Abstract
Natural history collections provide an immense record of biodiversity on Earth. These repositories have traditionally been used to address fundamental questions in biogeography, systematics, and conservation. However, they also hold the potential for studying evolution directly. While some of the best direct observations of evolution have come from long-term field studies or from experimental studies in the lab, natural history collections are providing new insights into evolutionary change in natural populations. By comparing phenotypic and genotypic changes in populations through time, natural history collections provide a window into evolutionary processes. Recent studies utilizing this approach have revealed some dramatic instances of phenotypic change over short time scales in response to presumably strong selective pressures. In some instances evolutionary change can be paired with environmental change, providing a context for potential selective forces. Moreover, in a few cases, the genetic basis of phenotypic change is well understood, allowing for insight into adaptive change at multiple levels. These kinds of studies open the door to a wide range of previously intractable questions by enabling the study of evolution through time, analogous to experimental studies in the laboratory, but amenable to a diversity of species over longer timescales in natural populations.
Keywords: natural history collections, evolution, environmental change, morphology, phenotypic change, genomics
Natural history collections as records of evolutionary change
Most of evolutionary biology is retrospective: we look at the present and attempt to make inferences about the past. For example, the distribution of a trait among species today can be used to infer the origin and evolution of that trait in the past. Similarly, patterns of genetic variation in present-day populations can be used to infer how allele frequencies have changed over the recent past. While powerful, these approaches are indirect, based on inference, and thus depend on assumptions of the underlying models. Observing evolutionary change directly is more difficult and typically comes from either laboratory studies of evolution or from very long-term field studies. Laboratory studies of experimental evolution over multiple generations are extremely powerful but are necessarily limited to organisms with short generation times (Barrick & Lenski 2013). These studies have mainly been conducted with microbes such as viruses (e.g. Wichman et al. 2005), bacteria (e.g. Wiser et al. 2013) and yeast (e.g. Lang et al. 2013) or with small multicellular organisms such as Drosophila (Burke et al. 2010). Since these studies rely on the relatively simple conditions of a laboratory environment it is often unclear how to extrapolate from them to the more complex conditions of natural populations. Long-term field studies have produced some spectacular successes with direct observations of evolutionary change in complex organisms (e.g. Coulsen et al. 2001, Grant & Grant 2002), but such studies often require decades of work in challenging field environments and are still relatively rare.
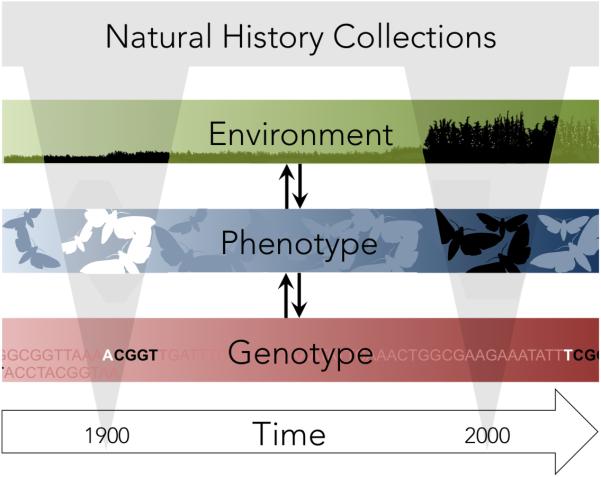
Natural history collections (NHCs) offer and alternative way to study evolution over reasonably long time scales (Figure 1). NHCs constitute an impressive record of life on earth with billions of specimens housed in museums around the world. These collections have provided a rich source of material for studies of phylogenetics, biogeography, conservation and many other areas of ecology and evolutionary biology (Suarez & Tsutsui 2004). However, the value of museum collections for studying the process of evolution directly–as genetic and phenotypic changes in populations through time–is less appreciated.
Figure 1.
Natural history collections provide specimens dating back, in some cases, over 100 years. These large repositories provide an opportunity to study evolution in response to changes in the environment. Whereas it has always been possible to study phenotypic change, new advances in genomic techniques are allowing researchers to also study the impacts of environmental change on genetic processes.
Collections provide a historical record of specimens against which modern samples can be compared, with many collections dating back 100 years or more. For organisms with short generation times, this represents a time scale over which substantial changes in allele frequency can be expected if selection is strong. Comparison of historical and modern samples can be used to study changes in phenotype, genotype, or both (Box I). If changes in the environment are well documented, in some cases it is even possible to ascribe observed changes to specific selective forces.
Here we review developments in this new field, highlighting the potential for NHCs to be used as resources to study evolutionary change directly. First, we document the extent of available collections in major museums and the associated data that enable this kind of study. Next, we describe some of the major anthropogenic environmental changes that are likely to have acted as agents of selection over the timescales in question. We then discuss the use of genetic and genomic tools to study evolutionary change using collections, focusing on empirical studies as well as theoretical approaches that utilize time-series data. Finally we discuss studies that have focused primarily on phenotypic changes, highlighting those with greatest potential to make links between genotypic and phenotypic change over time.
Natural history collections are large and diverse
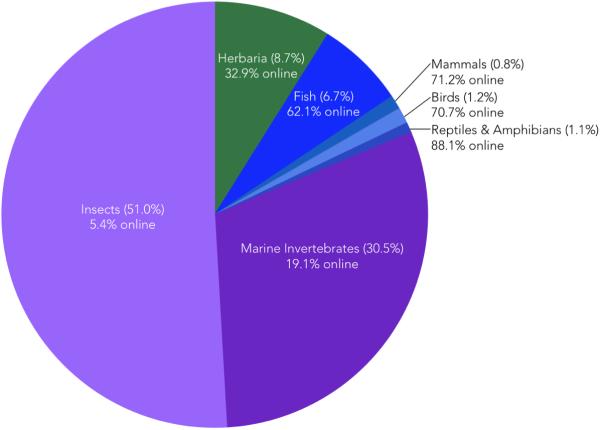
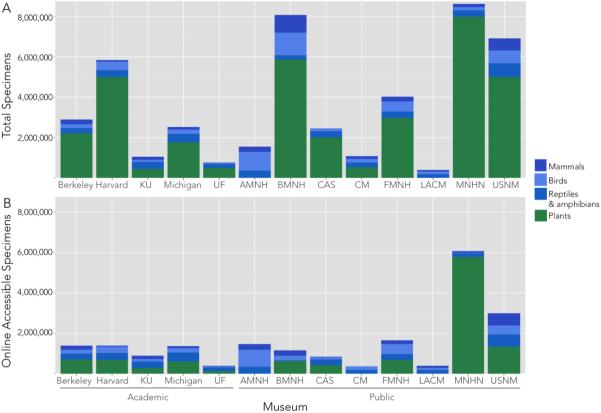
Natural history collections worldwide contain large numbers of plant and animal specimens (Figure 2). Nearly 90% of all specimens consist of insects, marine invertebrates, and fish, and these are usually preserved in “lots” consisting of groups of specimens. The holdings of select taxa in several of the best-known collections are summarized in Figure 3A. Specimens may include dry skins, skulls, skeletons, fluid-preserved whole organisms, frozen tissue samples, pressed plants, pinned insects, nests, eggs, seeds, scat and gut-contents, parasites, and cleared and stained organisms (Table 1). Different traits are preserved in this variety of specimens, permitting a wide range of studies. Since the 1970's, frozen tissues have often been preserved along with other kinds of specimens, facilitating molecular studies.
Figure 2.
Overview of holdings from a sample of thirteen well-known NHCs that together house approximately 392 million individual specimens. Holdings are colored by taxonomic group. The exact proportion of specimens accounted for by each group is listed in parenthesis; the percentage of each group that is digitally databased and online-accessible (“online”) is also listed. For fish, insects and other invertebrates, collections are typically made as lots, where individual lots may contain tens to thousands of specimens, thus, estimates for the number of specimens in these collections are approximate. The 13 NHCs summarized here are listed in the caption for Figure 3.
Figure 3.
Overview of available specimens in terrestrial vertebrate and herbaria collections in thirteen of the world's most diverse and well-known academic and public collections. (A) Total number of specimens in several major taxonomic groups. (B) Number of specimens that are digitally databased and available online. Numbers were acquired from museum websites, online databases, and/or from curators and are estimates,as collections and digitization efforts are constantly growing. Estimates for many herbaria include plants and fungi; we have not distinguished between these taxa here. This figure includes only non-lot-based taxonomic groups, which are easier to precisely quantify in a standardized manner. Abbreviations: Berkeley:Berkeley Natural History Museums, Berkeley; Harvard: Harvard University Herbaria and Museum of Comparative Zoology, Cambridge; KU: University of Kansas Biodiversity Institute, Lawrence; Michigan: University of Michigan Herbarium and Museum of Zoology, Ann Arbor; UF: Florida Museum of Natural History, Gainesville; AMNH: American Museum of Natural History, New York; BMNH: Natural History Museum, London; CAS: California Academy of Sciences, San Francisco; CM: Carnegie Museum, Pittsburgh; FMNH: Field Museum of Natural History, Chicago; LACM: Natural History Museum of Los Angeles County, Los Angeles; MNHN Muséum National d'Histoire Naturelle, Paris; USNM: National Museum of Natural History, Smithsonian Institution, Washington, D.C.
Table 1.
Types of data available in museum collections.
| Biological (storage method) | Common Taxa | Preserved traits |
|---|---|---|
| Fluid-preserved whole organisms (formalin, ethanol) | Fish, herpetofauna, invertebrates | Morphology, genetics |
| Fluid-preserved organism parts (e.g. gastrointestinal or reproductive tracts, stomach contents) (formalin, ethanol) | Mammals | Morphology, genetics, ecology |
| Frozen tissues (freezer, liquid nitrogen, ethanol, RNAlater) | Vertebrates | Genetics, isotopes |
| Pinned (dry) | Insects | Morphology, genetics |
| Pressings (dry) | Herbarium collections | Morphology, genetics |
| Seeds, spores (dry, frozen) | Herbarium collections | Morphology, genetics, isotopes, physiology, ecology, phenology |
| Skins (dry) | Birds, mammals | Morphology (e.g. color), isotopes, genetics, physiology, environmental contamination |
| Skeleton (dry) | Vertebrates | Morphology, genetics, isotopes |
| Nests (dry) | Birds | Behavior, ecology |
| Eggs, egg sacs (dry, formalin, ethanol) | Birds, insects, herpetofauna, | Morphology, ecology, environmental contamination |
In addition to the specimens themselves, collections often contain detailed supplementary data such as field notes, observations, geo-references, audio/video recordings and photographs (Table 2). Georeferences and photographs enable precise re-visitation of historically surveyed sites to provide contemporary points of reference. Field notes can be used to calculate sampling effort and measure population densities. Field notes and photographs may also provide insight into the changes in overall community structure over time. For example, along with collections of bird and mammal specimens from extensive surveys in the western United States, Grinnell and colleagues left detailed field notes and photographs of collecting localities (Grinnell 1943). These ancillary data allowed for precise site re-visitation and replication of survey methods in contemporary re-surveys, in addition to providing supplementary occurrence and non-occurrence data, all of which were essential to the documentation of extensive shifts in avian species richness over the last century in the Sierra Nevada mountains (Tingley et al. 2012, Tingley & Beissinger, 2013).
