Abstract
We report the design and characterization of UNC3866, a potent antagonist of the methyl-lysine (Kme) reading function of the Polycomb CBX and CDY families of chromodomains. Polycomb CBX proteins regulate gene expression by targeting Polycomb Repressive Complex 1 to sites of H3K27me3 via their chromodomains. UNC3866 binds the chromodomains of CBX4 and CBX7 most potently with a Kd of ∼100 nM for each, and is 6- to 18-fold selective versus seven other CBX and CDY chromodomains while being highly selective versus >250 other protein targets. X-ray crystallography revealed that UNC3866 closely mimics the interactions of the methylated H3 tail with these chromodomains. UNC4195, a biotinylated derivative of UNC3866, was used to demonstrate that UNC3866 engages intact PRC1 and that EED incorporation into PRC1 is isoform-dependent in PC3 prostate cancer cells. Finally, UNC3866 inhibits PC3 cell proliferation, a known CBX7 phenotype, while UNC4219, a methylated negative control compound, has negligible effects.
Methylation of lysine residues on histones critically contributes to chromatin regulation1, 2. This post-translational modification (PTM) provides a docking site for effector proteins, termed Kme readers, which recognize this PTM through an aromatic cage that accommodates the hydrophobic lysine side chain and engages the methylated amine through cation-π, electrostatic, and van der Waals interactions1. Selectivity between different methylation states (mono-, di-, and trimethylation) is conferred primarily by cage geometry and composition, while sequence specificity is achieved through interactions with the amino acids flanking the modified lysine1, 3.
Trimethyl-lysine 27 on histone 3 (H3K27me3) is a transcriptionally repressive signal that is installed by the methyltransferases EZH1 and EZH2 of Polycomb Repressive Complex 2 (PRC2). The other two core components of PRC2 are SUZ12 and the H3K27me3 reader, EED4. Under the ‘canonical’ Polycomb-signaling model, H3K27me3 is deposited by PRC2 and this mark is then recognized by the chromodomain-containing Kme reader component of the multiprotein repressor complex, Polycomb Repressive Complex 1 (PRC1). The PRC1 chromodomains (CBX2, -4, -6, -7 and -8) incorporate into PRC1 in a mutually exclusive manner5 and are thought to facilitate PRC1-mediated transcriptional repression by targeting the complex to H3K27me3, allowing for monoubiquitination of lysine 119 on H2A (H2AK119ub) by the E3 ubiquitin ligase subunits of PRC1, RING1A/B and BMI6. H2AK119ub is in turn associated with chromatin compaction, DNA methylation and repression of the underlying genes7, 8.
Despite this simplistic model of sequential PRC action, it has become clear that a large number of diverse PRC1 complexes exist. However, the role of this diversity in the context of specific gene regulation events or pathology is not understood. Further adding to the complexity of PRC1, it was recently reported that EED, the H3K27me3 reader once believed to only participate in PRC2, also exists as a component of CBX-containing PRC1 complexes4, giving PRC1 two potential Kme readers for the same mark. The variable CBX component is another point of diversity that potentially impacts the genomic targeting of PRC1. However, the ability of CBX proteins to target PRC1 to overlapping regions of the genome9 suggests that these proteins may be partially functionally redundant. Therefore, modulation of chromatin regulation through PRC1 Kme reader antagonism may require ligands that target multiple CBX proteins.
Of the CBX chromodomains, CBX7 is the best studied and has been implicated as contributing to numerous cancer types9-19. Overexpression of CBX7 confers a proliferative advantage in prostate, gastric, leukemia and lymphoma cell lines that can be inhibited through knockdown of CBX79, 10, 17, 18, while CBX2, -4, and -8 have also been shown to enhance proliferation in various cancer types20-22. Interestingly, CBX7 also functions as a tumor suppressor in lung, pancreatic, colon and thyroid tissues11, 14-16. In addition to CBX7, other developmental Polycomb factors have similarly been shown to exhibit contradictory functions in oncogenesis such as the H3K27 demethylase, JMJD323.
Modulation of H3K27me3 signaling via small molecules has recently garnered much pharmaceutical interest. Inhibitors of the PRC2 methyltransferase, EZH2, are effective at targeting aberrant H3K27me3 levels in lymphomas with activating mutations and are currently in clinical trials24. Inhibitors of the H3K27me3 demethylases, JMJD3 and UTX, also have growth-suppressive properties in acute lymphoblastic leukemia25 and robust antitumor activity in xenograft models of diffuse intrinsic pontine glioma26. Therefore, we hypothesized that antagonism of the H3K27me3 reader function of PRC1, thought to be immediately downstream of PRC2, may also have therapeutic potential. While there have been other reports of ligands for PRC1 chromodomains27, 28, no molecules have been reported that meet the stringent criteria29-31 for classification as a high-quality, cellularly active chemical probe. Chemical probes that inhibit the Kme3-reading capacity of PRC1 will be valuable tools in explicating PRC1 biology, determining the therapeutic potential of CBX chromodomains, investigating potential pharmacologic synergy with EZH2 and JMJD3/UTX inhibitors, and understanding the precise roles of CBX target genes in different cancer types. We report the design and characterization of UNC3866 as a potent and cellularly active antagonist of PRC1 chromodomains, and our initial investigation into the biological effects of antagonizing the H3K27me3 reading function of CBX chromodomains in PC3 prostate cancer cells.
Results
Molecular dynamics-based design of a CBX antagonist
Although monospecific chemical probes are often desirable, probes with well-characterized multi-target profiles are also of great utility and may, pragmatically, be the best tools achievable within highly homologous target families. For example, the initially reported bromodomain antagonists were not selective within the BET family and yet their impact has been extraordinary32. The high degree of similarity between the CBX chromodomains of PRC1, particularly in the regions that interact with H3, suggests that any molecule that binds to one of these domains is likely to have affinity for other Polycomb chromodomains as well. With this in mind, we selected CBX7 as an initial target for ligand discovery due to the existing structural information3, but with an understanding that quantitative profiling versus other chromodomains would be required for full ligand characterization.
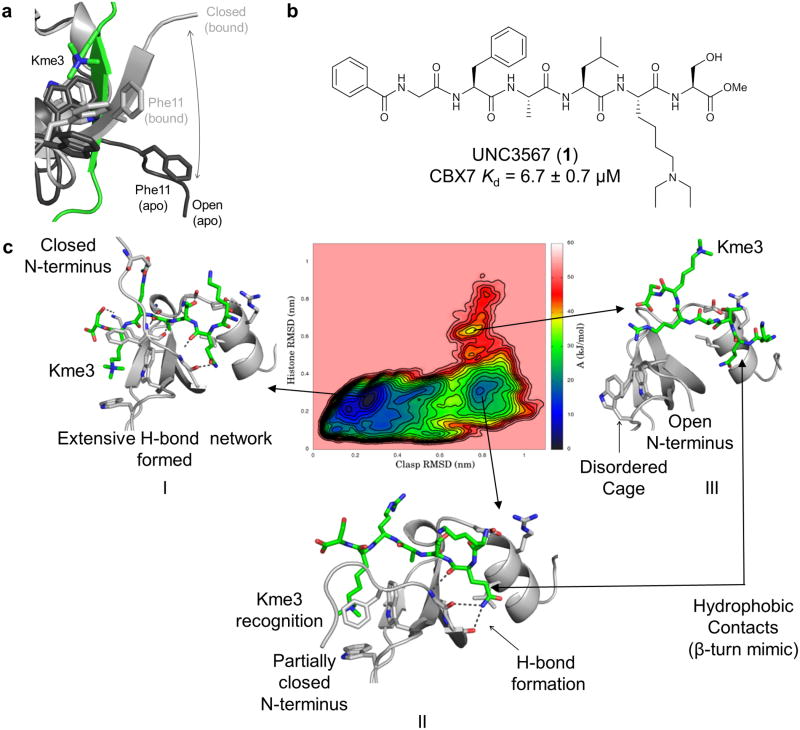
We first screened our epigenetic targeted library of small molecules designed largely for Kme reader domains and lysine methyltransferases using an established AlphaScreen® competition assay33. Additionally, we screened a set of commercially available ligands known to interact with other quaternary ammonium binding proteins at concentrations up to 30 μM (see Supplementary Results, Supplementary Fig. 1 for compound names and structures). This proved unsuccessful in identifying tractable hits capable of inhibiting the CBX7-H3 peptide interaction. Thus, we turned to the existing structural information to guide our hit discovery efforts. We compared the apo CBX7 NMR structure (PDB: 2K1B) with the H3K27me3-CBX7 NMR structure (PDB: 2L12) and observed that the aromatic cage of CBX7 is unformed in the apo structure (Fig. 1a). The absence of a preformed aromatic cage makes the discovery of small molecule Kme mimetics targeting this portion of CBX7 a significant challenge. Histone peptide substrates, in contrast, engage CBX7 via an induced fit, β-sheet-like interaction that results in formation of the Kme-reading aromatic cage3.
Figure 1. Molecular dynamics guided inhibitor design.
(a) Comparison of the aromatic cage of CBX7 in the NMR structure of the apo (PDB: 2K1B, dark grey) and H3K27me3 bound (PBD: 2L1B, light grey, Kme3 shown in green) conformations of CBX7. (b) Structure of UNC3567. The Kd represents the average of 3 replicates ± the standard deviation. (c) Free energy plot from the CBX7-H3K9me3 MDS studies. Arrows are drawn from potential energy wells to representative structures associated with that well. Frames I-III represent one possible dissociative path of H3 from CBX7, and the reverse of these frames represents the corresponding associative path of CBX7 and H3. The N-terminus label is referring to the N-terminus of CBX7.
