Abstract
Conversion of the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) to the secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA) is performed by a few species of intestinal bacteria in the genus Clostridium through a multistep biochemical pathway that removes a 7α-hydroxyl group. The rate-determining enzyme in this pathway is bile acid 7α-dehydratase (baiE). In this study, we report crystal structures of apo-BaiE and its putative product-bound (3-oxo-Δ4,6- lithocholyl-Coenzyme A (CoA)) complex. BaiE is a trimer with a twisted α+β barrel fold with similarity to the Nuclear Transport Factor 2 (NTF2) superfamily. Tyr30, Asp35 and His83 form a catalytic triad that is conserved across this family. Site-directed mutagenesis of BaiE from Clostridium scindens VPI 12708 confirmed that these residues are essential for catalysis and also confirmed the importance of other conserved residues, Tyr54 and Arg146, which are involved in substrate binding and affect catalytic turnover. Steady state kinetic studies revealed that the BaiE homologs are able to turn over 3-oxo-Δ4-bile acid and CoA-conjugated 3-oxo-Δ4-bile acid substrates with comparable efficiency questioning the role of CoA-conjugation in the bile acid metabolism pathway.
Keywords: Gut microbes, secondary bile acid synthesis, gut microbe mediated human metabolite, 7α-dehyroxylation, bile acid 7α-dehydratase, primary bile acid, secondary bile acid, nuclear transport factor-2 superfamily
Introduction
The human gut microbiome is a key aspect of normal physiology and metabolic homeostasis. A large number of normal host functions and disease states have been associated with intestinal microbiota 1–5. Understanding the interplay between host and microbe is a pivotal challenge since the underlying biochemical events are undefined. Metabolomics studies performed with germ-free and conventional mice have identified metabolites processed by microbes, which can influence human health 6–8. Among such metabolites, secondary bile acids, deoxycholate (DCA) and lithocholate (LCA), synthesized from primary bile acids cholate and chenodeoxycholate, respectively, are associated with an increase in the incidence of cancers of the colon, esophagus and biliary track as well as cholesterol gallstone disease in some patients 9–12. Furthermore, recent studies of obesity-associated hepatocellular carcinoma (HCC) in mice, implicated DCA with increased secretion of various inflammatory and tumor promoting factors in the liver. 13 This study showed that blocking DCA production could prevent HCC in obese mice.
Bile acids also act as hormones via activating specific nuclear receptors (FXR, PXR and Vitamin D Receptor) and plasma membrane G-protein coupled receptors (TGR-5, sphingosine-1-phosphate receptor 2, muscarinic M (2) receptor) in a physiologically relevant manner. Secondary bile acids are more powerful than primary bile acids as activators of PXR, Vitamin D receptor and TGR-5 in cell signaling pathways.14 In humans, unlike rodents, DCA can accumulate to very high levels (> 60%) in the bile acid pool as the human liver is unable to hydroxylate DCA at the 7α-position. 15 Therefore, by controlling the composition of the bile acid pool, specific gut bacteria may influence aspects of host physiology and influence pathophysiology. Hence, the microbial enzymes involved in secondary bile acid synthesis are potential drug targets for human disease.
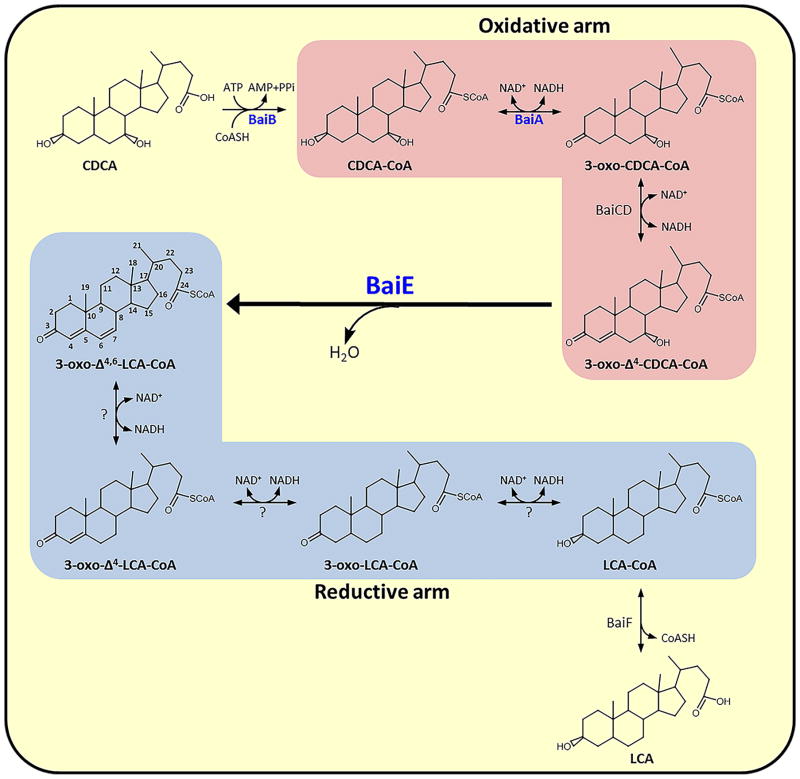
Microbial synthesis of secondary bile acids in the human gut involves a multistep biochemical process that results in the removal of C7-hydroxy group from primary bile acids (Fig. 1). The pathway, split into oxidative and reductive arms, involves a series of enzymes that are encoded by a bile acid inducible (bai) operon 15. The pathway is initiated by the uptake of primary bile acid via a H+-dependent active transporter (BaiG) 16 followed by thioesterification to coenzyme A (CoA) by a CoA-ligase (BaiB) 17. The oxidative arm of the pathway converts bile acid-CoA thioester to 3-oxo-7α-hydroxy bile acid-CoA and 3-oxo-Δ4-7α-hydroxy bile acid-CoA intermediates by BaiA18 and BaiCD19, respectively. The 7α-hydroxy group is removed from the latter through release of water by bile acid 7α dehydratase, BaiE, yielding a 3-oxo-Δ4,6-bile acid-CoA intermediate. 20 The subsequent three reductive steps catalyzed by yet unidentified enzymes constitute the reductive arm generating the secondary bile acid. Two CoA transferases, BaiF and BaiK, participate in the removal of the CoA moiety, although it remains unclear at which stage in the pathway this occurs. 21 Despite the significant influence of this pathway on human health and disease, limited structural and biochemical information is available for the key microbial enzymes involved. We recently reported the structural and steady-state kinetic characterization of BaiA2 from C. scindens VPI 12708, which represents the first structure-function characterization of enzymes in this pathway. 22
Figure 1. Proposed steps involved in synthesis of secondary bile acid in human gut anaerobe, Clostridium sp.
The oxidative and reductive arms of the pathway are enclosed within salmon and blue colored boxes, respectively. Enzymes with crystal structures solved using the JCSG pipeline are denoted in blue.
Here we report the structural and biochemical characterization of BaiE homologs from Clostridium sp. a key human gut microbe associated with secondary bile acid synthesis. The 7α-dehydroxylation step catalyzed by BaiE is rate limiting for this pathway and is the only irreversible step that directs the intermediate from the oxidative arm to the reductive arm generating the secondary bile acid. BaiE has <30% sequence identity with its nearest structural homolog. The BaiE crystal structure reveals a canonical twisted α+β barrel fold that is also observed in the Nuclear Transport Factor 2 (NTF2) family. Structure-guided site directed mutagenesis studies were carried out to identify and differentiate residues involved in substrate binding versus those specifically involved in catalysis. Furthermore, comparison of the co-crystal structure of BaiE from Clostridium hiranonis DSM13275 with the partially bound product (3-oxo-Δ4,6-Lithocholyl Coenzyme A (3-oxo-Δ4,6-LC-CoA)) with apo-structures of BaiE homologs revealed a dramatically different conformation of the loop formed by residues 48–63. This segment forms the roof of the active site pocket and suggests conformational flexibility that may be needed for substrate binding. Site-directed mutagenesis of Tyr54 indicated impaired substrate turnover indicating the importance of this loop in catalysis. A mechanism for bile acid dehydratase catalysis is proposed based on structural features and site-directed mutagenesis studies. The insight from these structures should facilitate the design of inhibitors which could modulate levels of secondary bile acids and thereby gene expression in pathways associated with human health and disease.
Materials and Methods
Gene Cloning and Site-directed Mutagenesis
Genes of the BaiE homologs were cloned from the corresponding genomic DNAs of C. scindens VPI12708, C. scindens ATCC35704, C. hylemonae DSM15053 and C. hiranonis DSM13275 by Polymerase Incomplete Primer Extension (PIPE) cloning method.23,24 C. scindens VPI12708 genomic DNA was isolated from anaerobically grown culture 25. All other genomic DNAs were obtained from the DSMZ (Braunschweig, Germany). The genes were amplified by polymerase chain reaction (PCR) and cloned into the expression vector, pSpeedET, which also encodes an N-terminal tobacco etch virus (TEV) protease-cleavable expression and purification (MGSDKIHHHHHHENLYFQ/G). Expression clones for PDB 4L8P, 4LEH, 4L8O, and 4N3V are available from the NIH PSI Materials Repository (http://psimr.asu.edu/MRLinks.html) Site-directed mutants were generated using the QuikChangeTM Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA). The expression vector pSport1-19k containing the baiE sequence was used as a template 20.