Table 2.
Types of metadata available in museum collections
| Metadata | Preserved information |
|---|---|
| Audio/Video Files | Vocalizations (e.g. specimen-linked bird/herp calls) Behavior Ecology (e.g. habitat associations, species interactions) |
| Field Notes, Observations | Trapping effort (population size/demographics), collecting method, distribution/biogeography, locality (time series site matching), habitat associations, species interactions, behavior |
| Geo-references/Maps | Distribution/biogeography, time series site-matching |
| Photographs | Time series site-matching Habitat change Color in life (especially herpetofauna) |
| Relationships | Genetic relationships, species interactions (e.g. parasite/host interactions, predator/prey interactions, mating behavior), habitat associations |
Importantly, NHCs can serve as repositories of evolutionary change because they often contain large samples from the same localities at different points in time. For example, Schroeder et al. (2009) examined specimens of the black-tailed godwit, Limosa limosa, from museums in the Netherlands and Denmark to reveal a decrease in the extent of male feather ornamentation over time, attributing this change to increased costliness of ornaments in human-dominated environments. Primack et al. (2004) showed that flowering date had advanced by an average of eight days in dozens of herbarium species over a hundred year period in Boston. Moritz et al. (2008) characterized changes in the distributions of mammals collected over 80 years in California, finding that half of the small mammal species in the Yosemite region have shifted their elevational ranges upslope. These kinds of studies highlight the importance of long-term data series from the same localities.
Data from many NHCs are increasingly available online (Figure 3B). Museum databases range from single- to multi-institutional and from taxon-specific (e.g. VertNet: Constable, 2010) to taxonomically broad (eg. GBIF: Flemons et al. 2007). In the best cases, online specimen records are detailed and can include morphological measurements, body condition indices, age and reproductive status, and georeferenced localities. Pressed plants often yield particularly complete digitized records, as many of their characteristics are easily captured online in an image or scan. Some current effort exists to create 3D scans of specimens that are available through databases such as iDigBio (Integrated Digitized Biocollections: https://www.idigbio.org/). Some collections also provide links to digitized field notes, audiovisual recordings, photographs, and, increasingly, genetic data via GenBank. Ecoinformatics, the field that integrates and increases accessibility of multiple lines of ecological and environmental data, will further advance the usefulness of online NHCs. Still, even the largest museums are not completely digitized. Terrestrial vertebrate collections, which are usually non-lot based and smaller than invertebrate and fish collections, are well-represented in online databases (Figure 3B), while huge efforts are currently underway to digitize the massive resources available in fish and invertebrate collections (e.g. CalBug, http://calbug.berkeley.edu), for which significant advances are being made (Fig. 2). Here we have focused on botanical and zoological collections of extant species, although we appreciate the wealth of data available in paleontological collections (Box 2).
Sampling Considerations
There are certain caveats to bear in mind when using NHCs to study evolutionary change (Boakes et al., 2010; Shaffer et al., 1998; Wandeler et al., 2007; Wehi et al., 2012). First, museum collecting is sometimes opportunistic and non-random and this may introduce sampling biases. In general, specimens are not proportionately distributed across taxonomy, geography, season, or age. For example, common or local species are more likely to be repeatedly and regularly sampled. In some cases, populations have been intensively sampled during a short period of time, but poorly sampled since that period; these populations could be targeted for current and future sampling to increase the pool of available time-series. Tropical areas are very diverse but are usually not proportionately represented in collections. For temperate areas, specimens are more likely to be collected during the summer than the winter. Adult specimens are generally more likely to be targeted than juveniles. If certain morphotypes of a species are more visible or easily captured they may be disproportionately represented in collections. These biases must be kept in mind when using specimens for demographic, distributional, and evolutionary studies (Wehi et al. 2012) and should help guide future collection efforts.
Some uses of museum specimens require “destructive sampling” in which a portion of a specimen is destroyed in order to obtain data. This applies to such things as frozen tissues or portions of specimens used for DNA studies or isotope analyses. Since specimens represent finite resources, most museums have strict policies governing destructive sampling and these may limit the availability of some samples.
Environmental drivers of change
Understanding the evolutionary process requires an understanding of links between genotype, phenotype, and environmental selective pressures (Figure 1). Though it can be difficult to attribute organismal change to a specific selective agent, recent studies suggest that five major anthropogenic causes of environmental change have been particularly important in the past century. Habitat destruction, including loss and fragmentation, is one of the most critical and immediate causes of biodiversity loss (Ewers & Didham 2006). Overexploitation, or exhaustive resource use via human hunting or harvesting has driven numerous recent extinctions (e.g. the great auk, Pinguinus impennis; Steller's sea cow, Hydrodamalis gigas; and the passenger pigeon, Ectopistes migratorius). Pollution has long been recognized as a major force of environmental change (Candelone et al. 1995) with extensive impacts on population structure (Blaustein et al. 2003) and evolution (e.g. Bradshaw & McNeilly 1981). Invasive species have similarly precipitated widespread ecological disruptions by outcompeting native species (Powell et al. 2013). These four agents can work together and in combination with the global phenomenon of anthropogenic climate change. Climate change has significantly altered Earth's ecosystems (Parmesan 2006) and undoubtedly set in motion a cascade of ecological and evolutionary events that await discovery. These drivers of environmental change are affecting biodiversity, altering everything from population genetic composition to community dynamics. By examining long-term collections from museum repositories we can elucidate the phenotypic and genotypic changes experienced by populations as they adapt to specific environmental changes.
Genetics and genomics
Over the last forty years, NHCs have shifted from almost exclusively housing traditional specimens (e.g. prepared skins) to an increasing focus on the preservation of tissues and genetic samples. This has opened the door to studies of molecular variation, including studies of changes in genotype frequencies through time. Most work to date has focused on documenting and understanding demographic changes, however, there is now great potential to make links between genotype and phenotype.
Allan Wilson and colleagues were the first to use museum specimens to make temporal comparisons of genetic variation (Thomas et al. 1990). They isolated mitochondrial DNA from 43 specimens of the Panamint kangaroo rat, Dipodomys panamintinus, from three populations collected between 1911 and 1937 and compared these to contemporary specimens collected in the same localities. The comparisons allowed them to ask whether populations were more connected spatially or temporally; they discovered that populations exhibited temporal continuity. This study was followed by many others that utilized museum specimens to examine genetic variation in historical and present Lozier & Cameron 2009; Pinsky et al. 2010; Spurgin et al. 2014, reviewed in Wandeler et al. 2007), including fossil and sub-fossil specimens (Box II). Many of these studies have documented declines in population size, although some have provided insight into more complex demographic processes (Groombridge et al. 2009; Lozier & Cameron 2009; Pinsky et al. 2010; Spurgin et al. 2014; Weber et al. 2000). For example, Pearse et al. (2011) suggested that migratory routes have been interrupted in steelhead trout, Oncorhynchus mykiss, from comparisons of modern-caught and historical specimens.
Most studies utilizing historical museum specimens have been limited to small fragments from a few loci. DNA from dried specimens is typically degraded and therefore difficult to PCR-amplify and sequence unless the target region is short (typically 100-200 bp). However, newer methods like Illumina sequencing are based on libraries with relatively short fragments and thus can be used on the degraded DNA in museum specimens (Bi et al. 2013; Rowe et al. 2011, Suchan et al. 2015, Hykin et al. 2015). For example, Bi et al. (2013) used exon-capture and Illumina sequencing to study patterns of genetic variation across the genome in historical and modern population samples of alpine chipmunks, Tamias alpinus T. alpinus has retracted its range in response to climate change (Moritz et al. 2008), and Bi et al. (2013) showed that modern samples exhibit more genetic subdivision compared to historical samples. This genome-scale work supported results from previous studies on T. alpinus based on fewer markers (Figure 4)(Rubidge et al. 2012).
Figure 4.
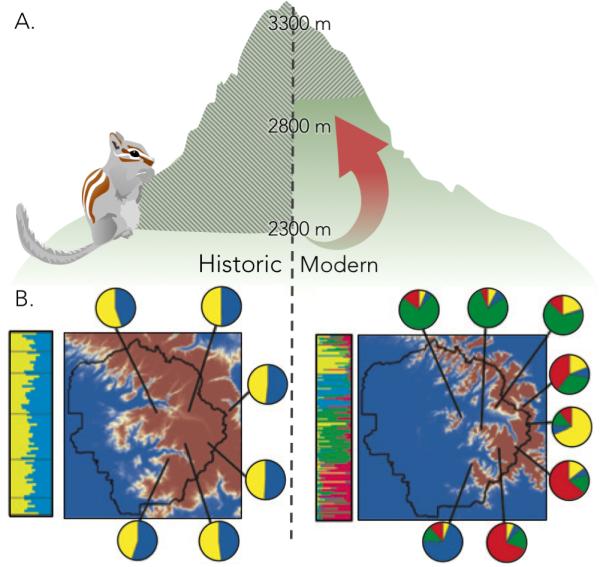
Ecological and genetic investigations on alpine chipmunks, Tamias alpinus, show the value of museum specimens for documenting ecological shifts and analyzing genetic change over time. (A) Cross-hatching indicates elevational range occupied by T. alpinus from the historical (left) to the modern (right) era. This species has retreated upslope as temperatures have warmed over the last century, isolating populations once open to gene flow. (B) Maps show Yosemite National Park (black outline) colored with the occupation probabilities (based on elevation) for T. alpinus (red: high probability, blue: low probability) in historical (left) and modern (right) eras. STRUCTURE barplots to the left of each map indicate population membership by individual; these data are split by site and represented in pie charts on the map. The greater diversity in pie charts in the modern samples compared to the historical samples is indicative of greater population subdivision. The availability of historical and modern specimens from the same localities was critical in revealing an increase in genetic subdivision of the alpine chipmunk populations over the past century (Rubidge et al. 2012). Figure based on data and figures from Rubidge et al., (2012) and Moritz et al.(2008).
