Abstract
The morphogenesis of midfacial processes requires the coordination of a variety of cellular functions of both mesenchymal and epithelial cells to develop complex structures. Any failure or delay in midfacial development as well as any abnormal fusion of the medial and lateral nasal and maxillary prominences will result in developmental defects in the midface with a varying degree of severity, including cleft, hypoplasia, and midline expansion. In spite of the advances in human genome sequencing technology, the causes of nearly 70 percent of all birth defects, which include midfacial development defects, remain unknown. Recent studies in animal models have highlighted the importance of specific signaling cascades and genetic-environmental interactions in the development of the midfacial region. This review will summarize the current understanding of the morphogenetic processes and molecular mechanisms underlying midfacial birth defects based on mouse models with midfacial developmental abnormalities.
Keywords: Midfacial development, neural crest cells, midfacial cleft, cell signaling, growth factor
Introduction
During embryogenesis, the midfacial region, defined as the nose, upper lip, maxilla, primary palate, and zygomatic bones, is derived from seven processes: the frontonasal process, the paired lateral and medial nasal processes, and the paired maxillary processes (Cox, 2004). This region is largely populated by mesenchymal cells derived from cranial neural crest (CNC) cells, which are covered with ectoderm (Helms et al., 2005). Interactions between the CNC cells and epithelial cells are crucial for normal midfacial development, but the cause of many defects remain unknown. Significant progress has been made in identifying both genetic and environmental risk factors for midfacial malformations such as midfacial cleft, hypoplasia, and hypertelorism (aka midline expansion). In this review, we will summarize the current understanding of the molecular mechanisms underlying midfacial birth defects.
Midfacial Development and Birth Defects
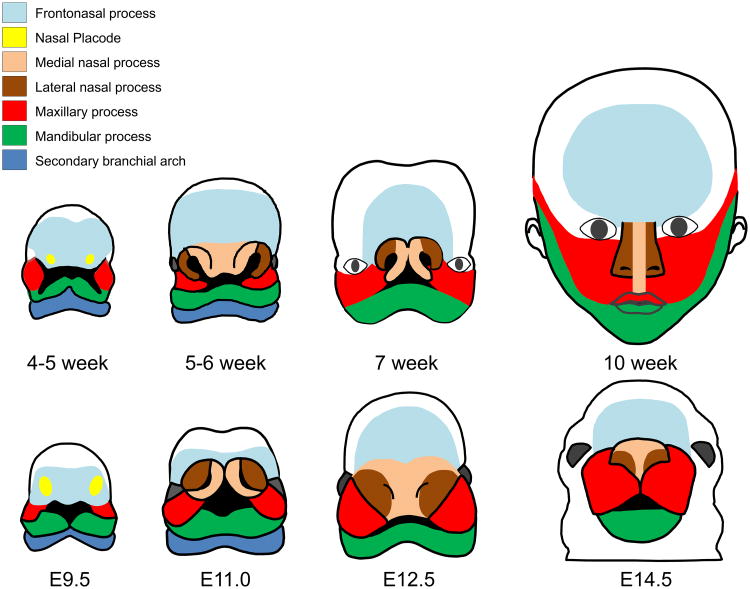
In humans and mice, early development of the midfacial region depends upon the growth and fusion of several processes, including the frontonasal process (aka frontonasal prominence), the paired lateral and medial nasal processes, and the paired maxillary processes. In the fifth week of gestation in humans and at embryonic day 10 (E10) in mice, the surface ectoderm of the frontonasal process thickens and develops into two bilateral nasal placodes. The lateral and medial nasal processes emerge from the nasal placodes, with the nasal pits forming in the center. In humans, the maxillary process of the first branchial arch (aka first pharyngeal arch) grows medially and approaches the lateral and medial nasal processes to form the upper lip and push the medial nasal process toward the midline. The merging of the two medial nasal processes also results in the formation of the primary palate carrying the incisors. In mice, the fusion between the medial and lateral nasal processes begins at E11. Subsequently, the maxillary processes grow toward each other and establish contact, fusing in the midline at E13 and forming one continuous structure. Prior to fusion, the epithelium on the opposing primordium is intact, with only occasional disruptions in the basal lamina, whereas during fusion, contact and adhesion, together with physical forces, trigger the degeneration of the basal lamina and change the fate of epithelial cells (induction of programmed cell death and activation of cell migration of medial edge epithelial cells) (Jiang et al., 2006; Shuler, 1995). At the seventh week in humans and E13.5 in mice, the fusion of the midfacial region is completed through the disappearance of the epithelial seams between the processes (Fig. 1). Failure of any of the steps of midfacial morphogenesis (defects in growth and patterning of the mesenchyme or epithelial fusion) results in midfacial cleft, hypoplasia, and/or hypertelorism in humans and mice.
Fig. 1.
Development of the midfacial primordium. Frontal views of the prominences that give rise to the main structures of the face in humans (upper panels) and mice (lower panels), namely the frontonasal process (light blue), the nasal placode (yellow), the medial nasal processes (light orange), the lateral nasal processes (brown), the maxillary processes (red), the mandibular process (green), and the secondary branchial arch (blue).
CNC cells are multipotent, proliferative cells originating from the dorsal aspect of the neural tube (Chai and Maxson, 2006). Facial processes arise during early embryogenesis from ventrally migrating CNC cells, in combination with ectoderm and mesoderm, and undergo directed growth and expansion around the nasal pits to fuse with one another (Jiang et al., 2006). CNC cells from the diencephalon and anterior mesencephalon give rise to the lateral and medial nasal processes, while those originating from the posterior mesencephalon and hindbrain give rise to the maxilla and mandible (Serbedzija et al., 1992). In addition, CNC cells acquire their anterior-posterior identities before migration. However, the patterning of CNC cells is determined by niches (i.e., reciprocal signals from the microenvironment), mainly at the post-migratory stage (Trainor et al., 2003). These findings suggest that each process may be regulated through temporal and spatial specific mechanisms to form each unique structure, and that CNC cells play a key role in the formation of the midfacial region.
Molecular Mechanisms of Midfacial Developmental Defects
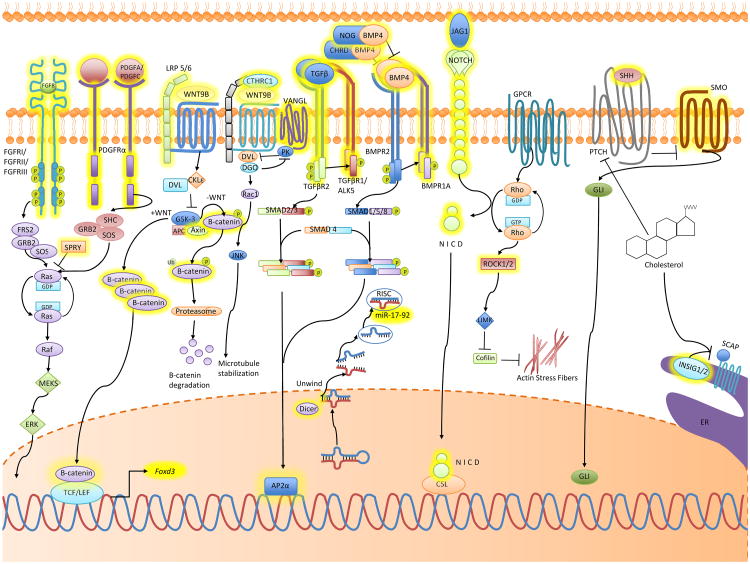
The identification of causal genes of midface anomalies in humans will help us understand their underlying molecular mechanisms. Mutations in genes involved in major signaling pathways (FGF, WNT, TGFβ, BMP, PDGF, and Notch signaling) and microRNAs (miRs) have been postulated as contributing genetic factors in midfacial birth defects in addition to environmental risk factors (Fig. 2 and Table 1).
Fig. 2.
Cell signaling in midfacial development. Depicted FGF, PDGF, WNT, TGFβ, BMP, NOTCH, Rho, and SHH signaling cascades. Molecules that have been shown to play direct roles in midfacial deformities are highlighted in yellow, but we cannot exclude the possibility of the molecules not highlighted being important in midfacial development because these may not have yet been reported or they may be compensated by other molecules. CSL: CBF1/Su(H)/Lag-1 transcription factor complex, ER: endoplasmic reticulum, ERK: extracellular signal-related kinase, GPRC: G protein-coupled receptor, JNK: Jun amino-terminal kinases, LIMK: LIM domain kinase, MAPK: mitogen-activated protein kinase, MEKS: MAPK/ERK kinases, NICD: Notch intracellular domain, RISC: RNA-induced silencing complex, SCAP: sterol regulatory element-binding protein cleavage-activating protein.
Table 1.
Selected mouse strains exhibited midfacial developmental defects.