Previous work utilizing peptide arrays revealed that the overall CBX7 consensus binding sequence is A(R/I/L/F/Y/V)Kme3(S/T)3. In fact, the sequence RGFALKme3STHG was shown to bind CBX7 with 22-fold greater affinity than the corresponding H3K27me3 peptide (Kd's of 5 μM and 110 μM, respectively)3. Therefore, we initiated optimization of peptide-based ligands with the goal of enhancing potency while decreasing charge and polarity to increase cell permeability, a known challenge for peptides. We first replaced the arginine preceding the Kme3 with leucine to facilitate cell permeability, and an N-terminal benzoyl cap was chosen to further increase the lipophilicity and aid in purification. Our next priority was to address the liabilities associated with Kme3: potential instability toward lysine demethylases34 and limited passive membrane permeability due to the permanent positive charge. A variety of peptides were prepared and screened in which tertiary and secondary amines of varying size, hydrophobicity and basicity were incorporated as quaternary amine replacements. We were encouraged to find that replacing the Kme3 in this context with a diethyl-lysine residue resulted in a peptide approximately equipotent for CBX7 as the original consensus peptide, UNC3567 (1) (Fig. 1b), Kd = 6.7 ± 0.7 μM as determined by isothermal titration calorimetry). This development represented encouraging progress toward our goal of cellularly active PRC1 antagonists and justified further optimization of UNC3567 to increase potency.
Because of the induced-fit recognition implied by prior structural studies3, we sought to better understand the critical CBX7-H3 interactions that drive the conformational rearrangement of CBX7, whereby residues 7-13 of CBX7 close around the histone and position Phe11 in close enough proximity to Trp32 and Trp35 to form the aromatic cage (Fig. 1a). We therefore turned to adaptively biased molecular dynamics simulations35 (MDS) using the previously published NMR structure of H3K9me3 bound to CBX7 (PDB: 2L12). We computed possible histone exit paths from the CBX7-H3K9me3 structure to understand the conformational dynamics of H3 recognition. The pathway shown in Fig. 1c is favored when we considered a truncated form of H3K9me3 (residues 4-10), which is most analogous to UNC3567. Starting from the bound state as shown in Fig. 1c, Frame I, the N-terminus of CBX7 opens by losing partial contact with H3 (transition from Frame I to II). Subsequently, the Kme3 loses contact with the aromatic cage (transition from Frame II to III), leaving H3 engaged with CBX7 primarily through hydrophobic contacts made with V13, D50, R52, and L53 (Frame III). Complete dissociation of H3 and CBX7 occurs following the loss of these contacts.
Association of CBX7 and H3 is simply the microscopic reversal of this exit path and is analogous to β-hairpin folding between two anti-parallel β-strands. In this case, one β-strand is formed by the histone; the other by residues 8-13 of CBX7 (Supplementary Fig. 2). Therefore, establishing the appropriate contacts between the N-terminal portion of the H3 peptide (residue Q5, Fig. 1c, Frame III) and CBX7 (namely residues V13, D50, R52 and L53) during association is analogous to forming the critical β-turn during hairpin folding. Once the turn is formed, backbone hydrogen bonds ‘zip’ the β-hairpin into the fully folded state36, 37. In the case of CBX7, in addition to the formation of an extensive hydrogen bond network, this also orients Phe11 of CBX7 to contact Kme3 and facilitates formation of the aromatic cage. We utilized this pathway for H3 binding as a model for the association of CBX7 with UNC3567 and hypothesized that optimizing the N-terminal interactions of UNC3567 with CBX7 would facilitate the necessary hydrogen bonding for closure of CBX7 around UNC3567, thereby increasing ligand affinity.
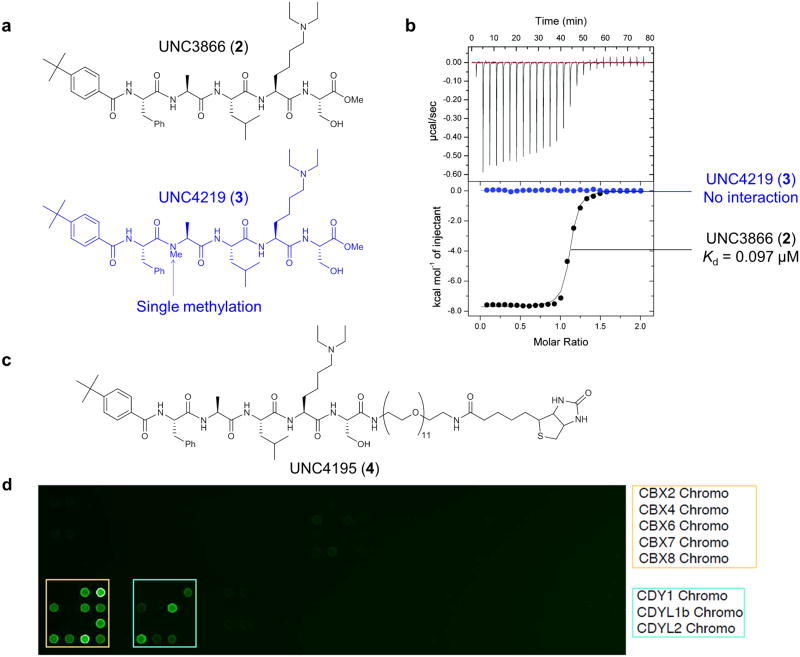
We synthesized a series of replacements for the N-terminal benzoyl-glycine of UNC3567 that were less flexible and predicted to interact favorably with V13, D50, R52 and L53 of CBX7. Structure activity relationship (SAR) studies ultimately led to the synthesis of UNC3866 (2) (Fig. 2a), which is to our knowledge, the most potent ligand reported for CBX7 with a Kd of 97 ± 2.4 nM (Fig. 2b, see Supplementary Table 1 for key SAR data that led to UNC3866). We also generated a negative control compound, UNC4219 (3), for use in cellular studies through methylation of the alanine residue at the Nα-position (Fig. 2a). This modification disrupts a key hydrogen bond, and consistent with our simulations, UNC4219 displays no interaction with CBX7 at concentrations up to 100 μM (Fig. 2b).
Figure 2. ITC studies of UNC3866 and UNC4219 with CBX7 and selectivity profiling of UNC3866.
(a) Structures of UNC3866 (black) and the structurally related negative control compound, UNC4219 (blue). (b) Comparison of UNC3866 (Kd = 97 ± 2.4 nM, average of 3 replicates ± the standard deviation) and UNC4219 (no interaction up to 100 μM) binding to CBX7 as measured by ITC. (c) Structure of UNC4195, a biotinylated derivative of UNC3866. (d) UNC4195 selectively interacts with 2 subfamilies of chromodomains on a microarray containing 96 epigenetic reader proteins across multiple domain types.
UNC3866 selectivity studies
Knowledge of the activity profile of a molecule is essential in order to associate its cellular effects with modulation of a specific molecular target(s) of interest. Thus, the use and classification of a molecule as a chemical probe is dependent upon an in-depth understanding of its activity against a broad panel of potential molecular targets29. Accordingly, we evaluated the selectivity of UNC3866 against a wide range of both epigenetic and non-epigenetic targets. The selectivity against a small set of Kme readers encompassing proteins from the MBT, PHD and Tudor families was first evaluated by AlphaScreen® or ITC (Supplementary Table 2). UNC3866 is a potent antagonist of the CBX7-H3 interaction as determined by AlphaScreen® (IC50 = 66 ± 1.2 nM) and is more than 100-fold selective for CBX7 over the other nine members of this Kme reader panel.
To facilitate further selectivity studies, we generated a biotinylated derivative of UNC3866, UNC4195 (4) (Fig. 2c), which retains high affinity for CBX7 (Kd = 220 ± 22 nM). We used this derivative to evaluate binding to 96 purified chromatin-associated effector proteins, including at least 19 known H3K9me3 or H3K27me3 binding proteins and 28 distinct chromodomains, spotted in duplicate onto a nitrocellulose coated membrane (Fig. 2c, Supplementary Fig. 3a). Binding interactions were visualized with fluorescently tagged streptavidin, revealing that UNC4195 binds to two families of chromodomains. As expected, a positive binding interaction was observed with CBX7 and the other PRC1 orthologues, CBX2, -4, -6 and -8. We did not detect binding to CBX1, -3 and -5 (also known as HP1β, -γ and -α, respectively) which are not associated with PRC1. The array also revealed binding to three members of the CDY family of chromodomains: CDY1, CDYL1b, and CDYL2. We expressed and purified all proteins that demonstrated positive binding to UNC4195 on the array and subsequently quantified the affinity for UNC3866 by ITC (Table 1 and Supplementary Fig. 3b). In agreement with the microarray data, we detected binding to each of these chromodomains. The affinity of UNC3866 for CBX2, -4, -6 and -8 is also surprisingly well correlated with the percent sequence identity of each chromodomain relative to that of CBX7 (Supplementary Fig. 3c). UNC3866 is equipotent for CBX4, which is most similar to CBX7, while it is 18-, 6- and 12-fold selective for CBX4/7 over CBX2, -6 and -8, respectively. Additionally, UNC3866 is 65-fold selective for CBX4/7 over CDY1 and 9-fold selective for CBX4/7 over CDYL1b and CDYL2. While some work has been done to explore the biochemistry of CDY proteins38, their biological function is less well characterized than Polycomb CBX proteins which makes assessing the potential consequences of antagonizing these proteins in a cellular context challenging. This is a “known unknown” in the use of UNC3866.
Table 1. Quantitative analysis of UNC3866 binding to CBX and CDY chromodomains by ITC.