Protein Expression and Purification
Proteins utilized for structure determination and kinetic studies were prepared as described previously. 22 Briefly, recombinant protein was expressed at 37°C by induction of the arabinose promoter with L-arabinose (0.13%) for 6 hours in a 0.5 L culture and purified to by a single step Ni-NTA chromatography on a 1.5 ml bed volume. LC-MS and SDS-PAGE analysis were consistent with the 22 kDa predicted molecular mass of the BaiE monomer with an N-terminal expression and purification tag. The oligomeric profile was determined by analytical size exclusion chromatography performed at room temperature on an Agilent HP1100 HPLC system utilizing a Shodex 8 x 300mm Protein KW-802.5 column in 20 mM Tris pH 7.5, 200 mM NaCl, 0.5 mM TCEP and 3 mM NaN3. The N-terminal expression and purification tags were not cleaved in this study. Selenomethionine (Se-Met) derivatized proteins were utilized for structure determination and native proteins were utilized for kinetic studies and co-crystallization trials.
Synthesis of Bile Acid Substrates and Products
CoA-thioesters of bile acids were prepared as described previously 22. 3-oxo-Δ4-chenodeoxycholic acid (3-oxo-Δ4-CDCA) 26 and 3-oxo-Δ4,6-lithocholic acid (3-oxo-Δ4,6-LCA) 27 were synthesized following published procedures. Commercially obtained chemicals used in the synthesis were utilized without further purification or drying.
Steady-state kinetics
Steady-state kinetic parameters were determined using a Varian Cary 100 Bio UV-Visible spectrophotometer equipped with a thermo-jacketed cuvette holder. The reactions at 25 °C in 20 mM Tris-HCl pH 7.3 monitored the formation of the product at 297 nm. Reaction rates were investigated under varying substrate concentrations below and above the respective KM values. The initial linear regions of the reaction progress curves were utilized for determining the reaction rates. The data were fitted to the Michelis-Menten equation by non-linear regression method using the enzyme kinetics module in GraphPad Prism (GraphPad Software, La Jolla, CA).
Detection of Substrate Turnover by 1H-NMR
1H-NMR spectra were acquired at 300 °K on a Bruker Avance instrument equipped with a 1H/13C/15N-TXI cryoprobe operating at 600 MHz with 16 scans and a recycle delay of 2 s employing excitation sculpting to suppress water signal (Hwang and Shaka). Isotope-labeled compounds were purchased from Sigma-Aldrich/Isotec. All reaction samples contained 3-(trimethylsilyl)-2,2′,3,3′-tetradeuteropropionic acid as an internal standard that was referenced to 0 ppm and 10% D2O. The reaction was initiated by adding enzyme to an ice cold solution containing 100 μM substrate prepared in deuterated DMSO. Samples were immediately transferred to the NMR instrument and data collection initiated after temperature equilibration as determined by the stability of the signal.
Substrate binding assay
Purified protein (100 βg) was suspended in 50 βl of reaction mixture containing 25 mM sodium acetate pH 7.5 with 3-[N-morpholino]propanesulfonic acid (MOPS) and bile acid substrate (18,000 dpm). The reaction was incubated for 1 min at 37 °C in 10K Nanosep centrifugal concentrators (Pall Filtron). The concentrators were centrifuged at 10,000xg for 2 min. Samples were washed with an additional 100 βl of reaction buffer and centrifuged again for 2 min. Bound counts and effluents were quantified by liquid scintillation spectrometry. Bound counts were corrected for non-specific binding against assays with heat-denatured protein and background binding to the filter.
Crystallization of BaiE
Selenomethionine derivatized BaiE protein from C. scindens ATCC35704, and C. hiranonis DSM13275 yielded crystals from the JCSG Core Suite (Qiagen Sciences, MD, USA), conditions I-D8 (20% w/v PEG3350, 0.2 M sodium sulfate, pH 6.6; space group P3121), and IV-D4 (50% w/v PEG200, 0.1M HEPES pH 7.5; space group P63), respectively. Standard JCSG protocols were utilized for all crystallization trials 28,29 using a drop volume of 0.2 μl and a 1:1 (vol/vol) ratio of reservoir to protein (18.6 mg/ml, C. scindens ATCC35704 and 20.6 mg/ml C. hiranonis DSM13275) in 20 mM HEPES pH 8.0 solution equilibrated against 100 μl of reservoir solution. The crystals were cryoprotected with addition of 10% 1,2 ethanediol (C. scindens ATCC35704) or 10% glycerol (C. hiranonis DSM13275) before harvesting and flash freezing with liquid nitrogen. Co-crystallization trials involving native preparations of the proteins were performed similarly with addition of 1 mM product, 3 -oxo-Δ4,6 -Lithocholyl-Coenzyme A (3-oxo-Δ4,6-LC-CoA). Partial binding of the product (see below) was observed in only one co-crystal of C. hiranonis DSM13275 (space group R32:h) obtained at a protein concentration of 21 mg/ml from JCSG Core Suite condition I-G5 (10 % PEG6000, 0.1 M citric acid pH 5.0). This crystal was cryoprotected with 15% 1,2 ethanediol. BaiE from C. hylemonae DSM15053 (8.5 mg/ml) yielded crystals [JCSG Core Suite condition III-E2 (0.01 M cobalt chloride, 1.8 M ammonium sulfate, 0.1M MES pH 6.5; space group R32:h)], but no ordered binding of the product was observed in the structure. Prior to harvesting, 10% ethanediol was added to the drop.
Structure Determination of BaiE
Experimental phases for BaiE homologs from C. scindens and C. hiranonis were generated by multiple-wavelength anomalous diffraction (MAD) and single-wavelength anomalous diffraction (SAD) methods, respectively. The X-ray diffraction data were collected at wavelengths corresponding to the peak (λ1), inflection (λ2) and high-energy remote (λ3) wavelengths of selenium for the MAD experiments and at the peak wavelength of selenium for the SAD experiments. The data were collected on beamline 11-1 at Stanford Synchrotron Radiation Lightsource at 100 K with a Rayonix Mar Mosaic MX-325 CCD detector using the Blu-Ice data-collection environment. 30 MOSFLM 31 and SCALA 32 were used for data integrating and scaling. Selenium positions were determined with SHELXD and the phases were refined using autoSHARP 33, with a mean figure of merit of 0.50 to 2.9 Å in space group P3121 for C. scindens ATCC 35704 and 0.28 to 1.6 Å in space group P63 for C. hiranonis DSM 13275. Automated model building was performed with ARP/wARP. 34 Data for the crystals grown from the co-crystallization trials were collected on beamline 8.2.2 at the Advanced Light Source with an ADSC Q315 CCD detector at 100 K. The data were reduced with XDS35 and scaled with XSCALE 36. The structure of BaiE, C. hiranonis DSM13275 was used as a search model for phasing the native BaiE from C.hylemonae DSM15053 and BaiE from C. hiranonis DSM13275 structures originating from the co-crystallization trials. Model completion and iterative cycles of refinement were performed with COOT37 and REFMAC 38 from the CCP4 program suite 39. The TLS refinement protocol included one TLS group per molecule in the asu. Coordinates for 3-oxo-Δ4,6-LC-CoA, polyethylene glycol (PEG) and ethylene glycol were generated using PRODRG 40. The library files containing the refinement restraints were generated using LIBCHECK within CCP4. The quality of the crystal structure was analyzed using the JCSG Quality Control server (http://smb.slac.stanford.edu/jcsg/QC). This server verifies: the stereochemical quality of the model using AutoDepInputTool41, MolProbity42, and Phenix43, the agreement between the atomic model and the data using RESOLVE44, the protein sequence using CLUSTALW45, the ADP distribution using Phenix, and differences in Rcryst/Rfree, expected Rfree/Rcryst and various other items including atom occupancies, consistency of NCS pairs, ligand interactions and special positions using in-house scripts to analyze refinement log file and PDB header. Data collection, model and refinement statistics are summarized in Table 1. Coordinates and the associated structure factors are deposited in the Protein Data Bank (PDB) with following accession codes: 4L8P - BaiE from C. hiranonis DSM 13275; 4L8O - BaiE from C. hylemonae DSM 15053; 4LEH – C. scindens ATCC 35704; 4N3V - BaiE from C. hiranonis DSM 13275 with 3-oxo-Δ4,6-LC-CoA.
Table I.