| Mouse Strain | Pathway/Component | Reference | Midfacial Phenotypes | Other Craniofacial Phenotypes |
|---|---|---|---|---|
| Sp8-/- | Upstream of FGF signaling | (Kasberg et al., 2013) | Midfacial cleft, maxillary hypoplasia, hypertelorism | Exencephaly, absence of frontal and parietal bones |
| Sp8F/F;Pax3-Cre | Upstream of FGF signaling | (Kasberg et al., 2013) | Midfacial cleft | Exencephaly, absence of frontal and parietal bones |
| Sp8F/F;Foxg1-Cre | Upstream of FGF signaling | (Kasberg et al., 2013) | Midfacial cleft | Exencephaly, absence of frontal and parietal bones |
| Fgfr1F/F;Wnt1-Cre | FGF receptor | (Wang et al., 2013a) | Cleft lip, premaxilla hypoplasia | Cleft palate, reduced frontonasal prominence and maxillary processes, micrognathia, tooth bud defect |
| Fgfr2+/P253R | FGF receptor | (Wang et al., 2010) | Midfacial cleft and hypoplasia | Cleft palate, skeletal system malformation (skull, palate, sternum, and long bones), craniosynostosis |
| Fgfr2+/S252W | FGF receptor | (Yu et al., 2000) | Midfacial cleft and hypoplasia | Skeletal system malformation, craniosynostosis |
| Fgf8null/Neo | FGF ligand | (Griffin et al., 2013) | Midfacial cleft | Cleft palate |
| Fgf8F/F;Nes-Cre | FGF ligand | (Trumpp et al., 1999) | Maxillomandibular malformation | Agnathia |
| Spry1TG;Wnt1-Cre | FGF inhibitor | (Yang et al., 2010) | Midfacial cleft, malformed maxilla | Cleft palate, failure of nasal and frontal bone formation, micrognathia |
| Spry4-/- | FGF inhibitor | (Taniguchi et al., 2007) | Midfacial hypoplasia | Micrognathia, malocclusion, increased incisors |
| Spry2-/-;Spry4-/- | FGF inhibitor | (Taniguchi et al., 2007) | Midfacial dysplasia | Micrognathia, malocclusion, increased incisors |
| Pbx1-/-;Pbx2+/- | Upstream of WNT signaling | (Ferretti et al., 2011) | Cleft lip (unilateral or bilateral), jaw hypoplasia | Cleft palate |
| Pbx1F/F;Pbx2+/-;Foxg1-Cre | Upstream of WNT signaling | (Ferretti et al., 2011) | Cleft lip (bilateral) | Abnormal nasal capsule, palatal and palatine processes of maxilla, absence of premaxillary process |
| Pbx1F/F;Pbx2+/-;Crect-Cre | Upstream of WNT signaling | (Ferretti et al., 2011) | Cleft lip (bilateral) | None |
| Wnt9b-/- | WNT ligands | (Jin et al., 2012) | Cleft lip (bilateral) | Cleft palate |
| Wnt9b-/-;Pbx1+/- | WNT ligands | (Ferretti et al., 2011) | Cleft lip (bilateral) | Cleft palate |
| Wnt9bF/F;Foxg1-Cre | WNT ligands | (Ferretti et al., 2011) | Cleft lip (bilateral) | Cleft palate |
| Lrp6βgeo/βgeo | WNT receptor | (Song et al., 2009) | Cleft lip (bilateral), midfacial hypoplasia | Defects in neural tube, eye, brain, limb and bone, cleft palate, midline cleft of the mandible |
| Axin1ΔC6/ΔC6;Ctnnb1+/- | WNT signaling mediator | (Chia et al., 2009) | Midfacial cleft, absence of nasal structures | Multiple brain malformations |
| Foxd3F/-;Wnt1-Cre | Downstream target of WNT signaling | (Teng et al., 2008) | Midfacial cleft, absence of frontal bone and nasal capsule | Small interparietal, parietal, basal occipital bones, and premaxilla, shortened and thickened mandible |
| Cthrc1-/-;Vangl2+/- | WNT inhibitor | (Yamamoto et al., 2008) | Midfacial hypoplasia | Exencephaly, neural tube closure defect in midbrain |
| Shh-/- | SHH ligand | (Chiang et al., 1996) | Severe craniofacial deformities, holoprosencephaly | Single optic vesicle, single nasal pit w/in a single olfactory placode |
| Shhc/n;K14-Cre | SHH ligand | (Lan and Jiang, 2009) | Nasal hypoplasia | Cleft palate |
| ShhF/F;K14-Cre | SHH ligand | (Iwata et al., 2014) | Nasal hypoplasia | Cleft palate |
| K14-Shh transgenic | SHH ligand | (Cobourne et al., 2009) | Cleft lip, hypertelorism | Cleft palate, arrested tooth development, absence of flat bones in skull vault, prominence of the frontal region, skeletal open bite, glossoptosis |
| SmoF/-;Wnt1-Cre | SHH receptor | (Jeong et al., 2004) | Midfacial deformities | Hypoplastic nasal bone, incomplete nasal septum, absence of lacrimal bone, mesethmoid and vomer |
| Cdon-/- | SHH receptor | (Cole and Krauss, 2003; Zhang et al., 2006) | Absence of primary palate | Hypoplasia of nasal septum cartilage, lack of maxillary incisors |
| HhatCreface/Creface | SHH ligand modifier | (Dennis et al., 2012) | Midfacial hypoplasia | Brain and eye abnormalities, acrania, agnathia |
| HhatCreface/Creface;Ptch1wiggable/wiggable | SHH ligand modifier/ SHH receptor | (Kurosaka et al., 2014) | Cleft lip, hypoplastic nasal process | Epithelial seam persistence |
| Ift144twt | SHH-Primary cilia | (Ashe et al., 2012) | Cleft lip (bilateral) | Exencephaly, skeletal and craniofacial anomalies |
| Kif3aF/F;Wnt1-Cre | SHH-Primary cilia | (Brugmann et al., 2010; Liu et al., 2014) | Midfacial deformities | Cleft palate, brain abnormalities, craniosynostosis, hypertelorism |
| Alk5F/F;Wnt1-Cre | TGFβ receptor | (Dudas et al., 2006) | Midfacial cleft, nasal cartilage cleft, hypoplastic premaxilla | Absence of frontal bones, cleft palate, reduced mandible |
| Alk5F/F;Nes-Cre | TGFβ receptor | (Li et al., 2008) | Cleft lip (unilateral or bilateral), hypoplastic maxillary process | Cleft palate |
| Bmpr1aDN/DN;Mpz-Cre | BMP receptor | (Saito et al., 2012) | Midfacial cleft | Hypertelorism, calvaria deformities, short faces, cleft palate, wide-open anterior fontanelles |
| Bmp4F/F;Nes-Cre | BMP ligand | (Liu et al., 2005) | Cleft lip (unilateral or bilateral) | Cleft palate |
| Nog-/- | BMP inhibitor | (Lana-Elola et al., 2011) | Enlarged nasal septum, holoprosencephaly | A solitary median maxillary incisor, larger hyoid bone, and absence of vomeronasal cartilage |
| Chrd-/-;Nog+/- | BMP inhibitor | (Anderson et al., 2002) | Midfacial hypoplasia, cleft lip (bilateral), single nostril with narrow proboscis | Agnathia, cebocephaly with cyclopic eye beneath proboscis |
| Ph-/- | PDGF receptor | (Morrison-Graham et al., 1992) | Midfacial cleft | Hemifacial microsomia, micrognathia, opened neural tube |
| Pdgfra-/- | PDGF receptor | (Soriano, 1997; Tallquist and Soriano, 2003) | Midfacial cleft | Hypoplastic brachial arches, wavy neural tube, skeletal defects |
| PdgfraF/F;Wnt1-Cre | PDGF receptor | (He and Soriano, 2013) | Midfacial cleft, absence of primary palate and philtrum | Shortening of nasal cartilage and premaxilla |
| Pdgfa-/-;Pdgfc-/- | PDGF ligand | (Ding et al., 2004) | Midfacial cleft | Loss of frontonasal tissues, open neural arches |
| Jag1F/F;Wnt1-Cre | Notch ligand | (Hill et al., 2014; Humphreys et al., 2012) | Short maxillary regions, midfacial hypoplasia | Malocclusion |
| RockDN/DN;Wnt1-Cre | Rho signaling mediator | (Phillips et al., 2012) | Hypoplasia of maxilla, midfacial cleft | Hypertelorism, hypoplasia of mandible, absence of frontonasal bones |
| Cdc42F/F;Wnt1-Cre | Rho GTPase | (Liu et al., 2013) | Cleft lip (unilateral or bilateral), midfacial hypoplasia, short snouts | Cleft palate, enlarged frontal cortex, encephalocele, micrognathia |
| Rac1F/F;Wnt1-Cre | Rho GTPase | (Fuchs et al., 2009) | Cleft lip (unilateral or bilateral), midfacial hypoplasia | Cleft palate, enlarged frontal cortex, short snouts, encephalocele, micrognathia |
| DicerF/F;Wnt1-Cre | MicroRNA | (Huang et al., 2010; Nie et al., 2011; Sheehy et al., 2010; Zehir et al., 2010) | Midfacial cleft and hypoplasia, short nose, microsomia | Malformation of midbrain, cerebellum and mandible, cleft palate |
| miR-17-92-/- | MicroRNA | (Wang et al., 2013b) | Cleft lip | Mandibular hypoplasia, incompletely penetrant cleft palate, microphthalmia |
| miR-17-92+/-;miR-106b-25+/- | MicroRNA | (Wang et al., 2013b) | Cleft lip | Mandibular hypoplasia, completely penetrant cleft palate |
| Alx3-/-;Alx4-/- | Transcription factor | (Beverdam et al., 2001) | Midfacial cleft and hypoplasia, split nose | Eye abnormalities, small skull vault |
| Foxc1-/- | Transcription factor | (Inman et al., 2013) | Maxillomandibular malformation, bony syngnathia | Alisphenoid and squamosal hypoplasia |
| Foxc1-/-;Fgf8+/- | Transcription factor/ FGF ligand | (Inman et al., 2013) | Maxillary hypoplasia, severe syngnathia | Alisphenoid and squamosal hypoplasia |
| Grhl21Nisw/1Nisw | Transcription factor | (Pyrgaki et al., 2011) | Midfacial cleft and hypoplasia | Exencephaly, defective optic fissure closure with complete penetrance, absence of skeletal components of the top of the skull and midface |
| Msx1-/- | Transcription factor | (Satokata and Maas, 1994) | Midfacial hypoplasia, small nasal bones, rounded shape of the frontal bones | Cleft palate, small mandible |
| Msx1-/-;Msx2-/- | Transcription factor | (Ishii et al., 2005; Satokata et al., 2000) | Defects in the fusion of the median lateral, nasal and maxillary prominences, midfacial hypoplasia | Exencephaly, cleft mandible, micrognathia |
| Sox11-/- | Transcription factor | (Sock et al., 2004) | Cleft lip (unilateral or bilateral) and upper jaw, maxillary hypoplasia | Cleft palate, eyelid closure defect |
| Tcfap2a-/- | Transcription factor | (Nottoli et al., 1998; Zhang et al., 1996) | Cleft lip, midline clefting of primary palate | Developmental defects in neural tube, head, and body wall, cleft palate |
| Tcfap2aChi/Chi | Transcription factor | (Nottoli et al., 1998) | Cleft lip, maxillary hypoplasia | Cleft palate, exencephaly, malocclusion |
| Tcof1+/- | Treacher Collins syndrome | (Dixon et al., 2006) | Maxillary hypoplasia, frontonasal dysplasia | Cleft palate, exencephaly, hypoplasia of the premaxilla, mandible, and temporal bones |
| Tw/Tw | Spontaneous | (Gong et al., 2000) | Cleft lip (unilateral or bilateral) | Cleft palate |
| Rdh10trex/trex | RA metabolism | (Rhinn et al., 2011; Sandell et al., 2007) | Midfacial cleft | Eye abnormalities, micrognathia |
| Insig1-/-;Insig2-/- | Cholesterol metabolism | (Engelking et al., 2006) | Midfacial cleft and hypoplasia | Cleft palate, exencephaly, unilateral or bilateral microphthalmia (or anophthalmia) |
1) FGF Signaling Pathway
Fibroblast growth factors (FGFs) regulate mesoderm establishment, neural crest (NC) patterning, myogenesis, and morphogenesis during embryogenesis (Nie et al., 2006a). FGF signaling activation is regulated by their receptors (five distinct membrane FGF receptors) and ligands (at least 22 known different FGF ligands) (Powers et al., 2000). Mutations in FGF signaling molecules result in craniofacial malformations, which include midfacial anomalies.
Upstream of FGF Signaling
The function of the zinc finger transcription factor SP8 in FGF signaling is essential for proper bone formation. Mice with a deficiency of Sp8 (Sp8-/- mice) exhibit severe midfacial defects via reduction of Fgf8 and Fgf17 expression (Kasberg et al., 2013). Sp8F/F;Pax3-Cre (Pax3-Cre is expressed in NC cells and in early metanephric mesenchymal lineages) and Sp8F/F;Foxg1-Cre (Foxg1-Cre is expressed in cells derived from the telencephalon) mice display midfacial clefts, exencephaly, and absence of frontal and parietal bones, as seen in Sp8-/- mice (Kasberg et al., 2013).
FGF Receptors
In humans, gain-of-function mutations in FGF receptors have been found in a series of midfacial hypoplasia and craniosynostosis (a premature suture closure) syndromes. Syndromes characteristic of midfacial malformations (e.g., hypertelorism and midfacial hypoplasia) and craniosynostosis include the Jackson–Weiss (OMIM 123150), Pfeiffer (OMIM 101600), Apert (OMIM 101200), and Crouzon syndromes (OMIM 123500) (Johnson and Wilkie, 2011; Kress et al., 2000; Roscioli et al., 2000; Senarath-Yapa et al., 2012). Pfeiffer syndrome is an autosomal dominant disorder caused by a gain-of-function mutation in the FGF receptor type 1 (FGFR1) gene [C>G transversion at position 755 of FGFR1, resulting in a proline to arginine substitution at amino acid 252 (FGFR1P252R)], whereas Jackson–Weiss syndrome, also an autosomal dominant disorder, is caused by a mutation in the FGF receptor type 2 (FGFR2) gene [C>G transversion at position 1043 of FGFR2, resulting in an alanine to glycine substitution at amino acid 344 (FGFR2A344G)]. Apert syndrome is also the result of gain-of-function mutations of FGFR2, over 99% of which are a serine to tryptophan substitution at amino acid 252 (FGFR2S252W) or a proline to arginine substitution at amino acid 253 (FGFR2P253R). In these syndromes, midfacial hypoplasia is often associated with a short cranial base, reduced pharyngeal height, narrow nasal cavity, and a diminished nasopharyngeal space leading to severe upper airway obstruction. In addition, other gain-of-function mutations in FGFR2 and FGFR3 have been identified in patients with Crouzon syndrome—an autosomal dominant condition characterized by midfacial hypoplasia, craniosynostosis, and ocular proptosis—and achondroplasia with midfacial hypoplasia [e.g., FGFR2C278F, a cysteine to phenylalanine substitution at amino acid 278, and FGFR2C342Y, a cysteine to tyrosine substitution at amino acid 342) (Nolan et al., 2013). Thus, particular mutations in FGF receptors seem to be strongly associated with the clinical features of these syndromes.
