Abstract
In epithelial tissues, cells constantly generate and transmit forces between each other. Forces generated by the actomyosin cytoskeleton regulate tissue shape and structure and also provide signals that influence cells’ decisions to divide, die, or differentiate. Forces are transmitted across epithelia because cells are mechanically linked through junctional complexes and forces can propagate through the cell cytoplasm. Here, we review some of the molecular mechanisms responsible for force generation, with a specific focus on the actomyosin cortex and adherens junctions. We then discuss evidence for how these mechanisms promote cell shape changes and force transmission in tissues.
Introduction
During development, epithelial tissues undergo dramatic shape changes to generate three-dimensional forms that are essential for organ function. For many of these processes, epithelial cells remain adhered to each other as they change shape and generate new tissue morphologies. Furthermore, many of the mechanisms used by tissues to shape developing organisms are also utilized to maintain and modify tissue properties in adulthood. For example, in developing embryos actomyosin-induced contractions drive cell shape changes to generate new tissue morphologies. In adult mammals, endothelial cells line blood vessels and function to provide a barrier between blood and surrounding tissues. In response to vasoactive compounds, like thrombin and histamine, activation of actomyosin induces endothelial cell contraction and increases endothelial tissue permeability (Lum and Malik, 1996). In both of these examples, the observed morphological response is the result of integration of an input signal that results in a mechanical, force-generating response that is transmitted over an individual cell or across a tissue.
Force transmission and mechanical signals are factors that influence cell survival and cell fate. The extent to which single cells spread influences the magnitude of force generated and a cell's decision to either undergo programmed cell death or to enter the cell cycle (Chen et al., 1997; Oakes et al., 2014). Accordingly, cells in an epithelial sheet respond to mechanical stress, and proliferation occurs in the regions of highest stress; these responses are dependent on tension generated by the actomyosin cytoskeleton and transmitted through cell-cell junctions (Nelson et al., 2005; Rauskolb et al., 2014). The degree of cell spreading can also influence cell differentiation in a manner that is dependent on cytoskeleton-dependent signals (McBeath et al., 2004). Stem cells can be biased to adopt certain cellular morphologies and transcriptional profiles by differences in substrate stiffness; this substrate-directed differentiation is dependent on myosin activity (Engler et al., 2006). Thus, intracellular and intercellular mechanical cues influence a variety of cell and tissue behaviors.
This review provides a “bottom-up” discussion of the principles of force transmission through a tissue. First, we discuss the molecular components essential for force transmission between and within cells, in this review we only discuss adherens junctions but tight junctions and desmosomes also play a role in force transmission (Nekrasova and Green, 2013; Van Itallie and Anderson, 2014; Bazellieres et al., 2015). We then discuss how cells organize the actomyosin cytoskeleton to transmit forces across the cytoplasm. Knowledge of how the actomyosin cytoskeleton promotes force generation in tissues is well advanced, and for this reason we focus on actomyosin-dependent force generation; however, intermediate filament meshworks (see (Kreplak and Fudge, 2007; Qin et al., 2010)) and microtubule networks (see (Brangwynne et al., 2006; Mofrad and Kamm, 2010)) also contribute to cell mechanics and thus influence force transmission.
We discuss measurements of the forces that cells can transmit in pairs and in tissues. We present evidence that suggests how adherens junctions and actomyosin are involved in transmitting forces across tissues in vivo. Finally, we discuss some challenges for the future and outstanding questions.
Molecular components critical to transmit force in tissues
Adherens Junctions
Effectively transmitting force across a tissue requires cells to be mechanically coupled; this coupling is achieved via junctional complexes. Adherens junctions (AJs) link neighboring cell membranes and their internal actin cytoskeletons. Cadherin-family proteins are the transmembrane elements of the AJ complex; the extracellular domains of cadherin proteins can interact with each other, linking neighboring cells (Figure 1A) (Oda et al., 1994; Yap et al., 1997). Because cadherin and its associated proteins connect to the cell's actin cytoskeleton, the AJ can be considered as a mechanical coupler, which connects the actin cytoskeleton of adjacent cells.
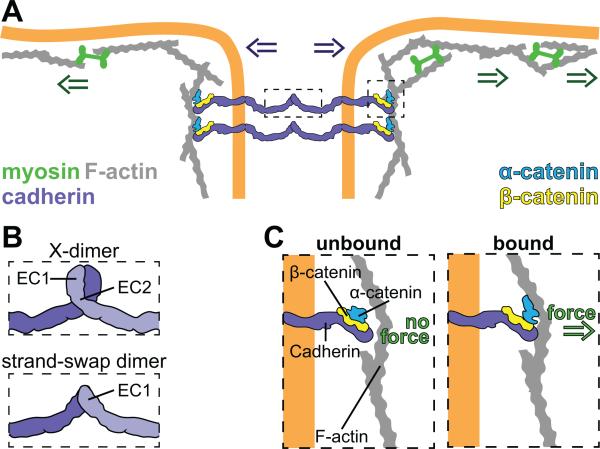
Figure 1. Adherens junctions mechanically couple adjacent cells.
(A) AJs are composed of cadherins, α-catenin, and β-catenin. The extracellular domains of cadherins can interact with each other to link adjacent cells. Together α-catenin and β-catenin form a scaffold that links the AJ to the actin cytoskeleton. In this way the intracellular force from actomyosin contraction (green arrows) is transmitted to adjacent cells (purple arrows). (B) Schematic of two different conformations for trans-interaction of vertebrate E-Cadherin highlighting the regions of interest. E-Cadherin can adopt the X-dimer form (top) or the strand-swapped dimer form (bottom). (C) Components of the AJs complex interact in a force dependent manner. β-catenin binds directly to the intracellular domain of E-cadherin in the presence or absence of force. In the absence of force, α-catenin adopts a folded conformation and does not bind strongly to actin filaments (F-actin) (left). When tensile force is applied to the junction, α-catenin adopts a new conformation and binds F-actin tightly (right).
Cadherin proteins are composed of extracellular (EC) domains arranged in tandem; ECadherin, the vertebrate epithelial cadherin isoform, has 5 EC domains, named EC1-EC5 (Shirayoshi et al., 1986; Ringwald et al., 1987; Shapiro and Weis, 2009). Structural analysis of the extracellular domains of cadherins demonstrated that these domains bind to each other in two distinct conformations, X-dimers (Figure 1B, top) or strand-swapped dimers (Figure 1B, bottom). X dimers were characterized by interactions between EC1 and EC2 domains of adhering partners (Harrison et al., 2010). Strand-swap dimers were mediated by insertion of a β-strand of the EC1 domain into a pocket on the EC1 domain of the trans-binding partner (Haussinger et al., 2004). In vitro force measurements of the different conformations provided insight into the functional difference of the two conformations. While X-dimers became longer-lived upon application of tensile force, behaving like catch bonds, strand-swap dimers were short-lived under tensile force, and thus behave like slip bonds (Rakshit et al., 2012). These results suggested that trans-cellular cadherin interaction conformations respond to mechanical forces, switching from X-dimers with catch-bond behavior under load, to more stable strand-swap dimers in the absence of load. Studies in epithelial cell culture demonstrated that conformational switching is a mechanism for cadherins to remodel junctions (Hong et al., 2011). Cells expressing a cadherin mutant locked in the X-dimer state displayed increased cadherin mobility at cellular junctions; conversely, cells expressing a mutant that only forms strand-swap dimers, stabilized cadherin at the junctions. Thus, force-sensitive conformational changes of trans-cellular cadherin interactions could be a mechanism to modulate AJ stability (Hong et al., 2011). Regardless of the conformation of trans-cellular cadherin interactions, these extracellular interactions are required to maintain tissue integrity (Kintner, 1992).