Data Collection and Refinement Statistics
| Crystal | C. hiranonis DSM 13275 | C. scindens ATCC 35704 | C. hylemonae DSM 15053 | C. hiranonis DSM 13275:3-oxo- Δ4,6-LC-CoA | ||
|---|---|---|---|---|---|---|
| PDB id | 4L8P | 4LEH | 4L8O | 4N3V | ||
| Space group | P63 | P3121 | R32:h | R32:h | ||
| Unit cell lengths (Å) | a=b=58.9,c=91.3 | a=b=109.3,c=118.3 | a=b=53.0,c=428.6 | a=b=84.3,c=311.8 | ||
| Data collection | ||||||
| Beamline | SSRL BL11-1 | SSRL BL11-1 | ALS 8.2.2 | ALS 8.2.2 | ||
| Data set | SAD | λ1 MAD | λ2 MAD | λ3 MAD | Native | Native |
| Wavelength (Å) | 0.9795 | 0.9790 | 0.9795 | 0.9184 | 1.000 | 1.000 |
| Resolution range (Å) | 29.45–1.60 | 29.57–2.90 | 29.55–2.98 | 29.54–2.90 | 28.58–2.20 | 47.40–1.89 |
| Highest resolution shell (Å) | 1.64–1.60 | 2.98–2.90 | 3.06–2.98 | 2.98–2.90 | 2.32–2.20 | 1.94–1.89 |
| No. of observations | 172,513 | 137,535 | 63,001 | 68,430 | 77,498 | 362,382 |
| No. of unique reflections | 23,715 | 18,559 | 17,071 | 18,502 | 12,340 | 34,714 |
| Completeness (%) | 100 (100)a | 99.9 (100) | 99.9 (100) | 99.9 (100) | 99.3 (96.0) | 99.9 (98.8) |
| Mean I/σ(I) | 11.1 (2.7)a | 13.1 (1.8) | 11.3 (2.0) | 11.2 (1.8) | 17.1 (2.7) | 19.5 (2.5) |
| Rmerge on Ib (%) | 9.5 (65.1)a | 9.4 (112.3) | 7.3 (61.8) | 7.1 (73.0) | 9.5 (55.7) | 6.9 (88.5) |
| Rmeas on Ic (%) | 10.2 (70.3)a | 10.1 (120.6) | 8.5 (72.3) | 8.3 (85.5) | 10.4 (61.9) | 7.3 (95.3) |
| Rpim on Id (%) | 3.8 (26.4)a | 3.7 (43.5) | 4.4 (36.8) | 4.2 (43.8) | 4.1 (26.4) | 2.2 (35.0) |
| Model and refinement statistics | ||||||
| Data used in refinement | λ1 MAD | |||||
| No. of reflections (total)g | 23,680 | 18,530 | 11,786 | 34,711 | ||
| No. of reflections (test) | 1,222 | 948 | 568 | 1,748 | ||
| Cutoff criteria | |F|>0 | |F|>0 | |F|>0 | |F|>0 | ||
| Rcryst (%)e | 15.1 | 17.7 | 19.4 | 18.8 | ||
| Rfree (%)f | 18.1 | 21.7 | 22.5 | 21.8 | ||
| Stereochemical parameters | ||||||
| Restraints (RMSD observed) | ||||||
| Bond lengths (Å) | 0.011 | 0.011 | 0.013 | 0.010 | ||
| Bond angles (°) | 1.49 | 1.49 | 1.32 | 1.38 | ||
| MolProbity statistics | ||||||
| Ramachandran plot (%)h | 97.6 (0) | 98.0 (0) | 98.8 (0) | 99.4 (0) | ||
| Rotamer outliers (%) | 2.1 | 0.7 | 0.0 | 2.1 | ||
| All atom clashscore | 1.4 | 1.0 | 0.4 | 3.2 | ||
| Average isotropic B-value (Å2)i | 24.8 (17.7) | 96.3 (104.1) | 35.2 (39.4) | 35.8 (26.2) | ||
| ESU based on Rfree (Å) | 0.08 | 0.31 | 0.18 | 0.13 | ||
| Protein residues/atoms | 168/1366 | 501/4091 | 168/1335 | 334/2722 | ||
| Non-protein entities | 124 H2O, 1 Ni+, 1 Na+, 2 PEG fragments |
3 H2O, 8 SO42−, 1 Zn2+ | 70 H2O, 1 SO42−, 1 Co2+, 1 Na+, 4 ethylene glycol | 189 H2O, 1 UNL, 2 Zn2+, 2 Na+, 2 citric acid, 1 ethylene glycol, 1 PEG fragment | ||
Values in the parentheses are for the highest resolution shell.
Rmerge=ΣhklΣi|Ii(hkl)−<I(hkl)>|/ΣhklΣiIi(hkl)
Rmeas(redundancy-independent Rmerge) =Σhkl[Nhkl/(Nhkl−1)]1/2Σi|Ii(hkl)−<I(hkl)>|/ΣhklΣiIi(hkl)
Rpim(precision-indicating Rmerge)=Σhkl[1/(Nhkl−1)]1/2Σi|Ii(hkl)−<I(hkl)>|/ΣhklΣiIi(hkl)
Rcryst =Σ| |Fobs|−|Fcalc| |/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5% of the total reflections chosen at random and omitted from refinement.
Typically, the number of unique reflections used in refinement is slightly less than the total number that were integrated and scaled. Reflections are excluded due to systematic absences, negative intensities and rounding errors in the resolution limits and cell parameters.
Percentage of residues in favored regions (Top8000) of Ramachandran plot (outliers in parenthesis).
This value represents the total B that includes TLS and residual B components. (Wilson plot B-value in parenthesis.)
Docking Simulations
Simulated substrate binding was performed by ASEDOCK program within MOE (Chemical Computing Group, Montreal, Canada) to predict possible enzyme:substrate interactions. Models of the substrates 3-oxo-Δ4-CDCA and 3-oxo-Δ4-CDC-CoA with expected protonation states were generated using the Molecule Builder of MOE and minimized using Merck Molecular Force Field 94×. Search for potential substrate-binding pockets were performed with the Site Finder module in MOE using the induced-fit protocol, which allows rotatable bonds during refinement of the docked molecules. The top 30 hits were retained for further analysis for relevance to catalysis.
Results and Discussion
Structure of BaiE, bile acid 7α-dehydratase
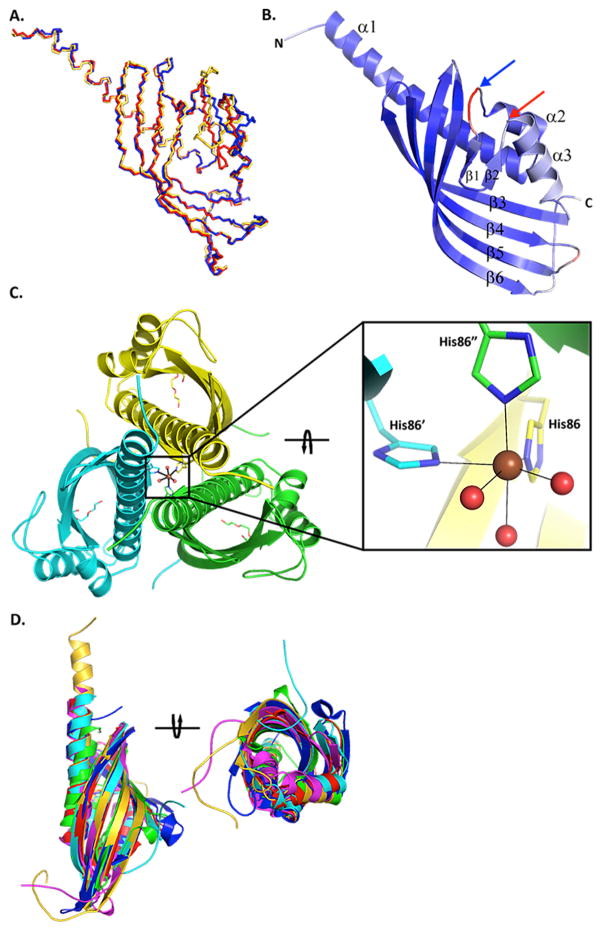
The final BaiE structures contained one (C. hiranonis DSM13275 in space group P63 and C. hylemonae DSM 15053 in space group R32:h), three (C. scindens ATCC 35704 in space group P3121) and two (C. hiranonis DSM13275 in space group R32:h) monomers in the asymmetric unit with Rcryst/Rfree ranging between 15.1/18.1 and 19.4/22.5, consistent with the respective resolution (Table 1). The apo-structures were superimposed using the Secondary Structure Matching (SSM) function in COOT 46 with pairwise rmsds ranging from 0.71 – 0.94 Å (Fig. 2A). The major difference in the structures is in the loop formed by residues 48–63, which is in a significantly distinct conformation in each structure. The electron density for the main chain and side chains is well defined throughout, except for the last two C-terminal residues of C. hiranonis DSM13275 and the last three residues of C. hylemonae DSM15053. In addition only Gly0 of the N-terminal expression and purification tag had clearly interpretable electron density. All models exhibited high-quality geometry with overall clash scores > 99th percentile, and Ramachandran plot with > 97% residues in favored regions as calculated by MolProbity (v4.02b-509) 47 The hexagonal crystals of BaiE, C. hiranonis DSM13275 diffracted to the highest resolution of 1.60 Å.