Kd's are reported as the average of at least 2 replicates ± the standard deviation.
| Chromodomain | UNC3866 Kd (μM) |
|---|---|
| CBX2 | 1.8 ± 0.21 |
| CBX4 | 0.094 ± 0.017 |
| CBX6 | 0.610 ± 0.0078 |
| CBX7 | 0.097 ± 0.0024 |
| CBX8 | 1.2 ± 0.021 |
| CDY1 | 6.3 ± 0.92 |
| CDYL1b | 0.91 ± 0.076 |
| CDYL2 | 0.85 ± 0.076 |
UNC3866 was also evaluated against a panel of 48 bromodomains, 33 protein methyltransferases, and 7 lysine demethylases and found to be inactive in each case (Supplementary Fig. 4 and Supplementary Tables 3 and 4). UNC3866 was tested against a general pharmacology panel (Cerep) consisting of 49 G protein-coupled receptors (GPCRs), 5 ion channels and 3 transporter proteins (Supplementary Table 5), as well as the National Institute of Mental Health's Psychoactive Drug Screening Program (NIMH PDSP) panel which includes 22 additional GPCRs. Follow-up functional assays39, 40 for targets displaying >50% inhibition of control radioligand binding at 10 μM UNC3866 were also performed with both UNC3866 and UNC4219 (negative control) (Supplementary Figures 5-8). Both UNC3866 and UNC4219 showed nearly identical profiles in these follow-up assays with both compounds showing very weak antagonism for the NK2 and V1a receptors (Supplementary Fig. 6 and Supplementary Fig. 8a). The similar profiles of UNC3866 and UNC4219 indicate that UNC4219 will be a valuable negative control in determining the biological consequences specific to chromodomain antagonism.
UNC3866 co-crystallization studies
We additionally solved an X-ray co-crystal structure of UNC3866 bound to CBX7 (PDB code 5EPJ) at a resolution of 1.6 Å to improve our understanding of its binding interactions. Each backbone amide of UNC3866 participates in at least one hydrogen bond with the backbone of CBX7 (Fig. 3a & 3b), while the N-terminal tert-butyl benzoyl cap of UNC3866 primarily contacts the side chains of D50, R52, and L53 (Fig. 3c). The guanidinium moiety of R52 forms a salt bridge with D50 and this rigidified R52 side-chain forms a hydrophobic groove with L53 within which the tert-butyl phenyl group of UNC3866 is embedded. Based on our MDS studies, we suspect that the 70-fold affinity increase observed upon substitution of the benzoyl-glycyl cap of UNC3567 for the tert-butyl benzoyl cap of UNC3866 is the result of an improvement in the ability of this moiety to nucleate closure of CBX7 around the molecule through interactions with D50, R52 and L53.
Figure 3. Structural studies of UNC3866 with CBX7 (PDB code 5EPJ) and CBX8 (PDB code 5EQ0).
(a) Interaction of CBX7 (surface shown) with the C-terminal portion of UNC3866 (green). (b) Interaction of CBX7 with the N-terminal portion of UNC3866. (c) Interactions of the tert-butyl benzoyl cap of UNC3866 with the hydrophobic groove formed by CBX7 residues D50, R52 and L53. (d) Comparison of the UNC3866 alanine binding pocket in CBX7 and CBX8 (top and bottom panels, respectively).
Additionally, we solved X-ray crystal structures of UNC3866 bound to CBX2 (PDB code 5EPK), CBX4 (PDB code 5EPL), and CBX8 (PDB code 5EQ0) in order to better understand the selectivity profile of UNC3866 (Fig. 3d and Supplementary Fig. 9). Alignment of these structures clearly illustrates that UNC3866 binds in a similar fashion to each chromodomain (Supplementary Fig. 9a). A closer inspection of the binding pocket for the alanine of UNC3866 in each protein revealed that the improved potency of UNC3866 for CBX4/7 over CBX2/8 might be the result of a subtle, yet significant difference in this pocket. The alanine pocket in CBX2 and -8 consists of a valine, leucine and alanine. The side chains of these residues form a relatively open hydrophobic pocket which binds the UNC3866 alanine methyl group. In contrast, this pocket in CBX4 and -7 is made up of two valines and a leucine (Fig. 3d, Supplementary Fig. 9b-f) which more tightly mold to the alanine, enhancing the van der Waals interactions with UNC3866. This single residue is likely the predominant factor that determines the higher affinity of UNC3866 for CBX4 and -7, as there are no other significant variations within the amino acids that directly interact with UNC3866.
In the absence of co-crystal structures, it is more challenging to explain the selectivity of UNC3866 for CBX4/7 over the CDY proteins. Alignment of the sequences (Supplementary Fig. 9f) of the CBX and CDY proteins that bind UNC4195 on the microarray (Fig. 3a) indicates that the key motif in the CBX family that engages the tert-butyl of UNC3866 (D-X-R) is replaced by an (N/H)CE motif in the CDY proteins. Given the demonstrated importance of this interaction in the binding of UNC3866 to CBX7, this sequence variation may contribute to the modest selectivity of UNC3866 for CBX4/7. Further structural and in-depth SAR studies are needed to obtain a more complete understanding to aid in the design of selective ligands.
We were also interested in the interactions between the Polycomb CBX chromodomains and the diethyl-lysine of UNC3866 revealed by our structures. While the H3K27me3 side-chain has been modeled as adopting a fully extended, all anti-conformation3, the lysine side chain of UNC3866 when bound to CBX proteins in our crystal structures is modeled as adopting a gauche conformation about the Cγ-Cδ bond in Fig. 3a. Although the corresponding electron density is comparatively weak for CBX7 (Supplementary Fig. 10a), we model the side chain in the gauche conformation based on the higher resolution CBX8-UNC3866 co-crystal structure (1.2 Å as compared to 1.6 Å for the CBX7-UNC3866 structure). While the electron density of the CBX8-UNC3866 structure more strongly supports the gauche conformation of this side chain in the bound state (Supplementary Fig. 10b), the ambiguity in the position of these atoms may reflect that the side chain in UNC3866 remains somewhat flexible when bound to CBX7. Further, this apparent flexibility in the bound state may explain the ability of CBX7 and the other Polycomb chromodomains to accommodate the diethyl functional group, which is more sterically demanding than the endogenous Kme3 recognized by these domains.
UNC4195 engages ‘canonical’ PRC1
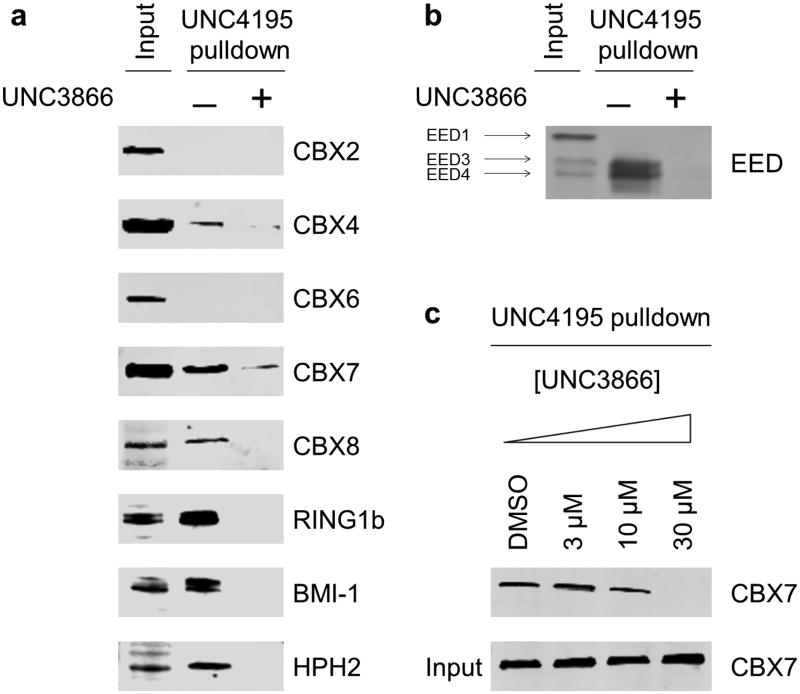
Chemiprecipitation experiments in PC3 prostate cancer cell lysates were performed with UNC4195 in order to investigate its utility as a CBX pull-down reagent. PC3 cells were chosen for the relatively high abundance of each CBX Polycomb protein and the prior utilization of this cell line to study CBX7 function10. Upon chemiprecipitation with UNC4195, we probed for the presence of CBX2, -4, -6, -7 and -8. We detected pull-down of CBX4, -7 and -8 in these experiments and this interaction could be inhibited in the presence of excess UNC3866 in the lysate (Fig. 4a), confirming that successful pull-down is dependent on the specific binding of the chromodomains to UNC4195. We failed to pull-down CBX2 and -6 in these experiments, suggesting that, in contrast to CBX4, -7 and -8, CBX2 and -6 participate in PRC1 complexes in such a way that their chromodomains become inaccessible to pull-down with UNC4195. This is supported by the fact that the purified, isolated chromodomains of CBX2 and -6 interact with UNC4195 on the microarrays (Fig. 2c). We also investigated the ability of UNC4195 to chemiprecipitate PRC1 as an intact complex by probing for representative proteins of the remaining three ‘canonical’ PRC1 subunits (Fig. 4b). We detected RING1b, BMI-1 and HPH2, indicating that UNC4195, and by extension, UNC3866, engages ‘canonical’ PRC1 without disrupting the complex's core composition.
Figure 4. UNC4195 pull-down studies in PC3 cells.