Fgfr1 and Fgfr2 are widely expressed in the facial mesenchyme and ectoderm, respectively, both in mice and humans (Bachler and Neubuser, 2001; Britto et al., 2001; Wilke et al., 1997). Ablation of Fgfr1 in CNC cells (Fgfr1F/F;Wnt1-Cre mice) results in cleft lip, due to a CNC migration defect in the frontonasal process, and cleft palate, which is caused by a proliferation defect in the palate (Wang et al., 2013a). Mice with a mesodermal deficiency of Fgfr2 (Fgfr2F/F;Dermo1-Cre mice) exhibit midfacial hypoplasia and domed-shaped skulls (Yu et al., 2003). By contrast, Fgfr2F/F;Wnt1-Cre and Fgfr3-/- mice show no midfacial defects (Pan et al., 2008; Valverde-Franco et al., 2004). These findings suggest that individual FGF receptors have different distribution and involvement in murine midfacial development. Gain-of-function mutations in FGFR2 in mice (Fgfr2+/P253R, Fgfr2+/S252W, Fgfr2+/S252W;Mesp1-Cre, and Fgfr2+/S252W;Wnt1-Cre mice) result in skull malformations including midfacial hypoplasia, as seen in patients with these mutations (Holmes and Basilico, 2012; Wang et al., 2010; Yu et al., 2000).
FGF Ligands
Mutations in the FGF8 gene have been found in patients with Kallmann-like idiopathic hypogonadotropic hypogonadism, midfacial hypoplasia, and cleft lip and palate (Stanier and Pauws, 2012). FGF8 mutations are also found in some cases of Crouzon syndrome (Li et al., 2013).
In mice, Fgf8 is broadly expressed in the midfacial ectoderm at E9.5; however, at later developmental stages, namely at E10.5 and E11.5, the expression is spatially restricted to the edge of the nasal pit and the oral edge of the medial nasal process (Bachler and Neubuser, 2001). Corresponding to the Fgf8 expression pattern, mice with inactivation of Fgf8 in the first branchial arch ectoderm (Fgf8F/F;Nes-Cre mice: Nes-Cre is expressed in the maxillary process mesenchyme, oral and dental epithelium, and epithelium of the medial and lateral nasal processes, between E10.5 and E11.5) are viable, but lack most first branchial arch-derived structures, including the maxilla and the mandible (Trumpp et al., 1999). In addition, the level of Fgf8 expression is strongly correlated with the phenotype in mutant mice: Fgf8null/null mice (0% of Fgf8 expression level) die from defective gastrulation at an early embryonic stage (Sun et al., 1999); Fgf8null/Neo mutant mice (20% of Fgf8 expression level) display midfacial cleft; Fgf8Neo/Neo newborn mice (40% of Fgf8 expression level) exhibit altered nasal capsule and optic capsule, as well as trabecular basal plate; and Fgf8+/null and Fgf8+/Neo mice are phenotypically comparable to Fgf8+/+ mice (50% of Fgf8 expression level in Fgf8+/null, 70% in Fgf8+/Neo, and 100% in Fgf8+/+ mice) (Griffin et al., 2013). These studies of mouse models demonstrate that FGF8 is a key mediator of proper orientation and polarity of facial primordia and subsequent frontonasal skeletal morphogenesis.
FGF Inhibitors
Sprouty (Spry) is one of the negative regulators of FGF signaling that plays crucial roles in embryogenesis (Hacohen et al., 1998). Mice with conditional expression of the Spry1 transgene in CNC cells (Spry1TG;Wnt1-Cre transgenic mice) exhibit midfacial cleft, cleft palate, and failure to form the nasal and frontal bones, as well as micrognathia and cardiovascular defects such as ventricular septal defects and outflow tract defects (Yang et al., 2010). In Spry1TG;Wnt1-Cre transgenic mice, these defects appear to be the result of decreased proliferation and increased apoptosis of NC and NC-derived cell populations. The expression of transcription factors Ap2 and Msx2, which are important to normal midfacial development, is reduced by inhibition of FGF signaling in Spry1TG;Wnt1-Cre mice. By contrast, Spry1-/- and Spry2-/- mice display no midfacial developmental defect, but Spry4-/- mice exhibit midfacial hypoplasia, micrognathia, malocclusion, and increased incisor size (Taniguchi et al., 2007). In addition, Spry2-/-;Spry4-/- double knockout mice exhibit a more severe midfacial dysplasia than Spry4-/- mice (Taniguchi et al., 2007), suggesting that Spry2 and Spry4 have both a redundant and non-redundant function in FGF signaling during midfacial development.
2) WNT Signaling Pathway
WNT ligands are secreted glycoproteins that compose a large family of 19 proteins in humans. Different combinations of WNT ligands and receptors can activate at least three distinct signaling pathways: the β-catenin-dependent (canonical) pathway, the β-catenin-independent planar cell polarity (PCP) pathway, and the β-catenin-independent Ca2+ pathway. Canonical WNT signaling is mediated by β-catenin through its stability and translocation into the nucleus. Without WNT ligands, β-catenin is phosphorylated and degraded by a β-catenin destruction complex, which includes AXIN, adenomatous polyposis coli (APC), and glycogen synthase kinase 3 (GSK-3). In presence of WNT ligands, β-catenin accumulates in the cytoplasm and eventually translocates into the nucleus to act as a transcriptional co-activator of transcription factors that belong to the T-cell factor/lymphoid enhancing factor (TCF/LEF) family.
The WNT signaling pathway is a highly conserved pathway across multiple species and regulates crucial aspects of embryonic development (Komiya and Habas, 2008). In craniofacial development, WNT signaling plays critical roles in the generation, migration, and proliferation of CNC cells in the facial processes (Mani et al., 2010).
Upstream of WNT Signaling
Pre-B-cell leukemia homeobox (PBX) proteins are homeodomain-containing homeobox (HOX) cofactors that increase HOX DNA-binding specificity and are required for organogenesis and skeletogenesis (Capellini et al., 2006). At E8.5-9.0, Pbx genes (Pbx1, Pbx2, and Pbx3) are highly expressed within the frontonasal process and first branchial arch, as well as in the surface of cephalic ectoderm and mesenchyme at E9.0; at E10.5, Pbx genes are highly expressed in the epithelium of the maxillary process and medial and lateral nasal processes. PBX proteins can directly regulate gene expression of Wnt9b and Wnt3, which in turn act upstream of Fgf8 (Ferretti et al., 2011). As a result, mice with a disrupted PBX pathway (Pbx1-/-;Pbx2+/-, Pbx1F/F;Pbx2+/-;Foxg1-Cre [Foxg1-Cre is expressed in facial and head ectoderm], and Pbx1F/F;Pbx2+/-;Crect-Cre [Crect-Cre, aka Tcfap2a-Cre, is induced by the ectoderm-specific Tcfap2a gene]) display cleft lip resulting from increased apoptosis at the medial edge of midfacial processes (Ferretti et al., 2011).
WNT Ligands
Wnt9b, a ligand for canonical WNT signaling, is expressed in the ectoderm of the facial processes and in the epithelial seam between the fusing facial processes during lip fusion in mice (Lan et al., 2006). Wnt9b null mice (Wnt9b-/- mice) show retarded outgrowth of the nasal (lateral and medial) and maxillary processes resulting from reduced proliferation of mesenchymal cells through tissue-tissue interactions, which subsequently leads to bilateral cleft lip and cleft palate (Jin et al., 2012). The cellular defects in Wnt9b-/- mice are mainly caused by reduced gene expression of members of the FGF family (Jin et al., 2012). As seen in Wnt9b-/- mice, deletion of Wnt9b in the cephalic ectoderm region (Wnt9bF/F;Foxg1-Cre mice) causes bilateral cleft lip through a mechanism similar to that of Wnt9b-/-mice. In addition, compound mutant mice Wnt9b-/-;Pbx1+/- (Pbx1 is upstream of WNT signaling) and Pbx1F/F;Pbx2+/-;Foxg1-Cre mice both exhibit cleft lip (Ferretti et al., 2011). These studies strongly suggest that the PBX-WNT pathway plays a crucial role in the induction of apoptosis in the medial edge epithelium during midfacial fusion.
WNT Receptor
In the canonical WNT pathway, WNT ligands bind to a receptor complex comprising members of the Frizzled family and a coreceptor of the low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6). Mice with ablation of Lrp6 (Lrp6βgeo/βgeo null mice) exhibit bilateral cleft lip, hypoplasia of the upper lip, cleft palate, and midline cleft of the mandible with complete penetrance (Song et al., 2009). Moreover, both proliferation and apoptosis are altered in Lrp6βgeo/βgeo lip epithelium, following downregulation of Msx1 and Msx2 expression.
WNT Signaling Mediators
AXIN (AXIN1 and AXIN2) proteins are negative regulators of canonical WNT signaling that promote the degradation of β-catenin (Zeng et al., 1997). Targeted disruption of Axin2 (Axin2-/- mice) promotes osteoblast proliferation and differentiation and induces skull structure malformations, including midfacial hypoplasia (Yu et al., 2005). Loss of Axin1 (Axin1-/-) or dominant negative knock-in of Axin1 allele, in which the regulator of G-protein signaling (RGS) domain or the six C-terminal amino acids (C6 motif) in Axin1 are deleted (Axin1ΔRGS/ΔRGS and Axin1ΔC6/ΔC6 mice), results in augmented (i.e., gain-of-function) β-catenin-dependent WNT signaling and early embryonic lethality at E9.5-E10.5 (Chia et al., 2009; Zeng et al., 1997). While the early embryonic lethality of Axin1ΔC6/ΔC6 mutant mice is rescued by haploinsufficiency of β-catenin (Ctnnb1+/-), Axin1ΔC6/ΔC6;Ctnnb1+/- mice still die within one day after birth presenting midfacial cleft (Chia et al., 2009). Similarly, loss-of-function of β-catenin in CNC cells (CtnnbF/F;Wnt1-Cre) leads to severe craniofacial abnormalities in mice (Brault et al., 2001). Thus, precise levels of WNT/β-catenin signaling activity seem to be crucial for midfacial development.
Downstream Targets of WNT Signaling
The TCF/LEF family regulates gene expression of WNT/β-catenin signaling downstream targets (Cadigan, 2012). For example, the TCF1 protein regulates gene expression of foxd3, a transcriptional repressor of the winged helix or “Forkhead” family of proteins, during Xenopus gastrulation (Janssens et al., 2013). Foxd3 is expressed in premigratory and migrating NC cells and is one of the earliest molecular markers of the NC lineage (Teng et al., 2008). Mice with a deficiency of Foxd3 in CNC cells (Foxd3F/-;Wnt1-Cre mice) die at birth with midfacial cleft (Teng et al., 2008).
Noncanonical WNT Signaling Pathway
Collagen triple helix repeat containing 1 (CTHRC1), a secreted glycoprotein, is involved in selective activation of the non-canonical PCP pathway induced by WNT ligands. The VANGL proteins, which are mammalian homologues of the Drosophila melanogaster protein Van Gogh (Vang/Strabismus) containing four transmembrane domains and intracellular N- and C- termini, can provide the scaffolding necessary for the assembly of PCP signaling complexes. Although Cthrc1 null mutant (Cthrc1-/-) mice appear normal, the introduction of a heterozygous mutation in a PCP gene, Vangl2 (Cthrc1-/-;Vangl2+/-), results in craniofacial deformities, including midfacial hypoplasia (Yamamoto et al., 2008). This indicates a compensative function of Vangl2 on a Cthrc1-/- background and the importance of the PCP pathway in midfacial development.