The ability to sequence much or all of the genome from historical samples opens the door to new avenues of research. For example, just as genome scans for selection have been applied to make spatial comparisons between different present-day populations, temporal comparisons can be made for a single population across multiple points in time. FST-outlier and other approaches can be used to identify specific genes with allele frequencies that have shifted, identifying candidate genes for responses to environmental change. Following this logic, Mikheyev et al. (2015) recently sequenced the genomes of population samples of 32 colonies of honeybees collected near Ithaca, New York. Using museum specimens from the Cornell University Insect Collection, they compared populations from 1977, sampled before the introduction of an ectoparasitic mite (Varroa destructor), to those collected in 2010. They documented phenotypic changes in these bees and also conducted scans for selection, identifying genes that may underlie resistance to this mite. Notably, some of the identified genes overlap with previously identified quantitative trait loci (QTL) for resistance to Varroa mites.
This kind of temporal genome scan using museum specimens is conceptually similar to “evolve-and-resequence” studies in which experimental laboratory populations of organisms are selected for a particular trait (e.g. Turner et al. 2011) or allowed to evolve in a new environment (e.g. Orozco-ter Wengel et al. 2012) and then sequenced at different time points to identify changes in allele frequency (reviewed in Schlötterer et al. 2015). Sophisticated analytic methods have recently been developed to explicitly analyze data from evolve-and-resequence studies (e.g. Illingworth et al. 2012; Terhorst et al. 2015).
Population genetic approaches utilizing time-series data
Inferring selection or demographic parameters from multiple time points has advantages over inference from single time points. For example, sampling from multiple time points can provide resolution on the strength of selection based on the rate of allele frequency shifts over time. This permits quantitative estimates of selection coefficients as well as more accurate estimates of demographic parameters (Bank et al. 2014).
A number of theoretical models have been developed for the analysis of time-series data; the first of these focused on estimating changes in effective population size over time (e.g. Anderson et al. 2000; Krimbas & Tsakas 1971; Nei & Tajima 1981; Waples 1989; Williamson & Slatkin 1999). More recently, methods have been developed to detect selection, estimate selection coefficients, estimate the age of alleles, or estimate migration rates (e.g. Bollback et al. 2008; Feder et al. 2014; Malaspinas et al. 2012; Mathieson & McVean 2013). For example, Malaspinas et al. (2012) developed a likelihood method to jointly estimate the strength of selection and the age of an allele, while Mathieson & McVean (2013) developed a method for jointly estimating selection coefficients and migration rates in structured populations. Jensen and colleagues have developed a Wright-Fisher approximate Bayesian computational (ABC) method to infer selection coefficients and have used this to study the evolution of drug resistance in influenza in cultured cells (Foll et al. 2014). Foll et al. (2015) extended this approach to estimate the selection and dominance coefficient of the medionigra allele in the scarlet tiger moth, sampled over a 60-year period.
These new theoretical models hold great promise for the analysis of time-series data from collections. Despite these advances, some limitations remain. It is still difficult to distinguish between the signatures of direct and indirect selection, as well as to correctly infer the joint effects of demographic changes and selection. While maximum likelihood-based methods are considered to be more accurate with small selection coefficients, they are computationally intensive. Newly developed ABC-approaches, in contrast, are relatively computationally efficient, and thus hold promise for surveys of large genomic datasets (reviewed in Bank et al. 2014).
From phenotype to genotype
Below we highlight some of the best-documented examples of phenotypic change in botanical and zoological museum collections. This discussion is not intended to be comprehensive, but to illustrate the ways in which NHCs can be used to study evolution directly. In some cases the documented changes have a clear genetic basis and likely represent an adaptive response to changes in the biotic or abiotic environment. In others, phenotypic change may be due in part to plasticity, occurring without corresponding changes in underlying genotypes. We attempt to highlight examples where connections between genotype and phenotype have either already been made or could be made because the traits are amenable to genetic analysis. This discussion is by no means taxonomically exhaustive; for example, we have not included some major groups, like fungi and fish, although they are well-represented in many collections, and excellent studies exist (Box 1).
Plants
Because plants provide a three-dimensional habitat for many other organisms and are the source of energy in terrestrial ecosystems, their evolutionary responses to environmental change can have cascading effects on the food web and community structure. Plant phenotypes are sensitive to both temperature and atmospheric CO2 concentrations, two aspects of recent anthropogenic environmental disturbances.
Environmental changes can affect both the microscopic and macroscopic morphologies of leaves. For instance, carbon dioxide concentrations have been linked to stomatal density in plants (Beerling & Woodward 1996; Woodward 1987). The relationship between stomatal density and CO2 concentration has also been examined by comparing modern and historical specimens from herbaria (Beerling & Woodward 1996). Woodward (1987) measured stomatal densities of seven tree species and one shrub species collected over a 200-year timespan beginning in the mid 18th century. Although the species differed in total stomatal density, a general trend emerged: a reduction in stomatal density by approximately 40% over the time period represented. Although it is not clear whether the changes seen by Woodward (1987) were genetically based or reflect phenotypic plasticity, a number of papers have investigated the genetic basis of stomatal density, revealing high heritability and individual QTL of large effect (Gailing et al. 2008; Muir et al. 2014). Moreover, a sign test for QTL underlying stomatal density provided strong evidence that genetic changes seen between species were driven by positive selection (Muir et al. 2014). Finally, individual genes contributing to stomatal density have now been identified, principally in Arabidopsis (e.g. Engineer et al. 2014; Meng & Yao 2015; Shimada et al. 2011). This suggests an intriguing arena for future work: candidate genes for stomatal density could be surveyed (or genome-wide resequencing approaches could be used) in historical and modern samples to look for evidence of changes in allele frequency over time. Given the importance of stomatal density in regulating CO2 uptake, this would provide a direct link between climate change and the genetic basis of organismal response.
Plant phenotypes can also respond to environmental change at the macroscopic level. Guerin & Lowe (2013) analyzed 255 specimens of narrow-leaf hopbush, Dodonaea viscosa angustissima, from collections spanning 127 years. Measuring leaf width, they found a decrease of 2mm in this species. Furthermore, they were able to show that this phenotype is negatively correlated with maximum temperature. Leger (2013) studied herbarium collections of several species of plants spanning nearly 120 years to examine links between climate change and leaf morphology. Among these species she found a general decrease in plant size with varying responses in leaf size and flower number to temperature changes over time. As with stomatal density, the extent to which these long-term changes in phenotype reflect plasticity or genetics is unclear. Nonetheless, the wealth of candidate genes underlying leaf morphology (Hay & Tsiantis 2006; Juenger et al. 2005; Tsuge et al. 1996) provides an opportunity to study changes in allele frequency at these genes using herbarium collections.
Changes in plant phenology are another important response to climate change (Richardson et al. 2013; Wolkovich et al. 2012). A recent study used a common garden experiment to rear cornflowers, Centaurea cyanus, from seeds that had been collected from the same locality 18 years apart (Thomann et al. 2015). They found that flowering time had shifted and floral display increased in both size and duration. QST-FST comparisons suggest that selection has been acting, likely in response to climate warming and a decrease in pollinators (Thomann et al. 2015). Another recent study by Robbirt et al. (2011) examined flowering times of 192 early spider orchids, Ophyrs sphegodes, with specimens spanning two long-term collection times, 1848-1958 and 1975-2006. They found that flowering times in both periods advanced by 6 days per 1° C increase in mean spring temperature. The genetic basis of flowering time has been intensively studied (eg. Brachi et al. 2010; Chiang et al. 2009; Ehrenreich et al. 2009; Irwin et al. 2012; Srikanth & Schmid 2011), providing many candidate genes that could be studied in NHC specimens. Moreover, the opportunity in some cases to perform crosses between plants grown from preserved and modern seeds opens the door to directly studying the genetic basis of changes that have occurred (Franks et al. 2007). In principle, this approach could be applied to any species that can be preserved in a dormant state (Orsini et al. 2013), a principle that has recently been applied to examine evolution of thermal tolerance in Daphnia (Geerts et al. 2015).
Invertebrates
An iconic example of evolution in response to anthropogenic environmental change comes from studies of melanism in the peppered moth, Biston betularia, in Britain. Following the Industrial Revolution, observers noted an increase in frequency of the previously rare melanic morph of of B. betularia. Tutt (1896) was the first to suggest the role of pollution in driving the increase in melanism. Kettlewell (1955; 1956) later confirmed this hypothesis with field experiments showing increased predation on non-melanic morphs. He strengthened his argument about the link between pollution, predation, and melanism by using museum records, private collections, and other published work to obtain data on the presence and frequency of the melanic morph in several areas throughout Britain. He concluded that a correlation exists between melanism and heavy levels of industrialization, and called for further work on the genetic mechanisms underlying the spread of melanic genes (Kettlewell 1958). Kettlewell's work laid the foundation for an extensive body of work on industrial melanism (summarized in Majerus & Brakefield 1998) including one study that used museum specimens and material from private collections to investigate the genetics underlying melanism (van't Hof et al. 2011). Using both laboratory-reared moths and specimens collected from 1925-2009, the authors concluded that industrial melanism in Britain is the result of a single mutation that spread rapidly in response to the strong selective force of pollution (van't Hof et al. 2011).
Another striking example of the use of NHCs comes from the soapberry bug, Jadera haematoloma, which feeds by inserting its needle-shaped beak into fruits. The soapberry bug feeds on several native host plants, but is capable of expanding its diet to include introduced species. In the southern United States, Carroll & Boyd (1992) used museum specimens to document changes in beak length over 20-50 years (40-150 generations) following the introduction of three new host plants to the area. They showed an increase in beak length of museum and field-caught specimens over time consistent with expectations based on fruit size. To test whether these changes were genetic or plastic, reciprocal rearing experiments were conducted that showed support for host-dependent genetic differentiation (Carroll et al. 1997). More recently, QTL mapping has suggested that beak length in J. haematoloma may be controlled by a relatively small number of loci of moderate effect (Yu & Andrés 2014).
Because invertebrates have short generation times and are generally abundant in NHCs, many other invertebrate studies have been able to utilize collections to examine evolutionary changes over time; phenotypic changes have been attributed to climate change (Anderson et al. 2008; Babin-Fenske et al. 2008; Ożgo & Schilthuizen 2012; Timms et al. 2013), habitat fragmentation (Anderson et al. 2008; Norberg & Leimar 2002), and invasive species (Carroll & Boyd 1992), among other drivers. As more insect genomes are sequenced and annotated, insects will become a particularly suitable group in which the exploration of candidate genes could be combined with the unique temporal and high-resolution spatial dimensions provided by NHCs.
