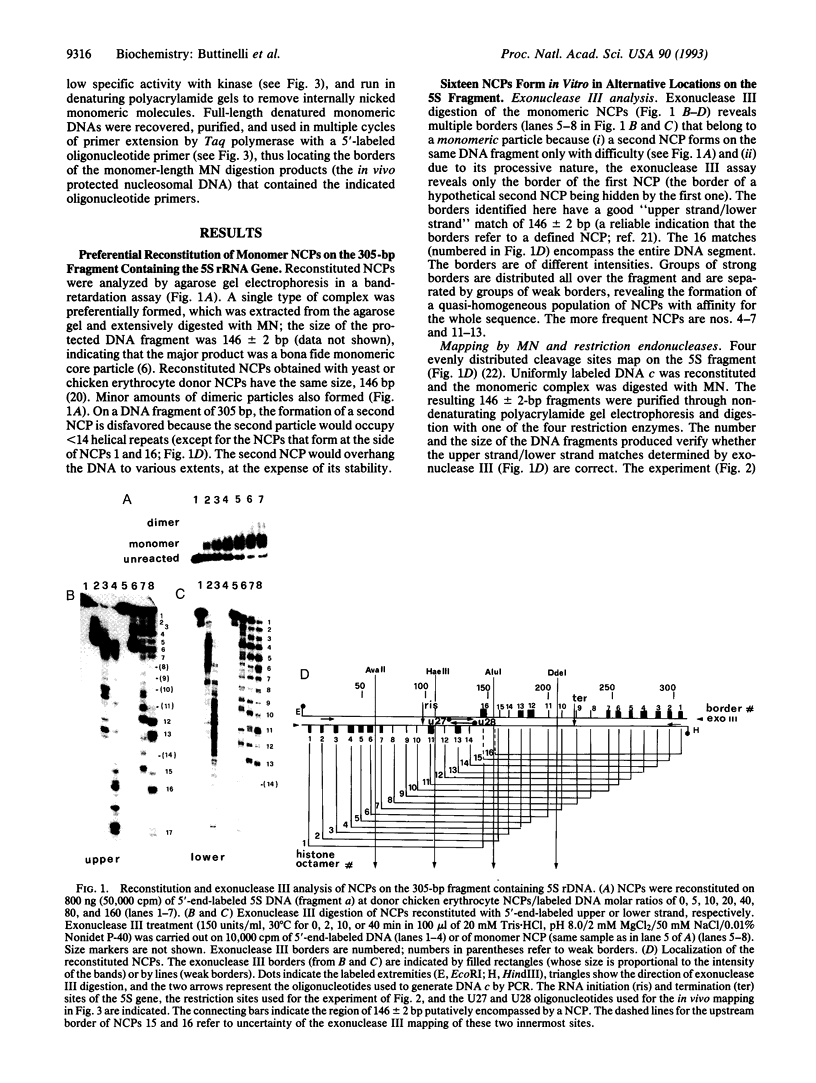
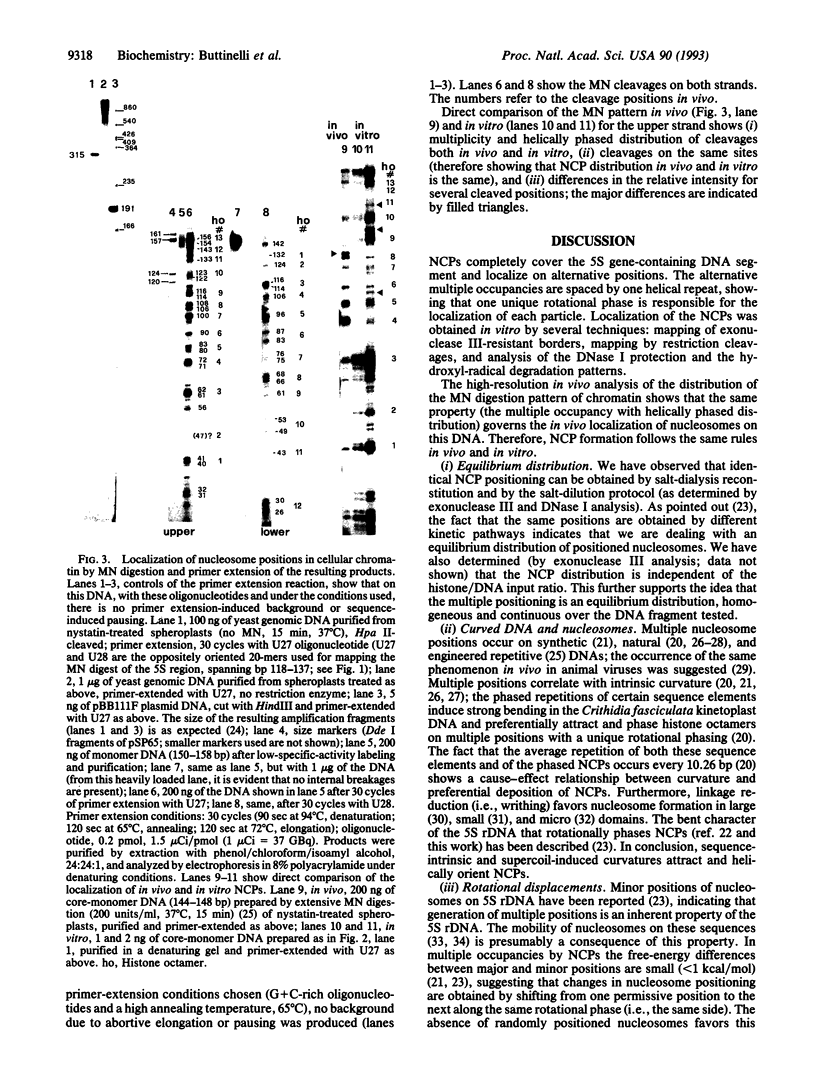
Abstract
A simple no-background assay was developed for high-resolution in vivo analysis of yeast chromatin. When applied to Saccharomyces cerevisiae 5S rRNA genes (5S rDNA), this analysis shows that nucleosomes completely cover this chromosomal region, occupying alternative positions characterized by a unique helical phase. This supports the notion that sequence-intrinsic rotational signals are the major determinant of nucleosome localization. Nucleosomal core particles reconstituted in vitro occupy the same positions and have the same helically phased distribution observed in vivo, as determined by mapping of exonuclease III-resistant borders, mapping by restriction cleavages, and by DNase I and hydroxyl-radical digestion patterns.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Kassavetis G. A., Geiduschek E. P. Bending of the Saccharomyces cerevisiae 5S rRNA gene in transcription factor complexes. J Biol Chem. 1992 Nov 5;267(31):22562–22569. [PubMed] [Google Scholar]
- Braun B. R., Riggs D. L., Kassavetis G. A., Geiduschek E. P. Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2530–2534. doi: 10.1073/pnas.86.8.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. F., FitzGerald P. C., Brubaker J. M., Simpson R. T. Sequence-specific interaction of histones with the simian virus 40 enhancer region in vitro. J Biol Chem. 1985 Oct 15;260(23):12394–12397. [PubMed] [Google Scholar]
- Costanzo G., Di Mauro E., Salina G., Negri R. Attraction, phasing and neighbour effects of histone octamers on curved DNA. J Mol Biol. 1990 Nov 20;216(2):363–374. doi: 10.1016/S0022-2836(05)80327-4. [DOI] [PubMed] [Google Scholar]
- Dong F., Hansen J. C., van Holde K. E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Forte P., Leoni L., Sampaolese B., Savino M. Cooperativity in nucleosomes assembly on supercoiled pBR322 DNA. Nucleic Acids Res. 1989 Nov 11;17(21):8683–8694. doi: 10.1093/nar/17.21.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L. A., Garrard W. T. DNA supercoiling in chromatin structure and gene expression. Crit Rev Eukaryot Gene Expr. 1992;2(2):165–209. [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. C., Ausio J., Stanik V. H., van Holde K. E. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989 Nov 14;28(23):9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- McMahon M. E., Stamenkovich D., Petes T. D. Tandemly arranged variant 5S ribosomal RNA genes in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Nov 12;12(21):8001–8016. doi: 10.1093/nar/12.21.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meersseman G., Pennings S., Bradbury E. M. Mobile nucleosomes--a general behavior. EMBO J. 1992 Aug;11(8):2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri R., Costanzo G., Venditti S., Di Mauro E. Linkage reduction allows reconstitution of nucleosomes on DNA microdomains. J Mol Biol. 1989 Jun 5;207(3):615–619. doi: 10.1016/0022-2836(89)90469-5. [DOI] [PubMed] [Google Scholar]
