Abstract
Intracellular bacterial pathogens have evolved many ways to manipulate host cells for successful infection. Many of these pathogens use specialized secretion systems to inject bacterial proteins into the host cytosol that manipulate cellular processes to favor infection. Autophagy is a eukaryotic cellular remodeling process with a critical role in many diseases, including bacterial clearance. A growing field of research highlights mechanisms used by intracellular bacteria to manipulate autophagy as a pro-survival strategy. This review focuses on a select group of bacterial pathogens with diverse intracellular lifestyles that exploit autophagy-derived nutrients and membrane for survival. This group of pathogens uses secretion systems and specific effectors to subvert distinct components of autophagy. By understanding how intracellular pathogens manipulate autophagy, we gain insight not only into bacterial pathogenesis but also host cell signaling and autophagolysosome maturation.
Introduction
Intracellular bacterial pathogens cause a spectrum of human disease and inflict significant morbidity and mortality on the human population. Increased antibiotic resistance and the emergence of new pathogens over the past few decades have alerted the scientific and healthcare communities to a need for new therapeutics and treatments. Intracellular pathogens must manipulate the host cell to acquire the nutrients necessary for proliferation and subsequent pathogenesis. Because this interaction is required for bacterial replication, altering the pathways that bacteria exploit for nutrients is a promising target for future therapeutics. Intracellular bacterial pathogens often use secretion systems to control host processes and promote infection and replication. Secretion systems are biological machines that secrete bacterial proteins either into the milieu or through host cell membranes into the cytosol. There are currently eight types of secretion systems (Type I to Type VIII) that bacteria use to translocate bacterial proteins to the extracellular space. Some of these secretion systems even puncture the membranes of target cells [1]. Translocated effectors control numerous infection events, including replication vacuole formation, apoptosis, cytokine responses, and autophagy. Sufficient nutrient and membrane supplies are key to the success of intracellular pathogens, and acquisition of these necessities involves manipulation of many host processes. In this review, we will focus on a group of bacterial pathogens with diverse intracellular activities that employ secreted effector proteins to exploit host autophagy and promote intracellular growth [2-6].
Autophagy
Macroautophagy (referred to as autophagy herein) is a eukaryotic process that maintains cellular homeostasis by degrading defunct organelles, protein aggregates, and, in cases of nutrient deprivation, bulk cytoplasm [7,8]. Additionally, recent discoveries show autophagy has many auxiliary roles, such as regulating immune signaling and clearing invading pathogens [9]. Briefly, autophagy initiates with formation of an isolation membrane that is generally derived from the endoplasmic reticulum (ER) but can originate from other organelles, such as mitochondria, or the plasma membrane [10,11]. Formation of an isolation membrane and phagophore is controlled by the Unc51-like kinase 1 (ULK1) complex that is activated when mTOR is inactive. The ULK1 complex activates Beclin-1, a critical protein in autophagosome nucleation. Beclin-1 forms a complex with Atg14, p150, and VPS34, a class III phosphoinositide 3-kinase (PI3K). Pharmacological inhibitors of autophagy, such as 3-methyladenine (3-MA), target VPS34 to abrogate the pathway [12]. Once autophagosome formation is initiated, a complex containing Atg5-Atg12 and Atg16 elongates the maturing phagophore. Atg4 cleaves cytoplasmic LC3, resulting in LC3-I while Atg3 and Atg7 to lipidate LC3-I, resulting in LC3-II [13]. LC3-II is then attached to the autophagosome and is commonly used as a marker of autophagosomes. LC3-II is involved in the final sealing steps that allow completion of an autophagosome. The newly formed autophagosome then matures to an acidified autophagolysosome that degrades harbored material into simple structures such as amino acids and fatty acids.
Many different intracellular components are degraded through autophagy. Targeted degradation of specific components is termed selective autophagy and is mediated by receptor proteins that target cargo for envelopment by autophagosomes. Common cargo receptors are p62 (sequestosome-1), nuclear dot protein 52 (NDP52), neighbor of BRCA1 gene 1 (NBR1), and optineurin (OPTN). These receptors interact with ubiquitinated substrates (p62, NBR1) or bacterial signatures (NDP52) and subsequently bind LC3-II, which directs autophagosome formation around selected cargo [14-16]. When the cargo is an invading pathogen, the process is referred to as xenophagy. Autophagy in the context of infection is often detrimental to intracellular bacteria and many species, such as Mycobacterium tuberculosis [17] and Legionella pneumophila [18], have developed mechanisms to avoid or prevent autophagy and interactions with autophagosomes. Several recent extensive review articles address bacterial avoidance or abrogation of autophagy. In contrast, several pathogens have evolved methods to utilize autophagy to promote infection. In this review, we highlight recent discoveries demonstrating intracellular bacterial pathogen manipulation of a host defense mechanism to benefit the pathogen and promote disease. We specifically focus on Anaplasma, Coxiella, Francisella, and Brucella species as examples of pathogens that uniquely interact with autophagy.
