio (OR) 2.02, p=0.022). The minor G allele for rs3802826 (ETS1) was found in 31% of those with anti-TNF associated psoriasis compared to 45% of controls (OR 0.55, p=0.025) while the distribution of rs1295685 (IL13, IL4) was borderline significant (p=0.07) with a frequency of 0.27 among those with anti-TNF associated psoriasis compared to 0.18 among controls. We then combined the two significant SNPs in a single risk score ranging from 0 to 4. The prevalence of anti-TNF psoriasis ranged from 2% in those with a score of 0 (no SNPs) to 33% with a score of 4 (homozygous at both loci) (p=0.005). None of the SNPs met independent significance on adjustment for multiple hypothesis testing.
On comparison of the genetic risk loci between anti-TNF associated psoriasis and idiopathic psoriasis, the lowest p-value was again observed for rs28998802 (NOS2) with a minor allele frequency of only 0.07 with idiopathic psoriasis compared to 0.22 in anti-TNF associated psoriasis (p=0.014, OR 3.65). Two other loci, rs9504361 (OR 0.51, p=0.05) and rs8016947 (OR 0.48, p=0.04) met nominal significance based on a univariate p-value < 0.05 (Supplemental Table 2). None retained significance on adjustment for multiple hypothesis testing.
DISCUSSION
Paradoxical immune-mediated diseases are a rare and unpredictable complication of anti-TNF therapy for IBD and much remains unknown about their pathogenesis and risk factors for occurrence8. Using data from a large and well-phenotyped cohort of patients with IBD, we demonstrate that patients who developed anti-TNF associated psoriasis have a weakly greater genetic predisposition towards psoriasis. Two psoriasis risk alleles were nominally associated with this increase in risk, though none remained robustly significant on adjustment for multiple hypothesis testing.
Most prior literature on anti-TNF associated psoriasis has been case series with few examining risk factors for onset, and none previously exploring genetic contribution8-14. In a review by Denadai et al., a total of 47 studies comprising 222 patients were identified, among which three-quarters were diagnosed with CD similar to our findings, reflecting the predominance of use of anti-TNF therapy for this disease12. While the most common inciting anti-TNF in our study and in prior studies has been infliximab, we identified several cases that occurred with adalimumab and certolizumab as well demonstrating the potential class-effect in the development of this complication rather than a drug specific adverse event. Plaque-type lesions were the most common reported types in our series as in prior reports9, 11-13. Two prior studies examined the role of genetics in anti-TNF associated psoriasis. Using the same risk loci as genotyped in this study, Cleynen et al. adopted an approach of performing initially an association analysis with all psoriasis risk loci, subsequently combining the significant SNPs into a risk score15. This was in distinction to our approach where the genetic risk score was constructed by using data from all known psoriasis risk loci. By utilizing this approach, we demonstrated a modest increase in genetic predisposition towards psoriasis among those who developed anti-TNF associated psoriasis, similar to their finding of a nominally higher frequency of psoriasis among those carrying more than 4 risk variants compared to those carrying between 1-4 such polymorphisms. Tillack et al. specifically examined the role of IL-23R, IL-23A, and IL-12B polymorphisms in explaining response to ustekinumab in such patients14.
The mechanism behind the development of anti-TNF associated psoriasis is unclear though several hypotheses have been proposed. The most widely accepted hypothesis for development of anti-TNF associated psoriasis is a reduction in tumor necrosis factor α levels in conjunction with an increase in interferon-α (IFNα) produced by plasmacytoid dendritic cells12, 25, 26. Supporting this hypothesis are studies demonstrating higher levels of IFNα in anti-TNF associated psoriasis lesions compared to idiopathic psoriasis and development of psoriasis after injection of IFN-α or inducers of IFN-α27, 28. However, the development of these lesions in only a small subset (fewer than 10%) of patients initiating anti-TNF therapy for various indications suggest a possible inherent tendency towards developing psoriasis. There is a considerable overlap in genetics between psoriasis, Crohn’s disease, and ulcerative colitis with several overlapping susceptibility genes including interleukin-12/interleukin-23 (IL12B, IL-23R), REL and TYK16-18. Indeed, our findings suggest that this likely plays some role as patients who developed anti-TNF associated psoriasis had a greater genetic predisposition towards psoriasis than those exposed to anti-TNF agents who did not develop psoriasis, and comparable to that with patients with idiopathic psoriasis.