While the extracellular domains of cadherins are required for cell-cell adhesion, they are not sufficient to mechanically couple cells. The intracellular domain of cadherin interacts with β-catenin and α-catenin, proteins that connect the cytoplasmic side of cadherins to the actin cytoskeleton, respectively (Figure 1A). In in vitro experiments, β-catenin bound the cytoplasmic domain of E-Cadherin with high affinity while α-catenin bound β-catenin and actin filaments but not E-Cadherin. These findings suggested the existence of a tetracomplex between E-Cadherin, β-catenin, α-catenin, and actin filaments; however, until recently only a complex between ECadherin, β-catenin, and α-catenin (tricomplex) could be isolated or reconstituted (Aberle et al., 1994; Rimm et al., 1995). Application of tensile force on the E-Cadherin, β-catenin, and α-catenin complex resulted in strong binding between the tricomplex and actin filaments; thus, tensile force is required for tetracomplex formation (Figure 1C) (Buckley et al., 2014). In vivo studies suggested that this tension-dependent interaction of the AJs components with actin filaments is due to force-sensitive conformational changes in α-catenin. Under low-tension α-catenin adopted an inhibited conformation that obstructs the binding sites of some its binding partners; when tension was applied to the junction, α-catenin adopted a different conformation that revealed these binding sites (Yonemura et al., 2010). Furthermore, measurement of cellular velocity, deformation rate, intercellular, and intracellular rates of cells in an epithelial sheet expressing siRNAs against α- or β-catenin demonstrated that both catenins were required to transmit tension through an epithelial sheet and to maintain epithelial cohesiveness (Bazellieres et al., 2015).
The components of AJs are important for generation and maintenance of tissues in vivo. During development of vertebrate and invertebrate organisms, mutations in cadherin genes caused defects in epithelial organization and morphogenetic movements (Larue et al., 1994; Tepass et al., 1996). Additionally, inhibition of trans-cellular cadherin interactions, via calcium chelation or antibodies, caused tissues to lose adhesion and dissociate (Takeichi et al., 1981; Damsky et al., 1983). Mutation of either α- or β-catenin resulted in tissue adhesion defects. In C. elegans, cadherin, α- or β-catenin (HMR-1, HMP-1, and HMP-2, respectively) are required for morphogenetic movements, including cell migration and tissue elongation (Costa et al., 1998). Structure-function analysis of α-catenin in Drosophila tissues found that both the β-catenin binding domain and the actin-binding domain are required for AJ formation (Desai et al., 2013). Furthermore, expression of an E-Cadherin–α-catenin fusion could rescue defects caused by α-catenin mutations in developing tissues as well as can rescue cell adhesion in cell culture systems (Nagafuchi et al., 1994; Sarpal et al., 2012). In addition to cadherins and catenins, other molecular components that link cellular adhesion molecules and the actin cytoskeleton have been implicated in driving cell shape changes. For example, depletion of the Drosophila homolog of Afadin, a protein can bind both actin filaments and α-catenin, that localizes to AJs, results in separation of the actomyosin cytoskeleton from AJs, disrupting cell shape changes during early Drosophila development (Pokutta et al., 2002; Sawyer et al., 2009; Sawyer et al., 2011). Thus, junctional complexes, including cadherins, catenins, Afadin, vinculin and the actin cytoskeleton, all function to not only adhere neighboring cells, but also, to mechanically couple the cytoskeletons of cells and thus transmit force.
The Actomyosin cytoskeleton
For a tissue to propagate force, force must be transmitted across the volumes of cells. The actomyosin cytoskeleton provides a means to generate and transmit force across a cell. Actin filaments (F-actin) are polymers composed of subunits that have an intrinsic polarity, such that filaments have a plus-end (barbed end) and a minus-end (pointed end) (Figure 2A). Assembly and disassembly occurs at both ends of a filament; however, actin monomers preferentially add to F-actin plus-ends and disassembly occurs preferentially at the minus-end (Pollard et al., 1982; Pollard and Borisy, 2003). F-actin is assembled in a meshwork just below the plasma membrane in a structure called the F-actin cortex (Bovellan et al., 2014). The F-actin cortex resists and generates force such that cells can maintain or change their shapes.
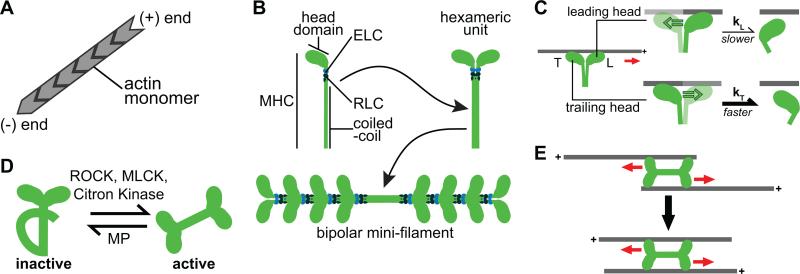
Figure 2. Properties and regulation of the actomyosin cytoskeleton.
(A) Actin monomers assemble into polar filaments with plus-ends and minus-ends. (B) Myosin is a hexameric complex composed of 2 myosin heavy chains (MHCs), 2 essential light chains (ELCs), and 2 regulatory light chains (RLCs). The MHC contains the head or motor domain, which binds and translocates F-actin, and a coiled-coil region that assembles with other coiled-coil tails to form bipolar minifilaments (bottom). (C) Leading and trailing heads experience different loads, which affect F-actin attachment lifetime. Unlike the trailing head (bottom), the leading head (top) experiences a resistive load, which slows the rate of ADP release (kL) and increases the lifetime of F-actin binding, compared to the trailing head (kT > kL). (D) Phosphorylation of the RLC by Rho-kinase (ROCK), myosin light chain kinase (MLCK), or Citron kinase promotes activation of motor activity and bipolar minifilament assembly. Dephosphorylation of the RLC by myosin phosphatase (MP) inactivates and disassembles myosin minifilaments. (E) Bipolar myosin minifilaments promote contraction of anti-parallel F-actin filaments because myosin heads walk towards the plus-ends of F-actin. In this schematic we have minimized the number of myosin heads in the bipolar minifilament for simplicity.