Figure 2. Structure of BaiE, bile acid 7α-dehydratase.
(A) Secondary structure match (SSM) superposition of BaiE crystal structures from different organisms. The main chain atoms are colored gold, red and blue respectively for C. hiranonis DSM13275, C. scindens ATCC35704 and C. hylemonae DSM15053. (B) Characteristic twisted α+β barrel fold observed in BaiE. The secondary structure elements are colored according to Cα atomic B-value from blue (lowest B-value: 20 Å2) to red (highest B-value: 65 Å2). The blue and red arrows respectively indicate the start and end of loop residues 48–63. (C) Trimer assembly interface as observed in the crystal packing of BaiE, C. hiranonis DSM13275. Bound PEG molecules in the active site are indicated as sticks. The three subunits of the trimer assembly are colored yellow, cyan and green. The six atom, (His)3– (H2O)3, octahedral coordination of the Ni ion is depicted in the square panel. The view in the square panel was obtained by ~90° clockwise rotation of the view of the trimer assembly in the primary figure around an axis perpendicular to the 3-fold symmetry. The coordinate bonds are depicted as solid black lines. N, O, and Ni atoms are colored blue, red and brown, respectively. The carbon atoms of the protein chain are colored yellow, cyan and green, respectively. (D) SSM superposition of BaiE, C. hiranonis DSM13275 (gold), onto closest structural homologues as determined by DALI. The proteins used in the superposition are two scytalone dehydrogenases (blue and cyan), LinA (magenta), an α+β barrel fold containing protein of unknown function (PDB ID: 3ROB) and Δ5,3-ketosteroid isomerase (green). Two views between the figures are related ~90° anticlockwise rotation.
The BaiE protomer is a single domain structure bearing the characteristic twisted α+β barrel fold of the NTF2-like protein superfamily. 48 The secondary structure elements consist of three α-helices and six β-strands (Fig. 2B). The six β-strands that form the antiparallel curved β-sheet are assembled on one side of the monomer with a β2-β1-β6- β5-β4-β3 strand order. The three α-helices flank the other side of the molecule and consist of a 34-residue N-terminal helix, α1, and two α-helices of 5 (α2) and 11 (α3) residues. Inspection of the main-chain B-values indicate that residues 48–63 tend to exhibit the highest values in all structures, suggesting conformational flexibility (Fig. 2B). A previously reported homology model of BaiE, C. scindens VPI 12708 15, superposes quite well with the crystal structures with a backbone rmsd of 1.7 Å for the core α+β barrel fold; however, as expected notable differences are observed for residues 48–63.
Analytical size exclusion chromatography indicated that all BaiE homologs here are trimers in solution. BaiE also assembles as a trimer in the crystal structure (Fig. 2C). The asymmetric unit of the C. scindens ATCC35704 BaiE crystal (space group P312) contains what is assumed to be the biological trimer. Similar trimer assemblies can be generated in the other crystal forms by space group symmetry. The trimer interface primarily involves α1 and strand-swapped interactions between β6 and the C-terminal “strand” of the neighboring monomer that stabilizes the bottom of the trimer, with addition minor contributions from β3, β4. Several key interactions are predicted to stabilize the inter-protomer interactions including an octahedral six-atom coordination of a divalent metal ion at the trimer interface. The metal ion coordinates with three H86-Nε2 atoms from each symmetry-related subunit of the trimer and three water molecules within 2.2 Å. This divalent ion-His coordination at the trimer interface is conserved across all known structures of BaiE homologs. Presence of divalent metal ion at the subunit interface has been previously reported and considered to stabilize the quaternary assembly. 49 . The anomalous difference Fourier electron density maps suggests a mixture of Ni2+ and Zn2+ ions in the trimer interface of BaiE, C. hiranonis DSM13275. The Ni2+ likely originated from the metal affinity column used for purification, whereas Zn2+ was likely acquired during protein. Neither of these metal ions was present in the purification or crystallization buffers. The peak intensity for Ni2+ in the structure is higher than Zn2+ as it likely that some Zn may have been exchanged with Ni during purification. Based on these observations, we modeled the divalent metal ion at the trimer interface in the final deposited model of BaiE, C. hiranonis DSM13275 as Ni2+. However, the divalent metal ion in the deposited model of the homolog from C. scindens ATCC35704 is modeled as Zn2+ based on the X-ray fluorescence spectroscopy 50 (data not shown). For the homolog from C. hylemonae DSM15053, X-ray fluorescence data were inconclusive and the divalent metal ion was modeled as Co2+ since the crystallization cocktail contained CoCl2. Many inter-subunit salt bridge interactions are also present at the trimer interface (E4-R6, R6-E11, E21-K24, R32-E89 and E150-R154) in addition to multiple hydrogen bond interactions. Similar trimer interface salt bridges and hydrogen bond interaction are also observed in related structures from this protein family, such as scytalone dehydratase (STD) 48 and LinA 51. However, to our knowledge, the metal-His interaction at the trimer interface in BaiE is the first such interaction observed in this family.
Analysis using DALI 52 and FATCAT 53 suggest BaiE is similar to members of the NTF2 superfamily (Fig. 2D). The closest structural homologs are scytalone dehydratases (DALI Z score: 16.8) and LinA (DALI Z score: 16.1). SSM superposition by COOT achieved a core backbone rmsd of 1.5 Å over 126 Cα atoms for comparison with scytalone dehydratase 54 (PDB ID: 3STD), 2.6 Å over 110 Cα atoms with Δ5,3-ketosteroid isomerase 55 (PDB ID: 1OPY) and 3.2 Å over 117 Cα atoms with LinA (PDB ID: 3A76). BaiE possesses < 10% sequence identity with the closest structurally related protein.
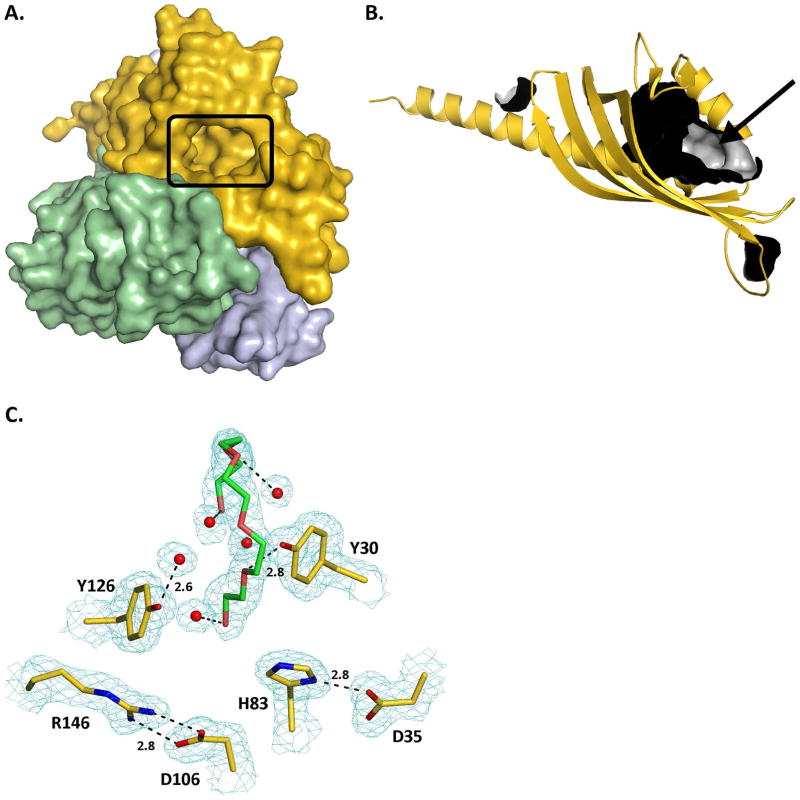
Substrate binding site
Analysis of both the monomer and the trimer BaiE structure with the Site Finder module of the Molecular Operating Environment package (MOE; Chemical Computing Group, Montreal, Canada) indicates that the substrate-binding pocket (Fig. 3A) is the largest surface cavity with a volume of 451 Å3 as determined by CASTp 56 (Fig. 3B). Secondary structure elements α1, β1 and β2 form the mouth, while α3, β3, β4, β5 and β6 form the floor of the binding pocket. Y30 (α1), D35 (α1) and H83 (β3) that were shown to participate in catalysis by site-directed mutagenesis study (see below) are indeed located in the substrate binding pocket. Crystal structures of the 4 BaiE homologs revealed the D35 and H83 side chains interact within a distance of 2.8 Å (Fig. 3C). However, no interaction is observed between Y30 and the other two catalytic residues. In BaiE, C. hiranonis DSM13275, Y30 interacts (2.8 Å) with an oxygen atom of a polyethylene glycol (PEG) molecule bound in the active site. D106 (β4) and R146 (β6) in the substrate binding pocket are conserved across all the BaiE homologs, and, interact via salt bridge (2.8 Å) that is conserved in all apo-structures of BaiE homologs.