(a) UNC4195 selectively chemiprecipitates CBX4 and -7, and -8 from PC3 cell lysates and chemiprecipitates additional PRC1 components. (b) UNC4195 chemiprecipitates EED3 and EED4 from PC3 cell lysates. (c) Incubation of intact PC3 cells with UNC3866 for 24 hours inhibits pull-down of CBX7 with UNC4195. (n=3 for all samples in a-c)
Isoform-specific incorporation of EED into PRC1
The recent finding that EED can participate in CBX-containing PRC1 complexes intrigued us4; therefore, we investigated the ability of UNC4195 to pull-down EED in PC3 cells. Three of the four mammalian isoforms of EED (EED1, -3 and -4)41 were detected in the PC3 lysate input; however, UNC4195 pulled-down only EED3 and EED4, both of which could be competed away through the addition of UNC3866 to the lysate (Fig. 4b). Additionally, because EED binds H3K27me3 via its WD40 Kme reading domain, we confirmed that UNC3866 does not bind directly to EED (Supplementary Fig. 11), indicating that pull-down is mediated through incorporation of EED into PRC1. We next examined the presence of EED in PRC2 using the EZH1 and EZH2 biotinylated pull-down reagent, UNC239924. Interestingly, we detected only EED4 in PRC2 (Supplementary Fig. 12). It has been shown that the incorporation of different isoforms of EED into PRC2 can dictate the in vitro histone substrate of PRC242, and hence our discovery that EED incorporates into PRC1 in an isoform-dependent fashion provides the basis for future work to examine the effects of these isoforms on PRC1 function. Furthermore, the absence of EED1 in PRC1 or PRC2 raises questions as to the binding partners and function of this isoform in PC3 cells.
UNC3866 antagonizes PRC1 chromodomains in cells
We next assessed the utility of UNC3866 in a cellular context. As peptide inhibitors often exhibit poor cellular activity, we assessed the permeability of UNC3866 using a Caco-2 assay and found it to be low (Supplementary Fig. 13), indicating a need for relatively high compound concentrations for cellular studies relative to the in vitro Kd. Additionally, we noted the potential for UNC3866 to serve as a pro-drug if the C-terminal methyl ester is susceptible to hydrolysis by intracellular esterases; therefore, we confirmed through a combination of pull-down and ITC experiments that the corresponding C-terminal acid, UNC4007 (5), is still capable of engaging PRC1 chromodomains (Supplementary Fig. 14). Next, we quantified the intracellular concentration of both UNC3866 and UNC4007 by LC-MS/MS (Supplementary Table 6). Owing to the low Caco-2 permeability, we incubated PC3 cells with 30 μM UNC3866 in order to ensure sufficient accumulation of intracellular UNC3866. Treatment for 24 hours resulted in an intracellular concentration of 1.4 ± 0.3 μM, indicating that the intracellular concentration of UNC3866 is ∼5% of the extracellular concentration. The UNC3866 intracellular concentration was approximately 7-fold higher than the intracellular concentration of UNC4007, indicating that hydrolysis of UNC3866 is limited under these conditions. Overall, UNC3866 is sufficiently cell permeable and stable to evaluate its ability to engage and antagonize the functions of its chromodomain targets in cells.
The non-enzymatic nature of chromodomains makes demonstrating cellular engagement of these proteins challenging. While PRC1 has E3 ligase activity, the presence of non-canonical PRC1 complexes that do not contain CBX proteins6 makes monoubiquitination of H2AK119 an unreliable readout. Further, it has been shown that neither mutation nor deletion of the chromodomains of CBX proteins has significant effects on the cellular mobility of these proteins as determined by Fluorescence Recovery After Photobleaching (FRAP) experiments43, ruling out this technique which has been previously applied to chromatin readers32, 33. We failed to detect thermostabilization of CBX7 by UNC3866 in cell lysates (Supplementary Fig. 15), likely because the chromodomain comprises a relatively small portion (∼25%) of the full-length protein, which precludes evaluation of cellular target engagement via the Cellular Thermal Shift Assay (CETSA)44. Therefore, we hypothesized that competition-based chemiprecipitation experiments could be utilized to demonstrate cellular target engagement, with the expectation that pretreatment with UNC3866 would block chemiprecipitation. Whole PC3 cells were incubated with varying concentrations of UNC3866 for 24 hours, after which excess media and compound were removed and the cells were washed, harvested with trypsin and lysed. Encouragingly, CBX7 chemiprecipitation with UNC4195 was completely inhibited in lysates from cells pretreated with 30 μM UNC3866 (intracellular concentration = 1.4 ± 0.3 μM, Fig. 4c), indicating that UNC3866 effectively engages CBX7 in cells at this concentration.
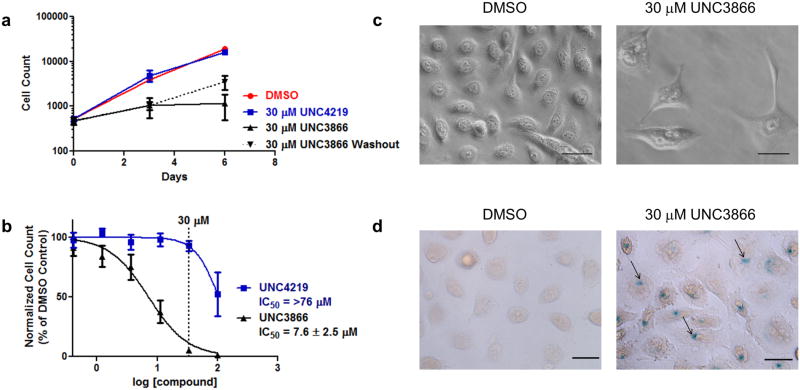
We next focused on establishing a biological consequence of treating cells with UNC3866. The high levels of CBX proteins in PC3 cells, coupled with the demonstrated ability of CBX7 overexpression to confer a growth advantage in these cells10, 45, encouraged us to investigate the effects of UNC3866 on PC3 cell proliferation. As a prelude to these studies, we confirmed that there were no toxic effects induced upon treatment with UNC3866 up to 100 μM as assessed by CellTiter-Glo® in PC3 and HEK293T cells (Supplementary Fig. 16). Similarly, UNC4219 exhibited no observable toxicity within the same concentration range. HEK293T cells were used to evaluate UNC3866 toxicity in a non-cancerous cell line. To assess effects on proliferation, PC3 cells were treated with UNC3866 and the negative control, UNC4219, at 30 μM, as this concentration was shown to effectively engage CBX7 in cells (Fig. 4c). Consistent with the known ability of CBX7 to enhance proliferative capacity in PC3 cells14, 45, we observed a dramatic decrease in cell proliferation upon UNC3866 treatment, resulting in almost no change in cell count after 3 and 6 days, while cells treated with UNC4219 or DMSO exhibited exponential cell growth (Fig. 5a). Further, we observed that cells resumed proliferating when washed and not replenished with UNC3866 after 3 days (Fig. 5a, dotted line), indicating that the compound effects are at least partially reversible. Dose dependency was next examined for UNC3866 and UNC4219, and a significant reduction in PC3 cell count was only observed for UNC3866 treated cells, resulting in an EC50 of 7.6 ± 2.5 μM (Fig. 5b). The intracellular concentration of UNC3866 at its EC50 in PC3 cells is therefore estimated to be 340 nM, which is adequate to fully engage CBX4 and -7 based on the measured ∼100 nM Kd's versus these targets (Fig. 3b). Although some contribution of other chromodomains bound by UNC3866 to this antiproliferative phenotype cannot be ruled out, the concordance of this result with previously published CBX7 genetic manipulation is striking17, 18.
Figure 5. Cellular effects of UNC3866.
(a) Treatment of PC3 cells with 30 μM UNC3866 inhibits cell proliferation. UNC4219 treatment has no effect on PC3 cell proliferation. For washout experiments (dotted line), UNC3866 was not replenished at day 3. Error bars represent the SD of that data point (n=3, single biological replicate with 3 technical replicates). The data is representative of at least 4 biological replicates with varying plating densities. (b) UNC3866 inhibits PC3 cell proliferation in a dose-dependent manner after exposure for 6 days (n=8, 2 biological replicates with 3 technical replicates and 1 biological replicate with 2 technical replicates). The EC50 is reported as the 95% confidence interval. UNC4219 was used to control for off-target effects, which were not evident at concentrations below 100 μM. The EC50 for UNC4219 is >76 μM, which is the lower bound of the 95% confidence interval (n=6, 3 biological replicates with 2 technical replicates each). Error bars represent the SEM of that data point. (c) UNC3866 induces a senescent-like morphology in PC3 cells and (d) expression of SA-β-gal. Arrows indicate examples of positive SA-β-gal expression. Cell morphology was assessed using phase contrast microscopy (40×). SA-β-gal expression was assessed using bright-field microscopy (40×). The images are representative of 3 biological replicates and the black scale bars are equal to 60 μm.
We also observed that PC3 cells treated with UNC3866 exhibited an enlarged and flattened morphology (Fig. 5c), suggesting that these cells had become senescent46. We checked for the presence of senescence-associated β-galactosidase (SA-β-gal) and observed a dramatic increase in the number of cells positive for SA-β-gal following treatment with 30 μM UNC3866 (Fig. 5d). This was an intriguing observation because CBX7 was originally identified in a genetic screen for genes capable of bypassing replicative senescence in Human Prostate Epithelial Cells (HPrEC)18. Further, the same study observed that CBX7 knockdown is capable of inducing senescence.