3) Rho GTPase Signaling Pathway
Rho-related small GTPases (e.g., RhoA, Rac1, and Cdc42) have emerged as key mediators of the WNT signaling pathway, most notably in the non-canonical pathways, including the PCP pathway, but also in the canonical pathway. Rho GTPases transduce extracellular signals into a variety of intracellular events controlling cell morphology and polarity (Marinissen et al., 2004). Key Rho effectors Rho kinase 1 (aka ROCK1) and ROCK2 are highly conserved serine/threonine kinases with multiple substrates and play essential roles in fundamental cellular processes, such as contraction, adhesion, migration, apoptosis, and proliferation (Julian and Olson, 2014).
Rho Signaling Mediators
The expression patterns of Rock1 and Rock2 are similar in developing embryos (Wei et al., 2001). Mice expressing a dominant negative (DN) form of ROCKs in CNC cells (RockDN/DN;Wnt1-Cre mice) exhibit hypoplasia of the frontonasal processes and first branchial arch, ultimately resulting in hypoplasia of the maxilla and mandible, midfacial cleft, and hypertelorism (Phillips et al., 2012). CNC cells in RockDN/DN;Wnt1-Cre mice show exacerbated cell death in the frontonasal processes (Phillips et al., 2012). In humans, a deletion on chromosome 18q, where ROCK1 resides (chromosome 18q deletion syndrome, OMIM 601808), causes a spectrum of abnormalities that include hypertelorism, cleft palate, micrognathia, and cardiac defects (Phillips et al., 2012).
Rho GTPases
Cdc42 and Rac1 are ubiquitously expressed small Rho GTPases that act as molecular switches, cycling between an active GTP-bound and an inactive GDP-bound state. Mice with deletion of either Cdc42 or Rac1 in the NC (Cdc42F/F;Wnt1-Cre and Rac1F/F;Wnt1-Cre) exhibit craniofacial deformities such as midfacial cleft and cleft palate resulting from a reduction in size of NC-derived structures. Increased cell-cycle exit is detected in these bony structures, while NC cells emigrating from the neural tube are not affected (Fuchs et al., 2009; Liu et al., 2013). In addition to these phenotypes, epithelial detachment, formation of acellular mesenchymal cavities, and elevated apoptosis have been found in CNC-derived mesenchymal regions of Rac1F/F;Wnt1-Cre mice (Thomas et al., 2010). In contrast to Cdc42F/F;Wnt1-Cre and Rac1F/F;Wnt1-Cre mice, RhoAF/F;Wnt1-Cre mice exhibit exencephaly but no obvious midfacial defects (Katayama et al., 2011). Although there is no conclusive evidence explaining these craniofacial phenotypic differences, they may reflect differential expression and regulation of RhoA in craniofacial development.
4) SHH Signaling Pathway
Sonic hedgehog (SHH) is a member of the hedgehog (HH) family of secreted proteins and plays crucial roles in diverse developmental processes, including left-right axis establishment, dorsoventral patterning of the neural tube, endoderm development, anteroposterior patterning of the developing limb, and brain development and patterning (Roessler and Muenke, 2003). The foregut endoderm, facial ectoderm, and neuroepithelium are all sources of SHH ligands (Echelard et al., 1993), which bind to their cell surface receptor Patched 1 (PTCH1), hence triggering SHH signaling, to relieve its basal repression of Smoothened (SMO) (Jiang et al., 2006; Pan et al., 2013). SHH signaling is critical for proliferation, survival, and patterning of CNC cells during craniofacial development (Hu and Marcucio, 2009; Iwata et al., 2014).
SHH Receptors
SHH ligands bind to PTCH1, a primary receptor for SHH ligands that works as a repressor of SHH signaling, in the absence of SHH ligand and co-receptors SMO, cell adhesion molecule-related/downregulated by oncogenes, aka CDO (CDON), brother of CDON (BOC), and growth arrest-specific 1 (GAS1) to induce SHH signaling activity. Mice with deletion of Smoothened in CNC cells (SmoF/-;Wnt1-Cre mice) exhibit severe craniofacial deformities such as midfacial hypoplasia, hypoplastic nasal bone, cleft palate, and micrognathia, owing to increased apoptosis and decreased cell proliferation of postmigratory CNC cells (Jeong et al., 2004). Cdon-/- mice show variability in the penetrance and expressivity of midfacial and holoprosencephaly phenotypes (e.g., absence of primary palate, hypoplasia of nasal septum cartilage, absence of maxillary incisors) in a strain-dependent manner (Cole and Krauss, 2003; Zhang et al., 2006). In fact, genetic variation in different inbred strains of mice might be sufficient to account for these differences in penetrance and expressivity, as seen in human populations.
SHH Ligands
Mutations in human SHH are responsible for holoprosencephaly (HPE), a disorder involving abnormalities of the forebrain and facial structures (Belloni et al., 1996). Shh null mutant mice (Shh-/- mice) show early embryonic lethality with severe craniofacial deformities (Chiang et al., 1996), but mice with a single allele of an epithelial cell-specific deletion of Shh in a Shh heterozygous null background (Shhc/n;K14-Cre mice), as well as mice with loss of Shh in the epithelium (ShhF/F;K14-Cre mice), exhibit nasal hypoplasia and cleft palate (Iwata et al., 2014; Lan and Jiang, 2009). Similarly, excessive SHH signaling activity in K14-Shh transgenic mice causes hypertelorism, wide cleft lip (a failure of growth of the maxillary processes), cleft palate, absence of flat bones within the skull vault, and arrested tooth development with decreased cell proliferation (Cobourne et al., 2009). Importantly, SHH ligands are post-translationally modified with both cholesterol and palmitic acid, which are required for HH protein multimerization, distribution and activity. Disrupting post-translational palmitoylation of SHH ligands by inhibiting the hedgehog acyltransferase (Hhat) gene disturbs SHH secretion and long-range activity and causes severe hypotelorism with midfacial bone defects in HhatCreface/Creface (The Creface [Ap2-Cre] transgene is integrated within intron 9 of Hhat to delete the Hhat gene) mice (Dennis et al., 2012). SHH-mediated WNT signaling activation is compromised in HhatCreface/Creface mice during the growth and fusion of the nasal processes (Kurosaka et al., 2014). Since disruption of Ptch1 results in an elevation of SHH signaling (Goodrich et al., 1997), severe midfacial defects in HhatCreface/Creface mice are partially restored by a deletion of the Ptch1 gene (HhatCreface/Creface;Ptch1wiggable/wiggable mice: Ptch1wiggable mice were generated through N-ethyl-N-nitrosourea [ENU] mutagenesis and the Ptch1 gene is mutated), although HhatCreface/Creface;Ptch1wiggable/wiggable mice still exhibit hypoplastic nasal process outgrowth, epithelial seam persistence, and cleft lip (Kurosaka et al., 2014). These studies indicate that both expression level and lipid modifications of SHH ligands are important for proper functioning of SHH ligands.
Primary Cilia
The primary cilium, a microtubule-based organelle, is required to induce HH signaling, a crucial pathway for embryonic patterning and organogenesis (Zaghloul and Brugmann, 2011). Two intraflagellar transport (IFT) complexes, IFT-A (6 subunits) and IFT-B (14 subunits), are required for the assembly and functioning of the primary cilia by mediating the activity of a number of developmental signaling pathways, including the HH, WNT, PDGF, FGF, and Notch cascades (Ashe et al., 2012; Liem et al., 2012). Of these signaling pathways, particularly HH signaling has been studied to date (Ashe et al., 2012; Brugmann et al., 2010).
In humans, mutations in IFT144 and IFT43 have been found in patients with Sensenbrenner and Jeune syndromes, which are characterized by craniofacial defects with hypertelorism, short ribs and limbs, and polydactyly, the result of increased SHH signaling (Arts et al., 2011; Ashe et al., 2012; Bredrup et al., 2011). Significantly, ENU-derived mutant mice [Random ENU mutagenesis is a complementary approach to obtain mouse models for human diseases (Nolan et al., 2000)] with a hypomorphic missense mutation in the Ift144 gene (Itf144twt mice) exhibit skeletal and craniofacial anomalies, including facial cleft, frontonasal dysplasia, cleft palate, and exencephaly, as seen in patients with human skeletal ciliopathies (Ashe et al., 2012). Recent studies also show that elimination of the kinesin family member 3A (KIF3A), another IFT protein, in CNC cells (Kif3aF/F;Wnt1-Cre) causes truncation of primary cilia on CNC cells and leads to expansion of the SHH-responsive area in the craniofacial mesenchyme (Brugmann et al., 2010; Liu et al., 2014). As a result, Kif3aF/F;Wnt1-Cre mice exhibit hypertelorism, cleft lip, frontonasal dysplasia, and cleft palate. Thus, molecules related to cilia formation play crucial roles in regulating multiple signaling cascades in midfacial development.
5) TGFβ Superfamily Signaling Pathway
The members of the transforming growth factor beta (TGFβ) superfamily form dimers and bind to heterodimeric receptor complexes consisting of two type I (TGFBR1/ALK5) and two type II receptors (TGFBR2) with serine/threonine kinase domains. Upon ligand binding, the type II receptor phosphorylates and activates the type I receptor and induces SMAD phosphorylation and translocation into the nucleus to regulate transcriptional activity of target genes. TGFβ signaling plays a critical role in a vast range of biological functions during craniofacial development, including cell proliferation, differentiation, and apoptosis (Iwata et al., 2011; Iwata et al., 2012).
Members of TGFβ signaling cascade are expressed in developing mouse facial primordia in a temporal and spatial specific manner (Dudas and Kaartinen, 2005). While the most established downstream signaling cascade involves the sequential phosphorylation of SMAD2/3 proteins (aka canonical pathway) in the formation of the frontal bone and some parts of other craniofacial structures, non-canonical (SMAD-independent) TGFβ signaling in CNC cells plays a crucial role in the formation of the palate and the majority of the craniofacial structures (Ho et al., 2015; Iwata et al., 2011).
The signaling pathway of bone morphogenetic proteins (BMPs), members of the TGFβ superfamily, regulates diverse developmental processes, including cell proliferation, differentiation, apoptosis, and tissue morphogenesis (Wan and Cao, 2005). BMPs signal through receptor complexes composed of type II (BMPR2, ACVR2A, and ACVR2B) and type I transmembrane receptors (ACVR1A/ALK2, BMPR1A/ALK3, and BMPR1B/ALK6); binding of BMPs to their receptors induces SMAD1/5/8 phosphorylation and translocation into the nucleus to regulate gene expression of the targets. Members of the BMP family are essential for the development of the mammalian orofacial region, due to their role in the regulation of cell proliferation, extracellular matrix synthesis, and cellular differentiation. Perturbation of BMP signaling results in orofacial clefting.
TGFβ Receptors
Deletion of TGFβ receptor type I (Tgfbr1, aka Alk5) in CNC cells (Alk5F/F;Wnt1-Cre mice) results in midfacial cleft with increased number of apoptotic cells within the mesenchyme of the first branchial arch (Dudas et al., 2006). Mice with loss of Alk5 in the first branchial arch ectoderm (Alk5F/F;Nes-Cre mice) die at E15 and display a hypoplastic maxillary process, unilateral or bilateral cleft lip, and cleft palate due to increased cell death in the medial nasal process and the maxillary process (Li et al., 2008). Interestingly, mice deficient for the other TGFβ receptors and ligands exhibit cleft palate, but no midfacial defects. This suggests that a combination of each ligand, receptor, and mediator differently activates a tissue-specific TGFβ cascade.