Anaplasma phagocytophilum Secretes Ats-1 to Initiate Autophagosome Formation
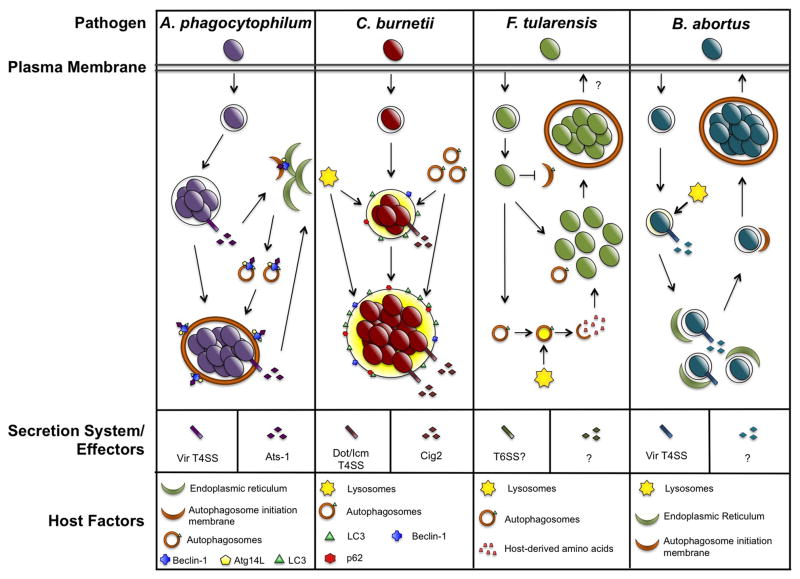
A. phagocytophilum is the causative agent of human granylocytic anaplasmosis, an acute febrile illness, and primarily infects granulocytes. The bacterium replicates within a host-derived vacuole that initially matures along the endocytic pathway, acquiring markers such as Rab5 and EEA1, but is halted by A. phagocytophilum before fusing with degradative lysosomes. Early studies showed this compartment has a double-lipid bilayer surrounding replicating organisms, a hallmark characteristic of an autophagosome [19]. LC3 and Beclin-1 are present on the replicative vacuole membrane, further demonstrating this compartment is autophagosome-derived [20]. Additionally, autophagy is induced in A. phagocytophilum-infected cells, and inhibition of autophagy using 3-MA prevents bacterial replication, indicating autophagy is necessary for productive infection. To control cellular infection events, A. phagocytophilum uses a Type IV secretion system (T4SS) to deliver effector proteins to the host cytosol. The bacterial effector Anaplasma translocated substrate-1 (Ats-1) co-localizes on A. phagocytophilum vacuoles with autophagy proteins typically found at the ER, where autophagosome nucleation often occurs [2]. Ats-1 directly binds Beclin-1 and recruits Atg14L to promote autophagosome nucleation at the vacuole membrane, and silencing Beclin-1 expression negatively impacts A. phagocytophilum replication. These findings are unique because pathogen-derived proteins that bind Beclin-1 typically inhibit, rather than promote, autophagy. The mechanism by which A. phagocytophilum triggers autophagosome formation is distinct from canonical autophagy due to the absence of mTOR dephosphorylation. Interestingly, Ats-1 also localizes to mitochondria and interferes with apoptotic signaling in a cell-density dependent manner [21]. This observation suggests A. phagocytophilum uses one effector to manipulate host cell survival and autophagy, with localization of the effector influenced by host cell conditions.