Figure 3. Substrate binding site of BaiE, bile acid 7α-dehydratase.
(A) Location of the active site of each protomer with respect to the trimer assembly. The protomers of the trimer are colored gold, blue and green. (B) Cavities detected in the monomer structure of BaiE. Inner surface of the cavities are colored gray and the outer surface are colored black. The substrate binding site is indicated by the black arrow. (C) Key interaction involving residues of the substrate binding site with PEG and water molecules in BaiE, C. hiranonis DSM13275. 2Fo-Fc electron density map colored cyan contoured at 1.0 sigma. Dashed black lines are hydrogen bond interactions with numbers being distances in Å between the interacting atoms. Carbon atoms of protein residues and PEG molecule are colored gold and green, respectively. O, N, P and S atoms are colored red, blue and orange, respectively.
Predicted enzyme:substrate interaction by simulated docking experiments
Docking experiments of the substrate 3-oxo-Δ4-CDCA into the BaiE binding pocket with the ASEDock module of MOE identified a potentially productive binding mode that is consistent with our site-directed mutagenesis experiments (Fig. 4A). In this binding mode, the C3-oxo and C7-hydroxyl groups are located 3.1 Å and 3.6 Å from Y30-Oη and H83-Nε2 atoms, respectively. H83-Nε2 is 2.7 Å from the 6α-hydrogen compared to 4.5 Å from the 6β-hydrogen. The H83-imidazole is therefore positioned to abstract the 6α hydrogen and protonate the leaving C7-hydroxy group complying with a cis- elimination of water (6αH, 7α-OH). However, previous studies with triply tritiated and singly C14 labeled cholic acid (6ε, 6ε, 8ε-H3, 24-C14) detected a diaxial trans- elimination of water (6βH, 7α-OH). 57 Further experiments with purified BaiE utilizing 3-oxo-Δ4 derivatives of both α- and β-muricholic acid (3α,6β,7α-trihydroxy-5β-cholanoic acid) and hyocholic acid (3α,6α,7α-trihydroxy-5β-cholanoic acid) should help resolve the stereochemistry of the 7α-hydroxyl elimination reaction.
Figure 4. Predicted enzyme:substrate interactions.
(A) Probable productive binding mode of 3-oxo-Δ4-Chenodeoxycholate (3-oxo-Δ4-CDCA). Blue dashed lines and adjacent numbers are predicted interaction of His83-Nε2 atom with C7-OH and C6 atoms and Y30-OH group with C3-oxo atom of 3-oxo-Δ4-CDCA. The 6α-H closest to H83-Nε2 atom colored magenta and 6β-H away from H83-Nε2 atom colored brown. (B) Predicted stacking interaction involving the adenine group of the Coenzyme (CoA) moiety of 3-oxo-Δ4-Chenodeoxycholyl CoA (3-oxo-Δ4-CDC-CoA) with Y115. The key interaction of the bile acid moiety of the docked CoA-bile acid ester with the active site residues is similar to what is predicted in (A). Carbon atoms of protein residues and product molecule are colored gold and green, respectively. H, O, N, P and S atoms are colored gray, red, blue, orange and olive, respectively.
Computational calculations with the substrate 3-oxo-Δ4 -CDC- CoA predicted probable binding interactions of the CoA moiety in addition to predicting a similar productive binding mode of the bile acid moiety (Fig. 4B). The calculation predicted a twisted conformation of CoA where the adenine moiety is involved in a π-π stacking interaction with Y115. In such a conformation, a significant portion of both the pantetheine and AMP moieties of CoA would be exposed to the bulk solvent without any direct interaction with the protein. This scenario has previously been observed in crystal structures of thiolase where the AMP portion projects out of the substrate binding pocket and interacts only with surface residues 58,59. Thus, it seems that only the bile acid moiety binds in the active site pocket with no space to accommodate the CoA moiety. The CoA moiety likely projects out in the solvent possibly interacting with surface residues. This topology is supported by the co-crystal structure of BaiE with 3-oxo-Δ4,6-LC-CoA where the bile acid moiety is observed to bind in the active site and the CoA moiety extends into the solvent exhibiting a poor electron density, as described below.
Crystal structure of BaiE 3-oxo-Δ4,6-Lithocholyl CoA
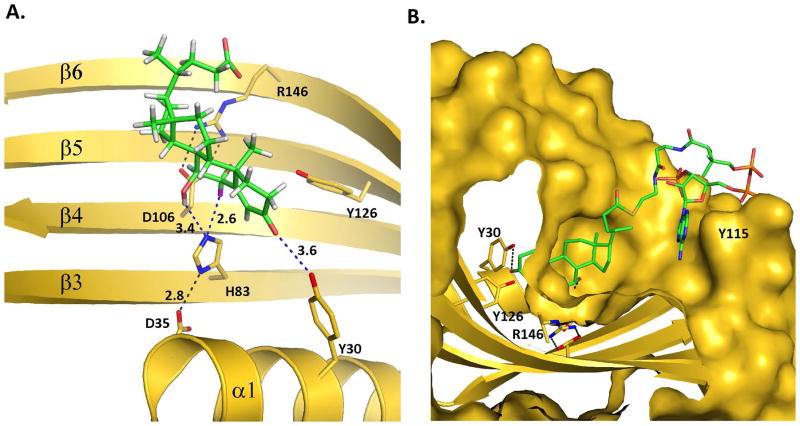
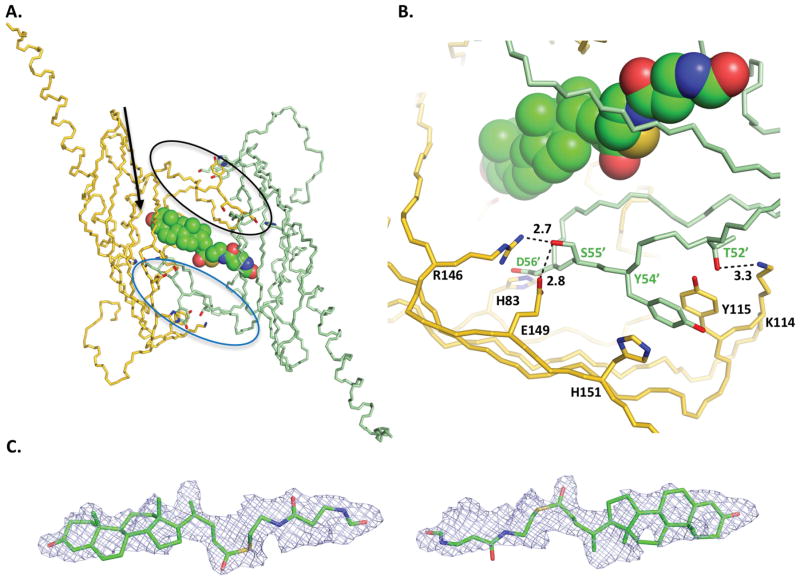
The co-crystal structure of BaiE with 3-oxo-Δ4,6-LC-CoA shows a novel extended pocket that was not predicted by the simulated docking experiments. This extended pocket is generated by the substrate binding sites of two protomers that originate from two different trimer assemblies related by an NCS 2-fold symmetry. A key loop comprising residues 48–63 in the substrate binding site of each protomer forms the extended pocket (black and blue circles in Fig. 5A). Interestingly, this loop in the co-crystal structure protrude into the substrate binding site of the protomer related by NCS 2-fold symmetry. This extended conformation is stabilized by hydrogen bond interaction with several residues from the NCS 2-fold related protomer (Fig. 5B). Importantly, the D56 carboxylate hydrogen bonds with the H83 imidazole in the NCS 2-fold related protomer, which is one of the three catalytic residues. An additional set of hydrogen bonds are observed with loop residues T52, Y54, S55, and G57, and their respective hydrogen partners located in the binding site of the NCS 2-fold related protomer: K114 (with T52), Y115 (with Y54), H151 (with Y54), R146 (with S55), E149 (with S55), and Y70 (with G57).
Figure 5. Binding of product, 3-oxo-Δ4,6- Lithocholyl-CoA (3-oxo-Δ4,6-LC-CoA, in BaiE, C. hiranonis DSM13275.