BMP Ligands
Embryonic orofacial tissues express BMPs and their receptors in a temporal and spatial specific manner, suggesting that BMPs play a crucial role in midfacial development. The role of BMP signaling is relatively well studied in the palate (Greene and Pisano, 2010; Nie et al., 2006b), but its involvement in midfacial development is still largely unknown. Mice with conditional inactivation of the BMP4 ligand (Bmp4) using the Nes-Cre transgenic line (Bmp4F/F;Nes-Cre mice) exhibit isolated bilateral cleft lip at E12.5, and a few (2/9 mice) exhibit unilateral cleft lip at E14.5 (Liu et al., 2005). However, the mechanism of cleft lip in Bmp4F/F;Nes-Cre mice is still largely unknown.
BMP Receptors
Mutations in BMP receptor type 1A (BMPR1A) are associated with facial dysmorphism in both juvenile polyposis and chromosome 10q23 deletion syndrome (Cao et al., 2006; Zhou et al., 2001). Mice harboring a DN form of Bmpr1a in NC-derived cells expressing the myelin protein zero (Mpz-Cre) transgene (Bmpr1aDN/DN;Mpz-Cre mice) exhibit wide-open anterior fontanelles with complete penetrance as a result of activated p53-dependent apoptosis (Saito et al., 2012). A total of 80% of Bmpr1aDN/DN;Mpz-Cre mice die from cleft face and palate soon after birth, and the other 20% survive and develop short faces, hypertelorism, and calvaria deformities.
BMP Inhibitors
The distribution and activity of BMPs are regulated by the antagonists Chordin (CHRD) and Noggin (NOG) in the extracellular space (Gazzerro and Canalis, 2006). Noggin null (Nog-/-) mice show early midfacial narrowing in the spectrum of holoprosencephaly, which is a heterogeneous craniofacial and neural developmental anomaly characterized in its most severe form by failure of the forebrain to divide (Lana-Elola et al., 2011). In addition, in Nog-/- mice, the expression area of Shh, as well as SHH target genes Ptch1 and Gli1, is decreased in the frontonasal region at E11.5 and E12.5, which are key developmental stages of early facial development. Chrd-/- mice die perinatally with an extensive array of malformations, including midfacial deformities, cleft palate, and a small jaw (Bachiller et al., 2003). Although Chrd-/-;Nog-/- double knockout mice show early lethality (Chrd and Nog are functionally redundant in embryogenesis), Chrd-/-;Nog+/- mice survive until the neonatal stage and present defects restricted to the craniofacial region, with a varying degree of severity (but more severe than Chrd-/- single knockout mice) ranging from a single nostril with narrow proboscis, anterior truncation, agnathia, and midfacial hypoplasia to bilateral cleft lip (Anderson et al., 2002). Gene expression of Shh is also decreased in Chrd-/-;Nog+/- mice, as seen in Nog-/- and Chrd-/- mice. Taken together, these findings indicate that BMP-mediated SHH signaling seems to play a crucial role in midfacial development.
6) PDGF Signaling Pathway
Platelet-derived growth factors (PDGFs) were originally identified as mitogenic factors for smooth muscle cells, fibroblasts, and glia cells. Taking into account recent findings, they are additionally known as paracrine growth factors that mediate epithelial-mesenchymal interactions in various tissues (Heldin and Westermark, 1999). PDGF ligands (PDGF-A to PDGF-D) exert their function by inducing dimerization and activation of the PDGF receptors (PDGFRα and PDGFRβ). Ligand binding leads to phosphorylation of tyrosine residues in their cytoplasmic domains, which in turn results in activation of a multitude of intracellular signaling cascades (Smith and Tallquist, 2010). PDGF signaling regulates progenitor cell proliferation and cell migration during early embryonic development and also controls extracellular matrix formation, tissue remodeling, and patterning determination during later stages of embryogenesis (Sun et al., 2000).
PDGF Receptors
PDGFRα is expressed in CNC cells and is required for their migration into the branchial arches (Smith and Tallquist, 2010). The Patch (Ph) mutation, originally identified as a distinctive pigmentation phenotype, includes a PDGFRα deletion. The homozygous null Patch mutation in mice (Ph-/- mice) results in pleiotropic defects and first branchial arch deformities, including facial clefting, hemifacial microsomia, micrognathia, opened neural tube, and heart deformities (Morrison-Graham et al., 1992; Gruneberg H and Truslove GM, 1960). Pdgfra-deficient mice (Pdgfra-/- mice) die by E15.5 and have a severe phenotype of midfacial cleft, spina bifida, subepidermal blistering, and skeletal and vascular defects (Soriano, 1997; Tallquist and Soriano, 2003). Similarly, loss of Pdgfra in CNC cells (PdgfraF/F;Wnt1-Cre) results in disruption of CNC cell migration and proliferation, which are essential to form the medial nasal processes as well as cranial bone and cartilage (He and Soriano, 2013). As a result, PdgfraF/F;Wnt1-Cre mice exhibit midfacial cleft, shortened nasal cartilage and premaxilla, and cleft palate.
PDGF Ligands
PDGF-A and PDGF-C are members of the PDGF family that signal through PDGF receptors. Pdgfa-/- mice exhibit no craniofacial defect, although Pdgfc-/- mice show cleft palate and subepidermal blood-filled blisters in the frontal and frontonasal regions, resulting from diminished filopodia formation of epithelial cells and cell proliferation defects in both epithelium and mesenchyme (Ding et al., 2004). Interestingly, Pdgfa-/-;Pdgfc-/- double knockout mice exhibit a midfacial cleft, spina bifida, subepidermal blistering, and skeletal and vascular defects, as seen in Pdgfra-/- mice (Ding et al., 2004). This suggests that both PDGF-A and PDGF-C signal through PDGFRα and that PDGF-C is dominant in the midfacial region, although it is in part functionally redundant with PDGF-A. These two genes also exhibit distinct expression patterns (He and Soriano, 2013).
7) Notch Signaling Pathway
The Notch signaling pathway is a cell-cell adhesion signaling mechanism evolutionarily conserved among species. A Notch transmembrane receptor interacts with a Notch transmembrane ligand on a contacting cell, initiating proteolytic cleavage of the receptor and the subsequent release of the Notch intracellular domain (NICD) of the receptor. The NICD then translocates into the nucleus to induce the transcription of Notch target genes. Several gene mutations of this pathway have been implicated in craniofacial birth defects.
Notch Ligands
Mutations in JAGGED1 (JAG1), a ligand of the Notch signaling pathway, cause Alagille syndrome characterized by biliary, cardiac, and craniofacial anomalies, including a prominent forehead, deep-set eyes, a straight nose with a flat tip, flattened midface, and prominent chin. Jag1F/F;Wnt1-Cre mice exhibit shortened maxillary regions and class III malocclusion but can survive until adulthood. The midfacial hypoplasia in Jag1F/F;Wnt1-Cre mice results from decreased cell proliferation and continuous reduction in hyaluronic acid production at E14.5 and later stages (Humphreys et al., 2012). In the maxilla, cell autonomous Jagged1 signaling in CNC-derived maxillary mesenchyme is required for osteoblast differentiation and mineralization (Hill et al., 2014).
8) MicroRNAs
MicroRNAs (miRs) are short (∼22 nucleotides) noncoding RNAs that regulate approximately 30% of protein-coding genes at the post-transcriptional level (Ross et al., 2007). miRs repress gene expression post-transcriptionally by Watson-Crick base pairing to the seed sites in the 3′ untranslated region (UTR) of target genes. Multiple processes are involved in the synthesis and transcription of miRs as long primary transcripts that are cleaved by Dicer, a type 3 ribonuclease, to produce mature miRs (Wilson and Doudna, 2013). Recent studies indicate that loss of all mature miRs in CNC cells (DicerF/F;Wnt1-Cre) in mice results in increased apoptosis and defects in osteogenic differentiation, causing severe craniofacial deformities including midfacial cleft and hypoplasia (Huang et al., 2010; Nie et al., 2011; Sheehy et al., 2010; Zehir et al., 2010). These findings suggest that miRs play crucial roles in the fate determination of CNC cells. By contrast, although Dicer was expected to play a role in palatal fusion, epithelial-specific Dicer knockout (DicerF/F;K14-Cre: K14-Cre is expressed after the fusion of the upper lip) mice exhibit no significant changes in craniofacial development (Otsuka-Tanaka et al., 2013). Future studies are thus necessary to understand the role of miRs during the fusion of facial processes.
miRs Associated with BMP Signaling
The miR-17-92 cluster encodes six miRs (miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92a-1) within a region on chromosome 13q, whose deletion is associated with cleft lip and/or palate, lung hypoplasia, microophthalmia, microcephaly, and small stature in humans (Kirchhoff et al., 2009). A recent study shows that the MiR-17-92 cluster is a direct target of BMP signaling and transcription factor AP2α (Wang et al., 2013b). Mice with miR-17-92 deficiency (miR-17-92-/- mice: the clefting phenotype is incompletely penetrant) and compound deletion of miR-17-92 and the related miR-106b-25 cluster (miR-17-92-/-;miR-106b-25-/+ mice: the clefting phenotype is more severe and completely penetrant) exhibit cleft lip, cleft palate, and mandibular hypoplasia owing to cell proliferation defects in the mesenchyme (Wang et al., 2013b).
9) Transcription Factors
ALX3/ALX4
Aristaless-like homeobox genes such as Alx3 and Alx4 form a distinct gene family, whose members are characterized by a paired type homeobox with partially redundant functions and a small conserved C-terminal domain. Although mice with deletion of Alx3 (Alx3-/- mice) or Alx4 (Alx4-/- mice) are indistinguishable in regard to midfacial development from wild-type control mice, Alx3/Alx4 double knockout (Alx3-/-;Alx4-/-) mice show severe craniofacial abnormalities, among them midfacial cleft, following an increase in apoptosis in the outgrowing frontonasal process at E10.0 (Beverdam et al., 2001). This indicates that Alx3 and Alx4 are functionally redundant in midfacial development.
FOXC1
Foxc1 is a member of the forkhead box winged helix transcription factor family and plays roles in a variety of tissue developmental processes. Foxc1 null (Foxc1-/-) mice display bony syngnathia, defects in maxillary and mandibular structures, and craniofacial muscular developmental defects (Inman et al., 2013). Interestingly, mice with haploinsufficiency of Fgf8 in a Foxc1-/- background (Foxc1-/-;Fgf8+/-) resemble Foxc1-/- mice but have more severe frontonasal shortening and no externally visible mandible. This indicates that Foxc1 and Fgf8 function synergistically in frontonasal and mandibular development (Inman et al., 2013).
GRHL2
Mice with an ENU-induced nonsense mutation in the grainyhead-like 2 (Grhl2) gene that encodes transcription factor GRHL2 (Grhl21Nisw/1Nisw mutant mice) display midfacial cleft and hypoplasia, exencephaly, body wall closure defects, and defective optic fissure closure with complete penetrance (Pyrgaki et al., 2011). In Grhl21Nisw/1Nisw mutant mice, expression of adhesion molecules is downregulated during neural fold fusion, suggesting that GRHL2 plays a crucial role in epithelial fusion.
MSX1
Msx1-/- mice die at birth and exhibit midfacial hypoplasia, small nasal bones, rounded shape of the frontal bones, cleft palate, and a small mandible (Satokata and Maas, 1994). Although Msx2-/- mice exhibit no midfacial deformities or very mild ones, Msx1-/-;Msx2-/- mice exhibit more severe midfacial developmental defects than Msx1-/- mice (Ishii et al., 2005; Satokata et al., 2000), which suggests that MSX1 and MSX2 are functionally redundant in midfacial development.
SOX11
Transcription factor SOX11 is expressed transiently during embryogenesis in many tissues that undergo inductive remodeling (Hong and Saint-Jeannet, 2005). Approximately 70% of all Sox11-/- embryos exhibit cleft lip (unilateral or bilateral) and cleft palate resulting from failure of the maxillary processes to fuse with the lateral nasal processes (Sock et al., 2004). Unfortunately, the cellular mechanism behind this phenotype has not been studied yet.