While it has been suggested that CBX7 controls cellular proliferation through regulation of the Ink4a/ARF locus18, its actual role in regulating this locus is controversial9, 11, 18. For example, CBX7 has been shown to enhance proliferative capacity without negatively regulating this locus9, and negative regulation of this locus does not necessarily enhance proliferative capacity14. Nonetheless, we analyzed the expression of this locus in response to knockdown of CBX7 by shRNA (>60% mRNA reduction) in PC3 cells and did not observe a statistically significant change in expression of the Ink4a/ARF locus under these conditions. Further, evaluation of the effects of UNC3866 on expression of this locus showed no change at 30 μM, ruling out Ink4a/ARF regulation in the antiproliferative effects of UNC3866 (Supplementary Fig. 17). This was not entirely surprising as there is considerable evidence indicating that this locus is deeply repressed in PC3 cells through DNA hypermethylation47-50. Further, methylation of this locus is unaffected by both DNMT knockdown and inhibition with 5-AzaC, suggesting that this gene may be irreversibly repressed in PC3 cells47-49.
Finally, we evaluated the stability of UNC3866 following intraperitoneal (IP) injection (10 mg/kg) in male swiss albino mice and found that UNC3866 is the predominant species in plasma at all time points tested relative to UNC4007 (Supplementary Tables 7-9 and Supplementary Fig. 18), and shows 25% bioavailability and moderate clearance. While these PK results are promising for a peptidic compound, the use of UNC3866 in vivo may be limited because of the high circulating levels required for intracellular target engagement due to its poor cell permeability. The potential utility of UNC3866 at higher doses for in vivo experiments is currently under investigation.
Discussion
Our investigations into the molecular dynamics of H3 recognition by CBX7 enabled the design of UNC3866, the most potent CBX7 antagonist reported to date27, 28. UNC3866 is an equipotent nanomolar inhibitor of CBX4 and -7 Kme recognition in vitro with a thoroughly characterized selectivity profile. Further, through the use of UNC4195, we demonstrated that UNC3866 binds in the context of intact, ‘canonical’ PRC1. Finally, UNC3866 exhibits micromolar cellular potency in competition pull-down and cellular proliferation assays with no associated toxicity. The replacement of the quaternary amine of the native peptide ligand with an unnatural tertiary amine mimetic was a key achievement in the development of a cellularly active ligand, and more broadly signifies that a quaternary amine is not required for inhibition of Kme3 readers.
Knowledge of the activity profile of a molecule is essential in order to associate its cellular effects with modulation of a specific molecular target(s) of interest. While UNC3866 is most potent for CBX4 and -7, it possesses affinity for other Polycomb CBX proteins and the CDY family of chromodomains. Its use as a chemical probe will require consideration of these activities and evaluation of dose dependency to assess possible affects from these other targets. This is not unusual for first-in-class chemical probes; for example, the initially reported bromodomain antagonists were not selective within the BET family and yet their utility is well established32. The thorough characterization of the in vitro selectivity of UNC3866 versus >250 proteins, the demonstrated cellular target engagement of UNC3866, and the use of UNC4219 as a negative control validates UNC3866 as the first high-quality chemical probe for both the Polycomb CBX family of Kme readers and, to our knowledge, any Kme3 reader.
Our work towards the development of chemical tools for Polycomb CBX Kme readers has revealed that EED is incorporated into PRC1 and PRC2 in an isoform-dependent manner in PC3 cells, providing the basis for future investigation into the biochemical role of the different EED isoforms in Polycomb-mediated signaling. The ability of UNC3866 to inhibit PC3 cell proliferation supports further investigation of the use of PRC1 chromodomain antagonists as therapeutics in oncology, alone or in combination with EZH2 inhibitors. In addition to their role in different cancers, PRC1 chromodomains play a pivotal role in regulating cellular differentiation9, and UNC3866 and its related derivatives provide a new set of tools that will assist in further probing the role of PRC1 chromodomains in cell-fate determination, and more generally, in understanding Polycomb signaling.
Online Methods
I. Experimental Methods
(i) Adaptively Biased Molecular Dynamics Simulations
Adaptive biasing potential35 was implemented in GROMACS 4.5.551. The bias parameters were b=0.8, c=0.0005/δt, where δt=2 fs is the molecular dynamics time step. The Gaussian width was 1/4 Angstrom. This choice of bias parameters floods stable states extremely slowly, to minimize adaptation of the bias near transition states. Thus, one may argue for approximate state-to-state dynamics as has recently been done for metadynamics52 following the principles of hyperdynamics53. Using the first frame of the CBX7+H3K9me3 NMR structure, two collective variables were defined: RMSD of clasp, and RMSD of peptide. We used the Kabsch algorithm to align the reference and trajectory during the simulation. All simulations were performed in a cubic box with a minimum of 12 Angstroms between the protein or peptide and the nearest cube face, resulting in 23710 atoms after adding water and salt. System construction included 5 nsec of isothermal–isobaric equilibration at 1 bar. Production simulations were carried out in the canonical ensemble with stochastic velocity rescaling54. Van der Waals and direct electrostatic interactions used a 10 Å cutoff. Long-range electrostatics were treated with the particle mesh Ewald approach with a grid spacing of 1.6 Å. The neighbor list was updated every five steps. We ran five 128 ns simulations to get some intuition that could inform ligand design. All images from the simulations were rendered using the PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.
(ii) ITC Experiments
All ITC measurements were recorded at 25 °C with an AutoITC200 microcalorimeter (MicroCal Inc., MA). All protein and compound stock samples were in the target buffer (25 mM Tris-HCl, pH 8, 150 mM NaCl, and 2 mM β-mercaptoethanol), and then diluted in the same buffer to achieve the desired concentrations: 50 μM protein and 0.5 mM compound. The concentration of the protein stock solution was established using the Edelhoch method, whereas compound stock solutions were prepared based on mass. A typical experiment included a single 0.2 μl compound injection into a 200 μl cell filled with protein, followed by 26 subsequent 1.5 μl injections of compound. Injections were performed with a spacing of 180 seconds and a reference power of 8 μcal/sec. Control experiments were performed titrating each compound into buffer under identical conditions to determine the heat signals, if any, that arise from diluting the compound. If applicable, the heats of dilution generated were then subtracted from the protein-compound binding curves. The initial data point was routinely deleted. The titration data was analyzed using Origin Software (MicroCal Inc., USA) by non-linear least squares, fitting the heats of binding as a function of the compound:protein ratio to a one site binding model.
(iii) Kme Reader AlphaScreen® Assays
The AlphaScreen® assay (Perkin Elmer) was generally performed as previously described.55 In brief, compound plates (1 μL at 10 mM highest concentration; 3-fold, 10-point dilutions in DMSO) were diluted in 1× assay buffer (20 mM TRIS pH 8.0, 25 mM NaCl, 2 mM DTT and 0.05% Tween-20) to 1 mM using a Multimek robotic pipettor (Nanoscreen) and 1 μL was spotted into the wells of 384-well low-volume Proxiplates (Perkin Elmer). To these plates 9 μL of protein-peptide mix in 1× assay buffer was added by Multidrop (Thermo) to bring the final compound concentration to 100 μM and incubated for 30 min at room temperature. Next, 2 μL of a 1:1 mixture of streptavidin-conjugate donor and nickel-chelate or α-GST acceptor beads (45 μg/mL in 1× assay buffer) were added and the plates were allowed to incubate for an additional 30 min in the dark at room temperature. After incubation, the plates were read on an EnVision multi-label reader equipped with an HTS AlphaScreen laser (Perkin Elmer). The expression and purification of the constructs used in this assay was described previously56.
(iv) Methyltransferase Selectivity Assays
The effect of UNC3866 on the methyltransferase activity of G9a, EHMT1, SUV39H1, SUV39H2, SETDB1, SETD8, SUV420H1, SUV420H2, SETD7, MLL1 trimeric complex, MLL3 tetrameric complex, EZH2 trimeric complex, PRMT1, PRMT3, PRMT5-MEP50 complex, PRMT6, PRMT7, PRMT8, PRDM9, SETD2, SMYD2, SMYD3, BCDIN3D and DNMT1 was assessed by monitoring the incorporation of tritium-labeled methyl group to lysine or arginine residues of peptide substrates using Scintillation Proximity Assay (SPA) as previously described57. Assays were performed in a 20 μL reaction mixture containing 3H-SAM (Cat. # NET155V250UC; Perkin Elmer; www.perkinelmer.com) at substrate concentrations close to the Km values for each enzyme. Three concentrations (1 μM, 10 μM, and 50 μM) of UNC3866 were used in all selectivity assays. To stop the enzymatic reactions, 7.5 M Guanidine hydrochloride was added, followed by 180 μl of buffer (20 mM Tris, pH 8.0). The reactions were mixed and then transferred to a 96-well FlashPlate (Cat. # SMP103; Perkin Elmer; www.perkinelmer.com). The reaction mixtures in Flash plates were incubated for 1 hour and the CPM were measured using a TopCount plate reader (Perkin Elmer, www.perkinelmer.com). The CPM counts in the absence of compound for each data set were defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set were defined as background (0%).
For DOT1L, NSD1, NSD2, NSD3, ASH1L, METTL21A, METTL21D, DNMT3A/3L, and DNMT3B/3L, a filter-based assay was used. In this assay, 20 μl of reaction mixtures were incubated at 22 °C for 1 hour, 100 μl of 10% trichloroacetic acid (TCA) was added, and the samples were mixed and transferred to filter-plates (Millipore; cat.# MSFBN6B10; www.millipore.com). Plates were centrifuged at 2000 rpm (Allegra X-15R - Beckman Coulter, Inc.) for 2 min, followed by two additional 10% TCA washes, one ethanol wash (180 μl), and centrifugation. Plates were dried and 100 μl of MicroO (MicroScint-O; Cat. # 6013611, Perkin Elmer; www.perkinelmer.com) was added to each well, centrifuged and removed. Another 70 μl of MicroO was added and the CPM was measured using a TopCount plate reader.