A) The extended binding pocket and binding of 3-oxo-Δ4,6-LC-CoA, in BaiE, C. hiranonis DSM13275. Main chain of protomers arising from different trimer assemblies is colored gold and green, respectively. The loop formed by residues 48–63 is bordered by black and blue ellipses in the gold and green monomers, respectively. The black arrow indicates the location of the C3-oxo atom. (B) Key inter-protomer interactions involved in generating the extended binding pocket. Numbers indicate distances in Å. (C) Unbiased mFo-DFc electron density map of 3-oxo-Δ4,6-LC-CoA contoured at +1.8 sigma (0.085 e−/Å3). For clarity, both orientations of the ligand are shown separately to illustrate how the partial product model fits the density. A model with the two orientations together at partial occupancy fits the density better than either orientation on its own. The pyrophosphate and AMP moiety of the product is not shown. The density fit for those regions is not as good (see text). Product 3-oxo-Δ4,6-LC-CoA in panels A and B is depicted as spheres.
Although the resolution (1.9 Å) of the structure is reasonably high, the electron density of the ligand is relatively poor compared to the protein. Analysis of the electron density map suggests partial occupancy and a dual conformation of the substrate across the NCS 2-fold symmetry axis (Fig. 5C). Attempts at modeling the entire product indicated that the ligand extends from the binding site of one protomer to the binding site of the other protomer with the CoA moiety presumably extending into the solvent. Therefore, the bile acid product was modeled in two different orientations. As shown in Fig. 5C in the top and bottom panels, the bile acid moiety is modeled respectively into each half of the electron density maps. Furthermore, the ligand comprising only the bile acid and pantetheine moieties fits well into the unbiased mFo – DFc electron density at relatively low contour level, +1.8 rmsd (0.085 e−/Å3) in either orientation. In addition, in either orientation, the C3-oxo group of the bile acid is within 2.7 Å from the side chain hydroxyl group of the active site Y30 within the same protomer. Hence, in the final deposited structure, the bile acid product was modeled in two overlapping orientations in each active site.
The 2mFo – DFc electron density map generated after addition of the entire product in either orientation revealed continuous electron density features over bile acid moiety and part of the pantetheine moiety at contour levels ranging from +0.8 rmsd (0.21 e−/Å3) to +1.0 rmsd (0.26 e−/Å3). However, the AMP, the dimethyl group, and the pyrophosphate linkage regions do not have electron density coverage. Although an additional electron density blob is observed adjacent to the bile acid moiety, attempts at modeling the AMP moiety into this blob shows that the shape is inconsistent. Therefore, due to difficulty in unequivocal assignment of the ligand arising from partial occupancy and dual binding mode across the NCS 2-fold, the ligand is modeled as an unknown ligand (UNL) with a dual conformation. In the UNL, the AMP moiety, pyrophosphate linkage and dimethyl group is omitted.
Detection of product formation by 1H-NMR
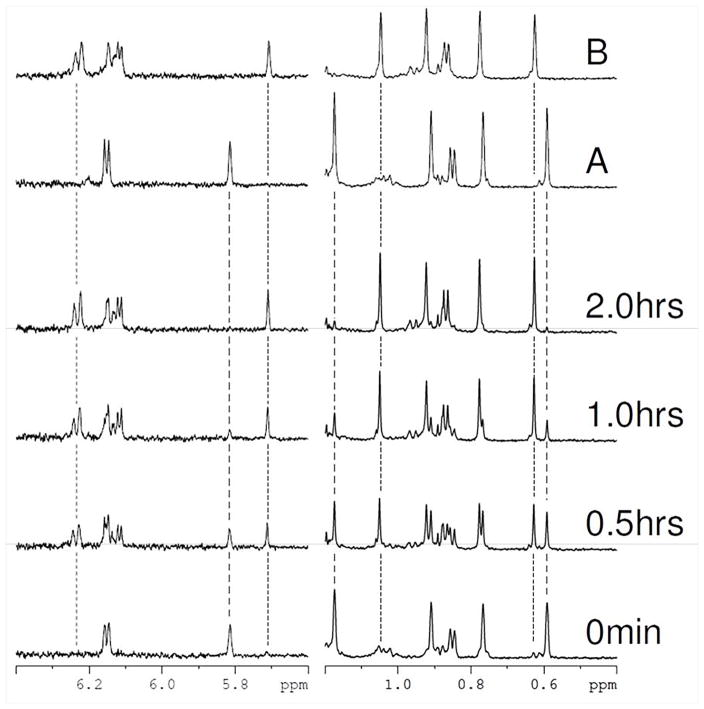
A time series of proton NMR spectra were collected to directly monitor the enzymatic removal of the C7α-hydroxy group and the generation of 3-oxo-Δ4,6 bile acid intermediate by BaiE. These data provided an unambiguous analytical probe to confirm the generated product. Selected regions of proton spectra for the BaiE mediated conversion of 3-oxo-Δ4-CDC-CoA to 3-oxo-Δ4,6-LC-CoA are shown in Figure 6. Aromatic and methyl regions (left and right panels respectively) of the spectra acquired at different time points after the addition of enzyme are shown. During the reaction, the intensities of the substrate peaks decrease with a concomitant appearance of a new set of peaks, as readily observed in Figure 6 where the intensity of selected substrate peaks at 5.81, 1.17, and 0.59 ppm (marked with dashed lines) decrease as the reaction proceeds. This intensity loss is associated with the appearance of a new set of peaks at 5.71, 1.05, 0.63 ppm (marked with dotted lines) that increase in intensity. The reaction appears almost complete after two hours with only minor evidence of substrate peaks remaining. The chemical shifts of the new peaks perfectly match a control spectrum acquired of 3-oxo-Δ4,6-LC-CoA in the same buffer (compound B in Fig. 6) and is observed for all peaks in the spectra and not isolated only to the regions displayed. Close examination of the aromatic region in Fig. 6 shows the appearance of two new doublets during the reaction as expected for the methene groups generated by removal of the 7α-hydroxyl group. One doublet is isolated and clearly observed at 6.23 ppm (marked with a dotted line) and the second is observed as part of the overlapped peak at approximately 6.14 ppm. These data confirm that BaiE is converting 3-oxo-Δ4-CDC-CoA to the expected product 3-oxo-Δ4,6-LC-CoA.
Figure 6. In vitro generation of 3-oxo-Δ4,6- lithocholyl-Coenzyme A from 3-oxo-Δ4-Chenodeoxycholyl CoA(3-oxo-Δ4-CDC-CoA) using purified BaiE from C. hiranonis DSM13275 as monitored by 1H-NMR.
A reaction mixture of 100μM 3-oxo-Δ4-CDC-CoA (3-oxo-Δ4,6-LC-CoA) and 0.001μM BaiE was prepared in 20 mM HEPES pH 7.4 and incubated at 300°K. Selected regions from one-dimensional 1H-NMR spectra that were acquired at the indicated times after mixing and temperature equilibration (bottom four spectra) are displayed. Reference spectra of 3-oxo-Δ4-CDC-CoA and 3-oxo-Δ4,6-LC-CoA acquired in the same buffer are shown at the top labeled A and B respectively. Some representative peaks associated with 3-oxo-Δ4-CDC-CoA and 3-oxo-Δ4,6-LC-CoA are marked with dashed and dotted lines respectively. The intensity of peaks associated with the substrate decrease with a concomitant increase in product peaks during the reaction. The spectrum of the product matches perfectly the spectra of3-oxo-Δ4,6-LC-CoA . Examination of the amide region of the spectra (right hand panel) shows the appearance of two new peaks during the reaction and is associated with removal of the 7α-hydroxyl group and the generation of methene groups.
Site-directed mutagenesis
Mutation of predicted key catalytic and substrate binding residues residues of BaiE from C. scindens VPI 12708 provide important insights into their involvement in substrate turnover and binding (Table 2). Y30F, D35N and H83N mutations completely abolished substrate turnover indicating the involvement of these residues in catalysis. However, increased substrate binding is observed for mutants Y30F (5.6-fold) and D35N (3.0-fold) compared to wild type, but no substrate binding is observed for the H83N mutant. Y30 of BaiE is equivalent to Y14 in 3-oxo-Δ5-ketosteroid isomerase (KSI) and is predicted to participate as a general acid protonating or forming a low barrier hydrogen bond (LBHB) with the dienolate intermediate during the isomerization reaction. 55 Y126 may stabilize the negative charge on the dienolate intermediate as confirmed by 98% reduced catalytic activity observed in Y126F. In KSI, D99 participates in catalysis in a similar manner. H83 and D35 are equivalent to the H85-D31 catalytic dyad in scytalone dehydratase (STD) that is predicted to participate as a general base. 60 Mutation of the H85-D31 pair in STD also impaired catalysis.
Table II.
Relative activity and substrate binding of several site-directed mutants of BaiE, C. scindens VPI12708
| BaiE Mutation | Relative activity a | Substrate Binding (%) |
|---|---|---|
| Wild Type | +++ | 1.5 |
| Y30F | − | 8.4 |
| D35N | − | 4.6 |
| Y54A | − | 2.8 |
| Y54F | ++ | 4.1 |
| H83N | − | <0.1 |
| D106N | − | 2.3 |
| Y126F | + | 10.7 |
| R146Q | − | 10.4 |
dash denotes activity assays where <0.1% of substrate converted to product.