Myosins are a superfamily of motors that utilize energy from ATP hydrolysis to move along F-actin tracks (Howard, 2001). A key force-generating protein in a cell's cortex is the molecular motor non-muscle myosin II (myosin), which has roles in cell migration, cell-cell adhesion, and morphogenesis (Vicente-Manzanares et al., 2009). Myosin is a hexameric complex that consists of two myosin heavy chains (MHC) and two pairs of light chains called the regulatory light chain (RLC) and the essential light chain (ELC) (Sellers and Knight, 2007) (Figure 2B). The N-terminal head domain of MHC binds F-actin and ATP. The head domain couples energy from ATP hydrolysis to myosin movement along F-actin. Each enzymatic cycle of the motor domain results in its displacement towards the plus-end of F-actin, (refer to (De La Cruz and Ostap, 2004) for a detailed review of the actin-activated ATPase cycle). Compared to other molecular motors, like kinesins and dyneins, myosin heads spend a relatively short time bound to actin during the ATPase cycle. This characteristic means that individual myosin heads have a low duty-ratio, defined as the proportion of the ATPase cycle a head domain is bound to F-actin. To promote processive movement along F-actin, myosin hexamers assemble into bipolar minifilaments via the C-terminal coiled-coil domain of the MHC. Bipolar minifilament assembly increases the effective duty ratio of the minifilament structure.
Myosin has to perform diverse functions in tissues, including dynamic cell shape changes and stable generation of tension (Vicente-Manzanares et al., 2011). In mammals there are three isoforms of myosin (myosin-IIA, myosin-IIB, myosin-IIC). These isoforms are differentially expressed during development and in different cell types, suggesting that differences in function are reflected in the molecular behavior and assembly of these motors (Maupin et al., 1994; Rochlin et al., 1995; Ma et al., 2010). Myosin-IIA has the lowest duty ratio of the three isoforms (Table 1) (Kovacs et al., 2003). In contrast, myosin-IIB has a significantly greater duty ratio and has the highest affinity for F-actin of the three isoforms, suggesting it is suited for long-term maintenance of tension instead of short-term force generation, like myosin-IIA (Table 1) (Wang et al., 2003; Billington et al., 2013). The less well-studied myosin-IIC has an intermediate duty ratio; however, in comparison to myosin-IIA, its minifilaments have fewer heads, and as such, myosin-IIC has the lowest probability of the three isoforms of binding F-actin at any given time (Heissler and Manstein, 2011; Billington et al., 2013). Interestingly, Drosophila only expresses one myosin isoform, which behaves similar to the fast-acting myosin-IIA isoform, with a low duty ratio (Kiehart and Feghali, 1986; Kiehart et al., 1989; Mansfield et al., 1996; Heissler et al., 2015). Given that the three mammalian myosin isoforms have different kinetic behaviors and thus provide varying functionality to the organism, how Drosophila prevails with one isoform is not understood. One possible mechanism is that myosin heads tune their actin-binding kinetics in response to load. An in vitro study of two-headed myosin motors exploited the fact that the leading head and trailing head experience different loads to demonstrate that ADP release, the rate-limiting step for the motility of myosin motors, is sensitive to load (Kovacs et al., 2007). A resistive load slows ADP release and thus increases the lifetime of F-actin binding, while an assisting load increases the rate of ADP release, resulting in decreased F-actin attachment lifetimes (Figure 2C). Thus, tension in the cytoskeleton would be expected to slow myosin dynamics and promote a more stable myosinactin association. There is some indication that the application of force or tension in tissues leads to myosin stabilization and accumulation (Fernandez-Gonzalez et al., 2009; Pouille et al., 2009). However, whether this build-up is due to direct mechanical regulation of the motor, modulation of upstream signaling pathways, or both is not clear.
Table 1.
Comparison of mammalian myosin isoforms and Drosophila myosin properties
| Myosin | D.R. of a head | <# of heads per filament> | Effective filament D.R.* | Actin-activated ATPase activity of a head (s−1) |
|---|---|---|---|---|
| myosin-IIA | ~0.1a | 58e | 0.95 | 0.17a |
| myosin-IIB | ~0.4b | 60e | 0.99 | 0.13b |
| myosin-IIC | ~0.26c | 28e | 0.98 | 0.23c |
| Drosophila myosin-II | ~0.1d | 28f | 0.81 | 0.54d |
per-communication with S. Heissler, N. Billington, and J. Sellers
effective filament D.R. = p(myosin filament is attached to F-actin), at any given time
effective filament
D.R. = duty ratio
n = total number of heads per myosin mini-filament
Myosin activity is primarily regulated by phosphorylation of the RLC (Bresnick, 1999; Heissler and Manstein, 2013). Dephosphorylated myosin is inactive and adopts a folded (10S) conformation that blocks productive association of the motor domain with F-actin (Wendt et al., 2001; Jung et al., 2008; Lowey and Trybus, 2010). Phosphorylation of the RLC, primarily on Serine-19 but secondarily on Threonine-18, shifts myosin equilibrium towards an extended conformation (6S), which allows myosin to oligomerize, form bipolar minifilaments, bind F-actin, and activates ATPase activity (Scholey et al., 1980; Craig et al., 1983) (Figure 2B and 2D). The kinases primarily responsible for phosphorylation of the RLC, and consequently myosin activation, are the Rho-associated and coiled-coil kinase (ROCK), myosin light chain kinase, and citron kinase (Ikebe and Hartshorne, 1985; Amano et al., 1996; Totsukawa et al., 2000; Yamashiro et al., 2003). Additionally, ROCK can activate myosin by phosphorylating and inhibiting the myosin binding subunit of myosin phosphatase, the phosphatase that dephosphorylates RLC (Kimura et al., 1996; Hartshorne et al., 1998).
The polarity of F-actin and assembly of myosin into bipolar minifilaments enables plus-end-directed movement of myosin along opposing F-actin to mediate filament sliding (Figure 2E). In skeletal and cardiac muscle, the contractile unit is a sarcomere, which is composed of overlapping arrays of F-actin and muscle myosin, where F-actin plus-ends are fixed at the boundaries of sarcomeres, which themselves are arranged in series. Myosin movement towards F-actin plus ends results in decreasing sarcomere length, and muscle contraction (Huxley, 1963). Models of non-muscle cell contraction do not rely on ordered arrays of F-actin and myosin, but rely on the physical and biochemical properties of myosin and F-actin (Soares e Silva et al., 2011; Murrell and Gardel, 2012). Using a reconstituted actin cortex attached to a membrane surface, it was shown that a network, composed of F-actin, myosin, and other actin cross-linking proteins, can contract in the absence of initial F-actin order (Murrell and Gardel, 2012). The initiation of network contraction depended on breaking the balance between compressive and tensile stresses exerted on F-actin by myosin motors. F-actin resists tensile stresses, with the force required to break a single F-actin via extension being approximately 100 pN (Kishino and Yanagida, 1988). Therefore, pulling and filament extension most often results in translocation of F-actin. In contrast, the force required to buckle and break F-actin under compressive stresses is 0.16 pN, almost 4 orders of magnitude less than that required to break F-actin via extension (Lenz et al., 2012). Thus, in a disordered network the compressive stresses on F-actin, generated by myosin, were asymmetrically relieved by F-actin buckling, while the remaining tensile stresses were retained and promoted network contraction via shortening of F-actin segments via F-actin sliding.
Importantly, contraction of this pseudo-cortex has no requirement for a precise or ordered actin filament organization, demonstrating that contraction could be an intrinsic property of actomyosin networks (Billington et al., 2013; Carvalho et al., 2013). In non-muscle cells, actomyosin networks most often do not demonstrate a high degree of order and yet still contract (Verkhovsky et al., 1997). Thus, this in vitro work provides a physical model for how actomyosin networks without defined polarity and organization contract in vivo.