TCFAP2A
AP2α, a member of a small family of “basic-helix-span-helix” transcription factors, is a retinoic acid (RA)-inducible transcriptional factor expressed in epithelial and NC cell lineages during development (Knight et al., 2005; Zhang et al., 1996). Encoded by Tcfap2a in mice and TFAP2A in humans, AP2α is expressed in the developing midfacial region (Feng et al., 2008; Nelson and Williams, 2004). Mice with a homozygous disruption of Tcfap2a (Tcfap2a-/- mice) die at or before birth because of severe developmental defects in the neural tube, head, and body wall (Nottoli et al., 1998; Zhang et al., 1996). A total of 16% of chimeric null mice composed of both wild type and Tcfap2a null embryonic stem cells (Tcfap2αChi/Chi mice) exhibit craniofacial dysmorphology, including cleft lip with or without cleft palate (Nottoli et al., 1998). The phenotypes observed in the chimeras display a significant overlap with those caused by teratogenic levels of RA (Nottoli et al., 1998). Altogether, these findings indicate that AP2α is an important component of the RA metabolism.
TWSG1
Mice with a deficiency of the twisted gastrulation gene (Twsg1-/- mice) with exposure to all-trans RA manifest HPE with 100% penetrance, compared to 17% without exposure to all-trans RA via induction of apoptosis following upregulated Bmp2 expression and oxidative stress (Billington et al., 2015).
10) Others
INSIG1 and INSIG2
Insulin-induced genes 1 and 2 (Insig1/2), a pair of endoplasmic reticulum (ER) membrane proteins, are essential for end product-mediated feedback inhibition of cholesterol synthesis (Goldstein et al., 2006). Insig1/2 restricts the cholesterol biosynthetic pathway by preventing proteolytic activation of transcription factors sterol regulatory element-binding proteins (SREBPs) and by enhancing degradation of HMG-CoA reductase (Dong and Tang, 2010). Mice with loss of Insig1/2 (Insig1-/-;Insig2-/-) die within one day after birth and present midfacial developmental defects ranging from cleft palate to complete midfacial cleft with hypoplastic processes of the midfacial region (Engelking et al., 2006). The liver and head of Insig1-/-;Insig2-/- embryos overproduce sterols, causing a marked buildup of cholesterol intermediates. These findings have implications for the pathogenesis of midfacial cleft and other human malformation syndromes in which mutations in enzymes catalyzing the steps in cholesterol biosynthesis produce a buildup of sterol intermediates (Engelking et al., 2006). Although the cellular and molecular mechanism is still largely unknown, recent studies suggest that proper cholesterol metabolism is crucial for SHH signaling activation (Porter, 2006; Stottmann et al., 2011).
TCOF1
Autosomal dominant mutations of the TCOF1 gene cause Treacher Collins syndrome, which is characterized by hypoplasia of the first branchial arch derivatives, including facial bones (particularly the zygomatic complex and the mandible), cleft palate, and middle and external ear defects (Dixon et al., 2007). Treacle, the protein encoded by TCOF1, is required for the cell-autonomously formation and proliferation of NC cells (Dixon et al., 2006). Mice haploinsufficient for Tcof1 (Tcof1+/- mice) exhibit frontonasal dysplasia and cleft palate as a result of increased apoptosis in neuroepithelial cells, a source of CNC cells (Dixon et al., 2006).
Spontaneous Mouse Models
The Twirler (Tw) mouse model has proven to be an excellent model in the study of midfacial development, with all Tw homozygotes exhibiting clefting of the midfacial region. The Tw mutation is semidominant, and all homozygous mice display either a unilateral or bilateral cleft lip with cleft palate and a disrupted nasal cavity with misshapen nasal turbinates that are reduced in size and number (Gong et al., 2000).
11) Environmental Risk Factors for Midfacial Developmental Defects
Environmental risk factors and nutritional deficiencies have been implicated in midfacial cleft and hypoplasia (Johnston and Bronsky, 1995; Krapels et al., 2004b). It has been shown that cigarette smoking, maternal alcohol consumption and abnormal diets (zinc, myoinositol, folic acid, RA, etc.) can cause midfacial defects (Krapels et al., 2004a; Mossey et al., 2009).
Fetal Alcohol Syndrome
Mouse models for fetal alcohol syndrome (treated with a high dosage of ethanol during pregnancy) show an anterior neural plate deficiency and cleft lip with increased cell death (Kotch and Sulik, 1992; Sulik, 2005). Disruption of the SHH signaling pathway has been implicated as an important molecular mechanism in the pathogenesis of fetal alcohol syndrome (Ahlgren et al., 2002; Kietzman et al., 2014). For example, a correlation between the severities of facial dysmorphology and ethanol exposure during pregnancy has been described in both Shh+/- and Gli2+/- mice (Kietzman et al., 2014). CDON, a cell surface SHH-binding protein, promotes SHH pathway activity as a co-receptor with PTCH1 (Sanchez-Arrones et al., 2012). Cdon-/- mice exposed to ethanol in utero exhibit more severe midfacial hypoplasia as well as a broad spectrum of holoprosencephaly phenotypes, compared to non-treated Cdon-/- mice (Hong and Krauss, 2012).
Retinoic Acid Metabolism
RA is derived from vitamin A (retinol) by retinal dehydrogenase. The first step of RA synthesis is the oxidation of retinol to retinal by alcohol dehydrogenases; subsequently, retinal is converted to RA by retinaldehyde dehydrogenases. Craniofacial dysmorphism has been observed in cases of disruption of RA synthesis, as well as of exposure to excessive RA (Johnston and Bronsky, 1995). RA originates from the ventral epithelium of the frontonasal process and targets CNC cells (Schneider et al., 2001). Mice with ENU-induced mutation in the retinol dehydrogenase 10 (Rdh10) gene (Rdh10trex/trex mutant mice) display craniofacial, eye, limb, and major organ abnormalities and die by approximately E13.0 (Sandell et al., 2007). Rdh10-/- mice rescued by maternal administration of the missing retinoid compound can survive postnatally (Rhinn et al., 2011). Deletion of the Raldh2 gene, which is involved in the second step of RA synthesis, in mice causes lethality at E10.5. Treatment with RA can extend survival until E18.5, and these surviving embryos show midfacial hypoplasia (Niederreither et al., 2002). Raldh3 null mice, on the other hand, present malformations restricted to the ocular and nasal regions, which cause choanal atresia (aka upper airway obstruction) following downregulation of Fgf8 in nasal fins (Dupe et al., 2003). Thus, RA metabolic defects at each step of synthesis cause midfacial birth defects.
Perspectives
Midfacial development involves a few morphogenic processes such as growth and fusion that are more complex than general morphogenic processes involved in the timing and balance of growth of midfacial primordia. This is well reflected by the fact that the etiology of midfacial birth defects is complicated by a variety of genetic and environmental risk factors, as listed in Table 1. During midfacial development, the size and pattern of the primordia dramatically change in a short time window. The expression of hundreds of genes changes dramatically at each embryonic stage during craniofacial development, as confirmed by global gene expression studies, including microarrays and RNA-seq. These changes may be regulated by extracellular cues such as growth factors and the consequent orchestrated mechanisms that ultimately regulate cell fate (proliferation, differentiation, apoptosis, etc.). Specifically, in this review we learned that the BMP-SHH-WNT-FGF signaling axis may play a crucial role in midfacial development although each cell signaling cascade may directly regulate gene expression of causative genes of midfacial developmental defects. In addition to gene mutations, environmental and epigenetic factors also affect the expression and phenotypes of hundreds of genes without altering the DNA sequence and cause a number of diseases. Recent studies suggest that the distribution and contribution of miRs, which are regulated by environmental factors, is altered in midfacial birth defects. Interestingly, some cellular metabolisms play crucial roles in midfacial development through the regulation of cell signaling activity, which constitutes another example of how environmental and epigenetic factors affect midfacial morphogenesis. As implicated in a wide range of physiological and pathological conditions, a key cell signaling pathway may be activated to regulate gene and miR expression in a temporal and spatial specific manner during midfacial development. Further genetic studies and animal models will advance our understanding of the roles of genetic and epigenetic factors in midfacial development and will hopefully enable us to design future therapeutic approaches to diagnose and prevent midfacial developmental defects.
Acknowledgments
We thank Drs. Vesa M. Kaartinen, Fenglei He, and YiPing Chen for their critical reading of the manuscript and discussion, and Anne Scruggs and Sarah Zhao for helping with the literature search. This study was supported by a grant from the National Institute of Dental and Craniofacial Research, NIH (DE024759 to J.I.) and a faculty start-up fund from the UTHealth School of Dentistry (to J.I).
Grant sponsors: The National Institute of Dental and Craniofacial Research, NIH (DE024759 to J.I.) The UTHealth School of Dentistry (a faculty start-up fund to J.I.).
Footnotes
Conflict of interests: Authors state no conflicts of interest.