Reptiles and Amphibians
Although environmental change affects all taxa, amphibians and reptiles may be among the most susceptible of the vertebrates. Both are ectothermic and have relatively poor dispersal ability, which sensitizes them to temperature fluctuations while limiting their capacity to respond through range shifts (Sinervo et al. 2010). Amphibians have long been considered “canaries in the coal mine” for environmental change due to their permeable skin and a life history involving both aquatic and terrestrial stages (reviewed in Kerby et al. 2013). Reptiles with temperature-dependent sex determination may not be able to compensate behaviorally for climate change and may therefore experience skewed sex ratios and even demographic collapse (Refsnider et al. 2013). Furthermore, both amphibians and reptiles are experiencing elevated rates of extirpation and extinction often attributable to anthropogenic effects (Barnosky et al. 2011; Böhm et al. 2013).
Though much work on the response of reptiles and amphibians to environmental change has focused on phenology and associated reproductive behaviors such as nest depth (Urban et al. 2014), studies of morphological variation have demonstrated its close links to the ecology and fitness of herpetofauna (eg. Caruso et al. 2014; Losos 1990; Vidal-García et al. 2014). For example, Caruso et al. (2014) measured museum specimens from 15 species of Plethodon salamander collected between 1950 and 1996 and compared them to specimens collected between 2009 and 2011 to show that several species have undergone a considerable decrease in adult body size, possibly due to increasingly warm and dry habitats. In another study, hind limb length of Sceloporus undulatus has increased over seventy years across sites in the United States; this change was positively correlated with time since occupation by invasive fire ants, which can be better avoided by individuals with longer hind limbs (Langkilde 2009). Work in well-studied groups such as Anolis lizards has used common garden and laboratory experiments to tease out the roles of plasticity and genetics in determining lim length, body size, and other morphological traits (eg. Eales et al. 2010; Losos et al. 2000; Losos et al. 2001; Thorpe et al. 2005). From anoles we know that limb length is a largely plastic trait (Losos et al. 2000), and although body size has high broad-sense heritability (Berger et al. 1998), the genetic architecture remains uncharacterized. As more genomic resources become available for amphibians and reptiles it may become possible to elucidate the genetic components of some of these ecologically important traits.
Perhaps the best-known example of the use of NHCs to examine change over time in herpetological specimens comes from studies of chytrid fungus, Batrachochytrium dendrobatidis (Bd) infection, in amphibians. Although the earliest studies of Bd involved identifying and characterizing the nature of the infection and subsequent extirpation of amphibian populations (Berger et al. 1998; Lips et al. 2006) more recent studies have used museum specimens and genetic techniques (PCR and qPCR) to detect and characterize historical chytrid infections from around the world (Bataille et al. 2013; Crawford et al. 2010; Lips et al. 2008; Shaw et al. 2013; Swei et al. 2011). For example, one study tested for Bd in museum specimens of 38 Central American amphibian species collected before their decline and during the year of their decline, and revealed many cases of coincident Bd emergence and population collapse (Cheng et al. 2011). Bd infection likely exerts a strong selective force on any population that it does not completely eliminate. Although the nature of the immune response to Bd infection is complicated and varies among individuals and species (Ellison et al. 2014a; Ellison et al. 2014b; Rosenblum et al. 2012), some work has found a potential role for MHC variability in resistance (Bataille et al. 2015; Savage & Zamudio 2011). Future work could use museum specimens to assess immunogenetic diversity in populations before and after Bd infection to test hypotheses about specific genes that may contribute to susceptibility or resistance.
Partially because of the limited number of herpetological genomes available, genomic studies of evolutionary change in these animals have been constrained. Nonetheless, due to their strong susceptibility to environmental change, work on amphibians and reptiles has the potential to be particularly useful, and in some cases NHCs may provide the only remaining records of populations that have recently gone extinct.
Birds
The visibility and phenotypic variability of birds has long made them a subject of particular interest among researchers and the public alike. This interest has driven a number of excellent resources for ornithological study, including long-term banding records, extensive citizen science datasets, and large, well-curated ornithological collections. In combination with the extensive genomic resources recently made available through the 50 genomes project (Jarvis et al. 2014; Zhang et al. 2014), there is potential to make direct phenotype-genotype links for birds. For example, ornithological museum specimens have been used to study environmentally-induced changes in color and plumage over time (Alström et al. 2015; Galeotti et al. 2009). In one elegant study, Karell et al. (2011) used museum specimens and field observations to show that in less snowy years, brown-morph tawny owls, Strix alco, have a survival advantage over grey morphs. Using field data, they demonstrated a high heritability of these plumage types. Finally, they found that as annual snow cover decreased over a thirty-year period due to increasing temperatures, the frequency of brown morph individuals increased in comparison to the grey morphs (Figure 5). An exciting direction for potential future study could be to assay the large number of previously identified candidate pigmentation genes to make direct links between genes, phenotypic response, and environmental change.
Figure 5.
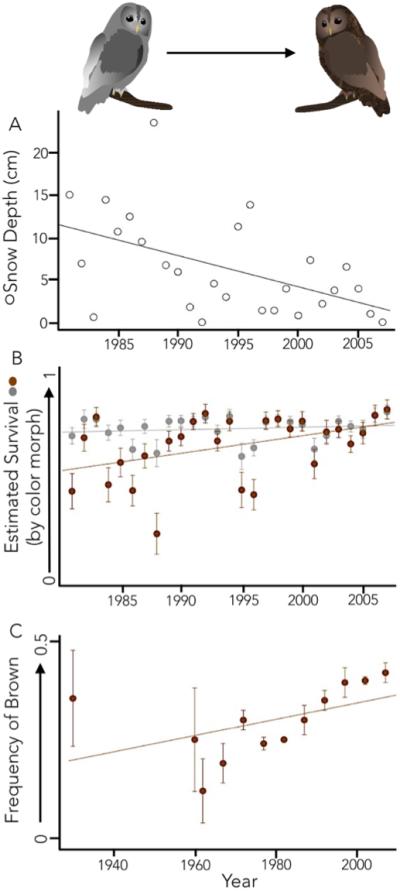
Work on tawny owls using museum and field-identified specimens indicates that plumage coloration can be affected by changes in climate. Brown owls are poorly camouflaged against snow, thus, selection against the brown morph is typically strongest in heavy-snow winters; this selection has been relaxed as snow levels have decreased due to climate change. (A) A decrease in snow depth (open circles) over a twenty-seven year period was documented. (B) Using mark-recapture modeling, survival estimates for grey and brown morphs were modeled. Graph shows estimated survival for grey (grey circles) and brown (brown circles) morphs, illustrating increased survival for brown morphs in years with less snow. (C) Using museum specimens and field-collected data, the frequency of the brown morph in the wild was found to have increased over time. Note that the x-axis for (C) differs from (A) and (B). Figure based on data from Karellet al. (2011).
Body size is another morphological trait in avian species that has been linked to environmental change. For example, Gardner et al. (2009) examined specimens of eight species of passerine birds collected from 1860 to 2001 and documented a decrease in body size of 1.8-3.6%, which the authors attribute to global warming. They suggest the observed decrease in body size represents evolutionary change and is not a result of diet-induced plasticity. These results are consistent with other studies showing a decrease in body sizes of birds that may be correlated with increased local temperatures (Yom-Tov 2001; Yom-Tov & Yom-Tov 2006).
Habitat modification has also been linked to phenotypic changes in birds studied in NHCs. Desrochers (2010) examined 21 species of temporal and boreal forest passerines collected between 1900 and 2008. He found a direct correlation between habitat fragmentation and an increase in the projections of outer wing feathers. Fragmented habitats lead to a more pointed wing shape, which is generally observed in birds with longer migratory distance and movement (Senar et al. 1994; Winkler & Leisler 1992). In contrast, birds that were found in habitats with less fragmentation showed a decrease in primary project length, suggesting reduced long-distance movement. Though the authors did not directly explore whether these changes were genetic or plastic, previous studies in other bird species have identified genetic contributions to wing length and shape (Schielzeth et al. 2012), including one study that found a single QTL that explained over one third of the phenotypic variation in wing length in great reed warblers (Tarka et al. 2010).
Anthropogenic environmental change, particularly climate change, can also be expected to affect migratory behaviors in birds (Guttal & Couzin 2010), and some changes in migratory patterns have already been observed (eg. Pulido & Berthold 2010). While migratory phenotypes are not directly discernible in museum specimens, collection dates assign individuals to particular locations at particular times and thus can provide indirect information about migratory patterns. Moreover, it may be possible to assay historical and contemporary NHC specimens for candidate migratory genes (Kuhn et al. 2013; Liedvogel et al. 2011; Mueller et al. 2011).
Mammals
Like birds, mammals tend to be highly represented and well studied in museum collections. For example, mammal specimens have recently been used to study changes in body size in response to climatic shifts (Eastman et al 2012; Sonne et al. 2013). In contrast to expectations based on thermal ecology, mammals do not show a consistent decrease in body size in response to increasing temperature. Pergams & Lawler (2009) examined rodents from 28 NHCs representing 25 species from four continents. Rapid phenotypic changes were detected in skull and body size, but changes varied by region and were uncorrelated with climate variables and human population impact. Whether these changes are genetically or plastically controlled is often unclear, and in general, exploring the genetic underpinnings of body size is challenging since it is typically a highly polygenic trait (Allen et al. 2010; Chan et al. 2012; Kemper et al. 2012; Perola 2011) and susceptible to environmental influences (Boutin & Lane 2014).
Changes in mammalian cranial morphology have recently been studied in a number of species using museum specimens (Snell-Rood & Wick 2013; Tomassini et al. 2014; Holmes et al. 2015). However, the genetic basis of skull shape is complex (Burgio et al. 2012; Maga et al. 2015), and skull shape is also known to show considerable plasticity during development (Gonzalez et al. 2011; Noback & Harvati 2015). Changes in dentition have also been studied using museum collections. For example, Szuma (2003) examined changes in dental traits of the red fox, Vulpes vulpes, collections of which spanned over 70 years. Significant changes in dentition were found in occlusal surfaces of premolars and incisors, which the authors suggested might be driven by an increase in the types of food available for V. vulpes due to historical land-use changes. Although tooth development appears to be polygenic (Thesleff 2006), studies in both humans and mice have associated anomalous dental phenotypes with specific mutations; for example WNT10A mutations result in supernumerary molars, molars with fewer cusps, and incisors with distinctive apical-lingual wedge shapes (Yang et al. 2015). Candidate gene approaches would be useful for linking changes in dental phenotypes over time to specific mutations.