Transmitting forces across a cell-cytoskeletal network
The ability to transmit force across a cell is essential for force generation on a cellular scale; transmission requires transduction of force across the cell's cytoplasm through a coherent mechanical network. Myosin inhibition caused the cell's cytoplasm to fragment as it lost circumferential actin bundles and stress fibers (Cai et al., 2010). Thus, myosin activity is essential to maintain a coherent actin network that spans an entire cell.
Advances in microscopy have uncovered a suborganization of cytoskeletal components required to transmit forces in a cell. Treatment of cells with Latrunculin A, a small-molecule that sequesters actin monomers, revealed “nodes” of actin structures throughout the cortex (Luo et al., 2013). Using super-resolution microscopy, nodes were visible without drug treatment, demonstrating that these node structures were not an artifact of drug treatment. Analysis of node movement demonstrated that nodes do not move by pure diffusion, but movement is best described by a biased random walk, suggesting that node movements are coupled via the actin cytoskeleton. In particular, nodes that were less than 2 μm apart moved towards each other. Myosin localized to actin nodes and myosin motor inhibition with blebbistatin caused nodes to move by pure diffusion, demonstrating that myosin motor activity is required to transmit force between nodes. Actomyosin nodes and their coalescence was also observed in contractile rings of fission yeast (Vavylonis et al., 2008). Here, high-speed microscopy of node movements uncovered a mechanism where myosin in nodes stochastically “captured” F-actin growing from a nearby node, exerted force on it, and then released it. These transient connections between nodes were sufficient to generate a contractile ring that could separate daughter cells during cytokinesis. These studies demonstrated that, while non-muscle cells may not display highly-ordered actomyosin organizations (like those observed in muscle cells), they do display supramolecular complexes that are essential for force transmission through a cell.
Supramolecular structures, such as nodes, are integrated into larger cellular structures that transmit forces across a cell. For example, stress fibers are 10-200 μm long bundles of cross-linked actin and myosin that can consist of a repeated sarcomere-like structure and can transmit force across a cell (Katoh et al., 1998). Ablation of a single stress fiber, in living cells growing on a compliant surface, resulted in viscoelastic retraction of the cut ends, compensatory relaxation of the extracellular surface, cell shape change, and actin cytoskeleton remodeling (Kumar et al., 2006). These results demonstrated that individual stress fibers play a role in stabilizing cell shape and exerting force on extracellular adhesions via contractile forces. Cells grown on soft matrices form contractile lamellar networks, characterized by myosin nodes dispersed in a randomly polarized branched F-actin meshwork and without any stress fibers (Verkhovsky et al., 1997; Yeung et al., 2005). These lamellar networks also generate and transmit force with lamellar networks contributing up to 60% of a cell's total traction stress (Aratyn-Schaus et al., 2011). These results demonstrated that a range of actomyosin organizations generate and transmit forces, providing mechanisms for cells to modulate force generation in the context of a tissue.
Magnitudes of forces transmitted in a tissue
As discussed above the actomyosin cortex can adopt a variety of different organizations. In this section we discuss approaches developed to quantitatively assess how cells exert force on each other and their substrates, and how much force they exert. One approach to measure forces transmitted between cells is to examine the force balance for a pair of adherent cells in which cell-substrate traction forces are known. Maruthamuthu et al. found that the tensile force between pairs of isolated epithelial cells was approximately 100 nN and oriented perpendicularly to the interface (Maruthamuthu et al., 2011). This magnitude of force would require on the order of thousands of myosin minifilaments (Finer et al., 1994). Using fabrication techniques to adhere endothelial cell pairs to each other by a single contact and measuring the traction forces exerted by these cells, Chen et al. determined that the tugging force at a single cell contact is approximately 40 nN (Liu et al., 2010). Additionally, the size of the cell-cell contact is determined by the magnitude of the tugging force, which is dependent on myosin activity. Activation of myosin contraction, by inhibition of myosin phosphatase or addition of vasoactive compounds, increased the intercellular tugging force and resulted in a larger cell-cell contact. Conversely, inhibition of myosin activity decreased tugging force and diminished the size of the cell-cell contact. Furthermore, contractile force induces F-actin assembly at the junction, providing a possible mechanism for tension-dependent cell-cell contact size (Leerberg et al., 2014). Thus, studies examining the forces exerted by cells in cell pairs, demonstrated that there is coordination between actomyosin-generated mechanical forces and cell-cell contacts, suggesting that cell-cell contacts could be functioning to mechanically integrate cell adhesion and actomyosin-generated contractility (Lecuit and Yap, 2015). The principles of cellular force balance can also be used to quantify forces in epithelial sheets (Tambe et al., 2013). Measurement of the traction forces exerted by individual cells in a spreading epithelial monolayer demonstrated that tensile stress increases from the sheet edge to the sheet center (~300 pN μm−2 to ~1000 pN μm−2) (Trepat et al., 2009). Thus in a cell sheet with 2 mm in diameter, an enormous amount of force is transmitted across cells and through junctions.
An important question is whether similar levels of force are experienced in the tissues of an organism. Previously, force inference in living tissues has relied on laser ablation techniques to infer tissue tension from the initial recoil velocity of the tissue following a cut (Kiehart et al., 2000; Hutson et al., 2003). These studies have provided insight into the relative tensile state of a tissue prior to the cut, and the directionality of tension-generation in a tissue; however, ablation techniques do not allow absolute measurement of force in a tissue. To study forces dynamically generated by a tissue during morphogenesis, Zhou et al. embedded embryonic dorsal tissue of Xenopus in an agarose gel of known mechanical properties (Zhou et al., 2015). By tracking the displacement of fluorescent beads embedded in the gel as the dorsal tissue underwent convergent extension (convergence along the mediolateral embryonic axis and extension along the anterio-posterior axis), the authors were able to map the stress field surrounding the tissue. Through this analysis, the authors found that the dorsal tissue produces approximately 5 pN μm−2 of stress during convergent extension.
Another group has developed an independent technique to quantify stresses generated by epithelial cells in culture, aggregate and living embryonic tissues. This technique involved fluorescently-labeled cell-sized droplets with known physical properties that can adhere to and integrate into tissues. By tracking the deformations of the droplet in time, the stresses that cells apply to the droplet at every time point can be calculated. Campàs et al. showed that cultured epithelial cells exert 3.4 pN μm−2 of stress while cells in embryonic mouse explants exert 1.6 nN μm−2 (Campas et al., 2014).
A recent study optically trapped cell-cell interfaces in the early Drosophila embryo and found that they generate 100 pN of tension, which could be generated by about fifteen myosin minifilaments (Bambardekar et al., 2015). Furthermore, by measuring the deflection of an interface after release from an optical trap, the authors found that the embryonic tissue behaves as a viscoelastic material, which can be modeled by a combination of elastic components (springs) and viscous components (dashpots) (for a review and detailed description of these mechanical components see (Davidson et al., 2009). These studies and others have begun to provide information on the magnitude of forces generated in living tissues and organisms.