References
- Ahlgren SC, Thakur V, Bronner-Fraser M. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proceedings of the National Academy of Sciencesof the United States of America. 2002;99(16):10476–10481. doi: 10.1073/pnas.162356199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Lawrence AR, Stottmann RW, Bachiller D, Klingensmith J. Chordin and noggin promote organizing centers of forebrain development in the mouse. Development. 2002;129(21):4975–4987. doi: 10.1242/dev.129.21.4975. [DOI] [PubMed] [Google Scholar]
- Arts HH, Bongers EM, Mans DA, van Beersum SE, Oud MM, Bolat E, et al. C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. Journal of medical genetics. 2011;48(6):390–395. doi: 10.1136/jmg.2011.088864. [DOI] [PubMed] [Google Scholar]
- Ashe A, Butterfield NC, Town L, Courtney AD, Cooper AN, Ferguson C, et al. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Human molecular genetics. 2012;21(8):1808–1823. doi: 10.1093/hmg/ddr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Shneyder N, Tran U, Anderson R, Rossant J, et al. The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development. 2003;130(15):3567–3578. doi: 10.1242/dev.00581. [DOI] [PubMed] [Google Scholar]
- Bachler M, Neubuser A. Expression of members of the Fgf family and their receptors during midfacial development. Mechanisms of development. 2001;100(2):313–316. doi: 10.1016/s0925-4773(00)00518-9. [DOI] [PubMed] [Google Scholar]
- Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nature genetics. 1996;14(3):353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128(20):3975–3986. doi: 10.1242/dev.128.20.3975. [DOI] [PubMed] [Google Scholar]
- Billington CJ, Jr, Schmidt B, Marcucio RS, Hallgrimsson B, Gopalakrishnan R, Petryk A. Impact of retinoic acid exposure on midfacial shape variation and manifestation of holoprosencephaly in Twsg1 mutant mice. Disease models & mechanisms. 2015;8(2):139–146. doi: 10.1242/dmm.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128(8):1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Bredrup C, Saunier S, Oud MM, Fiskerstrand T, Hoischen A, Brackman D, et al. Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. American journal of human genetics. 2011;89(5):634–643. doi: 10.1016/j.ajhg.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto JA, Evans RD, Hayward RD, Jones BM. From genotype to phenotype: the differential expression of FGF, FGFR, and TGFbeta genes characterizes human cranioskeletal development and reflects clinical presentation in FGFR syndromes. Plastic and reconstructive surgery. 2001;108(7):2026–2039. doi: 10.1097/00006534-200112000-00030. discussion 2040-2026. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Allen NC, James AW, Mekonnen Z, Madan E, Helms JA. A primary cilia-dependent etiology for midline facial disorders. Human molecular genetics. 2010;19(8):1577–1592. doi: 10.1093/hmg/ddq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM. TCFs and Wnt/beta-catenin signaling: more than one way to throw the switch. Current topics in developmental biology. 2012;98:1–34. doi: 10.1016/B978-0-12-386499-4.00001-X. [DOI] [PubMed] [Google Scholar]
- Cao X, Eu KW, Kumarasinghe MP, Li HH, Loi C, Cheah PY. Mapping of hereditary mixed polyposis syndrome (HMPS) to chromosome 10q23 by genomewide high-density single nucleotide polymorphism (SNP) scan and identification of BMPR1A loss of function. Journal of medical genetics. 2006;43(3):e13. doi: 10.1136/jmg.2005.034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, et al. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133(11):2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235(9):2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chia IV, Kim MJ, Itoh K, Sokol SY, Costantini F. Both the RGS domain and the six C-terminal amino acids of mouse Axin are required for normal embryogenesis. Genetics. 2009;181(4):1359–1368. doi: 10.1534/genetics.109.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383(6599):407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Xavier GM, Depew M, Hagan L, Sealby J, Webster Z, et al. Sonic hedgehog signalling inhibits palatogenesis and arrests tooth development in a mouse model of the nevoid basal cell carcinoma syndrome. Developmental biology. 2009;331(1):38–49. doi: 10.1016/j.ydbio.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Krauss RS. Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Current biology : CB. 2003;13(5):411–415. doi: 10.1016/s0960-9822(03)00088-5. [DOI] [PubMed] [Google Scholar]
- Cox TC. Taking it to the max: the genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clinical genetics. 2004;65(3):163–176. doi: 10.1111/j.0009-9163.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- Dennis JF, Kurosaka H, Iulianella A, Pace J, Thomas N, Beckham S, et al. Mutations in Hedgehog acyltransferase (Hhat) perturb Hedgehog signaling, resulting in severe acrania-holoprosencephaly-agnathia craniofacial defects. PLoS genetics. 2012;8(10):e1002927. doi: 10.1371/journal.pgen.1002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Wu X, Bostrom H, Kim I, Wong N, Tsoi B, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nature genetics. 2004;36(10):1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(36):13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Trainor P, Dixon MJ. Treacher Collins syndrome. Orthodontics & craniofacial research. 2007;10(2):88–95. doi: 10.1111/j.1601-6343.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- Dong XY, Tang SQ. Insulin-induced gene: a new regulator in lipid metabolism. Peptides. 2010;31(11):2145–2150. doi: 10.1016/j.peptides.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Dudas M, Kaartinen V. Tgf-beta superfamily and mouse craniofacial development: interplay of morphogenetic proteins and receptor signaling controls normal formation of the face. Current topics in developmental biology. 2005;66:65–133. doi: 10.1016/S0070-2153(05)66003-6. [DOI] [PubMed] [Google Scholar]
- Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, et al. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Developmental biology. 2006;296(2):298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75(7):1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Engelking LJ, Evers BM, Richardson JA, Goldstein JL, Brown MS, Liang G. Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. The Journal of clinical investigation. 2006;116(9):2356–2365. doi: 10.1172/JCI28988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Huang J, Zhang J, Williams T. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Molecular and cellular biology. 2008;28(1):315–325. doi: 10.1128/MCB.01168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Developmental cell. 2011;21(4):627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Herzog D, Sumara G, Buchmann-Moller S, Civenni G, Wu X, et al. Stage-specific control of neural crest stem cell proliferation by the small rho GTPases Cdc42 and Rac1. Cell stem cell. 2009;4(3):236–247. doi: 10.1016/j.stem.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Canalis E. Bone morphogenetic proteins and their antagonists. Reviews in endocrine & metabolic disorders. 2006;7(1-2):51–65. doi: 10.1007/s11154-006-9000-6. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Gong SG, White NJ, Sakasegawa AY. The Twirler mouse, a model for the study of cleft lip and palate. Archives of oral biology. 2000;45(1):87–94. doi: 10.1016/s0003-9969(99)00101-6. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Greene RM, Pisano MM. Palate morphogenesis: current understanding and future directions. Birth defects research Part C, Embryo today : reviews. 2010;90(2):133–154. doi: 10.1002/bdrc.20180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JN, Compagnucci C, Hu D, Fish J, Klein O, Marcucio R, et al. Fgf8 dosage determines midfacial integration and polarity within the nasal and optic capsules. Developmental biology. 2013;374(1):185–197. doi: 10.1016/j.ydbio.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg H, Truslove GM. Two closely linked genes in the mouse. Genet Res. 1960;1:69–90. [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92(2):253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- He F, Soriano P. A critical role for PDGFRalpha signaling in medial nasal process development. PLoS genetics. 2013;9(9):e1003851. doi: 10.1371/journal.pgen.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiological reviews. 1999;79(4):1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- Helms JA, Cordero D, Tapadia MD. New insights into craniofacial morphogenesis. Development. 2005;132(5):851–861. doi: 10.1242/dev.01705. [DOI] [PubMed] [Google Scholar]
- Hill CR, Yuasa M, Schoenecker J, Goudy SL. Jagged1 is essential for osteoblast development during maxillary ossification. Bone. 2014;62:10–21. doi: 10.1016/j.bone.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Iwata J, Ho HA, Grimes WC, Park S, Sanchez-Lara PA, et al. Integration of comprehensive 3D microCT and signaling analysis reveals differential regulatory mechanisms of craniofacial bone development. Developmental biology. 2015;400(2):180–190. doi: 10.1016/j.ydbio.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G, Basilico C. Mesodermal expression of Fgfr2S252W is necessary and sufficient to induce craniosynostosis in a mouse model of Apert syndrome. Developmental biology. 2012;368(2):283–293. doi: 10.1016/j.ydbio.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. Sox proteins and neural crest development. Seminars in cell & developmental biology. 2005;16(6):694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Hong M, Krauss RS. Cdon mutation and fetal ethanol exposure synergize to produce midline signaling defects and holoprosencephaly spectrum disorders in mice. PLoS genetics. 2012;8(10):e1002999. doi: 10.1371/journal.pgen.1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009;136(1):107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. Journal of molecular cell biology. 2010;2(3):152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- Humphreys R, Zheng W, Prince LS, Qu X, Brown C, Loomes K, et al. Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients. Human molecular genetics. 2012;21(6):1374–1383. doi: 10.1093/hmg/ddr575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman KE, Purcell P, Kume T, Trainor PA. Interaction between Foxc1 and Fgf8 during mammalian jaw patterning and in the pathogenesis of syngnathia. PLoS genetics. 2013;9(12):e1003949. doi: 10.1371/journal.pgen.1003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Han J, Yen HY, Sucov HM, Chai Y, Maxson RE., Jr Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development. 2005;132(22):4937–4950. doi: 10.1242/dev.02072. [DOI] [PubMed] [Google Scholar]
- Iwata J, Parada C, Chai Y. The mechanism of TGF-beta signaling during palate development. Oral diseases. 2011;17(8):733–744. doi: 10.1111/j.1601-0825.2011.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, Chai Y. Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. The Journal of clinical investigation. 2012;122(3):873–885. doi: 10.1172/JCI61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Suzuki A, Pelikan RC, Ho TV, Sanchez-Lara PA, Chai Y. Modulation of lipid metabolic defects rescues cleft palate in Tgfbr2 mutant mice. Human molecular genetics. 2014;23(1):182–193. doi: 10.1093/hmg/ddt410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Van DenBroek O, Davenport IR, Akkers RC, Liu F, Veenstra GJ, et al. The Wnt signaling mediator tcf1 is required for expression of foxd3 during Xenopus gastrulation. The International journal of developmental biology. 2013;57(1):49–54. doi: 10.1387/ijdb.120191kv. [DOI] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes & development. 2004;18(8):937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235(5):1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YR, Han XH, Taketo MM, Yoon JK. Wnt9b-dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development. 2012;139(10):1821–1830. doi: 10.1242/dev.075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Wilkie AO. Craniosynostosis. European journal of human genetics : EJHG. 2011;19(4):369–376. doi: 10.1038/ejhg.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston MC, Bronsky PT. Prenatal craniofacial development: new insights on normal and abnormal mechanisms. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists. 1995;6(4):368–422. doi: 10.1177/10454411950060040601. [DOI] [PubMed] [Google Scholar]
- Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small GTPases. 2014;5 doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasberg AD, Brunskill EW, Steven Potter S. SP8 regulates signaling centers during craniofacial development. Developmental biology. 2013;381(2):312–323. doi: 10.1016/j.ydbio.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K, Melendez J, Baumann JM, Leslie JR, Chauhan BK, Nemkul N, et al. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(18):7607–7612. doi: 10.1073/pnas.1101347108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzman HW, Everson JL, Sulik KK, Lipinski RJ. The teratogenic effects of prenatal ethanol exposure are exacerbated by Sonic Hedgehog or GLI2 haploinsufficiency in the mouse. PloS one. 2014;9(2):e89448. doi: 10.1371/journal.pone.0089448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M, Bisgaard AM, Stoeva R, Dimitrov B, Gillessen-Kaesbach G, Fryns JP, et al. Phenotype and 244k array-CGH characterization of chromosome 13q deletions: an update of the phenotypic map of 13q21.1-qter. American journal of medical genetics Part A. 2009;149A(5):894–905. doi: 10.1002/ajmg.a.32814. [DOI] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development. 2005;132(13):3127–3138. doi: 10.1242/dev.01879. [DOI] [PubMed] [Google Scholar]
- Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4(2):68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotch LE, Sulik KK. Experimental fetal alcohol syndrome: proposed pathogenic basis for a variety of associated facial and brain anomalies. American journal of medical genetics. 1992;44(2):168–176. doi: 10.1002/ajmg.1320440210. [DOI] [PubMed] [Google Scholar]
- Krapels IP, Rooij IA, Wevers RA, Zielhuis GA, Spauwen PH, Brussel W, et al. Myo-inositol, glucose and zinc status as risk factors for non-syndromic cleft lip with or without cleft palate in offspring: a case-control study. BJOG : an international journal of obstetrics and gynaecology. 2004a;111(7):661–668. doi: 10.1111/j.1471-0528.2004.00171.x. [DOI] [PubMed] [Google Scholar]
- Krapels IP, van Rooij IA, Ocke MC, West CE, van der Horst CM, Steegers-Theunissen RP. Maternal nutritional status and the risk for orofacial cleft offspring in humans. The Journal of nutrition. 2004b;134(11):3106–3113. doi: 10.1093/jn/134.11.3106. [DOI] [PubMed] [Google Scholar]
- Kress W, Collmann H, Busse M, Halliger-Keller B, Mueller CR. Clustering of FGFR2 gene mutations inpatients with Pfeiffer and Crouzon syndromes (FGFR2-associated craniosynostoses) Cytogenetics and cell genetics. 2000;91(1-4):134–137. doi: 10.1159/000056833. [DOI] [PubMed] [Google Scholar]