Coxiella burnetii Secretes T4SS Effectors to Promote Fusion with Autophagosomes
C. burnetii is an intracellular Gram-negative bacterium that causes human Q fever, an acute influenza-like illness that can progress to chronic endocarditis. Similar to A. phagocytophilum, C. burnetii encodes a Dot/Icm T4SS required for productive infection of macrophages, the pathogen’s favored host cells. C burnetii uniquely replicates within an acidic (pH ~ 4.5) lysosome-like compartment termed the parasitophorous vacuole (PV) that decorates with lysosome-associated membrane protein-1, -2, and -3 (LAMP-1, -2, and -3) and contains active proteases such as cathepsin D. Early studies indicated that PV incorporate autophagic proteins, including LC3, and PV generation is enhanced when autophagy is activated by amino acid and serum starvation [22]. Beclin-1 localizes to the PV and is beneficial for vacuole expansion, though the mechanism remains undefined [23]. Recruitment of LC3 to the PV is dependent on the T4SS in human alveolar macrophages, the target cell during human infection [4]. Interestingly, no change in autophagic flux occurs during infection, indicating autophagy is not actively triggered by C. burnetii. Thus, C. burnetti likely recruits existing autophagosomes. Additionally, the cargo receptor p62 is recruited to the PV in a T4SS-dependent manner. A separate human siRNA screen identified many host cell factors required for intracellular replication and PV generation [24]. When expression of the autophagy regulator Atg5 is silenced, C. burnetii infection efficiency is not altered, indicating canonical autophagy is not required for infection. Shortly after this study, an effector protein termed Cig2 was discovered that recruits LC3 to the PV and aids maintainance of typical PV structure [3]. Cig2-deficient C. burnetii does not form prototypical large PV or recruit autophagosomes, but replicates similar to wild type bacteria, albeit within small irregular vacuoles. Additional effectors, such as Coxiella plasmid effector B (CpeB) and CpeL, were identified in ectopic expression studies to co-localize with LC3 [25,26], suggesting C. burnetii uses multiple effectors to control interactions with autophagosomes.
Francisella tularensis Derives Nutrients from Autophagosomes
F. tularensis is the causative agent of the zoonotic disease tularemia. F. tularensis replicates in the cytosol of a wide range of cells including macrophages and epithelial cells [27]. In general, live F. tularensis bacteria evade autophagy [28]. The mechanism of evasion is still being characterized, but one critical component is the structure of the O-antigen [29]. Mutants deficient in O-antigen synthesis are viable but are destroyed by autophagy. In certain mouse cell types, Francisella can re-enter the endosomal compartment by entering an autophagosome [30]. However, F. tularensis is not destroyed within this structure and the importance of endosomal re-entry during infection is unclear. F. tularensis replicates efficiently in host cells with medium lacking amino acids, indicating that bacteria derive nutrients from the host [5]. This observation led to investigation of autophagy as a source of these nutrients. Interestingly, while Beclin-1 has a pro-bacterial effect, Atg5 is not required for F. tularensis intracellular growth. Indeed, non-canonical autophagy is activated during infection and contributes nutrients for replicating F. tularensis. F. tularensis harbors a pathogenicity island (FPI) that encodes genes belonging to a type VI secretion system. The FPI is critical for phagosome maturation and eventual phagosomal escape, and a secreted effector(s) likely mediates these events [31]. As with most intracellular pathogens, it is likely that a secreted effector(s) is also responsible for interactions with non-canonical autophagy. Future studies will provide insight into this novel area of study with a unique secretion system and use of non-canonical methods for intracellular replication.
Brucella spp. Use Autophagy to Replicate in and Exit Host Cells
Brucellosis is a chronic zoonosis caused by several bacterial species in the Brucella genus. Brucella encodes a VirB T4SS and has a unique and complex intracellular lifecycle involving transient interaction with several host organelles during growth in professional phagocytes. After internalization into a host cell, the Brucella-containing vacuole (BCV) traffics through the endosomal pathway, acquiring membrane markers such as Rab5, and briefly interacts with lysosomes in a “kiss and run” manner [32,33]. This event activates the VirB T4SS, which mediates interactions between the endosomal BCV (eBCV) and the ER. After fusion with the ER, the BCV is permissive for replication, deeming this niche the replicative BCV (rBCV). After replication, Brucella abortus rBCVs acquire a double membrane characteristic of autophagosomes [6]. Interestingly, LC3 is not present at the BCV. Formation of this newly discovered autophagic BCV (aBCV) does not require canonical autophagy elongation regulators, including Atg5, Atg16L, Atg4B, Atg7 and LC3B. However, aBCV biogenesis requires expression of Beclin-1 and ULK1, two proteins required for initiation of autophagy. Importantly, development of the aBCV may aid cell to cell spread. These results suggest Brucella activates autophagy at a late stage of infection to promote spread to bystander cells. This prediction presents a novel area of intracellular pathogenesis that could be important for other pathogens spreading from the primary infection site.