These results demonstrate that tissues can generate magnitudes of force varying in 3 orders of magnitude (pN to nN). These differences likely reflect differences in the mechanism of force measurement; however, they also suggest that tissues themselves have different force-generating properties. Some properties that may influence that magnitude of force a tissue generates could be differences in tissue size, the degree of morphological changes (like how much a tissue needs to move), and force-generating components. For example, tissue invagination in the sea urchin embryo is driven by apical extracellular matrix remodeling (Davidson et al., 1999), while actomyosin contractions drive tissue invagination in the early Drosophila embryo (Martin et al., 2009). Another possible explanation for the differences observed in force magnitudes could be due to the difference in requirements for survival in cell culture and in vivo environments. Cultured cells could need large stresses and signaling processes at focal adhesions to be able to continue proliferating; this requirement could contribute to the observation of a greater the magnitude of force exerted by cultured cells than cells in living tissues. More in vivo studies will elucidate the range of forces that tissues produce and furthermore will determine key properties of cells and their surrounding environments that are required to tune force generation in a tissue.
Transmitting forces across a tissue
To transmit force across a tissue cells must couple intracellular force transmission with mechanical coupling across intercellular contacts. Force transmission is required throughout many morphogenetic processes, for excellent reviews see (Lecuit et al., 2011; Heisenberg and Bellaiche, 2013). Here we discuss two model systems where the importance of force transmission on a tissue scale has been well studied. An excellent model of epithelial sheet contraction and folding is the formation of the Drosophila ventral furrow. At the onset of furrow formation, the ellipsoid embryo consists of a single epithelial sheet that surrounds the yolk. Cells of the prospective mesoderm constrict their apical (outside) ends, which promotes furrowing, or folding, of the epithelial sheet into a tube (Figure 3A). Another model for epithelial sheet contraction is Drosophila dorsal closure, whereby amnioserosa cells, a squamous epithelium on the dorsal face of the embryo, apically constrict, and the lateral epidermis moves dorsally such that they meet and fuse over the amnioserosa cells. The lateral epidermis forms a supracellular actomyosin cable that surrounds the amnioserosa and shortens throughout dorsal closure (Figure 3C) (Kiehart et al., 2000; Peralta et al., 2007). From laser cutting experiments, it has been inferred that both ventral furrow formation and dorsal closure involve tension transmitted across the tissue (Hutson et al., 2003; Martin et al., 2010).
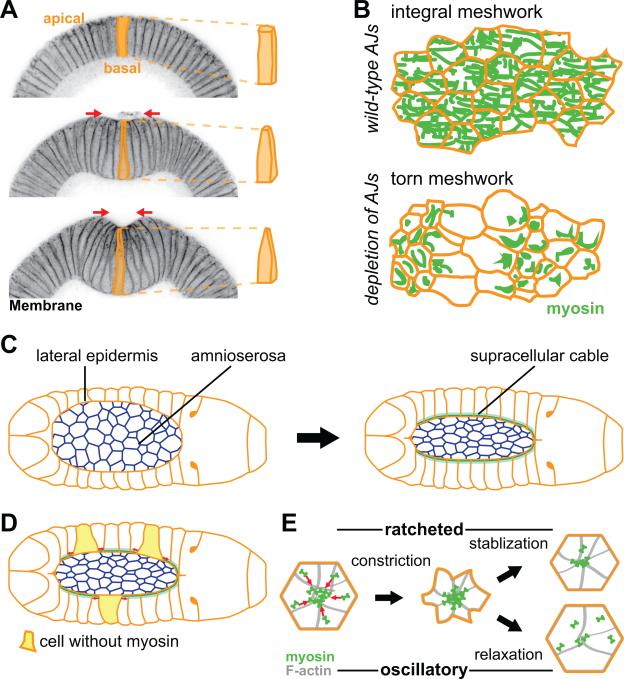
Figure 3. Evidence for AJ and actomyosin based force transmission in vivo.
(A) Apical constriction drives folding of the ventral tissue (ventral furrow formation) in the early Drosophila embryo. The left panels are cross-sections of staged embryos, stained for a membrane marker, undergoing ventral furrow formation. Cells are initially columnar (top) and then apically constrict to become wedged shape (middle and bottom), driving invagination of the tissue. (B) Illustration showing importance of AJs in transmitting tension across a constricting tissue. Myosin forms a supracellular meshwork that spans the constricting tissue (top). Depletion of any AJs component results in tears in the supracellular cytoskeletal meshwork (bottom). (C) Schematic of dorsal closure process. Amnioserosa cells apically constrict, while lateral epidermis forms a supracellular actomyosin cable (green) that also contracts the amnioserosa tissue. (D) In a mosaic animal, epidermal cells that express myosin stretch (red arrows) cells that do not express myosin (yellow cells). (E) Schematic of the apical surface of a cell undergoing ratcheted or oscillatory constriction. Initially, both cells constrict, however the decrease in cell area is stabilized and epithelial tension is facilitated by ratcheted constriction (top). In contrast, the cell shape is not stabilized and relaxes during oscillatory constrictions (bottom).
During ventral furrow formation and tissue folding, AJs are required to transmit tension across the tissue. Depletion of any component of the AJ, E-Cadherin, β-catenin, or α-catenin, during ventral formation resulted in tears in the normally intact supracellular myosin meshwork that spans the ventral tissue (Martin et al., 2010) (Figure 3B). Depletion of E-Cadherin or β-catenin during dorsal closure resulted in separations between the amnioserosa and the epidermis and also disrupts the actomyosin cable (Gorfinkiel and Arias, 2007). Tearing of the tissue-wide cytoskeletal meshwork in embryos with weakened AJs demonstrates that AJs are required to mechanically integrate the cytoskeleton across a tissue. Additionally, the connection between E-cadherin and the F-actin network is also necessary to transmit tension across the tissue. Depletion of alpha-catenin or Afadin, results in separation of neighboring actomyosin networks and contraction of the actomyosin fibers into spots, suggesting a loss of force transmission (Sawyer et al., 2009; Martin et al., 2010; Fernandez-Gonzalez and Zallen, 2011). Furthermore, during ventral furrow formation it has been shown that the connection between the apical F-actin network and AJs in ventral furrow cells is dynamic and F-actin turnover is required to maintain the cytoskeletal attachment to the junctional interface (Jodoin et al., 2015). It appears that during ventral furrow cell constriction the apical F-actin network often releases the junctions. F-actin turnover re-establishes the connection between the actomyosin network and the junction, allowing for stable force balance across intercellular junctions as the cells constrict. The function of this dynamic connection in the context of a wild-type tissue is not yet clear, but it is possible that such a dynamic coupling between actomyosin and the junction could allow cells to fine tune force transmission in the context of a tissue changing shape.
Myosin is required not only to generate contractile force, but also to maintain cell shape. Depletion of the myosin heavy chain during dorsal closure caused the apically constricting amnioserosa cells to separate from each other, as well as disrupted the organization of the supracellular cable (Franke et al., 2005). Generation of an animal with mosaic myosin expression resulted in the stretching of myosin-null cells adjacent to cells expressing myosin (Figure 3D). Thus, cells that do not express myosin cannot generate resistive tensile forces from the supracellular cable and are passively stretched by the neighboring cells contracting their sections of the supracellular cable.