(v) Bromodomain Selectivity Assays
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene). Proteins were buffered in 10 mM HEPES pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 μM in 20 μL volume. Compounds were added at a final concentration of 25 μM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C and fluorescence readings were taken at each interval.
(vi) Demethylase AlphaScreen® Assays
The demethylase AlphaScreen® assay was performed in 384-well plate format using white proxiplates (Perkin Elmer) and compound transfers (100 nl) were performed using an ECHO 550 Acoustic Dispenser (Labcyte). All subsequent steps were carried out in assay buffer (50 mM HEPES pH 7.5, 0.1% (w/v) Bovine Serum Albumin and 0.01 % (v/v) Tween-20). In brief, 5 ml of assay buffer containing demethylase enzyme at 2× final assay concentration (see Supplementary Table 13 for assay specifics) was pre-incubated for 15 minutes with dilutions of compound. The enzyme reaction was initiated by addition of substrate (5 ml) consisting of L-Ascorbic Acid (100 mM), 2-oxoglutarate (2× final assay concentration), Ferrous Ammonium Sulphate (2× final assay concentration) and Histone H3 substrate peptide (2× final assay concentration) and the enzyme reaction was allowed to proceed for the required time. The final assay DMSO concentration was 1%. The enzyme reaction was stopped by addition of 5 ml assay buffer containing EDTA (30 mM) and NaCl (800 mM). Streptavidin Donor beads (0.08 mg/ml) and Protein-A conjugated acceptor beads (0.08 mg/ml) were pre-incubated for 1 hour with anti-methyl mark antibody (4× final assay concentration) and the presence of Histone H3 product methyl mark was detected by addition of the pre-incubated AlphaScreen® beads (5 ml). Detection was allowed to proceed for 2 hour at room temperature and the assay plates were read in a BMG Pherastar FS plate reader (Excitation 680 nM / Emission 570 nM). Data were normalized to the no enzyme control and the IC50 determined from the nonlinear regression curve fit using GraphPad Prism 5.
(vii) NIMH PDSP Functional Assays
The NK1 and NK2 receptors; Δ-, κ-, and μ-opioid receptors; and V1A, V1B and V2 receptor functional assays were performed according to the NIMH PDSP Assay Protocol Book Version II found at https://pdspdb.unc.edu/pdspWeb/?site=binding
(viii) Cell culture and lysis
PC3 cells were obtained from ATCC® (CRL-1435™) through the UNC Lineberger Tissue Culture Facility. Cells were cultured using GIBCO® DMEM/F12 (Ham), [+] L-Glutamine, and [+] 15 mM HEPES media. Cells were trypsinized using 0.25% trypsin. Lysis was performed using Cytobuster™ protein extraction reagent supplemented with protease inhibitors and Benzonase® (used at 25 U/mL). Samples were incubated at 37°C for 10 minutes, followed by incubation at RT on a rotator for 20 minutes. The samples were spun down and the supernatant collected and transferred to a clean Eppendorf tube. Protein concentrations were quantified using the Bradford protein assay.
(ix) Pull-down studies
Cells were cultured on a T175 tissue culture flask until reaching ∼80-90% confluency. Following trypsinization, the pellet was washed twice with PBS and lysed with 500 μL of lysis buffer. When applicable, vehicle control or 100 μM UNC3866 was added to an aliquot of 1000 μg of total protein and diluted to 500 μL with 20 mM Tris (pH 8)/150 mM NaCl/0.1% Tween-20 (TBST). Pull-down reagents were bound to Streptavidin M-270 Dynabeads® by rotating 30 μL of beads with a 20-fold excess of pull-down reagent for 30 minutes at room temperature in. Unbound pull-down reagent was then removed and the beads were washed 3× with 200 μL of TBST. The lysate was then transferred to an Eppendorf tube containing 30 μL of Streptavidin M-270 Dynabeads® that had been pre-bound to UNC4195 or 2399. The mixture was rotated overnight at 4°C. The following morning, the depleted lysate was removed and the beads were washed 3× with 300-500 μL of TBST. The beads were re-suspended with 30 μL of 1× Laemmli sample buffer and heated at 95°C for 3 minutes. Fifteen microliters of the sample was then loaded into a gel and analyzed via western blotting using the appropriate primary antibody (Supplementary Table 14) and detected using a LI-COR Odyssey® instrument and the appropriate fluorescent secondary antibodies (diluted 1:10 ,000 in PBST). One percent of input was used for western blotting of all proteins except CBX8 and HPH2. For both of these proteins, 3% of sample input was used.
(x) Cell Proliferation Assay
PC3 cells were seeded at 200 cells/well into 24-well plates (Costar #3524). Cells were allowed to adhere overnight. The media (DMEM supplemented with 10 % FBS) was then exchanged with fresh media containing DMSO, UNC3866 or UNC4219. On day three, the media were exchanged with fresh media containing DMSO, UNC3866 or UNC4219. For dose-response studies, the EC50 was derived from a six-point titration ranging from 100 μM to 0.4 μM of UNC3866 or UNC4219. At day 0, 3 or 6, cells were fixed with ice-cold methanol for 30 sec. and rehydrated with PBS. Nuclei of the cells were stained with DAPI (0.05 μg/ml) and numerated using High Content Microscopy (Array Scan™ High Content Analysis, Thermo Fisher #NX10002L). For dose-response studies, the cell count of UNC3866- or UNC4219-treated cells was normalized to the average cell count of DMSO-treated cells. The EC50 was calculated using the “log[inhibitor] vs. the normalized response -- Variable slope” equation in GraphPad Prism 5.
(xi) CellTiter-Glo Luminescent Cell Viability Assay
The effect of UNC3866 and UNC4219 on cell viability was determined using a CellTiter-Glo™ ATP detection system (Promega #7573). Ten point, 1:3 dilution curves of compounds starting at 100 μM final concentration were diluted to 5× final concentration in PBS (vehicle control) and then 5 μL were added to 384-well white, clear bottom tissue culture plates (Corning #3707) with a Multimek automated liquid handling device (Nanoscreen, Charleston, SC). Twenty microliters of low passage, subconfluent HEK293T/PC3 cells grown in Dulbecco's Modified Eagle's Medium without phenol red (GIBCO® #31053) and supplemented with 10% Fetal Bovine Serum (GIBCO® #26140) were immediately added at a density of 5,000 cells per well using a Multidrop 384 (Titertek). Cell plates were incubated for 48 hours at 37°C and 5% C02, and then lysed with 25 microliters of CellTiter-Glo™ reagent. Luminescence was read on an Envision platereader (Perkin Elmer) after 15 minutes at room temperature in dim light.
(xii) Cellular Thermal Shift Assay (CETSA)
A PC3 cell pellet consisting of 107 cells was re-suspended in 1200 μL of PBS complete with protease inhibitors. The suspension was subjected to 3 consecutive freeze-thaw cycles performed with liquid N2 in order to lyse the cells. Following the final freeze-thaw cycle, the suspension was centrifuged at 20,000 g for 20 minutes at 4°C. The supernatant was collected and the resulting pellet was discarded. The lysate was then divided into two tubes (600 μL) each. The volume of each tube was brought up to 650 μL with PBS + protease inhibitors. DMSO or 30 μM UNC3866 was added to each tube. The tubes were incubated at RT for 30 minutes. The lysates were aliquoted into 12 thin-walled PCR tubes (50 μL/tube). The tubes were incubated for 3 minutes at temperatures ranging from 37°C to 73°C in 3° increments in a Mastercycle® nexus gradient qPCR machine. Tubes were then incubated at room temperature for 3 minutes followed by centrifugation at 20,000 g for 20 minutes at 4°C. The supernatant was collected. Ten microliters of 6× Laemmli buffer was added to each tube. The tubes were then heated to 95°C for 3 minutes. Twenty microliters was then loaded into an SDS page gel and analyzed by western blotting.
(xiii) CBX7 knockdown and quantitative PCR
PC3 cells were seeded at a density of 105 cells/well in a 6-well plate and allowed to adhere overnight. The media was then exchanged for 2 mL of media containing polybrene (8 μg/mL) and pre-packaged shRNA (200,000 Transducing Units/well) targeting CBX7 (Mission CBX7, SHCLNV (Sigma) TRCN0000019144RNA) or Non-targeting control (Mission pLKO.1-puro Non-Target shRNA, SHC016V (Sigma)). After 20 hours, the transduction mixture was exchanged with fresh media (GIBCO® DMEM/F12 (Ham), [+] L-Glutamine, and [+] 15 mM HEPES) and allowed to incubate for an additional 24 hours. The media was then replaced with media containing 2 μg/mL of puromycin and allowed to incubate until selection was complete as evidence by the death of all control cells that were not transduced. RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) and RNA was quantified on the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The reverse transcription reaction was performed with 500 ng RNA and the Superscript III First Strand Synthesis Supermix (Invitrogen #1172-050) according to manufacturer instructions. The resulting cDNA was diluted 1:10, and 2 μL was used for each PCR reaction. Quantitative PCR was performed using Roche FastStart Universal SYBR Green Master Mix (Rox) in a 384-well plate format on the ABI 2900HT instrument. Oligo sequences are available upon request.