Site-directed mutagenesis also revealed D106 and R146 in the substrate binding pocket as essential for catalysis. In all apo-structures of BaiE, side chains of D106 and R146 interact via salt bridge at a distance of 2.9 Å. Disruption of this salt bridge by mutation to Asn and Gln at positions 106 and 146, respectively, abolished catalysis. Site-directed mutagenesis of one residue of the conformationally flexible loop comprising residues 48–63 indicates the importance of this loop in catalysis. A Y54F mutation reduced enzyme activity by 74% compared to wild type, whereas Y54A abolished enzyme activity. This result suggests Y54 is probably important for maintaining the shape of the active site and recognition of the substrate. In the 3-oxo-Δ4,6-LC-CoA co-crystal structure of BaiE, Y54 seems to be involved in key stacking interactions with Y115 and H151 of the NCS 2-fold symmetry related monomer at the pocket interface.
Steady state kinetic studies
Steady-state kinetics (Table 3) analysis was performed by continuous UV assay monitoring of the formation of product utilizing an extinction coefficient, ε297 = 0.0188 μM −1 cm−1, measured at pH 7.3. All four homologs of BaiE revealed robust turnover of CoA-esters of bile acid and free bile acid substrates. The substrate affinity, KM, for the free bile acid substrate, 3-oxo-Δ4-CDCA is of comparable magnitude across all the four homologs. The KM values of the CoA-bile acid ester 3-oxo-Δ4-CDC-CoA are also of comparable magnitude across all homologs except for BaiE C. hiranonis DSM13275 where the KM is 208 μM, which is at most two orders of magnitude higher than the others. Furthermore, the turnover rate constants (kcat) of both substrates are comparable in all three homologs except C. scindens VPI12708, where the kcat is at most 7.7 times higher. Both BaiE homologs from C. scindens ATCC35704 and VPI12708 respectively exhibited higher substrate specificity (kcat/KM) towards the CoA-ester substrate by an order of a magnitude. In contrast, both BaiE homologs from C. hylemonae DSM15053 and C. hiranonis DSM13275 respectively exhibited kcat/KM values of equal magnitude towards both substrates. Overall, higher kcat/KM values are attained for the BaiE homologs from C. scindens strains in comparison to the homologs from the other two strains.
Table III.
Steady state kinetic parameters of different homologs of BaiE from key Clostridium sp. associated with secondary bile acid synthesis†
| Homolog | Kinetic Parameters | Substrates
|
|
|---|---|---|---|
| 3-oxo-Δ4-CDCA | 3-oxo-Δ4-CDC-CoA | ||
| BaiE_15053 | kcat (sec−1) | 18.6(0.6) | 12.6 (0.6) |
| KM (μM) | 21.0 (2.0) | 11.4 (1.9) | |
| kcat/KM (μM−1 sec−1) | 0.9 | 1.1 | |
|
| |||
| BaiE_13275 | kcat (sec−1) | 4.4 (0.1) | 22.2 (2.6) |
| KM (μM) | 25.0 (1.8) | 208.0 (40.0) | |
| kcat/KM (μM−1 sec−1) | 0.2 | 0.1 | |
|
| |||
| BaiE_35704 | kcat (sec−1) | 18.0 (0.9) | 15.0 (0.2) |
| KM (μM) | 11.0 (1.9) | 1.2 (0.2) | |
| kcat/KM (μM−1 sec−1) | 1.6 | 12.5 | |
|
| |||
| BaiE_12708 | kcat (sec−1) | 24.0(1.4) | 98.0 (2.7) |
| KM (μM) | 9.9 (2.1) | 4.0 (0.56) | |
| kcat/KM (μM−1 sec−1) | 2.4 | 24.5 | |
Steady state kinetic parameters were analyzed at substrate concentrations ranging beyond and below the respective KM values. Reaction was monitored by the formation of product 3-oxo-Δ4,6-lithocholic acid and 3-oxo-Δ4,6-lithocholyl-CoA for respective substrates at wavelength 297 nm. Values in parentheses indicate standard error.
BaiE_15053: BaiE from Clostridium hylemonae DSM15053
BaiE_13275: BaiE from Clostridium hiranonis DSM13275
BaiE_35704: BaiE from Clostridium scindens ATCC35704
BaiE_12708: BaiE from Clostridium scindens VPI12708
Mechanism of catalysis
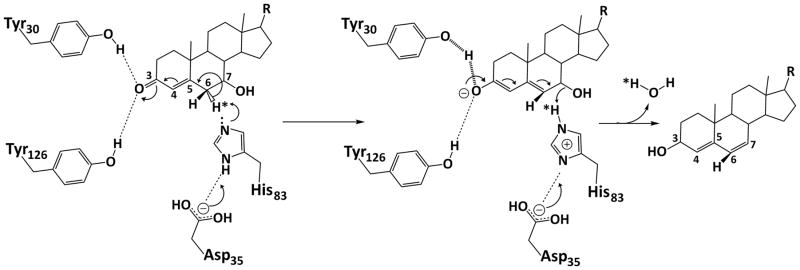
Based on the active site architecture as observed in the crystal structures and the site-directed mutagenesis study, we propose the following mechanism of catalysis by BaiE (Fig. 7). Y30 acts as a general acid (assisted by Y126) facilitating delocalization of the π-electrons across atoms C3, C4, C5 and C6. Based on the 3-oxo-Δ4-CDC-CoA co-crystal structure the terminal hydroxyl group of Y30 could possibly protonate the oxyanion generated on C3-oxo group to stabilize the negative charge. This would result in an electron shift destabilizing the C6-6αH bond. H83 is then positioned to abstract the 6αH atom and protonate the leaving C7α-hydroxy group. D35 maintains the pKa value of the H83-imidazole group for executing the deprotonation and ensuing protonation reaction with the subsequent release of a water molecule.
Figure 7.
Proposed mechanism of catalysis by BaiE highlighting the role of catalytically important residues in elimination of the 7α-hydroxy group through release of a water molecule.
Conclusion
Combined structural and biochemical analysis of BaiE provides key insights into substrate binding and turnover. The 1.9 Å resolution 3-oxo-Δ4,6-LC-CoA co- crystal structure of BaiE revealed an extended pocket between monomers from different trimer assemblies, although the biological relevance of this is unclear. Substrate and co-factor induced oligomerization is previously noted to be involved in catalysis of varied enzyme classes. 58,59,61 Moreover, using analytical size exclusion chromatography (AnSEC) p on the batch of BaiE C. hiranonis DSM13275 that yielded the co-crystal structure, we were able to detect higher-order species of the protein in presence of both bile acid substrates and products (data not shown). Although we have not been able to directly correlate these higher-order species with enzymatic activity, our data suggest possible substrate and ligand-induced oligomerization where the substrate/product binds in the extended pocket created between trimers by conformational flexibility of residues 48–63, as observed in the crystal structure. Comparison of the apo- structures across varied homologs reveals a dramatically different conformation of loop 48–63. The conformational flexibility of this loop at the substrate binding site may be essential for substrate binding. Site-directed mutagenesis studies with Y54Fand Y54A mutants of BaiE from C. hylemonae VPI12708 revealed respective impaired substrate binding and catalytic activity, suggesting the importance of this loop in catalysis.
The biochemical characterization reported herein provides key insights into the mechanism of catalysis of the enzyme, and also sheds new light onto the pathway. Steady-state kinetic studies reveal that all homologues of BaiE efficiently turnover both 3-oxo-Δ4-bile acid and CoA-3-oxo-Δ4-bile acid esters. This result is in stark contrast to our earlier reported characterization of BaiA enzymes that exhibited substantially high substrate specificity towards CoA-bile acid esters22. The substrate selectivity of bile acids is substantially lower for BaiA enzymes. It is unclear at what point in the pathway the CoA moiety is released. Since the BaiE homologues turnover bile acid and CoA-bile acid ester with comparable efficiency, it seems likely that a dual possibility exists in the pathway where the CoA moiety can be removed either before or after elimination of C7-hydroxy group. Strains of human bile acid 7 alpha/beta-dehydroxylating bacteria do not 7-dehydroxylate muricholic acids. This could be due to lack of uptake of muricholic acids (transporter specificity), or ligation to CoA, oxidation of 3-alpha-hydroxy group, or introduction of a C=C double bond (C4-C5). In other words, the initial reactions in the pathway may not allow the 3-oxo-delata 4- 6-beta, 7alpha-dihydroxy-cholenoic acid to be formed. Therefore, it is unclear if the bile acid 7alpha-dehydratase would recognize this as a substrate. However, muricholic acids are not found in humans and we have been unable to isolate bile acid 7-dehydroxylating bacteria from mice using the same techniques used to successfully isolate these bacteria from human feces. Furthermore, the DNA probes used to detect genes in human isolates give negative results using mice fecal samples. Evaluating the substrate specificity of yet-to-be identified enzymes in the reductive arm of the pathway for CoA-bile acid esters and bile acid is required to understand the role of CoA in the pathway. Site-directed mutagenesis with the homologue from C. scindens VPI12708 provides key insights into substrate binding and catalysis. Y30, D35 and H83, which are conserved in the family, were confirmed as active site residues participating in catalysis. In addition, Y54, D106 and R146 in the active site are crucial in catalysis, although they do not directly participate in the catalytic chemistry.