A surprising property of contraction in many systems, including the ventral furrow and dorsal closure, is that actomyosin activity is pulsatile (for reviews see (Martin and Goldstein, 2014) and (Gorfinkiel, 2015)). Myosin pulses involve cycles of myosin accumulation and subsequent remodeling, which can result in incremental cell constriction (Martin et al., 2009) (Figure 3E). Myosin pulses require dynamic regulation of the myosin motor by regulatory light chain phosphorylation, with cycles of phosphorylation and dephosphorylation (Vasquez et al., 2014; Munjal et al., 2015). Interestingly, abrogation of myosin pulses either via depletion of a subunit of myosin phosphatase (a negative regulator of myosin activity) or by expression of myosin regulatory light chain phosphomutants resulted in tears in the supracellular myosin meshwork (Vasquez et al., 2014). Thus, dynamic regulation of myosin activity is somehow required to maintain stable mechanical connections between cells, although the mechanism is not yet clear. One possibility is that coordinating contractile pulses between neighboring cells is required to set up stable force transmission. Dynamic regulation of myosin also appears to be required to efficiently reattach the actomyosin network to AJs after wounding, suggesting a cell autonomous role for myosin regulation in maintaining cytoskeletal attachment to junctions (Jodoin et al., 2015).
The observation of pulsatile contraction, where myosin levels periodically ebb, leads to the question, how do cells transmit forces between myosin pulses. Analysis of pulsatile behavior of ventral cells reveals that myosin pulses can generate different types of cellular responses: ratcheted pulses (cell constricts and stabilizes constricted area), oscillating or unratcheted pulses (cell constricts but relaxes area), and unconstricting pulses (cell does not constrict, or minimally constricts) (Figure 3E) (Xie and Martin, 2015). Myosin persists in the apical domain during ratcheted pulses, but does not persist during unratcheted pulses, suggesting that, specifically during ratcheted pulses, a cell's myosin network is stabilized (Martin et al., 2010; Xie and Martin, 2015). Interestingly, ratcheted pulses seem to be required for cells to cooperatively contract in a tissue (Xie and Martin, 2015). If a cell is undergoing an unratcheted pulse, neighboring pulses appear to slow the cell's constriction, suggesting competition between neighboring contractions. However, for a cell undergoing a ratcheted pulse, neighboring contractions enhance its constriction, suggesting that ratcheting allows cells to cooperate during tissue shape change. A model for this effect is that the persistence of myosin structures that follow ratcheted pulses bears tensile force, allowing forces in neighboring cells to propagate across a ratcheting cell, and thus add up, instead of dissipating. Consistent with this model, depletion of a gene that increases the frequency of unratcheted pulses dramatically reduced epithelial tension (Martin et al., 2010); thus, ratcheted pulses, but not unratcheted pulses, promote epithelial tension during tissue shape change. In addition, there is a transition from unratcheted to ratcheted pulses and the onset of ventral furrow formation (Xie and Martin, 2015).
Another example of pulsatile and ratchet-like apical constriction occurs in amnioserosa cells during dorsal closure. Early in dorsal closure, amnioserosa cell areas oscillate in a manner that correlates with pulsatile myosin behavior; as dorsal closure proceeds, area oscillations are dampened and apical constriction is more ratcheted (Figure 3E) (Solon et al., 2009; Blanchard et al., 2010; David et al., 2010). Thus, like ventral cells, amnioserosa cellular behavior transitions from a phase where there are more unratcheted or unconstricting pulses to a phase where ratcheted pulses are the dominant cellular behavior. David et al. identified a negative feedback loop with a delay that is responsible for cycles of myosin assembly and disassembly in amnioserosa cells (David et al., 2013). This delay is sufficient for amnioserosa cells to build up a persistent actomyosin network, inducing cellular constrictions to become more ratcheted (David et al., 2010; David et al., 2013). Thus, in both dorsal closure and the ventral furrow, effective force transmission across a tissue requires a transition from unratcheted or oscillatory cellular constrictions to ratcheted constrictions that sustain cell spanning cytoskeletal structures. Whether the transition from oscillatory to sustained contractions is a general feature of contractile systems requires live imaging and quantitative analysis of more contractile tissues, especially in vertebrates.
Conclusion
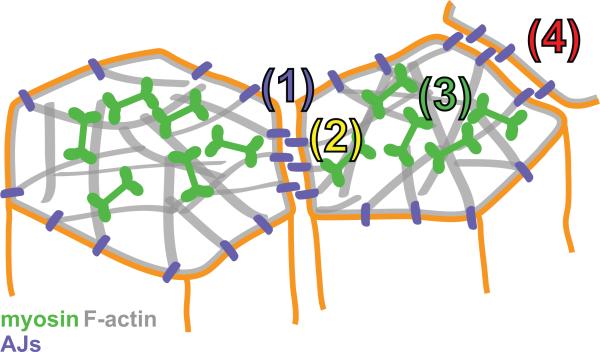
Effective transmission of force in between cells and through a tissue requires three properties: (1) Attachments between neighboring cells. In this review we only discussed cadherins but tight junctions and desmosomes also play a role in force transmission (Bazellieres et al., 2015). (2) A coherent intracellular meshwork, like the actomyosin cortex or an intermediate filament meshwork (Qin et al., 2010). (3) Coupling between the adhesion complex and the force-generating machinery (Figure 4). Together, these features mechanically couple cells to each other such that they can generate forces, react to forces, and ultimately change tissue shape.
Figure 4. Summary of components required to transmit forces across a tissue.
(1) Adherens junctions mediate cell-cell attachment.
(2) Adherens junctions couple the cell membrane to F-actin cytoskeleton.
(3) Actin and myosin form a coherent network across the cell, called the cortex. Myosin can contract F-actin networks to transmit force to the neighboring cell (4).
Recent advancements have lead to the development of various techniques that can measure mechanical forces in living organisms. Coupling of these quantitative tools with perturbations that affect specific molecular activities will be required to truly understand the molecular mechanisms of force generation, transmission, and efficiency in living organisms at the cellular and tissue scales. One question that can be answered using these new tools is the function and contribution of specific protein activities on force transmission in tissues. For example, determining whether myosin motor activity, myosin cross-linking, or myosin-driven F-actin depolymerization contribute to force generation in tissues, and furthermore, understanding how tuning of these different molecular activities influence cellular and tissue-scale force transmission and the ultimate morphogenetic output. Another important challenge will be to understand precisely what actomyosin pulsing does. Cycles of apical actomyosin network assembly and remodeling have not only been observed in apical constriction events in Drosophila embryonic development, but also during C. elegans development (Munro et al., 2004), Xenopus convergent extension (Skoglund et al., 2008), and during cell compaction in early mammalian development (Maitre et al., 2015). Thus, actomyosin pulsing appears to be a general mechanism to change cell shapes and understanding the underlying mechanisms and functions of pulsing will be critical to advance understanding of how cells generate and transmit force in a tissue.