Conclusions and future directions
Evolution is rarely directly observable, but comparisons of specimens collected at different points in time enable us to document and understand evolution in a way that is typically not possible in the lifetime of a single researcher. Understanding the genetic basis of evolutionary change requires that we make links from genotype to phenotype and from phenotype to environment. We envision a number of important future directions for making these links in the context of museum collections.
First, repeated sampling of populations that have been well sampled in the past will help identify specific phenotypic changes. Detailed historical records of biotic and abiotic conditions can be compared with current conditions to assess changes in the environment that may have driven changes in phenotype. For example, the PRISM climate data (http://prism.oregonstate.edu) contains climate records from 1895 to the present. Second, candidate genes can now be surveyed for many traits, and some of these will lead to specific genotype-phenotype links, which can then be followed through time. Although the relevance of genes identified from laboratory mutants to natural populations has been questioned (Palopoli & Patel 1996), it is clear that there have been many successes of this approach, especially for traits that are not highly polygenic (Barrett & Hoekstra 2011). Third, the ability to generate sequence data across the genome from historical specimens (Bi et al. 2013, Suchan et al. 2015) opens up many exciting areas. The obvious drawback of a candidate gene approach is that it focuses on a particular phenotype and limits discovery to genes that are already known. In contrast, genome scans are blind with respect to phenotype and can help identify genes that may have been under selection but were not previously suspected. Such genes may then point to phenotypic differences that may be important in evolution. In this way, museum specimens may provide a natural counterpart to the increasingly common “evolve-and-resequence” studies that have been carried out in the laboratory.
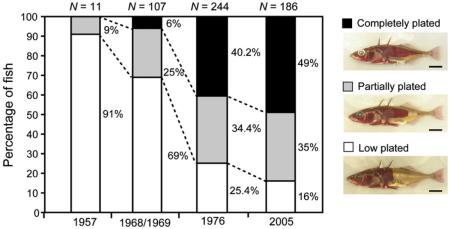
Box I. Case study: changes in stickleback armor over 50 years.
Kitano and colleagues examined threespine stickleback fish, Gasterosteus aculeatus, specimens from Lake Washington, WA to show that over a 48-year period (equivalent to 48 stickleback generations) the frequency of the completely plated morph had significantly increased (Kitano et al. 2008). The graph shows the proportion of completely plated (black) to partially plated (gray) and low plated (white) fish at four time points (1957, 1958-9, 1976, 2005); sample sizes are listed on the top of each bar. In combination with behavioral tests and lake transparency data collected over time, these data lead the authors to hypothesize that an increase in lake clarity due to the prevention of eutrophication had allowed trout to increase their predation on sticklebacks, acting as a selective force for increased plating. Genotyping revealed that the EDA gene largely controls this morphotype. This example, which combines morphological data from museum specimens, environmental data collected over time, and genetic analyses, illustrates the power of placing a time series of museum specimens in an ecological context to demonstrate adaptive evolutionary change. Figure reproduced from Kitano et al. (2008).
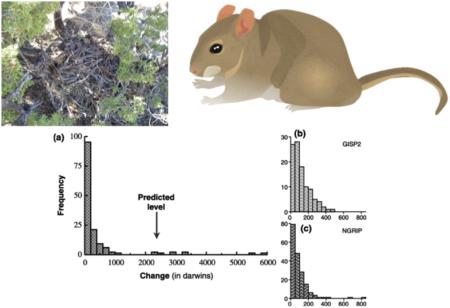
Box II. Woodrat body size and historical temperature.
Paleontological collections can also be used to study evolutionary change over time. The use of fossil and subfossil specimens allows for an assessment of phenotypic and genotypic change over long time periods and in response to dramatic and prolonged environmental transformations. Both morphological and genetic studies are possible with fossil and subfossil specimens.
The first study using ancient DNA (aDNA) sequenced a 229 bp fragment of mtDNA from a museum specimen of the extinct quagga (Higuchi et al. 1984). This sparked a flurry of studies utilizing aDNA from fossil and subfossil specimens. More recent studies have documented population genetic changes using heterochronous aDNA from temporal series of fossil and modern specimens (Chan et al. 2006; Hadly et al. 2004; Ramakrishnan et al. 2005; Ramakrishnan & Hadly 2009). Both exome capture (Bi et al. 2013) and targeted bait capture techniques (Carpenter et al. 2013; Mamanova et al. 2010; Peñalba et al. 2014) can generate large numbers of loci or SNPs from many individuals allowing for population-level studies in fossil organisms. The efficacy of targeted enrichment protocols for aDNA has been recently demonstrated with some samples yielding as many as 150k SNPs (Carpenter et al. 2013). These technologies provide the foundation for the nascent field of paleopopulation genetics (Wall & Slatkin 2012), adding to our knowledge of how species responded to environmental changes in recent geologic times (reviewed in Hofreiter and Stewart (2009) and de Bruyn et al. (2011)).
Morphology-based studies have been able to draw explicit links between body size proxies and environmental (climate) change for extant species (Blois et al. 2008; Hadly 1997; McGuire 2010; Smith et al. 1995). In a landmark study, Smith et al. (1995) examined the change in body size of woodrats (Neotoma spp.) over thirty thousand years. Woodrats, illustrated in the figure above (top right) next to a typical Neotoma midden (top left), show a tight negative correlation between body size and ambient temperature, a primary adaptation to climate change in Neotoma species. Broad-sense heritability estimates for body size are high (H2 > 0.8 (Smith & Betancourt 2006)), indicating that this trait is under substantial genetic control. Smith & Betancourt (2006) showed that body size change in woodrats from many geographic sites rapidly tracks climate change. The distribution of body size changes during the Holocene, measured in darwins, across 55 sites in the Western U.S. is shown in graph (a). The predicted rate of phenotypic change expected under current estimates of climate change for the next 100 years is shown with an arrow and falls within the range of rates of change observed during the Holocene. Graphs (b) and (c) depict histograms of changes in oxygen isotopes estimated from the GISP2 ice core and the NGRIP ice core respectively; higher oxygen isotope levels correspond to warmer temperatures. The distributions of inferred temperature changes (b and c) are similar to the distribution of body size changes (a). This series of studies on woodrats used fossilized fecal pellets and historical and modern museum specimens to create a highly integrative picture of environmentally-driven evolutionary change over time. Figure reproduced from Smith & Betancourt (2006).
Acknowledgements
We thank the curators and staff of the museums included in Figures 2 and 3 for providing us with information on collection sizes, Patrik Karell for sharing data, and Jim Patton, Bruce Baldwin, Kip Will, and two anonymous reviewers for helpful comments on the manuscript. Funding was provided by the Museum of Vertebrate Zoology, Department of Integrative Biology, and Graduate School at the University of California, Berkeley. Additional financial support was provided by an NSF GK-12 grant to MWH, REW, and LMB, a Chancellor's Fellowship from U.C. Berkeley to JCC, NSF Graduate Research Fellowships to TTH, KL, and JCC, NSF-DEB 1120356 to GOUW, and NIH RO1 GM074245 to MWN.
Works Cited
- Allen HL, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alström P, Jøønsson KA, Fjeldså J, et al. Dramatic niche shifts and morphological change in two insular bird species. Royal Society Open Science. 2015;2:140364. doi: 10.1098/rsos.140364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Williamson EG, Thompson EA. Monte Carlo evaluation of the likelihood for Ne from temporally spaced samples. Genetics. 2000;156:2109–2118. doi: 10.1093/genetics/156.4.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Conrad KF, Gillman MP, Woiwod IP, Freeland JR. Phenotypic changes and reduced genetic diversity have accompanied the rapid decline of the garden tiger moth (Arctia caja) in the UK. Ecological Entomology. 2008;33:638–645. [Google Scholar]
- Babin-Fenske J, Anand M, Alarie Y. Rapid morphological change in stream beetle museum specimens correlates with climate change. Ecological Entomology. 2008;33:646–651. [Google Scholar]
- Bank C, Ewing GB, Ferrer-Admettla A, Foll M, Jensen JD. Thinking too positive? Revisiting current methods of population genetic selection inference. Trends in Genetics. 2014;30:540–546. doi: 10.1016/j.tig.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Barnosky AD, Matzke N, Tomiya S, et al. Has the Earth's sixth mass extinction already arrived? Nature. 2011;471:51–57. doi: 10.1038/nature09678. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nature Reviews Genetics. 2013;14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille A, Cashins SD, Grogan L, et al. Susceptibility of amphibians to chytridiomycosis is associated with MHC class II conformation. Proceedings of the Royal Society of London B: Biological Sciences. 2015;282:20143127. doi: 10.1098/rspb.2014.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille A, Fong JJ, Cha M, et al. Genetic evidence for a high diversity and wide distribution of endemic strains of the pathogenic chytrid fungus Batrachochytrium dendrobatidis in wild Asian amphibians. Molecular Ecology. 2013;22:4196–4209. doi: 10.1111/mec.12385. [DOI] [PubMed] [Google Scholar]
- Beerling DJ, Woodward FI. Stomatal density responses to global environmental change. In: Stanhill G, editor. Advances in Bioclimatology. Springer-Verlag; Berlin: 1996. pp. 171–221. [Google Scholar]
- Berger L, Speare R, Daszak P, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proceedings of the National Academy of Sciences. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K, Linderoth T, Vanderpool D, et al. Unlocking the vault: next- generation museum population genomics. Molecular Ecology. 2013;22:6018–6032. doi: 10.1111/mec.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein AR, Romansic JM, Kiesecker JM, Hatch AC. Ultraviolet radiation, toxic chemicals and amphibian population declines. Diversity and Distributions. 2003;9:123–140. [Google Scholar]
- Blois JL, Feranec RS, Hadly EA. Environmental influences on spatial and temporal patterns of body-size variation in California ground squirrels (Spermophilus beecheyi). Journal of Biogeography. 2008;35:602–613. [Google Scholar]
- Boakes EH, McGowan PJ, Fuller RA, Chang-qing D, Clark NE, O'Connor K, Mace GM. Distorted views of biodiversity: spatial and temporal bias in species occurrence data. PLoS Biol. 2010;8(6):e1000385. doi: 10.1371/journal.pbio.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm M, Collen B, Baillie JEM, et al. The conservation status of the world's reptiles. Biological Conservation. 2013;157:372–385. [Google Scholar]