Two SNPs were nominally associated with risk of anti-TNF associated psoriasis. The first, ETS1, was inversely associated with a lower frequency of occurrence in cases. Ets1 is a transcription factor that is normally expressed in the stratified epithelium and has been associated with upregulation in squamous cell carcinomas16, 23, 29. It is important for CD8 T cell differentiation in the thymus, blocks differentiation of keratinocytes, and may have a potential role in immune regulation by inducing expression of matrix metalloproteases, defensins, chemokines, and cytokines29. Interestingly, a polymorphism near the NOS2 locus was found significantly more frequently among anti-TNF associated psoriasis cases compared to either idiopathic psoriasis or anti-TNF controls who did not develop psoriasis. NOS2 encodes inducible nitric oxide synthase (iNOS) which is expressed by TNF-α producing dendritic cells in psoriatic lesions30. NOS2 expression is absent in normal skin but is significantly upregulated in psoriatic lesions, associated by Th1 cytokines31. In addition, nitric oxide levels are elevated in active psoriasis and correlate with severity of disease32. Borderline significance was also observed for a locus proximate to genes for IL13 and IL4 which have been associated with asthma and atopic diseases in addition to psoriasis33, 34. Interleukin-4/IL-13 has been shown to inhibit development of Th17 cells, promote Th2 differentiation at lower levels and Th1 differentiation at higher levels35.
We readily acknowledge several limitations to our study. Despite being a large and one of the first studies to examine the overall genetic predisposition towards psoriasis in patients developing anti-TNF associated psoriasis, the number of cases limited robust genetic analysis to identify relevant SNPs. Post-hoc analysis revealed 81% power to detect a significant difference based on the mean genetic risk score estimates in each group. External validation cohorts are essential to confirm our findings and expand analysis to other immune mediated diseases where anti-TNF agents are used as treatments though our findings remained significant on interval validation with bootstrapping. Second, a referral cohort from a tertiary IBD center may be skewed towards more severe disease though one would not expect that to affect generalizability of our cohort of anti-TNF associated psoriasis, and no differences were noted in severity between the different groups. Third, owing to the retrospective design of the study, complete phenotypic characterization of all patients with psoriasis or histological evaluation was not performed. Moreover, retrospective association of anti-TNF medication use and the subsequent development of psoriasis is an observed correlation and causation cannot be inferred. Finally, establishing a diagnosis of anti-TNF associated may sometimes be clinically challenging. Patients where this diagnosis could not be made with high probability were excluded from our analysis. Since anti-TNF associated psoriasis remains a clinical diagnosis, our reliance on a dermatologist with expertise in this field in conjunction with treating gastroenterologists, represents the best available method to establish this diagnosis. We also examined only anti-TNF associated psoriasis and did not include other skin lesions such as eczema or lupus.
In conclusion, using a large well phenotyped cohort of patients with IBD, we demonstrate that patients who developed anti-TNF associated psoriasis had a greater genetic predisposition towards psoriasis than control anti-TNF users who did not develop these lesions, and a similar predisposition as those with idiopathic psoriasis. Two SNPs at NOS2 and ETS1 were nominally associated with the development of this complication. Larger studies are required to robustly replicate our findings and further inform our understanding of the pathogenesis of this paradoxical immune-mediated complication of anti-TNF therapy.
Supplementary Material
Acknowledgments
Source of funding: Ananthakrishnan is supported in part by a grant from the National Institutes of Health (K23 DK097142). This work is also supported by the National Institutes of Health (NIH) (P30 DK043351) to the Center for Study of Inflammatory Bowel Diseases.
Footnotes
Conflicts of Interest: Ananthakrishnan has served on the scientific advisory boards for Abbvie, Exact Sciences, and Cubist pharmaceuticals and has received grant support from Cubist and Amgen.