Although Brucella clearly interacts with autophagosomes, it is less clear whether autophagy alters overall intracellular replication. A study using B. melitensis showed that pharmacological inhibition of autophagy negatively impacts bacterial replication [34]. However, a recent study comparing B. abortus and B. melitensis demonstrated that the latter species does not require canonical Atg5-dependent autophagy for intracellular replication [35]. Thus, the overall impact of autophagy on replication is still under investigation. Additionally, secreted effectors likely control interactions with autophagosomes. Although there is no definitive effector yet linked to aBCV generation or interactions with autophagy, the VirB T4SS is required for completion of the Brucella life cycle [33], suggesting the system secretes effectors that impact all stages of infection.
Manipulation of the Host Unfolded Protein Response to Promote Autophagy
Recent studies have opened a new area of potential autophagy control during intracellular pathogen infection: ER stress and the unfolded protein response (UPR). The UPR consists generally of three ER-associated arms that combat stress responses through tightly controlled transcriptional programs. UPR pathways are controlled by one of three ER-associated receptors that sense stress: inositol-requiring enzyme-1 (IRE-1α), protein kinase RNA-like ER kinase (PERK), and activating transcription factor 6 (ATF6). IRE1α regulates autophagosome formation in response to ER stress and other stimuli. Following activation and autophosphorylation, IRE-1α cleaves X-box binding protein-1 (XBP-1) mRNA, resulting in XBP-1s that translocates to the nucleus to regulate transcription of numerous genes involved in lipid synthesis and ER-associated degradation (ERAD), a process by which proteins are degraded by the proteasome or autophagy [36]. The transcription factors ATF4 and ATF6 activate autophagy by up regulating expression of chaperones associated with the ERAD pathway. Multiple bacterial pathogens, including Listeria monocytogenes, M. tuberculosis, Brucella spp., and Helicobacter pylori trigger the UPR by secreting toxins or effector proteins [37]. For example, HP0175 is an antigen secreted by H. pylori that triggers UPR-dependent autophagy to manipulate host cell apoptosis [38]. Brucella activates the UPR using the host protein Yip1a to bind phosphorylated IRE-1α and trigger XBP-1-dependent transcription. Additionally, B. melitensis produces a protein termed TcpB that triggers activation of all three arms of the UPR. Although no reports have yet demonstrated activation of the UPR in cells infected with C. burnetii, A. phagocytophilum, or F. tularensis, it is intriguing to predict these pathogens also exploit the UPR to trigger autophagy-related events.
Conclusions
Intracellular pathogens have clearly evolved mechanisms to exploit autophagy, a typically antibacterial cellular process. The pathogens highlighted in this review replicate within distinct intracellular environments, using autophagosomes as convenient nutrient sources and, as predicted for Brucella spp., a method for escaping from infected cells. This area of intracellular pathogenesis has expanded in the past decade and a new appreciation has been gained for autophagy as a critical component of infection. However, much remains to be learned about the secreted bacterial proteins that control interactions with autophagy. Future research will undoubtedly discover many novel pathogen proteins that usurp autophagy for membrane and nutrients needed for growth within eukaryotic cells.
Figure 1.
Subversion of autophagy by intracellular bacterial pathogens. A. phagocytophilum, C. burnetii, F. tularensis, and Brucella spp. replicate within distinct compartments in host cells. Each pathogen promotes interaction with autophagosomes and these events are predicted to deliver nutrients to replicating bacteria. Additionally, secreted bacterial effectors are critical for autophagosome interactions and Brucella triggers the UPR to promote autophagy.
Highlights.
Intracellular bacterial pathogens must confront host cell autophagy to control infection.
A subset of pathogens actively engages autophagy to promote infection and survival.
Secreted bacterial proteins are responsible for many host cell exploitations.
New mechanisms of bacterial interplay with autophagy have been discovered.
Novel autophagy-related events provide insight into host cell responses and bacterial pathogenesis.