(xiv) Phase contrast microscopy images
PC3 cells were seeded at 200 cells/well into 24-well plates (Costar #3524). Cells were allowed to adhere overnight. The media (DMEM supplemented with 10 % FBS) was then exchanged with fresh media containing DMSO or 30 μM UNC3866. On day three, the media were exchanged with fresh media containing DMSO or 30 μM UNC3866. At day 6, cells were fixed with ice-cold methanol for 30 sec. and rehydrated with PBS. Samples were imaged using Volocity software and an Olympus IX81 microscope equipped with an Orca ER digital camera.
(xv) Senescence-associated β-Gal staining
PC3 cells were seeded in a 96-well plate at a density of 100-200 cells/well and allowed to adhere overnight. The media was then replaced with fresh media containing 30 μM UNC3866 or DMSO. After 3 days, the media was removed and the cells were fixed and stained using the Senescence β-Galactosidase Kit (#9860) from Cell Signaling Technology® according to the manufacturer's recommendations. Wells were imaged using bright-field microscopy with an Olympus IX81 microscope equipped with an Orca ER digital camera.
II. Protein Expression and Purification
(i) Expression constructs
The chromodomains of CBX2 (residues 9-66 of NP_005180), CBX4 (residues 8-65 of NP_003646), CBX6 (residues 8-65 of NP_055107) and CBX8 (residues 8-61 of NP_065700) were expressed with N-terminal His-tags in pET28 expression vectors. The chromodomain of CBX7 (residues 8-62 of NP_783640) was expressed with a C-terminal His-tag in a pET30 expression vector. The chromodomains of CDY1 (residues 1-101 of NP_733841), CDYL1b (residues 1-78 of NP_004815) and CDYL2 (residues 1-75 of NP_689555) were expressed with N-terminal GST-tags in pGEX4T expression vector.
(ii) Protein expression and purification
All expression constructs were transformed into Rosetta BL21(DE3)pLysS competent cells (Novagen, EMD Chemicals, San Diego, CA). Protein expression was induced by growing cells at 37°C with shaking until the OD600 reached ∼0.6-0.8 at which time the temperature was lowered to 18°C and expression was induced by adding 0.5mM IPTG and continuing shaking overnight. Cells were harvested by centrifugation and pellets were stored at -80°C.
His-tagged proteins were purified by re-suspending thawed cell pellets in 30ml of lysis buffer (50mM sodium phosphate pH 7.2, 50mM NaCl, 30mM imidazole, 1× EDTA free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN)) per liter of culture. Cells were lysed on ice by sonication with a Branson Digital 450 Sonifier (Branson Ultrasonics, Danbury, CT) at 40% amplitude for 12 cycles with each cycle consisting of a 20 second pulse followed by a 40 second rest. The cell lysate was clarified by centrifugation and loaded onto a HisTrap FF column (GE Healthcare, Piscataway, NJ) that had been preequilibrated with 10 column volumes of binding buffer (50mM sodium phosphate pH 7.2, 500mM NaCl, 30mM imidazole) using an AKTA FPLC (GE Healthcare, Piscataway, NJ). The column was washed with 15 column volumes of binding buffer and protein was eluted in a linear gradient to 100% elution buffer (50mM sodium phosphate pH 7.2, 500mM NaCl, 500mM imidazole) over 20 column volumes. Peak fractions containing the desired protein were pooled and concentrated to 2ml in Amicon Ultra-15 concentrators 3,000 molecular weight cut-off (Merck Millipore, Carrigtwohill Co. Cork IRL). Concentrated protein was loaded onto a HiLoad 26/60 Superdex 75 prep grade column (GE Healthcare, Piscataway, NJ) that had been preequilibrated with 1.2 column volumes of sizing buffer (25mM Tris pH 7.5, 250mM NaCl, 2mM DTT, 5% glycerol) using an ATKA Purifier (GE Healthcare, Piscataway, NJ). Protein was eluted isocratically in sizing buffer over 1.3 column volumes at a flow rate of 2ml/min collecting 3ml fractions. Peak fractions were analyzed for purity by SDS-PAGE and those containing pure protein were pooled and concentrated using Amicon Ultra-15 concentrators 3,000 molecular weight cut-off (Merck Millipore, Carrigtwohill Co. Cork IRL).
GST-tagged proteins were purified by re-suspending thawed cell pellets in 30ml of lysis buffer (1×PBS, 5mM DTT, 1× EDTA free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN)) per liter of culture. Cells were lysed on ice by sonication as described for His-tagged proteins. Clarified cell lysate was loaded onto a GSTrap FF column (GE Healthcare, Piscataway, NJ) that had been pre-equilibrated with 10 column volumes of binding buffer (1×PBS, 5mM DTT) using a AKTA FPLC (GE Healthcare, Piscataway, NJ). The column was washed with 10 column volumes of binding buffer and protein was eluted in 100% elution buffer (50mM Tris pH 7.5, 150mM NaCl, 10mM reduced glutathione) over 10 column volumes. Peak fractions containing the desired protein were pooled and concentrated to 2ml in Amicon Ultra-15 concentrators, 10,000 molecular weight cut-off (Merck Millipore, Carrigtwohill Co. Cork IRL). Concentrated protein was loaded onto a HiLoad 26/60 Superdex 200 prep grade column (GE Healthcare, Piscataway, NJ) that had been preequilibrated with 1.2 column volumes of sizing buffer (25mM Tris pH 7.5, 250mM NaCl, 2mM DTT, 5% glycerol) using an ATKA FPLC (GE Healthcare, Piscataway, NJ). Protein was eluted isocratically in sizing buffer over 1.3 column volumes at a flow rate of 2ml/min collecting 3ml fractions. Peak fractions were analyzed for purity by SDS-PAGE and those containing pure protein were pooled and concentrated using Amicon Ultra-15 concentrators 10,000 molecular weight cut-off (Merck Millipore, Carrigtwohill Co. Cork IRL).
(iii) Affinity tag removal
The N-terminal affinity tag was removed from CBX2, CBX6, CDY1, CDYL1b and CDYL2 by thrombin cleavage according to manufacturer's recommendations (Novagen, EMD Chemicals, San Diego, CA). Briefly, purified protein was incubated with biotinylated thrombin at a final concentration of 1 unit thrombin per milligram tagged protein for 16 hours at 4°C. The cleavage reaction was then passed over either a HisTrap FF column (His-tagged proteins) or a GSTrap FF column (GST-tagged proteins) to remove any protein that still retained the tag. The column flow through was collected and incubated with streptavidin agarose to remove thrombin from the sample. Streptavidin agarose was removed from the sample by centrifugation and the protein was concentrated using Amico Ultra-15 concentrators, 3,000 molecular weight cut-off (Merck Millipore, Carrigtwohill Co. Cork IRL). Protein was exchanged into a buffer containing 25mM Tris pH 7.5, 150mM NaCl, 2mM β-mercaptoethanol prior to use in ITC.
III. X-Ray Crystallography
(i) Preparation of proteins for crystallization
Expression and purification followed a previous description3. The coding DNA fragments of single chromodomains of CBX2 (residues 9-62), CBX4 (residues 8-65), CBX7 (residues 8-62) and CBX8 (residues 8-61) were subcloned into modified pET28-MHL or pET28a-Lic vectors to encode N-terminal His-tagged fusion proteins. Recombinant proteins were overexpressed in E. coli BL21 (DE3) Codon plus RIL cells (Stratagene) at 15 °C, purified by affinity chromatography on Ni-nitrilotriacetate resin (Qiagen), and treated with TEV or thrombin protease to remove the tag. The proteins were further purified by gel filtration over a Superdex75 column (GE Healthcare, Piscataway, NJ). For crystallization, purified proteins were concentrated to 10 mg/mL in a buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl and 1 mM DTT.
(ii) Crystallization
Purified protein was mixed with compound at a 1:2 molar ratio and crystallized using the sitting drop vapor diffusion method at 18–20°C by mixing 0.5 μL of the protein with 0.5 μL of the reservoir solution. Reservoir solutions were 20% PEG 3350, 0.2 M KSCN for CBX2, 30% PEG 3350, 0.2 M NaCl, 0.1 M sodium cacodylate, pH 5.5 for CBX4, 20% PEG 3350, 0.2 M ammonium formate for CBX7, and 1.4 M sodium citrate, 0.1 M Hepes, pH 7.5, 5% glycerol for CBX8. Crystals were soaked in a cryoprotectant consisting of 85% reservoir solution and 15% glycerol, and flash-frozen in liquid nitrogen.
(iii) Data collection/structure determination and refinement
Due to space restrictions, literature references to selected computer programs are provided in the Supplementary Information, references 4-16). Details on diffraction data and model refinement are given in Supplementary Table 15. Diffraction data for model refinement were collected at a copper rotating anode (CBX2, CBX7) or APS beam line 19ID (CBX4, CBX8; wavelength 0.979 Å). Diffraction intensities were integrated with XDS58 and merged with AIMLESS59. Molecular replacement was performed with the program PHASER60. Geometry restraints for the modified lysyl residue as well as the 4-t-butylbenzoyl protecting group were prepared with eLBOW/Mogul, restraints for selected peptide links with jLigand. Current crystallographic models underwent iterative re-building, refinement, validation in COOT, REFMAC, MOLPROBITY, respectively. Favored regions of the respective model's MOLPROBITY Ramachandran plot included at least 98% of the residues. Anisotropic displacement parameters were analyzed on the PARVATI server. PDB_EXTRACT as well as PHENIX, IOTBX and CCP4 software were used to compile data for model deposition in the Protein Data Bank and for publication.
Specifically:
CBX2. Coordinates from PDB entry 3H913 were used for molecular replacement.
CBX4. The structure of an isomorphous crystal was solved by molecular replacement with CBX2 coordinates. ARP/WARP was used in “map improvement” and “automated model building” modes. The model refinement was further refined against data from the current crystal.