- Pennings S., Meersseman G., Bradbury E. M. Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol. 1991 Jul 5;220(1):101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- Piña B., Barettino D., Truss M., Beato M. Structural features of a regulatory nucleosome. J Mol Biol. 1990 Dec 20;216(4):975–990. doi: 10.1016/S0022-2836(99)80015-1. [DOI] [PubMed] [Google Scholar]
- Ponder B. A., Crawford L. V. The arrangement of nucleosomes in nucleoprotein complexes from polyoma virus and SV40. Cell. 1977 May;11(1):35–49. doi: 10.1016/0092-8674(77)90315-4. [DOI] [PubMed] [Google Scholar]
- Ramsay N. Deletion analysis of a DNA sequence that positions itself precisely on the nucleosome core. J Mol Biol. 1986 May 5;189(1):179–188. doi: 10.1016/0022-2836(86)90389-x. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C., Travers A. A. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 1989 Jan;8(1):229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild C., Claret F. X., Wahli W., Wolffe A. P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993 Feb;12(2):423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka C., Hörz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991 Feb;10(2):361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Zatchej M. Chromatin folding modulates nucleosome positioning in yeast minichromosomes. Cell. 1988 Dec 23;55(6):945–953. doi: 10.1016/0092-8674(88)90240-1. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Masiarz F. R., DeGennaro L. J., Rutter W. J. Nucleotide sequence of the yeast 5S ribosomal RNA gene and adjacent putative control regions. Nature. 1977 Jun 16;267(5612):641–643. doi: 10.1038/267641a0. [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Hörz W. Nucleosomes are positioned on mouse satellite DNA in multiple highly specific frames that are correlated with a diverged subrepeat of nine base-pairs. J Mol Biol. 1984 Jun 15;176(1):105–129. doi: 10.1016/0022-2836(84)90384-x. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Goulet I., Revet B., Le Bret M., Prunell A. Chromatin reconstitution on small DNA rings. II. DNA supercoiling on the nucleosome. J Mol Biol. 1988 Mar 20;200(2):267–290. doi: 10.1016/0022-2836(88)90239-2. [DOI] [PubMed] [Google Scholar]
- van Holde K. Transcription. The omnipotent nucleosome. Nature. 1993 Mar 11;362(6416):111–112. doi: 10.1038/362111a0. [DOI] [PubMed] [Google Scholar]