- Bollback JP, York TL, Nielsen R. Estimation of 2Nes from temporal allele frequency data. Genetics. 2008;179:497–502. doi: 10.1534/genetics.107.085019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin S, Lane JE. Climate change and mammals: evolutionary versus plastic responses. Evolutionary Applications. 2014;7:29–41. doi: 10.1111/eva.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Faure N, Horton M, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genetics. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw AD, McNeilly T. Evolution and Pollution Edward Arnold, London. 1981 [Google Scholar]
- Burke MK, Dunham JP, Shahrestani P, Thornton KR, Rose MR, et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature. 2010;467:587–590. doi: 10.1038/nature09352. [DOI] [PubMed] [Google Scholar]
- Burgio G, Baylac M, Heyer E, Montagutelli X. Nasal bone shape is under complex epistatic genetic control in mouse interspecific recombinant congenic strains. PloS One. 2012;7:e37721. doi: 10.1371/journal.pone.0037721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelone J-P, Hong S, Pellone C, Boutron CF. Post-Industrial Revolution changes in large-scale atmospheric pollution of the northern hemisphere by heavy metals as documented in central Greenland snow and ice. Journal of Geophysical Research: Atmospheres. 1995;100:16605–16616. [Google Scholar]
- Carpenter ML, Buenrostro JD, Valdiosera C, et al. Pulling out the 1%: Whole-Genome Capture for the Targeted Enrichment of Ancient DNA Sequencing Libraries. The American Journal of Human Genetics. 2013;93:852–864. doi: 10.1016/j.ajhg.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SP, Boyd C. Host race radiation in the soapberry bug: natural history with the history. Evolution. 1992;46:1052–1069. doi: 10.1111/j.1558-5646.1992.tb00619.x. [DOI] [PubMed] [Google Scholar]
- Carroll SP, Dingle H, Klassen SP. Genetic differentiation of fitness-associated traits among rapidly evolving populations of the soapberry bug. Evolution. 1997;51:1182–1188. doi: 10.1111/j.1558-5646.1997.tb03966.x. [DOI] [PubMed] [Google Scholar]
- Caruso NM, Sears MW, Adams DC, Lips KR. Widespread rapid reductions in body size of adult salamanders in response to climate change. Global Change Biology. 2014;20:1751–1759. doi: 10.1111/gcb.12550. [DOI] [PubMed] [Google Scholar]
- Chan YF, Jones FC, McConnell E, et al. Parallel selection mapping using artificially selected mice reveals body weight control loci. Current Biology. 2012;22:794–800. doi: 10.1016/j.cub.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Chan YL, Anderson CNK, Hadly EA. Bayesian estimation of the timing and severity of a population bottleneck from ancient DNA. PLoS Genetics. 2006;2:e59. doi: 10.1371/journal.pgen.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TL, Rovito SM, Wake DB, Vredenburg VT. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proceedings of the National Academy of Sciences. 2011;108:9502–9507. doi: 10.1073/pnas.1105538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2009;106:11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constable H, Guralnick R, Wieczorek J, et al. VertNet: a new model for biodiversity data sharing. PLoS Biology. 2010;8:e1000309. doi: 10.1371/journal.pbio.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson T, Catchpole EA, Albon SD, Morgan BJT, Pemberton JM, Clutton-Brock TH, Crawley MJ, Grenfell BT. Age, sex, density, winter weather, and population crashes in Soay sheep. Science. 2001;292:1528–1531. doi: 10.1126/science.292.5521.1528. [DOI] [PubMed] [Google Scholar]
- Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proceedings of the National Academy of Sciences. 2010;107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn M, Hoelzel AR, Carvalho GR, Hofreiter M. Faunal histories from Holocene ancient DNA. Trends in Ecology & Evolution. 2011;26:405–413. doi: 10.1016/j.tree.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Desrochers A. Morphological response of songbirds to 100 years of landscape change in North America. Ecology. 2010;91:1577–1582. doi: 10.1890/09-2202.1. [DOI] [PubMed] [Google Scholar]
- Eales J, Thorpe RS, Malhotra A. Colonization history and genetic diversity: adaptive potential in early stage invasions. Molecular Ecology. 2010;19:2858–2869. doi: 10.1111/j.1365-294X.2010.04710.x. [DOI] [PubMed] [Google Scholar]
- Eastman LM, Morelli TL, Rowe KC, Conroy CJ, Moritz C. Size increase in high elevation ground squirrels over the last century. Global Change Biology. 2012;18:1499–1508. [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, et al. Candidate gene association mapping of Arabidopsis flowering time. Genetics. 2009;183:325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison AR, Savage AE, DiRenzo GV, et al. Fighting a losing battle: vigorous immune response countered by pathogen suppression of host defenses in the chytridiomycosis-susceptible frog Atelopus zeteki. Genes| Genomes| Genetics. 2014a;4:1275–1289. doi: 10.1534/g3.114.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison AR, Tunstall T, DiRenzo GV, et al. More than skin deep: functional genomic basis for resistance to amphibian chytridiomycosis. Genome Biology and Evolution. 2014b;7:286–298. doi: 10.1093/gbe/evu285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer CB, Ghassemian M, Anderson JC, et al. Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature. 2014;513:246–250. doi: 10.1038/nature13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers RM, Didham RK. Confounding factors in the detection of species responses to habitat fragmentation. Biological Reviews. 2006;81:117–142. doi: 10.1017/S1464793105006949. [DOI] [PubMed] [Google Scholar]
- Feder AF, Kryazhimskiy S, Plotkin JB. Identifying signatures of selection in genetic time series. Genetics. 2014;196:509–522. doi: 10.1534/genetics.113.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemons P, Guralnick R, Krieger J, et al. A web-based GIS tool for exploring the world's biodiversity: The Global Biodiversity Information Facility Mapping and Analysis Portal Application (GBIF:MAPA). Ecological Informatics. 2007;2:49–60. [Google Scholar]
- Foll M, Poh Y-P, Renzette N, et al. Influenza virus drug resistance: a time-sampled population genetics perspective. PLoS Genetics. 2014;10:e1004185. doi: 10.1371/journal.pgen.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Shim H, Jensen JD. WFABC: a Wright–Fisher ABC-based approach for inferring effective population sizes and selection coefficients from time746 sampled data. Molecular Ecology Resources. 2015;15:87–98. doi: 10.1111/1755-0998.12280. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proceedings of the National Academy of Sciences. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailing O, Langenfeld-Heyser R, Polle A, Finkeldey R. Quantitative trait loci affecting stomatal density and growth in a Quercus robur progeny: implications for the adaptation to changing environments. Global Change Biology. 2008;14:1934–1946. [Google Scholar]
- Galeotti P, Rubolini D, Sacchi R, Fasola M. Global changes and animal phenotypic responses: melanin-based plumage redness of scops owls increased with temperature and rainfall during the last century. Biology Letters. 2009;11:532–534. doi: 10.1098/rsbl.2009.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JL, Heinsohn R, Joseph L. Shifting latitudinal clines in avian body size correlate with global warming in Australian passerines. Proceedings of the Royal Society B: Biological Sciences. 2009;276:3845–3852. doi: 10.1098/rspb.2009.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts A, Vanoverbeke J, Vanschoenwinkel B, et al. Rapid evolution of thermal tolerance in the water flea Daphnia. Nature Climate Change. 2015;5:665–668. [Google Scholar]
- Gonzalez PN, Hallgrímsson B, Oyhenart EE. Developmental plasticity in covariance structure of the skull: effects of prenatal stress. Journal of Anatomy. 2011;218:243–257. doi: 10.1111/j.1469-7580.2010.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin's finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- Grinnell J. Joseph Grinnell's Philosophy of Nature. University of California Press; 1943. p. 237. [Google Scholar]
- Groombridge JJ, Dawson DA, Burke T, et al. Evaluating the demographic history of the Seychelles kestrel (Falco araea): genetic evidence for recovery from a population bottleneck following minimal conservation management. Biological Conservation. 2009;142:2250–2257. [Google Scholar]
- Guerin GR, Lowe AJ. Leaf morphology shift: new data and analysis support climate link. Biology Letters. 2013;9:882–886. doi: 10.1098/rsbl.2012.0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttal V, Couzin ID. Social interactions, information use, and the evolution of collective migration. Proceedings of the National Academy of Sciences. 2010;107:16172–16177. doi: 10.1073/pnas.1006874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadly EA. Evolutionary and ecological response of pocket gophers (Thomomys talpoides) to late-Holocene climatic change. Biological Journal of the Linnean Society. 1997;60:277–296. [Google Scholar]
- Hadly EA, Ramakrishnan U, Chan YL, et al. Genetic response to climatic change: insights from ancient DNA and phylochronology. PLoS Biology. 2004;2:e290. doi: 10.1371/journal.pbio.0020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nature Genetics. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. DNA sequences from the quagga, an extinct member of the horse family. Nature. 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Stewart J. Ecological change, range fluctuations and population dynamics during the Pleistocene. Current Biology. 2009;19:R584–R594. doi: 10.1016/j.cub.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Holmes MW, Boykins GKR, Bowie RCK, Lacey EA. Cranial morphological variation in Peromyscus maniculatus over nearly a century of environmental change in three areas of California. Journal of Morphology. 2015 doi: 10.1002/jmor.20482. DOI:10.1002/jmor.20482. [DOI] [PubMed] [Google Scholar]
- Hykin SM, Bi K, Mcguire JA. Fixing formalin: a method to recover genomic-scale DNA sequence data from formalin-fixed museum specimens using high-throughput sequencing. PLoS One. 2015 doi: 10.1371/journal.pone.0141579. DOI:10.137/journal.pone.0141579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth CJR, Parts L, Schiffels S, Liti G, Mustonen V. Quantifying selection acting on a complex trait using allele frequency time series data. Molecular Biology and Evolution. 2012;29:1187–1197. doi: 10.1093/molbev/msr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JA, Lister C, Soumpourou E, et al. Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC Plant Biology. 2012;12:21. doi: 10.1186/1471-2229-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Pérez-Pérez JM, Bernal S, Micol JL. Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: evidence for modular genetic architecture. Evolution & Development. 2005;7:259–271. doi: 10.1111/j.1525-142X.2005.05028.x. [DOI] [PubMed] [Google Scholar]