Acknowledgments
Funding information - Sponsor: NIH, Grant #: R01GM105984
References
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107(Pt 12):3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Aratyn-Schaus Y, Oakes PW, Gardel ML. Dynamic and structural signatures of lamellar actomyosin force generation. Mol Biol Cell. 2011;22:1330–1339. doi: 10.1091/mbc.E10-11-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambardekar K, Clement R, Blanc O, Chardes C, Lenne PF. Direct laser manipulation reveals the mechanics of cell contacts in vivo. Proc Natl Acad Sci. 2015;112:1416–1421. doi: 10.1073/pnas.1418732112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazellieres E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, Munoz JJ, Sales-Pardo M, Guimera R, Trepat X. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, Gorfinkiel N. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 2010;137:2743–2752. doi: 10.1242/dev.045872. [DOI] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, Jegou A, Thrasher AJ, Romet-Lemonne G, Roux PP, Paluch EK, Charras G. Cellular control of cortical actin nucleation. Curr Biol. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol. 2006;173:733–741. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick AR. Molecular mechanisms of nonmuscle myosin-II regulation. Curr Opin Cell Biol. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Rossier O, Gauthier NC, Biais N, Fardin MA, Zhang X, Miller LW, Ladoux B, Cornish VW, Sheetz MP. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123:413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campas O, Mammoto T, Hasso S, Sperling RA, O'Connell D, Bischof AG, Maas R, Weitz DA, Mahadevan L, Ingber DE. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat Methods. 2014;11:183–189. doi: 10.1038/nmeth.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho K, Tsai FC, Lees E, Voituriez R, Koenderink GH, Sykes C. Cell-sized liposomes reveal how actomyosin cortical tension drives shape change. Proc Natl Acad Sci. 2013;110:16456–16461. doi: 10.1073/pnas.1221524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- Damsky CH, Richa J, Solter D, Knudsen K, Buck CA. Identification and purification of a cell surface glycoprotein mediating intercellular adhesion in embryonic and adult tissue. Cell. 1983;34:455–466. doi: 10.1016/0092-8674(83)90379-3. [DOI] [PubMed] [Google Scholar]
- David DJ, Tishkina A, Harris TJ. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development. 2010;137:1645–1655. doi: 10.1242/dev.044107. [DOI] [PubMed] [Google Scholar]
- David DJ, Wang Q, Feng JJ, Harris TJ. Bazooka inhibits aPKC to limit antagonism of actomyosin networks during amnioserosa apical constriction. Development. 2013;140:4719–4729. doi: 10.1242/dev.098491. [DOI] [PubMed] [Google Scholar]
- Davidson L, von Dassow M, Zhou J. Multi-scale mechanics from molecules to morphogenesis. Int J Biochem Cell Biol. 2009;41:2147–2162. doi: 10.1016/j.biocel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA, Oster GF, Keller RE, Koehl MA. Measurements of mechanical properties of the blastula wall reveal which hypothesized mechanisms of primary invagination are physically plausible in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 1999;209:221–238. doi: 10.1006/dbio.1999.9249. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric alpha-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Zallen JA. Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys Biol. 2011;8:045005. doi: 10.1088/1478-3975/8/4/045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Franke JD, Montague RA, Kiehart DP. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol. 2005;15:2208–2221. doi: 10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N. From actomyosin oscillations to tissue-level deformations. Dev Dyn. 2015 doi: 10.1002/dvdy.24363. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N, Arias AM. Requirements for adherens junction components in the interaction between epithelial tissues during dorsal closure in Drosophila. J Cell Sci. 2007;120:3289–3298. doi: 10.1242/jcs.010850. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, Shapiro L. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne DJ, Ito M, Erdodi F. Myosin light chain phosphatase: subunit composition, interactions and regulation. J Muscle Res Cell Motil. 1998;19:325–341. doi: 10.1023/a:1005385302064. [DOI] [PubMed] [Google Scholar]
- Haussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Heissler SM, Chinthalapudi K, Sellers JR. Kinetic characterization of the sole nonmuscle myosin-2 from the model organism Drosophila melanogaster. FASEB J. 2015;29:1456–1466. doi: 10.1096/fj.14-266742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler SM, Manstein DJ. Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J Biol Chem. 2011;286:21191–21202. doi: 10.1074/jbc.M110.212290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler SM, Manstein DJ. Nonmuscle myosin-2: mix and match. Cell Mol Life Sci. 2013;70:1–21. doi: 10.1007/s00018-012-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Cadherin exits the junction by switching its adhesive bond. J Cell Biol. 2011;192:1073–1083. doi: 10.1083/jcb.201006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Inc.; Sunderland, MA: 2001. [Google Scholar]
- Hutson MS, Tokutake Y, Chang MS, Bloor JW, Venakides S, Kiehart DP, Edwards GS. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- Huxley HE. Electron Microscope Studies on the Structure of Natural and Synthetic Protein Filaments from Striated Muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- Ikebe M, Hartshorne DJ. Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem. 1985;260:10027–10031. [PubMed] [Google Scholar]
- Jodoin JN, Coravos JS, Chanet S, Vasquez CG, Tworoger M, Kingston ER, Perkins LA, Perrimon N, Martin AC. Stable Force Balance between Epithelial Cells Arises from F Actin Turnover. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.11.018. http://dx.doi.org/10.1016/j.devcel.2015.11.018. [DOI] [PMC free article] [PubMed]
- Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kano Y, Masuda M, Onishi H, Fujiwara K. Isolation and contraction of the stress fiber. Mol Biol Cell. 1998;9:1919–1938. doi: 10.1091/mbc.9.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophila melanogaster. J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Lutz MS, Chan D, Ketchum AS, Laymon RA, Nguyen B, Goldstein LS. Identification of the gene for fly non-muscle myosin heavy chain: Drosophila myosin heavy chains are encoded by a gene family. EMBO J. 1989;8:913–922. doi: 10.1002/j.1460-2075.1989.tb03452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kintner C. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs M, Wang F, Hu A, Zhang Y, Sellers JR. Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J Biol Chem. 2003;278:38132–38140. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- Kreplak L, Fudge D. Biomechanical properties of intermediate filaments: from tissues to single filaments and back. Bioessays. 2007;29:26–35. doi: 10.1002/bies.20514. [DOI] [PubMed] [Google Scholar]
- Kumar S, Maxwell IZ, Heisterkamp A, Polte TR, Lele TP, Salanga M, Mazur E, Ingber DE. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys J. 2006;90:3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- Leerberg JM, Gomez GA, Verma S, Moussa EJ, Wu SK, Priya R, Hoffman BD, Grashoff C, Schwartz MA, Yap AS. Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr Biol. 2014;24:1689–1699. doi: 10.1016/j.cub.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz M, Gardel ML, Dinner AR. Requirements for contractility in disordered cytoskeletal bundles. New J Phys. 2012;14 doi: 10.1088/1367-2630/14/3/033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S, Trybus KM. Common structural motifs for the regulation of divergent class II myosins. J Biol Chem. 2010;285:16403–16407. doi: 10.1074/jbc.R109.025551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum H, Malik AB. Mechanisms of increased endothelial permeability. Can J Physiol Pharmacol. 1996;74:787–800. doi: 10.1139/y96-081. [DOI] [PubMed] [Google Scholar]