- Kurosaka H, Iulianella A, Williams T, Trainor PA. Disrupting hedgehog and WNT signaling interactions promotes cleft lip pathogenesis. The Journal of clinical investigation. 2014;124(4):1660–1671. doi: 10.1172/JCI72688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, et al. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235(5):1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009;136(8):1387–1396. doi: 10.1242/dev.028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana-Elola E, Tylzanowski P, Takatalo M, Alakurtti K, Veistinen L, Mitsiadis TA, et al. Noggin null allele mice exhibit a microform of holoprosencephaly. Human molecular genetics. 2011;20(20):4005–4015. doi: 10.1093/hmg/ddr329. [DOI] [PubMed] [Google Scholar]
- Li WY, Dudas M, Kaartinen V. Signaling through Tgf-beta type I receptor Alk5 is required for upper lip fusion. Mechanisms of development. 2008;125(9):874–882. doi: 10.1016/j.mod.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Young NM, Tropp S, Hu D, Xu Y, Hallgrimsson B, et al. Quantification of shape and cell polarity reveals a novel mechanism underlying malformations resulting from related FGF mutations during facial morphogenesis. Human molecular genetics. 2013;22(25):5160–5172. doi: 10.1093/hmg/ddt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr, Ashe A, He M, Satir P, Moran J, Beier D, et al. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. The Journal of cell biology. 2012;197(6):789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen S, Johnson C, Helms JA. A ciliopathy with hydrocephalus, isolated craniosynostosis, hypertelorism, and clefting caused by deletion of Kif3a. Reproductive toxicology. 2014 doi: 10.1016/j.reprotox.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, et al. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132(6):1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jin Y, Li J, Seto E, Kuo E, Yu W, et al. Inactivation of Cdc42 in neural crest cells causes craniofacial and cardiovascular morphogenesis defects. Developmental biology. 2013;383(2):239–252. doi: 10.1016/j.ydbio.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, Atit R. Visualizing canonical Wnt signaling during mouse craniofacial development. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239(1):354–363. doi: 10.1002/dvdy.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen MJ, Chiariello M, Tanos T, Bernard O, Narumiya S, Gutkind JS. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Molecular cell. 2004;14(1):29–41. doi: 10.1016/s1097-2765(04)00153-4. [DOI] [PubMed] [Google Scholar]
- Morrison-Graham K, Schatteman GC, Bork T, Bowen-Pope DF, Weston JA. A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development. 1992;115(1):133–142. doi: 10.1242/dev.115.1.133. [DOI] [PubMed] [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- Nelson DK, Williams T. Frontonasal process-specific disruption of AP-2alpha results in postnatal midfacial hypoplasia, vascular anomalies, and nasal cavity defects. Developmental biology. 2004;267(1):72–92. doi: 10.1016/j.ydbio.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. FGF signalling in craniofacial development and developmental disorders. Oral diseases. 2006a;12(2):102–111. doi: 10.1111/j.1601-0825.2005.01176.x. [DOI] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. BMP signalling in craniofacial development. The International journal of developmental biology. 2006b;50(6):511–521. doi: 10.1387/ijdb.052101xn. [DOI] [PubMed] [Google Scholar]
- Nie X, Wang Q, Jiao K. Dicer activity in neural crest cells is essential for craniofacial organogenesis and pharyngeal arch artery morphogenesis. Mechanisms of development. 2011;128(3):200–207. doi: 10.1016/j.mod.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dolle P. Embryonic retinoic acid synthesis is required for forelimb growth and anteroposterior patterning in the mouse. Development. 2002;129(15):3563–3574. doi: 10.1242/dev.129.15.3563. [DOI] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nature genetics. 2000;25(4):440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- Nolan YM, Sullivan AM, Toulouse A. Parkinson's disease in the nuclear age of neuroinflammation. Trends in molecular medicine. 2013;19(3):187–196. doi: 10.1016/j.molmed.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, Williams T. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13714–13719. doi: 10.1073/pnas.95.23.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka-Tanaka Y, Oommen S, Kawasaki M, Kawasaki K, Imam N, Jalani-Ghazani F, et al. Oral lining mucosa development depends on mesenchymal microRNAs. Journal of dental research. 2013;92(3):229–234. doi: 10.1177/0022034512470830. [DOI] [PubMed] [Google Scholar]
- Pan A, Chang L, Nguyen A, James AW. A review of hedgehog signaling in cranial bone development. Frontiers in physiology. 2013;4:61. doi: 10.3389/fphys.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Carbe C, Powers A, Zhang EE, Esko JD, Grobe K, et al. Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development. 2008;135(2):301–310. doi: 10.1242/dev.014829. [DOI] [PubMed] [Google Scholar]
- Phillips HM, Papoutsi T, Soenen H, Ybot-Gonzalez P, Henderson DJ, Chaudhry B. Neural crest cell survival is dependent on Rho kinase and is required for development of the mid face in mouse embryos. PloS one. 2012;7(5):e37685. doi: 10.1371/journal.pone.0037685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD. Cholesterol precursors and facial clefting. The Journal of clinical investigation. 2006;116(9):2322–2325. doi: 10.1172/JCI29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocrine-related cancer. 2000;7(3):165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Developmental biology. 2011;353(1):38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Schuhbaur B, Niederreither K, Dolle P. Involvement of retinol dehydrogenase 10 in embryonic patterning and rescue of its loss of function by maternal retinaldehyde treatment. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(40):16687–16692. doi: 10.1073/pnas.1103877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Muenke M. How a Hedgehog might see holoprosencephaly. Human molecular genetics. 2003:R15–25. doi: 10.1093/hmg/ddg058. 12 Spec No 1. [DOI] [PubMed] [Google Scholar]
- Roscioli T, Flanagan S, Kumar P, Masel J, Gattas M, Hyland VJ, et al. Clinical findings in a patient with FGFR1 P252R mutation and comparison with the literature. American journal of medical genetics. 2000;93(1):22–28. doi: 10.1002/1096-8628(20000703)93:1<22::aid-ajmg5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Ross JS, Carlson JA, Brock G. miRNA: the new gene silencer. American journal of clinical pathology. 2007;128(5):830–836. doi: 10.1309/2JK279BU2G743MWJ. [DOI] [PubMed] [Google Scholar]
- Saito H, Yamamura K, Suzuki N. Reduced bone morphogenetic protein receptor type 1A signaling in neural-crest-derived cells causes facial dysmorphism. Disease models & mechanisms. 2012;5(6):948–955. doi: 10.1242/dmm.009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Arrones L, Cardozo M, Nieto-Lopez F, Bovolenta P. Cdon and Boc: Two transmembrane proteins implicated in cell-cell communication. The international journal of biochemistry & cell biology. 2012;44(5):698–702. doi: 10.1016/j.biocel.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, et al. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes & development. 2007;21(9):1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nature genetics. 1994;6(4):348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nature genetics. 2000;24(4):391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Hu D, Rubenstein JL, Maden M, Helms JA. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development. 2001;128(14):2755–2767. doi: 10.1242/dev.128.14.2755. [DOI] [PubMed] [Google Scholar]
- Senarath-Yapa K, Chung MT, McArdle A, Wong VW, Quarto N, Longaker MT, et al. Craniosynostosis: molecular pathways and future pharmacologic therapy. Organogenesis. 2012;8(4):103–113. doi: 10.4161/org.23307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116(2):297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Sheehy NT, Cordes KR, White MP, Ivey KN, Srivastava D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development. 2010;137(24):4307–4316. doi: 10.1242/dev.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuler CF. Programmed cell death and cell transformation in craniofacial development. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists. 1995;6(3):202–217. doi: 10.1177/10454411950060030301. [DOI] [PubMed] [Google Scholar]
- Smith CL, Tallquist MD. PDGF function in diverse neural crest cell populations. Cell adhesion & migration. 2010;4(4):561–566. doi: 10.4161/cam.4.4.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Molecular and cellular biology. 2004;24(15):6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Li Y, Wang K, Wang YZ, Molotkov A, Gao L, et al. Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development. 2009;136(18):3161–3171. doi: 10.1242/dev.037440. [DOI] [PubMed] [Google Scholar]
- Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124(14):2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- Stanier P, Pauws E. Development of the lip and palate: FGF signalling. Frontiers of oral biology. 2012;16:71–80. doi: 10.1159/000337618. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Turbe-Doan A, Tran P, Kratz LE, Moran JL, Kelley RI, et al. Cholesterol metabolism is required for intracellular hedgehog signal transduction in vivo. PLoS genetics. 2011;7(9):e1002224. doi: 10.1371/journal.pgen.1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik KK. Genesis of alcohol-induced craniofacial dysmorphism. Experimental biology and medicine. 2005;230(6):366–375. doi: 10.1177/15353702-0323006-04. [DOI] [PubMed] [Google Scholar]
- Sun T, Jayatilake D, Afink GB, Ataliotis P, Nister M, Richardson WD, et al. A human YAC transgene rescues craniofacial and neural tube development in PDGFRalpha knockout mice and uncovers a role for PDGFRalpha in prenatal lung growth. Development. 2000;127(21):4519–4529. doi: 10.1242/dev.127.21.4519. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes & development. 1999;13(14):1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130(3):507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Ayada T, Ichiyama K, Kohno R, Yonemitsu Y, Minami Y, et al. Sprouty2 and Sprouty4 are essential for embryonic morphogenesis and regulation of FGF signaling. Biochemical and biophysical research communications. 2007;352(4):896–902. doi: 10.1016/j.bbrc.2006.11.107. [DOI] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135(9):1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PS, Kim J, Nunez S, Glogauer M, Kaartinen V. Neural crest cell-specific deletion of Rac1 results in defective cell-matrix interactions and severe craniofacial and cardiovascular malformations. Developmental biology. 2010;340(2):613–625. doi: 10.1016/j.ydbio.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Melton KR, Manzanares M. Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. The International journal of developmental biology. 2003;47(7):541–553. [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes & development. 1999;13(23):3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Franco G, Liu H, Davidson D, Chai S, Valderrama-Carvajal H, Goltzman D, et al. Defective bone mineralization and osteopenia in young adult FGFR3-/- mice. Human molecular genetics. 2004;13(3):271–284. doi: 10.1093/hmg/ddh034. [DOI] [PubMed] [Google Scholar]
- Wan M, Cao X. BMP signaling in skeletal development. Biochemical and biophysical research communications. 2005;328(3):651–657. doi: 10.1016/j.bbrc.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Wang C, Chang JY, Yang C, Huang Y, Liu J, You P, et al. Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. The Journal of biological chemistry. 2013a;288(30):22174–22183. doi: 10.1074/jbc.M113.463620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bai Y, Li H, Greene SB, Klysik E, Yu W, et al. MicroRNA-17-92, a direct Ap-2alpha transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS genetics. 2013b;9(9):e1003785. doi: 10.1371/journal.pgen.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun M, Uhlhorn VL, Zhou X, Peter I, Martinez-Abadias N, et al. Activation of p38 MAPK pathway in the skull abnormalities of Apert syndrome Fgfr2(+P253R) mice. BMC developmental biology. 2010;10:22. doi: 10.1186/1471-213X-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Roberts W, Wang L, Yamada M, Zhang S, Zhao Z, et al. Rho kinases play an obligatory role in vertebrate embryonic organogenesis. Development. 2001;128(15):2953–2962. doi: 10.1242/dev.128.15.2953. [DOI] [PubMed] [Google Scholar]
- Wilke TA, Gubbels S, Schwartz J, Richman JM. Expression of fibroblast growth factor receptors (FGFR1, FGFR2, FGFR3) in the developing head and face. Developmental dynamics : an official publication of the American Association of Anatomists. 1997;210(1):41–52. doi: 10.1002/(SICI)1097-0177(199709)210:1<41::AID-AJA5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual review of biophysics. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Nishimura O, Misaki K, Nishita M, Minami Y, Yonemura S, et al. Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Developmental cell. 2008;15(1):23–36. doi: 10.1016/j.devcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Yang X, Kilgallen S, Andreeva V, Spicer DB, Pinz I, Friesel R. Conditional expression of Spry1 in neural crest causes craniofacial and cardiac defects. BMC developmental biology. 2010;10:48. doi: 10.1186/1471-213X-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HM, Jerchow B, Sheu TJ, Liu B, Costantini F, Puzas JE, et al. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development. 2005;132(8):1995–2005. doi: 10.1242/dev.01786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Herr AB, Waksman G, Ornitz DM. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proceedings of theNational Academy of Sciences of the United States of America. 2000;97(26):14536–14541. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130(13):3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- Zaghloul NA, Brugmann SA. The emerging face of primary cilia. Genesis. 2011;49(4):231–246. doi: 10.1002/dvg.20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Developmental biology. 2010;340(2):459–467. doi: 10.1016/j.ydbio.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, 3rd, et al. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90(1):181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381(6579):238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Kang JS, Cole F, Yi MJ, Krauss RS. Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Developmental cell. 2006;10(5):657–665. doi: 10.1016/j.devcel.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Zhou XP, Woodford-Richens K, Lehtonen R, Kurose K, Aldred M, Hampel H, et al. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. American journal of human genetics. 2001;69(4):704–711. doi: 10.1086/323703. [DOI] [PMC free article] [PubMed] [Google Scholar]