CBX7. The structure was first solved in the lower symmetry P41 setting by molecular replacement with coordinates from PDB entry 4MN327. After merging of data in space group P41212, molecular replacement was performed with preliminarily refined CBX7 coordinates.
CBX8. The structure of an isomorphous crystal was solved by molecular replacement with CBX7 coordinates. ARP/WARP was used in “map improvement” and “automated model building” modes. The model was further refined against data from an additional crystal and, subsequently, the current crystal.
Supplementary Material
Acknowledgments
We thank Kenneth Pearce (UNC Chapel Hill) for useful discussions and careful reading of the manuscript, Oleg Fedorov (SGC Oxford) for support with the bromodomain selectivity screening, Anthony Tumber (SGC Oxford) for support with the lysine demethylase selectivity screening and John R. Walker for the review of the crystal structures. We thank Greg Wang (UNC) for providing PHF1, PHF19, PHF23 and JARID1A protein constructs. We thank the labs of Terry Magnuson (UNC) and David Margolis (UNC) for providing EED and BMI-1 antibodies, respectively. We also thank Noah Sciaky (UNC) for help with cell counting using the high content imaging microscope. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The research described here was supported by the National Institute of General Medical Sciences, US National Institutes of Health (NIH, grant R01GM100919), the Carolina Partnership and the University Cancer Research Fund, University of North Carolina at Chapel Hill, and the Welch Foundation (G-1847). The SGC is a registered charity (no. 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation (CFI), the Canadian Institutes of Health Research (CIHR), Genome Canada, Ontario Genomics Institute Grant OGI-055, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and Wellcome Trust Grant 092809/Z/10/Z. M.T.B directs the Protein Array Core, which is supported by the Cancer Prevention Research Institute of Texas (RP130432), and by the Centre for Environmental and Molecular Carcinogenesis at MD Anderson.
Footnotes
Author contributions: J.I.S. designed and synthesized all compounds and related analogs, performed ITC studies, pull-down studies, rendered structural images, assisted with AlphaScreen® studies and performed all cell culture; B.D.M. performed adaptively biased molecular dynamics studies and assisted in compound design; N.C. performed CellTiter-Glo® assays, and cellular proliferation assays; Y.L., W.T., S.Q. solved the X-ray crystal structures of UNC3866 with the various CBX proteins; J.L.N. and S.H.C. expressed purified proteins; K.G.H performed pull-down studies; C.S. and K.B. performed protein array experiments; F.L. performed methyltransferase assays; X.P.H. performed GPCR functional assays. B.M.B. performed and analyzed AlphaScreen® assays; G.S. performed EED ITC experiments; J.I.S., B.D.M., S.G.P., J.M., B.L.R., M.V., P.J.B., M.T.B., C.H.A, L.I.J., and S.V.F. designed studies and discussed results; J.I.S., L.I.J. and S.V.F. wrote the paper
Competing financial interests: The authors declare no competing financial interests.
Additional Information: Any supplementary information, chemical compound information and source data are available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to L.I.J. and S.V.F.
References
- 1.Wagner T, Robaa D, Sippl W, Jung M. Mind the methyl: methyllysine binding proteins in epigenetic regulation. ChemMedChem. 2014;9:466–483. doi: 10.1002/cmdc.201300422. [DOI] [PubMed] [Google Scholar]
- 2.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 3.Kaustov L, et al. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem. 2011;286:521–529. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Q, et al. The central role of EED in the orchestration of polycomb group complexes. Nature communications. 2014;5:3127. doi: 10.1038/ncomms4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandamme J, Volkel P, Rosnoblet C, Le Faou P, Angrand PO. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol Cell Proteomics. 2011;10:M110 002642. doi: 10.1074/mcp.M110.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell reports. 2013;3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 7.Jin B, et al. Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. Cell reports. 2012;2:1411–1424. doi: 10.1016/j.celrep.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klauke K, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 10.Bernard D, et al. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- 11.Forzati F, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinjo K, et al. Expression of chromobox homolog 7 (CBX7) is associated with poor prognosis in ovarian clear cell adenocarcinoma via TRAIL-induced apoptotic pathway regulation. Int J Cancer. 2014;135:308–318. doi: 10.1002/ijc.28692. [DOI] [PubMed] [Google Scholar]
- 13.Mansueto G, et al. Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes Cancer. 2010;1:210–224. doi: 10.1177/1947601910366860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallante P, et al. Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Res. 2008;68:6770–6778. doi: 10.1158/0008-5472.CAN-08-0695. [DOI] [PubMed] [Google Scholar]
- 15.Pallante P, et al. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer. 2010;46:2304–2313. doi: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Karamitopoulou E, et al. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur J Cancer. 2010;46:1438–1444. doi: 10.1016/j.ejca.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XW, et al. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J Exp Clin Cancer Res. 2010;29:114. doi: 10.1186/1756-9966-29-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 19.Scott CL, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan J, et al. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Annals of surgical oncology. 2013;20(Suppl 3):S684–692. doi: 10.1245/s10434-013-3171-7. [DOI] [PubMed] [Google Scholar]
- 22.Clermont PL, et al. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. Br J Cancer. 2014;111:1663–1672. doi: 10.1038/bjc.2014.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauvageau M, Sauvageau G. Polycomb group genes: keeping stem cell activity in balance. PLoS Biol. 2008;6:e113. doi: 10.1371/journal.pbio.0060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konze KD, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS chemical biology. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simhadri C, et al. Chromodomain antagonists that target the polycomb-group methyllysine reader protein chromobox homolog 7 (CBX7) J Med Chem. 2014;57:2874–2883. doi: 10.1021/jm401487x. [DOI] [PubMed] [Google Scholar]
- 28.Ren C, et al. Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain. Chemistry & biology. 2015;22:161–168. doi: 10.1016/j.chembiol.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frye SV. The art of the chemical probe. Nature chemical biology. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 30.Bunnage ME, Chekler EL, Jones LH. Target validation using chemical probes. Nature chemical biology. 2013;9:195–199. doi: 10.1038/nchembio.1197. [DOI] [PubMed] [Google Scholar]
- 31.Workman P, Collins I. Probing the probes: fitness factors for small molecule tools. Chemistry & biology. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.James LI, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nature chemical biology. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkinson RJ, et al. Is JmjC oxygenase catalysis limited to demethylation? Angew Chem Int Ed Engl. 2013;52:7709–7713. doi: 10.1002/anie.201303282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickson BM. Approaching a parameter-free metadynamics. Physical Review E. 2011;84 doi: 10.1103/PhysRevE.84.037701. [DOI] [PubMed] [Google Scholar]
- 36.Muñoz V, Thompson PA, Hofrichter J, Eaton WA. Folding dynamics and mechanism of β-hairpin formation. Nature. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Y, Chen C, He Y. Folding mechanism of beta-hairpin trpzip2: heterogeneity, transition state and folding pathways. International journal of molecular sciences. 2009;10:2838–2848. doi: 10.3390/ijms10062838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J Biol Chem. 2008;283:19626–19635. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Besnard J, et al. Automated design of ligands to polypharmacological profiles. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montgomery ND, Yee D, Montgomery SA, Magnuson T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. Journal of molecular biology. 2007;374:1145–1157. doi: 10.1016/j.jmb.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-Containing Comlexes Target Methylation of Histone H1 or Nucleosomal Histone H3. Molecular Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 43.Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, et al. Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PloS one. 2010;5:e13732. doi: 10.1371/journal.pone.0013732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong Y, Cui H, Ramkumar C, Zhang H. Regulation of senescence in cancer and aging. J Aging Res. 2011;2011:963172. doi: 10.4061/2011/963172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarrard DF, et al. Deletional, Mutational, and Methylation Analyses of CDKN2 (p16/MTS1) in Primary and Metastatic Prostate Cancer. GENES, CHROMOSOMES & CANCER. 1997;19:90–96. [PubMed] [Google Scholar]
- 48.Yaqinuddin A, Qureshi SA, Qazi R, Abbas F. Down-regulation of DNMT3b in PC3 cells effects locus-specific DNA methylation, and represses cellular growth and migration. Cancer Cell Int. 2008;8:13. doi: 10.1186/1475-2867-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaqinuddin A, Qureshi SA, Qazi R, Farooq S, Abbas F. DNMT1 silencing affects locus specific DNA methylation and increases prostate cancer derived PC3 cell invasiveness. J Urol. 2009;182:756–761. doi: 10.1016/j.juro.2009.03.082. [DOI] [PubMed] [Google Scholar]
- 50.Mirochnik Y, et al. Androgen receptor drives cellular senescence. PloS one. 2012;7:e31052. doi: 10.1371/journal.pone.0031052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 52.Tiwary P, Parrinello M. From Metadynamics to Dynamics. Physical Review Letters. 2013;111:230602. doi: 10.1103/PhysRevLett.111.230602. [DOI] [PubMed] [Google Scholar]
- 53.Voter AF. Hyperdynamics: Accelerated Molecular Dynamics of Infrequent Events. Physical Review Letters. 1997;78:3908–3911. [Google Scholar]
- 54.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 55.Wigle TJ, et al. Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins. Journal of biomolecular screening. 2010;15:62–71. doi: 10.1177/1087057109352902. [DOI] [PubMed] [Google Scholar]
- 56.Perfetti MT, et al. Identification of a Fragment-like Small Molecule Ligand for the Methyl-lysine Binding Protein, 53BP1. ACS chemical biology. 2015;10:1072–1081. doi: 10.1021/cb500956g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barsyte-Lovejoy D, et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1407358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.