- Karell P, Ahola K, Karstinen T, Valkama J, Brommer JE. Climate change drives microevolution in a wild bird. Nature Communications. 2011;2:1–7. doi: 10.1038/ncomms1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper KE, Visscher PM, Goddard ME. Genetic architecture of body size in mammals. Genome Biology. 2012;13:1–13. doi: 10.1186/gb-2012-13-4-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerby JL, Richards-Hrdlicka KL, Storfer A, Skelly DK. An examination of amphibian sensitivity to environmental contaminants: are amphibians poor canaries? Ecology Letters. 2010;13:60–67. doi: 10.1111/j.1461-0248.2009.01399.x. [DOI] [PubMed] [Google Scholar]
- Kettlewell HBD. Selection experiments on industrial melanism in the Lepidoptera. Heredity. 1955;9:323–342. [Google Scholar]
- Kettlewell HBD. Further selection experiments on industrial melanism in the Lepidoptera. Heredity. 1956;10:287–301. [Google Scholar]
- Kettlewell HBD. A survey of the frequencies of Biston betularia (L.)(Lep.) and its melanic forms in Great Britain. Heredity. 1958;12:51–72. [Google Scholar]
- Kitano J, Bolnick DI, Beauchamp DA, et al. Reverse evolution of armor plates in the threespine stickleback. Current Biology. 2008;18:769–774. doi: 10.1016/j.cub.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Krimbas CB, Tsakas S. The genetics of Dacus oleae. V. changes of esterase polymorphism in a natural population following insecticide control-selection or drift? Evolution. 1971;25:454–460. doi: 10.1111/j.1558-5646.1971.tb01904.x. [DOI] [PubMed] [Google Scholar]
- Kuhn K, Schwenk K, Both C, et al. Differentiation in neutral genes and a candidate gene in the pied flycatcher: using biological archives to track global climate change. Ecology and Evolution. 2013;3:4799–4814. doi: 10.1002/ece3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langkilde T. Invasive fire ants alter behavior and morphology of native lizards. Ecology. 2009;90:208–217. doi: 10.1890/08-0355.1. [DOI] [PubMed] [Google Scholar]
- Leger EA. Annual plants change in size over a century of observations. Global Change Biology. 2013;19:2229–2239. doi: 10.1111/gcb.12208. [DOI] [PubMed] [Google Scholar]
- Liedvogel M, Åkesson S, Bensch S. The genetics of migration on the move. Trends in Ecology & Evolution. 2011;26:561–569. doi: 10.1016/j.tree.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Lips KR, Brem F, Brenes R, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proceedings of the National Academy of Sciences. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips KR, Diffendorfer J, Mendelson Iii JR, Sears MW. Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biology. 2008;6:441–454. doi: 10.1371/journal.pbio.0060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. The evolution of form and function: morphology and locomotor performance in West Indian Anolis lizards. Evolution. 1990;44:1189–1203. doi: 10.1111/j.1558-5646.1990.tb05225.x. [DOI] [PubMed] [Google Scholar]
- Losos JB, Creer DA, Glossip D, et al. Evolutionary implications of phenotypic plasticity in the hindlimb of the lizard Anolis sagrei. Evolution. 2000;54:301–305. doi: 10.1111/j.0014-3820.2000.tb00032.x. [DOI] [PubMed] [Google Scholar]
- Losos JB, Schoener TW, Warheit KI, Creer D. In: Microevolution Rate, Pattern, Process. Springer; 2001. Experimental studies of adaptive differentiation in Bahamian Anolis lizards. pp. 399–415. [PubMed] [Google Scholar]
- Lozier JD, Cameron SA. Comparative genetic analyses of historical and contemporary collections highlight contrasting demographic histories for the bumble bees Bombus pensylvanicus and B. impatiens in Illinois. Molecular Ecology. 2009;18:1875–1886. doi: 10.1111/j.1365-294X.2009.04160.x. [DOI] [PubMed] [Google Scholar]
- Maga AM, Navarro N, Cunningham ML, Cox TC. Quantitative trait loci affecting the 3D skull shape and size in mouse and prioritization of candidate genes in-silico. Frontiers in Physiology. 2015;6:1–13. doi: 10.3389/fphys.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus MEN, Brakefield PM. Melanism: Evolution in Action. Oxford University Press; Oxford: 1998. [Google Scholar]
- Malaspinas A-S, Malaspinas O, Evans SN, Slatkin M. Estimating allele age and selection coefficient from time-serial data. Genetics. 2012;192:599–607. doi: 10.1534/genetics.112.140939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, et al. Target-enrichment strategies for next-generation sequencing. Nature Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- Mathieson I, McVean G. Estimating selection coefficients in spatially structured populations from time series data of allele frequencies. Genetics. 2013;193:973–984. doi: 10.1534/genetics.112.147611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire JL. Geometric morphometrics of vole (Microtus californicus) dentition as a new paleoclimate proxy: Shape change along geographic and climatic clines. Quaternary International. 2010;212:198–205. [Google Scholar]
- Meng LS, Yao SQ. Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA). Plant Biotechnology Journal. 2015;38:1–10. doi: 10.1111/pbi.12324. [DOI] [PubMed] [Google Scholar]
- Mikheyev AS, Tin MMY, Arora J, Seeley TD. Museum samples reveal rapid evolution by wild honey bees exposed to a novel parasite. Nature Communications. 2015;6:1–8. doi: 10.1038/ncomms8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Patton JL, Conroy CJ, et al. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science. 2008;322:261–264. doi: 10.1126/science.1163428. [DOI] [PubMed] [Google Scholar]
- Mueller JC, Pulido F, Kempenaers B. Identification of a gene associated with avian migratory behaviour. Proceedings of the Royal Society B: Biological Sciences. 2011;278:2848–2856. doi: 10.1098/rspb.2010.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir CD, Pease JB, Moyle LC. Quantitative genetic analysis indicates natural selection on leaf phenotypes across wild tomato species (Solanum sect. Lycopersicon; Solanaceae). Genetics. 2014;198:1629–1643. doi: 10.1534/genetics.114.169276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Tajima F. Genetic drift and estimation of effective population size. Genetics. 1981;98:625–640. doi: 10.1093/genetics/98.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noback ML, Harvati K. The contribution of subsistence to global human cranial variation. Journal of Human Evolution. 2015;80:34–50. doi: 10.1016/j.jhevol.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Norberg U, Leimar O. Spatial and temporal variation in flight morphology in the butterfly Melitaea cinxia (Lepidoptera: Nymphalidae). Biological Journal of the Linnean Society. 2002;77:445–453. [Google Scholar]
- Orozco-ter Wengel P, Kapun M, Nolte V, et al. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Molecular Ecology. 2012;21:4931–4941. doi: 10.1111/j.1365-294X.2012.05673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini L, Schwenk K, De Meester L, et al. The evolutionary time machine: using dormant propagules to forecast how populations can adapt to changing environments. Trends in Ecology & Evolution. 2013;28:274–282. doi: 10.1016/j.tree.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oźgo M, Schilthuizen M. Evolutionary change in Cepaea nemoralis shell colour over 43 years. Global Change Biology. 2012;18:74–81. [Google Scholar]
- Palopoli MF, Patel NH. Neo-Darwinian developmental evolution: can we bridge the gap between pattern and process? Current Opinion in Genetics & Development. 1996;6:502–508. doi: 10.1016/s0959-437x(96)80074-8. [DOI] [PubMed] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution, and Systematics. 2006;37:637–669. [Google Scholar]
- Pearse DE, Martinez E, Garza JC. Disruption of historical patterns of isolation by distance in coastal steelhead. Conservation Genetics. 2011;12:691–700. [Google Scholar]
- Peñalba JV, Smith LL, Tonione MA, et al. Sequence capture using PCR-generated probes: a cost-effective method of targeted high-throughput sequencing for nonmodel organisms. Molecular Ecology Resources. 2014;14:1000–1010. doi: 10.1111/1755-0998.12249. [DOI] [PubMed] [Google Scholar]
- Pergams ORW, Lawler JJ. Recent and widespread rapid morphological change in rodents. PloS One. 2009;4:e6452. doi: 10.1371/journal.pone.0006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perola M. Genome-wide association approaches for identifying loci for human height genes. Best Practice & Research Clinical Endocrinology & Metabolism. 2011;25:19–23. doi: 10.1016/j.beem.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Pinsky ML, Newsome SD, Dickerson BR, et al. Dispersal provided resilience to range collapse in a marine mammal: insights from the past to inform conservation biology. Molecular Ecology. 2010;19:2418–2429. doi: 10.1111/j.1365-294X.2010.04671.x. [DOI] [PubMed] [Google Scholar]
- Powell KI, Chase JM, Knight TM. Invasive plants have scale-dependent effects on diversity by altering species-area relationships. Science. 2013;339:316–318. doi: 10.1126/science.1226817. [DOI] [PubMed] [Google Scholar]
- Primack D, Imbres C, Primack RB, Miller-Rushing AJ, Del Tredici P. Herbarium specimens demonstrate earlier flowering times in response to warming in Boston. American Journal of Botany. 2004;91:1260–1264. doi: 10.3732/ajb.91.8.1260. [DOI] [PubMed] [Google Scholar]
- Pulido F, Berthold P. Current selection for lower migratory activity will drive the evolution of residency in a migratory bird population. Proceedings of the National Academy of Sciences. 2010;107:7341–7346. doi: 10.1073/pnas.0910361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan U, Hadly EA, Mountain JL. Detecting past population bottlenecks using temporal genetic data. Molecular Ecology. 2005;14:2915–2922. doi: 10.1111/j.1365-294X.2005.02586.x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan UMA, Hadly EA. Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Molecular Ecology. 2009;18:1310–1330. doi: 10.1111/j.1365-294X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Refsnider JM, Bodensteiner BL, Reneker JL, Janzen FJ. Nest depth may not compensate for sex ratio skews caused by climate change in turtles. Animal Conservation. 2013;16:481–490. [Google Scholar]
- Richardson AD, Keenan TF, Migliavacca M, et al. Climate change, phenology, and phenological control of vegetation feedbacks to the climate system. Agricultural and Forest Meteorology. 2013;169:156–173. [Google Scholar]
- Robbirt KM, Davy AJ, Hutchings MJ, Roberts DL. Validation of biological collections as a source of phenological data for use in climate change studies: a case study with the orchid Ophrys sphegodes. Journal of Ecology. 2011;99:235–241. [Google Scholar]