Disclosures
Priyanka Vedak – no conflict of interest exists
Daniela Kroshinsky – no conflict of interest exists
Jessica St. John – no conflict of interest exists
Ramnik Xavier – no conflict of interest exists
Vijay Yajnik - no conflict of interest exists
Ashwin Ananthakrishnan - Scientific advisory board for Cubist pharmaceuticals, Abbvie, Exact Sciences
Author Contributions:
Vedak, Kroshinsky, Xavier: study design, data collection, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content
St. John: data collection, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content
V Yajnik study design, data collection, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, supervision for the study
A Ananthakrishnan study design, data collection, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content
REFERENCES
- 1.Cosnes J, Gower-Rousseau C, Seksik P, et al. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–94. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Horin S, Kopylov U, Chowers Y. Optimizing anti-TNF treatments in inflammatory bowel disease. Autoimmun Rev. 2014;13:24–30. doi: 10.1016/j.autrev.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 3.D’Haens GR, Panaccione R, Higgins PD, et al. The London Position Statement of the World Congress of Gastroenterology on Biological Therapy for IBD with the European Crohn’s and Colitis Organization: when to start, when to stop, which drug to choose, and how to predict response? Am J Gastroenterol. 2011;106:199–212. doi: 10.1038/ajg.2010.392. quiz 213. [DOI] [PubMed] [Google Scholar]
- 4.Peyrin-Biroulet L. Anti-TNF therapy in inflammatory bowel diseases: a huge review. Minerva Gastroenterol Dietol. 2010;56:233–43. [PubMed] [Google Scholar]
- 5.Afif W, Loftus EV., Jr. Safety profile of IBD therapeutics: infectious risks. Gastroenterol Clin North Am. 2009;38:691–709. doi: 10.1016/j.gtc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Bewtra M, Lewis JD. Safety profile of IBD: lymphoma risks. Gastroenterol Clin North Am. 2009;38:669–89. doi: 10.1016/j.gtc.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Fiorino G, Allez M, Malesci A, et al. Review article: anti TNF-alpha induced psoriasis in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2009;29:921–7. doi: 10.1111/j.1365-2036.2009.03955.x. [DOI] [PubMed] [Google Scholar]
- 8.Fiorino G, Danese S, Pariente B, et al. Paradoxical immune-mediated inflammation in inflammatory bowel disease patients receiving anti-TNF-alpha agents. Autoimmun Rev. 2014;13:15–9. doi: 10.1016/j.autrev.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Afzali A, Wheat CL, Hu JK, et al. The association of psoriasiform rash with anti-tumor necrosis factor (anti-TNF) therapy in inflammatory bowel disease: a single academic center case series. J Crohns Colitis. 2014;8:480–8. doi: 10.1016/j.crohns.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Barthel C, Biedermann L, Frei P, et al. Induction or exacerbation of psoriasis in patients with Crohn’s disease under treatment with anti-TNF antibodies. Digestion. 2014;89:209–15. doi: 10.1159/000358288. [DOI] [PubMed] [Google Scholar]
- 11.Cullen G, Kroshinsky D, Cheifetz AS, et al. Psoriasis associated with anti-tumour necrosis factor therapy in inflammatory bowel disease: a new series and a review of 120 cases from the literature. Aliment Pharmacol Ther. 2011;34:1318–27. doi: 10.1111/j.1365-2036.2011.04866.x. [DOI] [PubMed] [Google Scholar]
- 12.Denadai R, Teixeira FV, Steinwurz F, et al. Induction or exacerbation of psoriatic lesions during anti-TNF-alpha therapy for inflammatory bowel disease: a systematic literature review based on 222 cases. J Crohns Colitis. 2013;7:517–24. doi: 10.1016/j.crohns.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Rahier JF, Buche S, Peyrin-Biroulet L, et al. Severe skin lesions cause patients with inflammatory bowel disease to discontinue anti-tumor necrosis factor therapy. Clin Gastroenterol Hepatol. 2010;8:1048–55. doi: 10.1016/j.cgh.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Tillack C, Ehmann LM, Friedrich M, et al. Anti-TNF antibody-induced psoriasiform skin lesions in patients with inflammatory bowel disease are characterised by interferon-gamma-expressing Th1 cells and IL-17A/IL-22-expressing Th17 cells and respond to anti-IL-12/IL-23 antibody treatment. Gut. 2014;63:567–77. doi: 10.1136/gutjnl-2012-302853. [DOI] [PubMed] [Google Scholar]