This study has provided key insights into the mechanism of C7α-hydroxy group removal from the bile acid moiety catalyzed by BaiE. Previous studies alluded to the possibility of trans- elimination as the stereochemical route of the reaction. 57 However, from the crystal structures and simulated docking study, cis- elimination is a likely possibility. Further experiments involving 3-oxo-Δ4 intermediate of α-muricholic acid may ascertain the stereochemical route.
The reaction catalyzed by BaiE is pivotal in the pathway as it catalyzes the only irreversible reaction whereby the intermediate is channeled onto the reductive arm generating the secondary bile acids. Hence, the biochemical and the structural characterization of this enzyme will be crucial in generating key modulators controlling the level of secondary bile acids in disease.
Acknowledgments
We thank the members of the JCSG high-throughput structural biology pipeline for their contribution to this work. This work was in part supported by the National Institutes of Health (NIH), National Institute of General Medical Sciences (NIGMS) Protein Structure Initiative (U54 GM094586). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL) and the Advanced Light Source (ALS). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division of the US Department of Energy under Contract No. DE-AC02-05CH11231 at Lawrence Berkeley National Laboratory. The Berkeley Center for Structural Biology is supported in part by NIH, NIGMS and the Howard Hughes Medical Institute. The work was also supported, in part, by a VA Merit Award BX001328 (to PBH).
Abbreviations used
- NTF2
nuclear transport factor 2
- IBD
inflammatory bowel disease
- TEV
tobacco etch virus
- CoA
Coenzyme A
- LCA
lithocholic acid
- 3-oxo-Δ4-CDCA
3-oxo-Δ4-Chenodeoxycholate
- 3-oxo-Δ46-LCA
3-oxo-Δ4,6-Lithocholate
- 3-oxo-Δ4-CDC-CoA
3-oxo-Δ4-Chenodeoxycholyl Coenzyme A
- 3-oxo-Δ46-LC-CoA
3-oxo-Δ4,6-Lithocholyl Coenzyme A
Footnotes
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or the NIH.
References
- 1.Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 2.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 4.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134:479–482. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 6.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Holmes E, Nicholson JK, Cloarec O, Chollet J, Tanner M, Singer BH, Utzinger J. Metabonomic investigations in mice infected with Schistosoma mansoni: An approach for biomarker identification. Proc Natl Acad Sci USA. 2004;101:12676–12681. doi: 10.1073/pnas.0404878101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap IKS, Li JV, Saric J, Martin F-P, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger, Marchesi JR, Holmes E. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 9.Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutation Research/Reviews in Mutation Research. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Berr F, Kullak-Ublick GA, Paumgartner G, Munzing W, Hylemon PB. 7 α-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology. 1996;111:1611–1620. doi: 10.1016/s0016-5085(96)70024-0. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 14.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Mallonee DH, Hylemon PB. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallonee DH, Adams JL, Hylemon PB. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J Bacteriol. 1992;174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallonee DH, Lijewski MA, Hylemon PB. Expression in Escherichia coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol. 1995;30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 19.Kang D-J, Ridlon JM, Moore DR, II, Barnes S, Hylemon PB. Clostridium scindens baiCD and baiH genes encode stereo-specific 7α/7β-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductases. Biochim Biophys Acta. 2008;1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson JA, Mallonee DH, Björkhem I, Hylemon PB. Expression and characterization of a C24 bile acid 7 α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 21.Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J Lipid Res. 2012;53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhowmik S, Jones DH, Chiu HP, Park IH, Chiu HJ, Axelrod HL, Farr CL, Tien HJ, Agarwalla S, Lesley SA. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins. 2013;82:216–229. doi: 10.1002/prot.24353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 24.Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 25.Mallonee DH, White WB, Hylemon PB. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J Bacteriol. 1990;172:7011–7019. doi: 10.1128/jb.172.12.7011-7019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iida T, Momose T, Nambara T, Chang FC. Potential bile acid metabolites. X. : Syntheses of stereoisomeric 3, 7-dihydroxy-5α-cholanic acids. Chem Pharm Bull (Tokyo) 1986;34:1929–1933. [Google Scholar]
- 27.Leppik RA. Improved synthesis of 3-keto, 4-ene-3-keto, and 4,6-diene-3-keto bile acids. Steroids. 1983;41:475–484. doi: 10.1016/0039-128x(83)90087-9. [DOI] [PubMed] [Google Scholar]
- 28.Elsliger M-A, Deacon AM, Godzik A, Lesley SA, Wooley J, Wüthrich K, Wilson IA. The JCSG high-throughput structural biology pipeline. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1137–1142. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, Uber DC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. An approach to rapid protein crystallization using nanodroplets. J Appl Crystallogr. 2002;35:278–281. [Google Scholar]
- 30.McPhillips TM, McPhillips SE, Chiu H-J, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 31.Leslie AW, Powell H. Processing diffraction data with mosflm. In: Read R, Sussman J, editors. Evolving methods for macromolecular crystallography. Vol. 245. Netherlands: Springer; 2007. pp. 41–51. NATO Science Series. [Google Scholar]
- 32.Evans P. Scaling and assessment of data quality. Acta Crystallographica Section D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 33.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 34.Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, Lamzin VS, Kleywegt GJ, Perrakis A. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 41.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 42.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2. 0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Davis IW, Murray LW, Richardson JS, Richardson DC. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundqvist T, Rice J, Hodge CN, Basarab GS, Pierce J, Lindqvist Y. Crystal structure of scytalone dehydratase — a disease determinant of the rice pathogen, Magnaporthe grisea. Structure. 1994;2:937–944. doi: 10.1016/s0969-2126(94)00095-6. [DOI] [PubMed] [Google Scholar]
- 49.Auld D. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 50.Bearden JA. X-Ray wavelengths. Rev Mod Phys. 1967;39:78–124. [Google Scholar]
- 51.Okai M, Kubota K, Fukuda M, Nagata Y, Nagata K, Tanokura M. Crystal structure of γ-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26. J Mol Biol. 2010;403:260–269. doi: 10.1016/j.jmb.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 52.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Ye Y, Godzik A. Flexible Structural Neighborhood—a database of protein structural similarities and alignments. Nucleic Acids Res. 2006;34:D277–D280. doi: 10.1093/nar/gkj124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen JM, Xu SL, Wawrzak Z, Basarab GS, Jordan DB. Structure-based design of potent inhibitors of scytalone dehydratase: Displacement of a water molecule from the active site. Biochemistry. 1998;37:17735–17744. doi: 10.1021/bi981848r. [DOI] [PubMed] [Google Scholar]
- 55.Kim SW, Cha S-S, Cho H-S, Kim J-S, Ha N-C, Cho M-J, Joo S, Kim KK, Choi KY, Oh B-H. High-resolution crystal structures of Δ5-3-ketosteroid isomerase with and without a reaction intermediate analogue. Biochemistry. 1997;36:14030–14036. doi: 10.1021/bi971546+. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Samuelsson B. Bile acids and steroids: 96. On the mechanism of the biological formation of deoxycholic acid from cholic acid. J Biol Chem. 1960;235:361–366. [Google Scholar]
- 58.Sinha SC, Chaudhuri BN, Burgner JW, Yakovleva G, Davisson VJ, Smith JL. Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold. J Biol Chem. 2004;279:15491–15498. doi: 10.1074/jbc.M312733200. [DOI] [PubMed] [Google Scholar]
- 59.Sennett NC, Kadirvelraj R, Wood ZA. Cofactor binding triggers a molecular switch to allosterically activate human UDP-alpha-D-glucose 6-dehydrogenase. Biochemistry. 2012;51:9364–9374. doi: 10.1021/bi301067w. [DOI] [PubMed] [Google Scholar]
- 60.Basarab GS, Steffens JJ, Wawrzak Z, Schwartz RS, Lundqvist T, Jordan DB. Catalytic Mechanism of Scytalone Dehydratase: Site-Directed Mutagenisis, Kinetic Isotope Effects, and Alternate Substrates. Biochemistry. 1999;38:6012–6024. doi: 10.1021/bi982952b. [DOI] [PubMed] [Google Scholar]
- 61.Simanshu DK, Savithri HS, Murthy MR. Crystal structures of Salmonella typhimurium biodegradative threonine deaminase and its complex with CMP provide structural insights into ligand-induced oligomerization and enzyme activation. J Biol Chem. 2006;281:39630–39641. doi: 10.1074/jbc.M605721200. [DOI] [PubMed] [Google Scholar]