- Rosenblum EB, Poorten TJ, Settles M, Murdoch GK. Only skin deep: shared genetic response to the deadly chytrid fungus in susceptible frog species. Molecular Ecology. 2012;21:3110–3120. doi: 10.1111/j.1365-294X.2012.05481.x. [DOI] [PubMed] [Google Scholar]
- Rowe KC, Singhal S, Macmanes MD, et al. Museum genomics: low-cost and high-accuracy genetic data from historical specimens. Molecular Ecology Resources. 2011;11:1082–1092. doi: 10.1111/j.1755-0998.2011.03052.x. [DOI] [PubMed] [Google Scholar]
- Rubidge EM, Patton JL, Lim M, et al. Climate-induced range contraction drives genetic erosion in an alpine mammal. Nature Climate Change. 2012;2:285–288. [Google Scholar]
- Savage AE, Zamudio KR. MHC genotypes associate with resistance to a frog-killing fungus. Proceedings of the National Academy of Sciences. 2011;108:16705–16710. doi: 10.1073/pnas.1106893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schielzeth H, Forstmeier W, Kempenaers B, Ellegren H. QTL linkage mapping of wing length in zebra finch using genome-wide single nucleotide polymorphisms markers. Molecular Ecology. 2012;21:329–339. doi: 10.1111/j.1365-294X.2011.05365.x. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Kofler R, Versace E, Tobler R, Franssen SU. Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity. 2015;114:431–440. doi: 10.1038/hdy.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J, Lourenço PM, Hooijmeijer JCEW, Both C, Piersma T. A possible case of contemporary selection leading to a decrease in sexual plumage dimorphism in a grassland-breeding shorebird. Behavioral Ecology. 2009;20:797–807. [Google Scholar]
- Senar JC, Lleonart J, Metcalfe NB. Wing-shape variation between resident and transient wintering siskins Carduelis spinus. Journal of Avian Biology. 1994;25:50–50. [Google Scholar]
- Shaffer HB, Fisher RN, Davidson C. The role of natural history collections in documenting species declines. Trends in Ecology & Evolution. 1998;13(1):27–30. doi: 10.1016/s0169-5347(97)01177-4. [DOI] [PubMed] [Google Scholar]
- Shaw SD, Skerratt LF, Haigh A, et al. The distribution and host range of Batrachochytrium dendrobatidis in New Zealand, 1930-2010. Ecology. 2013;94:2108–2111. [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I. Positive and negative peptide signals control stomatal density. Cellular and Molecular Life Sciences. 2011;68:2081–2088. doi: 10.1007/s00018-011-0685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinervo B, Mendez-De-La-Cruz F, Miles DB, et al. Erosion of lizard diversity by climate change and altered thermal niches. Science. 2010;328:894–899. doi: 10.1126/science.1184695. [DOI] [PubMed] [Google Scholar]
- Smith FA, Betancourt JL. Predicting woodrat (Neotoma) responses to anthropogenic warming from studies of the palaeomidden record. Journal of Biogeography. 2006;33:2061–2076. [Google Scholar]
- Smith FA, Betancourt JL, Brown JH. Evolution of body size in the woodrat over the past 25,000 years of climate change. Science. 1995;270:2012–2014. [Google Scholar]
- Snell-Rood EC, Wick N. Anthropogenic environments exert variable selection on cranial capacity in mammals. Proceedings of the Royal Society B: Biological Sciences. 2013;280:1–9. doi: 10.1098/rspb.2013.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonne C, Bechshøft TØ, Riget FF, et al. Size and density of East Greenland polar bear (Ursus maritimus) skulls: valuable bio-indicators of environmental changes? Ecological Indicators. 2013;34:290–295. [Google Scholar]
- Spurgin LG, Wright DJ, Velde M, et al. Museum DNA reveals the demographic history of the endangered Seychelles warbler. Evolutionary Applications. 2014;7:1134–1143. doi: 10.1111/eva.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez AV, Tsutsui ND. The value of museum collections for research and society. BioScience. 2004;54:66–74. [Google Scholar]
- Suchan T Pitteloud C, Gerasimova N, Kostikova A, Arrigo N, Pajkovic M, Ronikier M, Alvarez N. Hybridization capture using RAD probes (hyRAD), a new tool for performing genomic analyses on museum collection specimens. bioRxiv. 2015 doi: 10.1371/journal.pone.0151651. DOI: http://dx.doi.org/10.1101/025551. [DOI] [PMC free article] [PubMed]
- Swei A, Rowley JJL, Rödder D, et al. Is Chytridiomycosis an emerging infectious disease in Asia? PloS One. 2011;6:e23179. doi: 10.1371/journal.pone.0023179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuma E. Microevolutionary trends in the dentition of the red fox (Vulpes vulpes). Journal of Zoological Systematics and Evolutionary Research. 2003;41:47–56. [Google Scholar]
- Tarka M, Åkesson M, Beraldi D, et al. A strong quantitative trait locus for wing length on chromosome 2 in a wild population of great reed warblers. Proceedings of the Royal Society of London B: Biological Sciences. 2010;277:2361–2369. doi: 10.1098/rspb.2010.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J, Schlötterer C, Song YS. Multi-locus analysis of genomic time series data from experimental evolution. PLoS Genetics. 2015;11:e1005069. doi: 10.1371/journal.pgen.1005069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I. The genetic basis of tooth development and dental defects. American Journal of Medical Genetics Part A. 2006;140:2530–2535. doi: 10.1002/ajmg.a.31360. [DOI] [PubMed] [Google Scholar]
- Thomann M, Imbert E, Engstrand RC, Cheptou PO. Contemporary evolution of plant reproductive strategies under global change is revealed by stored seeds. Journal of Evolutionary Biology. 2015;28:766–778. doi: 10.1111/jeb.12603. [DOI] [PubMed] [Google Scholar]
- Thomas WK, Pääbo S, Villablanca FX, Wilson AC. Spatial and temporal continuity of kangaroo rat populations shown by sequencing mitochondrial DNA from museum specimens. Journal of Molecular Evolution. 1990;31:101–112. doi: 10.1007/BF02109479. [DOI] [PubMed] [Google Scholar]
- Thorpe RS, Reardon JT, Malhotra A. Common garden and natural selection experiments support ecotypic differentiation in the Dominican anole (Anolis oculatus). The American Naturalist. 2005;165:495–504. doi: 10.1086/428408. [DOI] [PubMed] [Google Scholar]
- Timms LL, Bennett AMR, Buddle CM, Wheeler TA. Assessing five decades of change in a high Arctic parasitoid community. Ecography. 2013;36:1227–1235. [Google Scholar]
- Tingley MW, Koo MS, Moritz C, Rush AC, Beissinger SR. The push and pull of climate change causes heterogeneous shifts in avian elevational ranges. Global Change Biology. 2012;18(11):3279–3290. [Google Scholar]
- Tingley MW, Beissinger SR. Cryptic loss of montane avian richness and high community turnover over 100 years. Ecology. 2013;94(3):598–609. doi: 10.1890/12-0928.1. [DOI] [PubMed] [Google Scholar]
- Tomassini A, Colangelo P, Agnelli P, Jones G, Russo D. Cranial size has increased over 133 years in a common bat, Pipistrellus kuhlii: a response to changing climate or urbanization? Journal of Biogeography. 2014;41:944–953. [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development. 1996;122:1589–1600. doi: 10.1242/dev.122.5.1589. [DOI] [PubMed] [Google Scholar]
- Turner TL, Stewart AD, Fields AT, Rice WR, Tarone AM. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genetics. 2011;7:e1001336. doi: 10.1371/journal.pgen.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt JW. British Moths. Routledge; London: 1896. [Google Scholar]
- Urban MC, Richardson JL, Freidenfelds NA. Plasticity and genetic adaptation mediate amphibian and reptile responses to climate change. Evolutionary Applications. 2014;7:88–103. doi: 10.1111/eva.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Hof AE, Edmonds N, Dalíková M, Marec F, Saccheri IJ. Industrial melanism in British peppered moths has a singular and recent mutational origin. Science. 2011;332:958–960. doi: 10.1126/science.1203043. [DOI] [PubMed] [Google Scholar]
- Vidal-García M, Byrne PG, Roberts JD, Keogh JS. The role of phylogeny and ecology in shaping morphology in 21 genera and 127 species of Australo-Papuan myobatrachid frogs. Journal of Evolutionary Biology. 2014;27:181–192. doi: 10.1111/jeb.12292. [DOI] [PubMed] [Google Scholar]
- Wall JD, Slatkin M. Paleopopulation genetics. Annual Review of Genetics. 2012;46:635–649. doi: 10.1146/annurev-genet-110711-155557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandeler P, Hoeck PEA, Keller LF. Back to the future: museum specimens in population genetics. Trends in Ecology & Evolution. 2007;22:634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Waples RS. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics. 1989;121:379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DS, Stewart BS, Garza JC, Lehman N. An empirical genetic assessment of the severity of the northern elephant seal population bottleneck. Current Biology. 2000;10:1287–1290. doi: 10.1016/s0960-9822(00)00759-4. [DOI] [PubMed] [Google Scholar]
- Wehi PM, Whaanga H, Trewick SA. Artefacts, biology and bias in museum collection research. Molecular Ecology. 2012;21:3103–3109. doi: 10.1111/j.1365-294x.2012.05589.x. [DOI] [PubMed] [Google Scholar]
- Williamson EG, Slatkin M. Using maximum likelihood to estimate population size from temporal changes in allele frequencies. Genetics. 1999;152:755–761. doi: 10.1093/genetics/152.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, Millstein J, Bull JJ. Adaptive molecular evolution for 13,000 phage generations a possible arms race. Genetics. 2005;170:19–31. doi: 10.1534/genetics.104.034488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Leisler B. On the ecomorphology of migrants. Ibis. 1992;134:21–28. [Google Scholar]
- Wiser MJ, Ribeck N, Lenski RE. Long-term dynamics of adaptation in asexual populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- Wolkovich EM, Cook BI, Allen JM, et al. Warming experiments underpredict plant phenological responses to climate change. Nature. 2012;485:494–497. doi: 10.1038/nature11014. [DOI] [PubMed] [Google Scholar]
- Woodward FI. Stomatal numbers are sensitive to increases in CO2 from pre-industrial levels. Nature. 1987;327:617–618. [Google Scholar]
- Yang J, Wang SK, Choi M, et al. Taurodontism, variations in tooth number, and misshapened crowns in Wnt10a null mice and human kindreds. Molecular Genetics & Genomic Medicine. 2015;3:40–58. doi: 10.1002/mgg3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yom-Tov Y. Global warming and body mass decline in Israeli passerine birds. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2001;268:947–952. doi: 10.1098/rspb.2001.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yom-Tov Y, Yom-Tov S. Decrease in body size of Danish goshawks during the twentieth century. Journal of Ornithology. 2006;147:644–647. [Google Scholar]
- Yu Y, Andrés JA. Genetic architecture of contemporary adaptation to biotic invasions: quantitative trait locus mapping of beak reduction in soapberry bugs. Genes| Genomes| Genetics. 2014;4:255–264. doi: 10.1534/g3.113.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]