- 15.Cleynen I, Moerkercke WV, Billiet T, et al. Characteristics of Skin Lesions Associated With Anti-Tumor Necrosis Factor Therapy in Patients With Inflammatory Bowel Disease: A Cohort Study. Ann Intern Med. 2015:10–22. doi: 10.7326/M15-0729. [DOI] [PubMed] [Google Scholar]
- 16.Ellinghaus D, Ellinghaus E, Nair RP, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–47. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lees CW, Barrett JC, Parkes M, et al. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–53. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 18.Parkes M, Cortes A, van Heel DA, et al. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 19.Ananthakrishnan AN, Huang H, Nguyen DD, et al. Differential effect of genetic burden on disease phenotypes in Crohn’s disease and ulcerative colitis: analysis of a North American cohort. Am J Gastroenterol. 2014;109:395–400. doi: 10.1038/ajg.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ananthakrishnan AN, Nguyen DD, Sauk J, et al. Genetic polymorphisms in metabolizing enzymes modifying the association between smoking and inflammatory bowel diseases. Inflamm Bowel Dis. 2014;20:783–9. doi: 10.1097/MIB.0000000000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler SA, Ananthakrishnan AN, Gardet A, et al. SMAD3 gene variant is a risk factor for recurrent surgery in patients with Crohn’s disease. J Crohns Colitis. 2014;8:845–51. doi: 10.1016/j.crohns.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsoi LC, Spain SL, Knight J, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collamer AN, Battafarano DF. Psoriatic skin lesions induced by tumor necrosis factor antagonist therapy: clinical features and possible immunopathogenesis. Semin Arthritis Rheum. 2010;40:233–40. doi: 10.1016/j.semarthrit.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Collamer AN, Guerrero KT, Henning JS, et al. Psoriatic skin lesions induced by tumor necrosis factor antagonist therapy: a literature review and potential mechanisms of action. Arthritis Rheum. 2008;59:996–1001. doi: 10.1002/art.23835. [DOI] [PubMed] [Google Scholar]
- 27.de Gannes GC, Ghoreishi M, Pope J, et al. Psoriasis and pustular dermatitis triggered by TNF-{alpha} inhibitors in patients with rheumatologic conditions. Arch Dermatol. 2007;143:223–31. doi: 10.1001/archderm.143.2.223. [DOI] [PubMed] [Google Scholar]
- 28.Funk J, Langeland T, Schrumpf E, et al. Psoriasis induced by interferon-alpha. Br J Dermatol. 1991;125:463–5. doi: 10.1111/j.1365-2133.1991.tb14774.x. [DOI] [PubMed] [Google Scholar]
- 29.Nagarajan P, Chin SS, Wang D, et al. Ets1 blocks terminal differentiation of keratinocytes and induces expression of matrix metalloproteases and innate immune mediators. J Cell Sci. 2010;123:3566–75. doi: 10.1242/jcs.062240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart PE, Nair RP, Ellinghaus E, et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet. 2010;42:1000–4. doi: 10.1038/ng.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garzorz N, Eyerich K. NOS2 and CCL27: clinical implications for psoriasis and eczema diagnosis and management. Expert Rev Clin Immunol. 2015;11:167–9. doi: 10.1586/1744666X.2015.996549. [DOI] [PubMed] [Google Scholar]
- 32.Gokhale NR, Belgaumkar VA, Pandit DP, et al. A study of serum nitric oxide levels in psoriasis. Indian J Dermatol Venereol Leprol. 2005;71:175–8. doi: 10.4103/0378-6323.16232. [DOI] [PubMed] [Google Scholar]
- 33.Bottema RW, Nolte IM, Howard TD, et al. Interleukin 13 and interleukin 4 receptor-alpha polymorphisms in rhinitis and asthma. Int Arch Allergy Immunol. 2010;153:259–67. doi: 10.1159/000314366. [DOI] [PubMed] [Google Scholar]
- 34.Bottema RW, Reijmerink NE, Kerkhof M, et al. Interleukin 13, CD14, pet and tobacco smoke influence atopy in three Dutch cohorts: the allergenic study. Eur Respir J. 2008;32:593–602. doi: 10.1183/09031936.00162407. [DOI] [PubMed] [Google Scholar]
- 35.Elder JT, Bruce AT, Gudjonsson JE, et al. Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol. 2010;130:1213–26. doi: 10.1038/jid.2009.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.