Acknowledgments
This research was supported by funding to D. E. V. from the NIH/NIAID (R01AI087669), the Arkansas Biosciences Institute, and the Center for Microbial Pathogenesis and Host Inflammatory Responses (NIH/NIGMS P20GM103625), and to T. K. from the NIH/NIAID (R01AI082870).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest within the period of review have been highlighted as:
* of special interest
** of outstanding interest
- 1.Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 2009;9 (Suppl 1):S2. doi: 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niu H, Rikihisa Y. Ats-1: a novel bacterial molecule that links autophagy to bacterial nutrition. Autophagy. 2013;9:787–788. doi: 10.4161/auto.23693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 2014;10:e1004286. doi: 10.1371/journal.ppat.1004286. The authors identified Cig2, a T4SS effector involved in autophagosome recruitment and PV generation. This is the first C. burnetii effector shown to influence host autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winchell CG, Graham JG, Kurten RC, Voth DE. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun. 2014;82:2229–2238. doi: 10.1128/IAI.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 2013;9:e1003562. doi: 10.1371/journal.ppat.1003562. The authors show that non-canonical autophagy provides a major source of amino acid nutrients to F. tularensis during infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. A new component of the Brucella life cycle was discovered and termed the autophagic Brucella-containing vacuole. This compartment resembles an autophagosome and potentially aids cell to cell spread. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo EK, Yuk JM, Shin DM, Sasakawa C. Roles of autophagy in elimination of intracellular bacterial pathogens. Front Immunol. 2013;4:97. doi: 10.3389/fimmu.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 11.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;9:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Wandel MP, Li F, Liu Z, He C, Wu J, Shi Y, Randow F. Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Sci Signal. 2013;6:ra9. doi: 10.1126/scisignal.2003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Colombo MI, Gutierrez MG, Romano PS. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy. 2006;2:162–164. doi: 10.4161/auto.2827. [DOI] [PubMed] [Google Scholar]
- 18.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol. 2008;10:593–605. doi: 10.1111/j.1462-5822.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 20**.Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A. 2012;109:20800–20807. doi: 10.1073/pnas.1218674109. The authors discovered an auxillary role for the secreted A. phagocytophilum protein Ats-1. This effector directly binds to Beclin-1 and induces autophagosome formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 2010;6:e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 23.Vazquez CL, Colombo MI. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ. 2010;17:421–438. doi: 10.1038/cdd.2009.129. [DOI] [PubMed] [Google Scholar]
- 24.McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. MBio. 2013;4:e00606–00612. doi: 10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maturana P, Graham JG, Sharma UM, Voth DE. Refining the plasmid-encoded type IV secretion system substrate repertoire of Coxiella burnetii. J Bacteriol. 2013;195:3269–3276. doi: 10.1128/JB.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol. 2011;193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect Immun. 2008;76:5843–5852. doi: 10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, Celli J. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy. 2012;8:1342–1356. doi: 10.4161/auto.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Case ED, Chong A, Wehrly TD, Hansen B, Child R, Hwang S, Virgin HW, Celli J. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell Microbiol. 2014;16:862–877. doi: 10.1111/cmi.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broms JE, Sjostedt A, Lavander M. The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol. 2010;1:136. doi: 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celli J. The changing nature of the Brucella-containing vacuole. Cell Microbiol. 2015;17:951–958. doi: 10.1111/cmi.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lacerda TL, Salcedo SP, Gorvel JP. Brucella T4SS: the VIP pass inside host cells. Curr Opin Microbiol. 2013;16:45–51. doi: 10.1016/j.mib.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Guo F, Zhang H, Chen C, Hu S, Wang Y, Qiao J, Ren Y, Zhang K, Wang Y, Du G. Autophagy favors Brucella melitensis survival in infected macrophages. Cell Mol Biol Lett. 2012;17:249–257. doi: 10.2478/s11658-012-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamer I, Goffin E, De Bolle X, Letesson JJ, Jadot M. Replication of Brucella abortus and Brucella melitensis in fibroblasts does not require Atg5-dependent macroautophagy. BMC Microbiol. 2014;14:223. doi: 10.1186/s12866-014-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kario E, Amar N, Elazar Z, Navon A. A new autophagy-related checkpoint in the degradation of an ERAD-M target. J Biol Chem. 2011;286:11479–11491. doi: 10.1074/jbc.M110.177618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol. 2015;13:71–82. doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Halder P, Datta C, Kumar R, Sharma AK, Basu J, Kundu M. The secreted antigen, HP0175, of Helicobacter pylori links the unfolded protein response (UPR) to autophagy in gastric epithelial cells. Cell Microbiol. 2015;17:714–729. doi: 10.1111/cmi.12396. This study links a bacterial protein, autophagy, and the UPR, suggesting other pathogens may also use the UPR to direct autophagic events. [DOI] [PubMed] [Google Scholar]