- Luo W, Yu CH, Lieu ZZ, Allard J, Mogilner A, Sheetz MP, Bershadsky AD. Analysis of the local organization and dynamics of cellular actin networks. J Cell Biol. 2013;202:1057–1073. doi: 10.1083/jcb.201210123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Jana SS, Conti MA, Kawamoto S, Claycomb WC, Adelstein RS. Ablation of nonmuscle myosin II-B and II-C reveals a role for nonmuscle myosin II in cardiac myocyte karyokinesis. Mol Biol Cell. 2010;21:3952–3962. doi: 10.1091/mbc.E10-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre JL, Niwayama R, Turlier H, Nedelec F, Hiiragi T. Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat Cell Biol. 2015;17:849–855. doi: 10.1038/ncb3185. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, al-Shirawi DY, Ketchum AS, Newbern EC, Kiehart DP. Molecular organization and alternative splicing in zipper, the gene that encodes the Drosophila non-muscle myosin II heavy chain. J Mol Biol. 1996;255:98–109. doi: 10.1006/jmbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Goldstein B. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development. 2014;141:1987–1998. doi: 10.1242/dev.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci. 2011;108:4708–4713. doi: 10.1073/pnas.1011123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci. 1994;107(Pt 11):3077–3090. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Mofrad MRK, Kamm RD. Cellular mechanotransduction: Diverse perspectives from molecules to tissues. Cambridge University Press; New York: 2010. pp. 234–249. [Google Scholar]
- Munjal A, Philippe JM, Munro E, Lecuit T. A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature. 2015 doi: 10.1038/nature14603. [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Murrell MP, Gardel ML. F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc Natl Acad Sci. 2012;109:20820–20825. doi: 10.1073/pnas.1214753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A, Ishihara S, Tsukita S. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-alpha catenin fusion molecules. J Cell Biol. 1994;127:235–245. doi: 10.1083/jcb.127.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova O, Green KJ. Desmosome assembly and dynamics. Trends Cell Biol. 2013;23:537–546. doi: 10.1016/j.tcb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Banerjee S, Marchetti MC, Gardel ML. Geometry regulates traction stresses in adherent cells. Biophys J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- Peralta XG, Toyama Y, Hutson MS, Montague R, Venakides S, Kiehart DP, Edwards GS. Upregulation of forces and morphogenic asymmetries in dorsal closure during Drosophila development. Biophys J. 2007;92:2583–2596. doi: 10.1529/biophysj.106.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J Biol Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Aebi U, Cooper JA, Fowler WE, Tseng P. Actin structure, polymerization, and gelation. Cold Spring Harb Symp Quant Biol 46 Pt. 1982;2:513–524. doi: 10.1101/sqb.1982.046.01.048. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pouille PA, Ahmadi P, Brunet AC, Farge E. Mechanical signals trigger Myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci Signal. 2009;2:ra16. doi: 10.1126/scisignal.2000098. [DOI] [PubMed] [Google Scholar]
- Qin Z, Buehler MJ, Kreplak L. A multi-scale approach to understand the mechanobiology of intermediate filaments. J Biomech. 2010;43:15–22. doi: 10.1016/j.jbiomech.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014;158:143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Schuh R, Vestweber D, Eistetter H, Lottspeich F, Engel J, Dolz R, Jahnig F, Epplen J, Mayer S, et al. The structure of cell adhesion molecule uvomorulin. Insights into the molecular mechanism of Ca2+-dependent cell adhesion. EMBO J. 1987;6:3647–3653. doi: 10.1002/j.1460-2075.1987.tb02697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Itoh K, Adelstein RS, Bridgman PC. Localization of myosin II A and B isoforms in cultured neurons. J Cell Sci. 1995;108(Pt 12):3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- Sarpal R, Pellikka M, Patel RR, Hui FY, Godt D, Tepass U. Mutational analysis supports a core role for Drosophila alpha-catenin in adherens junction function. J Cell Sci. 2012;125:233–245. doi: 10.1242/jcs.096644. [DOI] [PubMed] [Google Scholar]
- Sawyer JK, Choi W, Jung KC, He L, Harris NJ, Peifer M. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol Biol Cell. 2011;22:2491–2508. doi: 10.1091/mbc.E11-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Harris NJ, Slep KC, Gaul U, Peifer M. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J Cell Biol. 2009;186:57–73. doi: 10.1083/jcb.200904001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Taylor KA, Kendrick-Jones J. Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature. 1980;287:233–235. doi: 10.1038/287233a0. [DOI] [PubMed] [Google Scholar]
- Sellers JR, Knight PJ. Folding and regulation in myosins II and V. J Muscle Res Cell Motil. 2007;28:363–370. doi: 10.1007/s10974-008-9134-0. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayoshi Y, Hatta K, Hosoda M, Tsunasawa S, Sakiyama F, Takeichi M. Cadherin cell adhesion molecules with distinct binding specificities share a common structure. EMBO J. 1986;5:2485–2488. doi: 10.1002/j.1460-2075.1986.tb04525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Rolo A, Chen X, Gumbiner BM, Keller R. Convergence and extension at gastrulation require a myosin IIB-dependent cortical actin network. Development. 2008;135:2435–2444. doi: 10.1242/dev.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares e Silva M, Depken M, Stuhrmann B, Korsten M, MacKintosh FC, Koenderink GH. Active multistage coarsening of actin networks driven by myosin motors. Proc Natl Acad Sci. 2011;108:9408–9413. doi: 10.1073/pnas.1016616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- Takeichi M, Atsumi T, Yoshida C, Uno K, Okada TS. Selective adhesion of embryonal carcinoma cells and differentiated cells by Ca2+-dependent sites. Dev Biol. 1981;87:340–350. doi: 10.1016/0012-1606(81)90157-3. [DOI] [PubMed] [Google Scholar]
- Tambe DT, Croutelle U, Trepat X, Park CY, Kim JH, Millet E, Butler JP, Fredberg JJ. Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS One. 2013;8:e55172. doi: 10.1371/journal.pone.0055172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. Physical forces during collective cell migration. Nature Physics. 2009;5:426–430. [Google Scholar]
- Van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol. 2014;36:157–165. doi: 10.1016/j.semcdb.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez CG, Tworoger M, Martin AC. Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. J Cell Biol. 2014;206:435–450. doi: 10.1083/jcb.201402004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Polarity sorting of actin filaments in cytochalasin-treated fibroblasts. J Cell Sci. 1997;110(Pt 15):1693–1704. doi: 10.1242/jcs.110.15.1693. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Newell-Litwa K, Bachir AI, Whitmore LA, Horwitz AR. Myosin IIA/IIB restrict adhesive and protrusive signaling to generate front-back polarity in migrating cells. J Cell Biol. 2011;193:381–396. doi: 10.1083/jcb.201012159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Martin AC. Intracellular signalling and intercellular coupling coordinate heterogeneous contractile events to facilitate tissue folding. Nat Commun. 2015;6:7161. doi: 10.1038/ncomms8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Totsukawa G, Yamakita Y, Sasaki Y, Madaule P, Ishizaki T, Narumiya S, Matsumura F. Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol Biol Cell. 2003;14:1745–1756. doi: 10.1091/mbc.E02-07-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Zhou J, Pal S, Maiti S, Davidson LA. Force production and mechanical accommodation during convergent extension. Development. 2015;142:692–701. doi: 10.1242/dev.116533. [DOI] [PMC free article] [PubMed] [Google Scholar]